Abstract
Prenylated flavonoids are natural compounds that often represent the active components in various medicinal plants and exhibit beneficial effects on human health. Prenylated flavonoids are hybrid products composed of a flavonoid core mainly attached to either 5-carbon (dimethylallyl) or 10-carbon (geranyl) prenyl groups derived from isoprenoid (terpenoid) metabolism, and the prenyl groups are crucial for their biological activity. Prenylation reactions in vivo are crucial coupling processes of two major metabolic pathways, the shikimate-acetate and isoprenoid pathways, in which these reactions are also known as a rate-limiting step. However, none of the genes responsible for the prenylation of flavonoids has been identified despite more than 30 years of research in this field. We have isolated a prenyltransferase gene from Sophora flavescens, SfN8DT-1, responsible for the prenylation of the flavonoid naringenin at the 8-position, which is specific for flavanones and dimethylallyl diphosphate as substrates. Phylogenetic analysis shows that SfN8DT-1 has the same evolutionary origin as prenyltransferases for vitamin E and plastoquinone. The gene expression of SfN8DT-1 is strictly limited to the root bark where prenylated flavonoids are solely accumulated in planta. The ectopic expression of SfN8DT-1 in Arabidopsis thaliana resulted in the formation of prenylated apigenin, quercetin, and kaempferol, as well as 8-prenylnaringenin. SfN8DT-1 represents the first flavonoid-specific prenyltransferase identified in plants and paves the way for the identification and characterization of further genes responsible for the production of this large and important class of secondary metabolites.
The prenylation of aromatic compounds is a major contributor to the diversity of plant secondary metabolites due to differences in prenylation position on the aromatic ring, various lengths of prenyl chain, and further modifications of the prenyl moiety, e.g. cyclization and hydroxylation, resulting in the occurrence of more than 1,000 prenylated compounds in plants (Tahara and Ibrahim, 1995; Barron and Ibrahim, 1996). In particular, prenylated flavonoids in higher plants protect them by exhibiting strong antibacterial and antifungal activities (Sohn et al., 2004). Many prenylated flavonoids have been identified as active components in medicinal plants with biological activities, such as anticancer, anti-androgen, anti-leishmania, and anti-nitric oxide production (De Naeyer et al., 2004; Ahmed-Belkacem et al., 2005; Han et al., 2006). Due to the beneficial effects for human health, prenylated flavonoids are of particular interest as lead compounds for producing new drugs and functional foods. The prenylation of the flavonoid core increases the lipophilicity and the membrane permeability, which is one of the proposed reasons for the enhanced biological activities of prenylated flavonoids (Wang et al., 1997; Maitrejean et al., 2000; Murakami et al., 2000). However, none of the genes responsible for the prenylation reactions has been identified despite more than 30 years of research in this field.
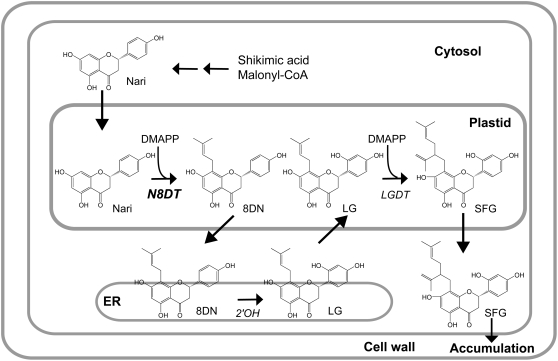
Cell cultures of Sophora flavescens produce the prenylated flavonoid sophoraflavanone G (SFG) in a large quantity. The biosynthesis of SFG involves two prenylation reactions that have been biochemically determined to be associated with the crude membrane fraction of cultured cells (Yamamoto et al., 2000; Zhao et al., 2003). Naringenin is first prenylated at the 8-position with one dimethylallyl diphosphate (DMAPP; Fig. 1). This intermediate, 8-dimethylallyl naringenin (8DN), is further hydroxylated to form leachianone G (LG) by 8DN 2′-hydroxylase (Yamamoto et al., 2001), and the second prenylation takes place at the prenyl side chain of LG catalyzed by LG 2″-dimethylallyltransferase (Zhao et al., 2003). Both prenylation reactions are Mg2+ dependent, plastid localized, and involve membrane-bound proteins.
Figure 1.
Biosynthetic pathway from naringenin to SFG in S. flavescens. Two dimethylallylations and a 2′-hydroxylation are involved. Nari, Naringenin; 2′OH, 8DN 2′-hydroxylase; LGDT, LG 2″-dimethylallyltransferase.
In this article, we describe the identification of a plant flavonoid prenyltransferase gene encoding naringenin 8-dimethylallyltransferase (SfN8DT-1) that catalyzes the primary dimethylallylation step in SFG biosynthesis, as well as functional expression of the gene products in yeast (Saccharomyces cerevisiae). We also demonstrated the enzymatic function of SfN8DT-1 in planta using transgenic Arabidopsis (Arabidopsis thaliana) plants.
RESULTS
Cloning of SfN8DT-1 cDNA and Characterization of Recombinant SfN8DT-1
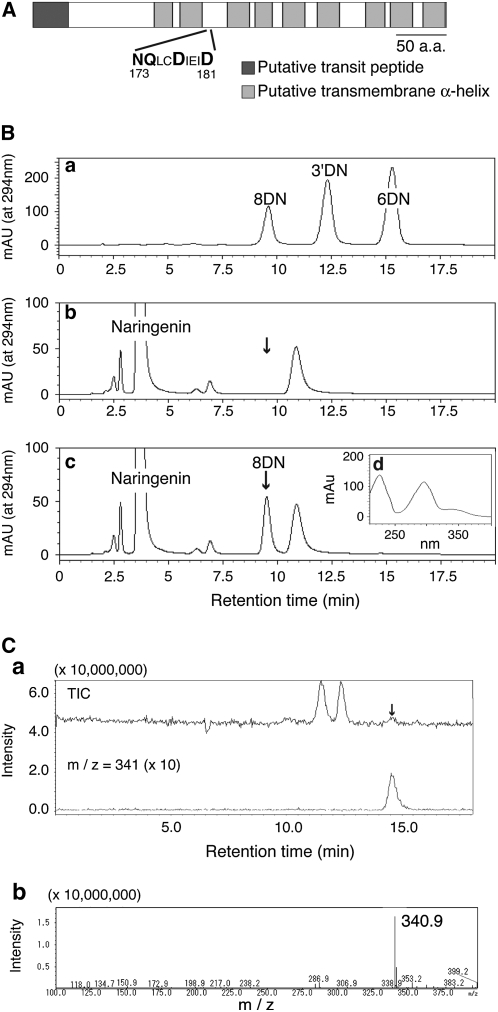
From cDNA library sequence data, we selected clones for flavonoid prenyltransferases via three criteria: possessing at least one transmembrane domain, having a putative plastid transit peptide, and the presence of conserved amino acid sequences responsible for prenyl diphosphate recognition among Mg2+-dependent prenyltransferases: NDxxDxxxD, NQxxDxxxD, NQxxExxxD, DDxxDxxxD, DQxxDxxxD, or DQxxExxxD (Table I). From 11,874 ESTs, 208 clones were found to contain one of these conserved sequences, from which membrane-bound proteins were selected using the SOSUI program, and three subcellular localization prediction programs (ChloroP, PSORT, WoLF PSORT) were used to predict transit peptides in the putative polypeptide sequences. Seven clones fulfilled all three criteria, and full-length clones were obtained for expression in yeast. The enzymatic activity was measured using the microsomal fraction of yeast transformants with naringenin and DMAPP, or LG and DMAPP as substrates, and one clone (SfN8DT-1; DNA Data Bank of Japan [DDBJ] accession no. AB325579) showed clear naringenin dimethylallyltransferase activity (1.38 ± 0.3 nmol h−1 mg protein−1) that was dependent on Mg2+ and DMAPP (Fig. 2B; Supplemental Fig. S1A). The SfN8DT-1 clone had a stop codon upstream of the initiation codon in the same frame, representing the full-length clone. The truncated protein, in which N-terminal SfN8DT-1 sequence for 30 amino acids was removed, showed almost the same activity as the full-length clone when expressed in yeast (data not shown). The predicted polypeptide possessed nine transmembrane α-helices and a prenyltransferase motif, NQxxDxxxD (Fig. 2A), and, accordingly, the activity of the recombinant enzyme was concentrated in the membrane fraction (Supplemental Fig. S1B), which could not be solubilized by high salt treatment. We also screened the prenyltransferase activity of the remaining 201 clones containing a conserved prenyltransferase motif by preparation of each recombinant protein in yeast, but no other clone showed prenyltransferase activity. The enzymatic reaction product (8DN) had an identical retention time and molecular mass representing the parent ion in liquid chromatography/electrospray ionization mass spectrometry (LC/ESI-MS) analysis with a standard specimen (Fig. 2C). The final identification of the reaction product was achieved using LC-NMR.
Table I.
Narrow down of candidate clones for flavonoid prenyltransferase
Flavonoid prenyltransferases in S. flavescens have three critical features: containing a conserved motif of prenyltransferase, a putative transit peptide, and membrane-spanning domains. The summary is in Table I, i.e. 208 clones out of 11,874 ESTs possessed d-rich amino acid sequences conserved in Mg2+-dependent prenyltransferases. Among 208 clones, 20 clones have at least one transmembrane α-helix in the sequenced stretch (average length, 559 nucleotide for 186 amino acids), whereas subcellular localization prediction programs detected 30 clones that possessed plastidial targeting signal. SfN8DT-1 was found in 34 clones possessing the NQxxDxxD motif and in 21 clones predicted by WoLF PSORT.
| Screening | Criteria | No. |
|---|---|---|
| First screening | Conserved motif | |
| NDxxDxxxD | 28 | |
| NQxxDxxxD | 34 | |
| NQxxExxxD | 17 | |
| DDxxDxxxD | 89 | |
| DQxxDxxxD | 23 | |
| DQxxExxxD | 17 | |
| Total 208/11,874 | ||
| Second screening | Transit peptide | |
| ChloroP | 7 | |
| PSORT | 6 | |
| WoLF PSORT | 21 | |
| Total 30/208 | ||
| Third screening | Transmembrane domain | Total 20/208 |
Figure 2.
Enzymatic characterization of recombinant SfN8DT-1 expressed in yeast. A, Structural features of the SfN8DT-1 polypeptide. B, HPLC chromatograms of standard specimens, 8DN, 3′-dimethylallyl naringenin (3′DN), and 6-dimethylallyl naringenin (6DN; a); ethyl acetate extract of incubation mixture of naringenin and DMAPP with empty-vector transformed cells (b); and recombinant SfN8DT-1 (c). Arrowheads indicate the retention time of 8DN (9.5 min). d, UV spectrum of 8DN formed from naringenin by recombinant SfN8DT-1. C, LC/ESI-MS analysis of enzymatic reaction products. a, Total ion chromatogram (top) and selected ion monitoring for specific m/z 341 for 8DN [M + H]+ (bottom) and mass spectrum of 8DN (b).
SfN8DT-1 coded for a polypeptide of 410 amino acids with significant similarity to homogentisate (HG) prenyltransferases (21%–55%; Collakova and DellaPenna, 2001; Savidge et al., 2002; Cahoon et al., 2003; Sadre et al., 2006) involved in vitamin E or plastoquinone biosynthesis, while relatively low similarity was seen with ubiquinone/shikonin synthesizing p-hydroxybenzoate (PHB) prenyltransferase (approximately 16%; Yazaki et al., 2002; Okada et al., 2004).
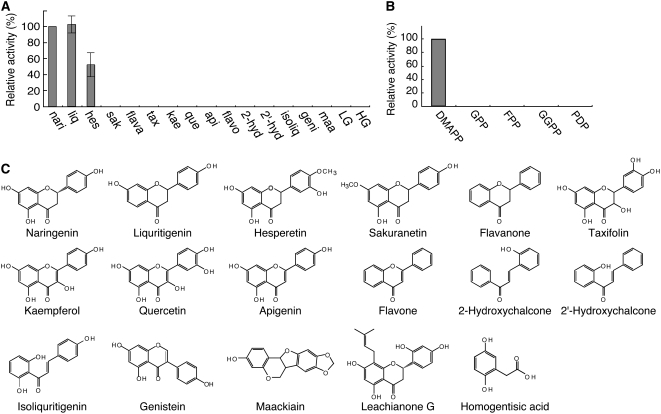
The substrate specificity of SfN8DT-1 was analyzed with various plant flavonoids as prenyl acceptors (Fig. 3, A and C) and compared to HG, where [14C]DMAPP was used as the prenyl donor. Recombinant SfN8DT-1 could accept only naringenin (100%), liquiritigenin (102%), and hesperetin (52%) as substrates, whereas flavonols or isoflavones are not prenylated.
Figure 3.
Substrate specificity of recombinant SfN8DT-1. A, Relative enzyme activity with various aromatic substrates as prenyl acceptors, naringenin (nari), liquiritigenin (liq), hesperetin (hes), sakuranetin (sak), flavanone (flava), taxifolin (tax), kaempferol (kae), quercetin (que), apigenin (api), flavone (flavo), 2-hydroxy chalcone (2-hyd), 2′-hydroxy chalcone (2′-hyd), isoliquiritigenin (isoliq), genistein (geni), maackiain (maa), LG, and HG. B, Relative enzyme activity with various prenyl diphosphates of different chain length as substrates, DMAPP, GPP, FPP, GGPP, and PDP. C, Chemical structures of flavonoids used for substrate specificity analysis.
The prenyl-donor substrate specificity of SfN8DT-1 is shown in Figure 3B. In addition to the native substrate DMAPP, geranyl diphosphate (GPP), farnesyl diphosphate (FPP), geranylgeranyl diphosphate (GGPP), and phytyl diphosphate (PDP) were tested, with naringenin as the prenyl acceptor. LC/ESI-MS analysis showed that the SfN8DT-1 protein had a strict substrate preference for DMAPP, and no detectable reaction products were obtained with any other prenyl diphosphates. The apparent Km values for DMAPP and naringenin were calculated to be 106 μm and 55 μm, respectively. These values coincided with those of the native protein (Yamamoto et al., 2000) and are in a similar range of many other secondary metabolic enzymes (Fujiwara et al., 1998; Choi et al., 2002; Suzuki et al., 2002; Fukuchi-Mizutani et al., 2003; Kim et al., 2006).
Subcellular Localization of SfN8DT-1
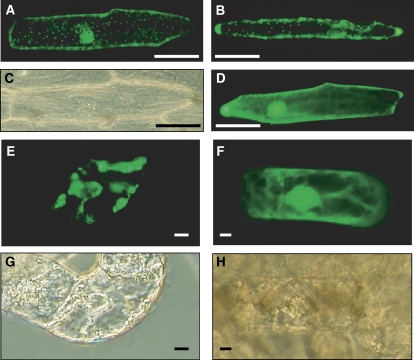
The N-terminal partial sequence of SfN8DT-1 obtained from EST analysis was predicted to have a transit peptide during in silico screening, but when the full sequence of SfN8DT-1 was submitted to prediction programs (TargetP, ChloroP, PSORT, WoLF PSORT, iPSORT), only iPSORT showed high scores for a transit peptide at the N terminus. To confirm the subcellular localization of SfN8DT-1, full-length SfN8DT-1 or its N-terminal SfN8DT-1 sequence (30 amino acids) was independently fused to GFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter and was introduced into onion (Allium cepa) peels and S. flavescens cultured cells by particle bombardment (Fig. 4). Following transient expression, the fluorescence of SfN8DT1-GFP was localized to dotted organelles in both cell types, whose size and pattern were highly similar to that of isoprene synthase, a typical plastid protein, used as a positive control (Sasaki et al., 2005). These results suggested that SfN8DT-1 was localized to plastids as the native enzyme in S. flavescens (Zhao et al., 2003) as well as prenyltransferase of other plant species (Dhillon and Brown, 1976; Biggs et al., 1990; Fellermeier et al., 2001).
Figure 4.
Transient expression of the SfN8DT1-GFP fusion protein. The plasmid containing SfN8DT1-GFP was introduced into onion peels (A–D) and cultured S. flavescens cells (E–H) by particle bombardment. Scale bars show 100 μm for onion and 10 μm for S. flavescens cells. A, SfN8DT1∷GFP; B, PaIspS-TP∷GFP, as a plastid-targeted control; D, 35S-GFP (control); E, SfN8DT1-TP∷GFP; F, 35S-GFP (control). C, G, and H show bright field images of A, E, and F, respectively.
Expression of SfN8DT-1 Genes in S. flavescens
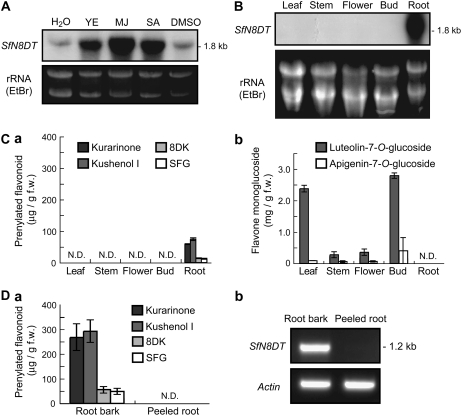
The prenyltransferase activity in cultured S. flavescens cells was inducible by the application of methyl jasmonate (MJ), which mimics defense reactions against insect and fungal attack. SfN8DT-1 expression in cultured cells was also strongly induced by yeast extract, MJ, and salicylic acid when monitored by RNA gel-blot analysis (Fig. 5A), suggesting that the induction of prenyltransferase activity detected in cultured cells was regulated at the transcriptional level. In intact S. flavescens plants, SfN8DT-1 mRNA was solely detected in root tissues (Fig. 5B), where many prenylated flavonoids, such as SFG, kurarinone, kushenol I, and 8-dimethylallyl kaempferol (8DK, des-O-methylanhydroicaritin), are localized (Yamamoto et al., 1992), and no detectable expression of SfN8DT-1 was seen in aerial tissues, where flavone monoglucosides such as luteolin-7-O-glucoside are abundant (Fig. 5C). In the root, prenylated flavonoids exclusively accumulated in the bark and were not detectable in peeled roots (Fig. 5D). In accordance with this accumulation pattern of prenylated compounds, SfN8DT-1 was specifically expressed in root bark (Fig. 5D).
Figure 5.
Accumulation of SfN8DT-1 mRNA and prenylated flavonoids in S. flavescens. A, Effects of yeast extract, MJ, and salicylic acid on SfN8DT-1 expression in cultured S. flavescens cells monitored by RNA gel-blot analysis. B, Organ-specific accumulation of SfN8DT-1 mRNA in intact S. flavescens plants detected by RNA gel-blot analysis. C, Organ-specific accumulation of flavonoids in S. flavescens. a, Contents of prenylated flavonoids in each plant organ. b, Contents of flavone monoglucosides in each plant organ. Values are the mean ± sd of three replicates. D, Correlation of accumulation pattern of prenylated flavonoids with SfN8DT-1 gene expression. a, Content of prenylated flavonoids in the roots of S. flavescens. b, SfN8DT-1 mRNA expression level in the roots of S. flavescens monitored by RT-PCR analysis.
Introduction of SfN8DT-1 cDNA into Arabidopsis Plants
Arabidopsis does not show flavonoid prenyltransferase activity, and accordingly no prenylated flavonoid was detected. Thus, we transformed Arabidopsis with the full-length SfN8DT-1 cDNA, and the enzymatic function of N8DT in planta was observed in the Arabidopsis transformant, in which SfN8DT-1 was under the control of a CaMV 35S promoter. In the T2 generation, the expression of SfN8DT-1 mRNA was confirmed by reverse transcription (RT)-PCR (Supplemental Fig. S2). In the aqueous acetone extract of transformed seedlings, 8DK was detected by LC/MS (4.4 ± 0.43 μg/g dry weight [DW]; Table II). Then we tested the effect of naringenin addition (100 μm) to the growth medium on the production of prenylated flavonoids. The supplementation of naringenin resulted in the production of 8DN (5.1 ± 0.93 μg/g DW), which provided the evidence that SfN8DT-1 was enzymatically functional in planta to form prenylated flavonoids in a heterologous host plant. It was noteworthy that, in addition to 8DN and 8DK (5.1 ± 1.05 μg/g DW), dimethylallylated derivatives of apigenin and quercetin were also detected in the Arabidopsis SfN8DT-1 transformants. The contents of these dimethylallylated apigenin and quercetin were expressed as 8DN equivalence according to their peak intensities in LC/MS (Table II), because we do not have authentic samples. In contrast, prenylated flavonoids were not detected in wild-type Arabidopsis seedlings, nor did the addition of naringenin produce 8DN.
Table II.
Production of prenylated flavonoids in transgenic Arabidopsis
Freeze-dried 2-week-old seedlings (approximately 100 mg) of either SfN8DT-1-expressing or wild-type Arabidopsis plants (approximately 300–500 plants) were extracted with 80% acetone, and the extract was subjected to LC/MS analysis. The effect of naringenin addition (100 μm) to the agar medium was also tested. Prenylated flavonoids were not detectable in wild-type Arabidopsis with or without naringenin addition. n.d., Not detectable; DA, dimethylallylated apigenin; DQ, dimethylallylated quercetin. With and without addition of naringenin is shown as + and −, respectively.
| Content | Naringenin Addition
|
|||
|---|---|---|---|---|
| Wild Type, − | Transgenic, − | Wild Type, + | Transgenic, + | |
| μg/g DW | ||||
| 8DN | n.d. | n.d. | n.d. | 5.1 ± 0.93 |
| 8DK | n.d. | 4.4 ± 0.43 | n.d. | 5.1 ± 1.05 |
| DA | n.d. | n.d. | n.d. | 4.4 ± 0.92 |
| DQ | n.d. | n.d. | n.d. | 2.7 ± 0.65 |
DISCUSSION
Many researchers have studied biochemical features of aromatic prenyltransferases and biological activities of prenylated aromatic compounds for a long time, as these compounds are attractive resources in food and pharmaceutical industry. There are several reports on the identification of prenyltransferase cDNAs of simple phenolic intermediates, such as PHB and HG involved in ubiquinone, plastoquinone, and vitamin E biosynthesis; however, there has been no report on the identification of a gene that encodes plant prenyltransferase producing valuable prenylated flavonoids or coumarins.
In this study, we isolated the first cDNA of flavonoid-specific plant prenyltransferase SfN8DT-1 involved in SFG biosynthesis of S. flavescens. Heterologous expression in yeast showed that the gene product exhibited enzyme activity to transfer the dimethylallyl moiety to flavonoid substrate naringenin. The result of the fractionation of the recombinant crude enzyme showed that SfN8DT-1 was a membrane-bound protein in accordance with prediction by the TMHMM program (Supplemental Fig. S1B). In addition, the apparent Km values and divalent cation requirements of recombinant SfN8DT-1 (Supplemental Fig. S1C) coincided with those of native protein (Yamamoto et al., 2000), and so SfN8DT-1 was confirmed to be the first flavonoid-specific prenyltransferase identified in any plant species, leading to a native secondary metabolic product.
SfN8DT-1 accepts flavanones as specific substrates, whereas flavonols or isoflavones are not prenylated. Judging from the preference for liquiritigenin as a substrate, the OH group at the 5-position of the A-ring does not play an important role in the enzymatic function of SfN8DT-1. In contrast, sakuranetin, a methyl derivative of naringenin, did not give any detectable reaction product, i.e. one methoxy group at the 7-position of the A-ring seemed to strongly hamper the recognition as a substrate, probably due to the vicinal position of prenylation. On the other hand, substitution of the B-ring did not strongly influence the enzymatic activity of SfN8DT-1, as hesperetin was clearly prenylated (Fig. 3A). In addition, SfN8DT-1 cannot accept LG, the substrate of the second prenylation step involved in SFG biosynthesis. A gene encoding LG 2″-dimethylallyltransferase was not found in the S. flavescens cDNA library either. However, we obtained three homologous genes by RT-PCR using internal sequences of SfN8DT-1 as primers, i.e. SfN8DT-2, SfL17a, and SfL17b (DDBJ accession nos. SfN8DT-2, AB370330; SfL17a, AB371287; SfL17b, AB370329). SfN8DT-2, sharing 93% amino acid identity with SfN8DT-1, exhibited the same enzymatic properties as SfN8DT-1, including the substrate specificity, which suggested the functional redundancy of these enzymes, but SfL17a and SfL17b, sharing 68% and 69% amino acid identities with SfN8DT-1, did not exhibit prenyltransferase activity as far as we tested using DMAPP and various flavonoids (naringenin, LG, liquiritigenin, hesperetin, kaempferol, apigenin, taxifolin, 2-hydroxychalcone, 2′-hydroxychalcone, isoliquiritigenin, and genistein).
In S. flavescens plants, prenylated flavonoids such as kurarinone, kushenol I, SFG, and 8DK were mainly localized in root bark and detected in neither aerial parts nor peeled roots (Yamamoto et al., 1992; Fig. 5, C and D). The first prenylation step in biosynthesis of these prenylated flavonoids is presumed to be 8-dimethylallylation of naringenin, and thus SfN8DT-1, which is expressed in only the root bark, will be responsible for the common prenylation reaction (Fig. 5, B and D). Although some secondary metabolites are translocated from source cells to sink organs, these results indicated that prenylated flavonoids are biosynthesized only in the root bark of S. flavescens, and are not transferred to peeled root and aerial parts. Furthermore SfN8DT-1 expression in cultured cells was strongly induced by yeast extract, MJ and salicylic acid, which mimics insect and pathogen attacks (Martin et al., 2002; Martin et al., 2003; Fig. 5A), indicative that this gene plays an important role as a defense reaction against insect and pathogen attack in S. flavescens root.
It was demonstrated that transgenic Arabidopsis overexpressing SfN8DT-1 accumulated a prenylated flavonoid, 8DK, whereas wild type did not (Table II). It is not clear if SfN8DT-1 catalyzed the prenylation of kaempferol in planta, or if naringenin, a precursor of kaempferol, was first prenylated by SfN8DT-1 and then converted to 8DK in vivo. However, because kaempferol is the major flavonoid in Arabidopsis and the in vivo level of naringenin is very low, the occurrence of 8DK may be reasonable (Peer et al., 2001). The detection of an appreciable amount of 8DN in naringenin-fed transgenic Arabidopsis indicated that SfN8DT-1 was enzymatically functional in planta and contributed to the formation of prenylated flavonoids in a heterologous plant. These findings demonstrated that nonproducing plants could be hosts for the production of valuable prenylated flavonoids by expressing prenyltransferases.
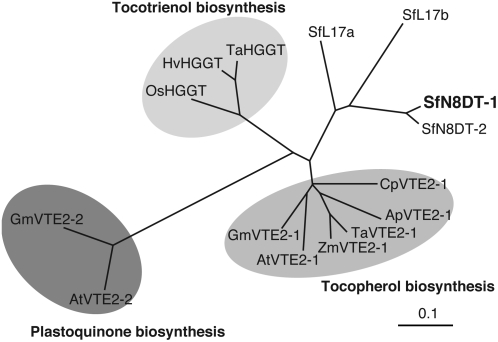
Biochemical properties of prenyltransferases involved in biosynthesis of vitamin E and plastoquinone are very similar to those of SfN8DT-1, i.e. these catalyze an aromatic proton substitution with prenyl moiety, have eight or nine putative transmembrane α-helices, and are localized in plastids (Fig. 4). Flavonoids are biosynthesized in cytosol and endoplasmic reticulum, but flavonoid prenyltransferases seem to be localized to plastids as far as characterized (Biggs et al., 1990; Zhao et al., 2003), suggesting that the subcellular movement of flavonoid molecules between plastids and endoplasmic reticulum is important for the biosynthesis of prenylated flavonoids. Phylogenetic analysis reveals that members of the HG prenyltransferases are divided into three groups, HG phytyltransferases for tocopherol synthesis, HG geranylgeranyltransferases responsible for tocotrienol synthesis, and HG solanesyltransferases for plastoquinone formation (Fig. 6). However, SfN8DT-1, SfN8DT-2, SfL17a, and SfL17b do not belong to any of these clusters despite their overall polypeptide similarities. Several amino acid residues in the conserved domains are present in SfN8DT-1, while divergent residues characteristic only to SfN8DT-1 are observed as well (Supplemental Fig. S3; Venkatesh et al., 2006). Accumulation of sequence data of further flavonoid prenyltransferases will enable us to predict which amino acids are responsible for the recognition of the flavonoid core as a prenyl accepter instead of HG.
Figure 6.
Phylogenetic relationship of prenyltransferases accepting aromatic substrates. A rooted phylogram was generated using a ClustalW alignment. Ap, Allium porrum; At, Arabidopsis; Cp, Cuphea pulcherrima; Gm, Glycine max; Hv, Hordeum vulgare; Os, Oryza sativa; Ta, Triticum aestivum; Zm, Zea mays. HG phytyltransferases (VTE2-1) are involved in tocopherol biosynthesis, HG geranylgeranyltransferases (HGGT) are involved in tocotrienol biosynthesis, and HG solanesyltransferases (VTE2-2) are involved in plastoquinone biosynthesis. Accession numbers: ApVTE2-1, DQ231057; AtVTE2-1, AY089963; AtVTE2-2, DQ231060; CpVTE2-1, DQ231058; GmVTE2-1, DQ231059; GmVTE2-2, DQ231061; HvHGGT, AY222860; OsHGGT, AY222862; SfN8DT-1, AB325579; SfN8DT-2, AB370330; SfL17a, AB371287; SfL17b, AB370329; TaHGGT, AY222861; TaVTE2-1, DQ231056; ZmVTE2-1, DQ231055.
SfN8DT-1 and SfN8DT-2 isolated in this study are the first cDNAs that encode plant membrane-bound prenyltransferases catalyzing the transfer of the prenyl moiety to flavonoid. These cDNAs are expected to be useful for genetic engineering of SFG production. Cell-free extract from S. flavescens-cultured cells catalyzed the prenylation of other flavonoids, such as genistein, taxifolin, kaempferol, and quercetin, so this cDNA could be very powerful tool for isolating prenyltransferase cDNAs of other flavonoids. In fact, several bands were observed in the genomic Southern blot using full-length SfN8DT-1 as a probe (data not shown). Controlled expression of SfN8DT-1 in plants will provide new resources to supply prenylated flavonoids, which have beneficial effects for human health, such as prevention of cancer and nonantibiotic antibacterial agents that have become more important to suppress multiple drug resistant microorganisms in the medical field (Stevens and Page, 2004; Botta et al., 2005). Similar metabolic engineering will be also applicable for the agricultural field to confer disease resistance in crop plants, for instance, against soil-borne pathogens like Fusarium spp. (Zhang and Smith, 1983; Hahn et al., 1985). Further use of SfN8DT genes for the generation of wood biomass with increased resistance for rot fungi can also be expected in sustainability sciences.
MATERIALS AND METHODS
Plant Materials and Reagents
Cultured cells of Sophora flavescens were maintained in Murashige and Skoog medium with 1 μm 2,4-dichlorophenoxyacetic acid and 1 μm kinetin as described previously (Yamamoto et al., 1991). DMAPP, GPP, FPP, and GGPP were chemically synthesized as described previously (Cornforth and Popjak, 1969). PDP was a kind gift from Dr. T. Koyama, Tohoku University.
Construction of cDNA Library
Cultured S. flavescens cells were treated with 20 μL of 0.1 m MJ dissolved in dimethyl sulfoxide (DMSO; final concentration 0.1 mm) for 36 h to induce the expression of flavonoid prenylation enzymes. Total RNA was prepared from the cultured cells, from which poly(A+) RNA was isolated using an Oligotex-MAG mRNA purification kit (TAKARA Bio). A cDNA plasmid library was constructed using a cDNA synthesis kit (Stratagene) with a yeast (Saccharomyces cerevisiae) shuttle vector, pDR196, for the purpose of functional expression of membrane-bound proteins. In brief, the first strand of cDNA was synthesized using 7 μg of poly(A+) RNA, and oligo(dT)18 anchor primer containing an XhoI restriction site. After second-strand cDNA synthesis, a blunt-ended adaptor containing an EcoRI restriction site was ligated onto the double-stranded DNA, and the fragments were then ligated into the pDR196 vector downstream of the strong constitutive promoter PMA1 (Rentsch et al., 1995).
EST Sequences
The cDNA library was introduced into Escherichia coli DH10B, and 11,874 independent clones of the primary library were randomly picked and sequenced from the 5′-end using a primer that annealed to the PMA1 promoter region (single-pass sequencing). The average size of the sequenced ESTs was 559 bp, and the insert size of the cDNA library was approximately from 500 bp to 5 kb. They were clustered into 1,519 contiguous sequences, 1,086 cluster singletons, and 4,270 singletons. These EST data are available on the Web site of the Research Institute for Sustainable Humanosphere, Kyoto University (http://database.rish.kyoto-u.ac.jp/arch/plantdb/index.html).
By motif searching with 11,874 clones, it was found that 208 clones contained one of the following amino acid sequences conserved among aromatic substrate prenyltransferases, NDxxDxxxD, NQxxDxxxD, NQxxExxxD, DDxxDxxxD, DQxxDxxxD, or DQxxExxxD, which appeared normally at 80 to 170 amino acids from the N terminus in PHB and HG prenyltransferases. These cDNA clones were analyzed using the SOSUI program to find that 20 clones out of 208 candidates contained a transmembrane domain, at least in the sequenced region. In PHB and HG prenyltransferases, the first transmembrane domain exists upstream of the conserved motif of prenyltransferases. Transit peptide sequences were searched for with three frequently used prediction programs, ChloroP, PSORT, and WoLF PSORT, which suggested that 30 out of 208 clones had putative plastid targeting signals. Taken together, a single-pass sequence covering approximately 170 amino acids from the N terminus enabled us to check three criteria. In fact, among these 208 clones, seven clones contained both plastid targeting signal and transmembrane α-helices. These clones were respectively introduced by the lithium acetate method into the yeast strain W303-1A-Δcoq2, in which the PHB:prenyltransferase gene COQ2 was disrupted (Yazaki et al., 2002).
Screening for Flavonoid Prenyltransferases
All 208 candidate clones were expressed as yeast transformants (W303-1A-Δcoq2) by culturing in synthetic dextrose (−ura) liquid media (180 mL) to reach mid-log phase and from which the microsomal fraction was prepared, as described previously (Yazaki et al., 2002). The membrane fraction from each transformant was resuspended in 500 μL of 0.1 mm Tris-HCl buffer, pH 9.0, which was then used as an enzyme solution for the flavonoid prenyltransferase assay using naringenin and DMAPP, or LG and DMAPP as substrates, as described previously (Yamamoto et al., 2000). The reaction product, 8DN, was analyzed quantitatively by HPLC with a Shimadzu LC-10A system: column, 4.6- × 250-mm YMC-Pack Pro C18 RS; solvent system, methanol:water:acetic acid (70:30:0.3); 230 to 320 nm with a SPD6A photodiode array detector.
Identification of 8DN
8DN was identified using LC/ESI-MS and LC-NMR by comparison with a chemically synthesized standard specimen (Tahara et al., 1994). LC/ESI-MS was carried out on a Shimadzu model 2010A system liquid chromatograph and mass spectrometer using an LC-10AD Solvent Delivery system: column, 4- × 250-mm LiChrospere 100RP-18 (Merck); solvent system, ethanol:water:formic acid (90:10:0.3); flow rate, 0.2 mL min−1. Samples were ionized by positive-ion mode from m/z 50 to 600. The enzymatic reaction product gave the same retention time and MS spectrum as those of standard 8DN.
LC-NMR data was acquired using a Varian UNITY-INOVA-500 spectrometer (1H: 499.83 MHz) equipped with a 60-μL triple-resonance microflow NMR probe (Iwasa et al., 2005). 1H-1D NMR spectra were obtained in stopped-flow mode. The HPLC system consisted of a Varian Pro Star model 230 solvent delivery system: CH3CN:D2O:CF3COOD (50:50:0.3); flow rate, 1 mL min−1; detection, Varian Pro Star model 310 variable-wavelength UV-vis detector. 1H NMR (500 MHz) δ: 7.31 (2H, d, J = 7.5 Hz, H-2′, 6′), 6.84 (2H, d, J = 8 Hz, H-3′, 5′), 5.99 (1H, s, H-6), 5.37 (1H, br d, J = 12.5 Hz, H-2″), 5.09 (1H, br t, J = 4Hz, H-2), 3.13 (2H, br d, J = 7.5 Hz, H-1″), 3.08 (1H, dd, J = 17 Hz, 13 Hz, H-3), 2.76 (1H, br d, J = 16.5 Hz, H-3), 1.59 (3H, s, H-5″), 1.56 (3H, s, H-4″).
Enzymatic Characterization of SfN8DT-1
The enzymatic features of recombinant SfN8DT-1 were mostly analyzed by HPLC. Substrate specificity for the prenyl donor was examined using LC/MS analysis, which covered the elution period of the predicted reaction products. Substrate specificity for various flavonoid compounds was examined using a radioactive assay.
Radioactive Assay of Prenyltransferase by Thin-Layer Chromatography
The same assay mixture was used as described above except that 4.5 μm [1-14C]DMAPP (specific activity 2.0 GBq/mmol; American Radiolabeled Chemicals) was used instead of cold DMAPP. After incubation, the ethyl acetate-soluble portion was evaporated to dryness, dissolved in 20 μL of methanol, and spotted onto a silica gel thin-layer chromatography plate (20 × 20 cm; Silica Gel 60 F254; Merck). Thin-layer chromatography was developed with toluene:ethyl acetate (8:2) and subjected to autoradiography to detect the reaction products. The radioactivity of the products was quantified using a BAS1800 Bio-image Analyzer (Fuji Film).
Construction of GFP Fusion Proteins
The nucleotide sequence for full-length SfN8DT-1 or its N-terminal sequence was amplified by PCR (SfN8DTfull_Fw: 5′-GCGGTACCATGGGTTCTATGCTTCTTGCATCTTT-3′ and SfN8DTfullnostop_Rv: 5′-CAGCGGCCGCTCTAAACAAAGGTATGAGGAAGTACTCTGC-3′ or SfN8DTtpnostop_Rv: 5′-CAGCGGCCGCGGATTCTTGGCATATTGTTTACTCCTCAAGC-3′; KpnI and NotI site underlined). The PCR product was subcloned into pENTR1A to give pENTR-SfN8DT1, pENTR-SfN8DT1-TP. For transient expression of the GFP fusion proteins, the entry vector constructs were subjected to GATEWAY system transfer of the full-length SfN8DT-1 or its N-terminal sequence into modified pGWB5, so that either SfN8DT1-GFP or SfN8DT1-TP-GFP was expressed from the CaMV 35S promoter. The resulting plasmids (10 μg) were introduced into onion (Allium cepa) peels or cultured S. flavescens cells using a particle gun (PDS-1000; Bio-Rad), and the GFP fluorescence was analyzed as described previously (Sasaki et al., 2005). Pictures were taken with more than 10 cells for each plant material, in which GFP fluorescence was observed, and all cells show nearly the identical pattern representing the plastidal localization of the fusion protein.
RNA Gel-Blot Analyses
For the expression analysis of cultured cells, filter-sterilized DMSO solutions of 100 mm MJ or salicylic acid (20 μL each), or autoclaved 500 mg/mL yeast extract solution in 200 μL of water were aseptically added to the cell cultures (final concentrations, 0.1 mm, 0.1 mm, and 5 mg/mL, respectively), and cultured for 1 d. Twenty microliters of DMSO and 200 μL of water were used as negative controls. For organ-specific expression analysis, an adult plant of S. flavescens (approximately 150–170 cm in height) was used, and total RNA was extracted with an RNeasy Plant Mini kit (Qiagen). Each RNA sample (10 μg) was separated by formamide-containing 1% agarose gel electrophoresis and then capillary blotted onto a nylon membrane (Hybond N+; GE Healthcare). The membrane was hybridized with a 32P-labeled probe of full-length SfN8DT-1 cDNA, washed, and exposed by a standard procedure. Signals of ribosomal RNA stained with ethidium bromide were used as a loading control.
RT-PCR Analysis
The tissue-specific expression of SfN8DT-1 in root was analyzed by RT-PCR. Total RNA (5 μg) extracted from the root bark and the peeled root of S. flavescens adult plants was reverse-transcribed with SuperScript III RNase H− reverse transcriptase (Invitrogen). PCR was carried out with Go Taq DNA polymerase (Promega) and the following primer pair (SfN8DTfull_Fw; SfN8DTfull_Rv: 5′-CAGCGGCCGCTCATCTAAACAAAGGTATGAGGAAGTACTCTGC-3′). For normalization, actin was used as an external standard (actin-Fw, 5′-CAACTGGGACGACATGGAGA-3′ and actin-Rv, 5′-GATCCACATCTGCTGGAAGG-3′).
Quantitative Analysis of Secondary Metabolites
Sample extraction was basically carried out according to the method of Wiesman et al. (2002). Methanol extracts from each S. flavescens sample were analyzed by HPLC analysis, and each compound was identified by direct comparison with standard specimens.
Construction of the Plant Expression Cassette and Arabidopsis Transformation
The nucleotide sequence for full-length SfN8DT-1 was amplified by PCR using primer pair SfN8DTfull_Fw and SfN8DTfull_Rv. The PCR product was subcloned into pENTR1A. The entry vector was subjected to the GATEWAY recombination to transfer the full-length cDNA of SfN8DT-1 into the binary destination vector pGWB2 for constitutive expression. Agrobacterium tumefaciens GV3101 (pMP90) was transformed with the binary vector and was used to transform Arabidopsis (Arabidopsis thaliana) Columbia wild-type plants by the floral dip method. T1 generation transformants were grown on germination medium supplemented with 50 μg/mL kanamycin for selection before transfer to soil. Expression of SfN8DT-1 mRNA in Arabidopsis was checked by RT-PCR using seedlings of the T2 generation. Total RNA prepared from 100 mg seedlings was subjected to RT-PCR, in which Go Taq DNA polymerase (Promega) was employed to amplify the full-length SfN8DT-1 (primer pair, SfN8DTfull_Fw and SfN8DTfull_Rv). Actin was used as an external standard (primer pair, actin-Fw and actin-Rv).
Measurements of Prenylated Flavonoids in Transgenic Arabidopsis
Arabidopsis plants (2-week-old T2 seedling, approximately 100 mg DW) were grown on plates with or without 0.1 mm naringenin, flavonoids were extracted with 80% aqueous acetone, and the acetone extract was evaporated and partitioned with water and diethyl ether. The ether soluble portion was evaporated to dryness, and the residue was dissolved with methanol. Methanol extracts from transgenic and wild-type plants were analyzed by LC/MS analysis (microTOF-Q; Bruker Daltonics), and 8DN and 8DK were identified by direct comparison with standard specimens. Other prenylated compounds were identified by accurate MS.
Isolation of cDNAs Homologous to SfN8DT-1 from S. flavescens Cultured Cells
Total RNA prepared from S. flavescens cell cultures treated with MJ was reverse-transcribed using GeneRacer kit (Invitrogen), and RT products were subjected to RACE according to the manufacturer's protocol. SfN8DT-2, SfL17a, and SfL17b were obtained by use of primers specific for SfN8DT-1. Their full-length clones were re-isolated by RT-PCR using primer pairs specific for each clone.
DDBJ accession numbers for the genes isolated in this article are AB325579 (SfN8DT-1 cDNA), AB370330 (SfN8DT-2 cDNA), AB371287 (SfL17a cDNA), and AB370329 (SfL17b cDNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Enzymatic characterization of recombinant SfN8DT-1 expressed in yeast.
Supplemental Figure S2. Detection of SfN8DT-1 mRNA in transgenic Arabidopsis plants.
Supplemental Figure S3. Multiple alignment of the amino acid sequences among HG substrate prenyltransferases including SfN8DT-1.
Supplementary Material
Acknowledgments
We thank Dragon Genomics Center (Takara Bio), Bruker Daltonics K.K., and Akita Prefectural University for in silico screening of ESTs, analysis of prenylated flavonoids in Arabidopsis, and DNA sequencing, respectively. We are grateful to Drs. W. Frommer (Carnegie Institution), T. Nakagawa (Shimane University), T. Shikanai (Kyushu University), and T. Koyama (Tohoku University) for experimental materials. Drs. K. Iwasa and M. Sugiura (Kobe Pharmaceutical University), and T. Umezawa and A. Oka (Kyoto University) provided technical assistance. S. flavescens plants were from the Kyoto Botanical Garden.
This work was supported in part by a Grant-in-Aid for Scientific Research (no. 17310126 to K.Y.) and by a research fellowship from the Japan Society for the Promotion of Science for Young Scientists (no. 183424 to K.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kazufumi Yazaki (yazaki@rish.kyoto-u.ac.jp).
The online version of this article contains Web-only data.
References
- Ahmed-Belkacem A, Pozza A, Munoz-Martinez F, Bates SE, Castanys S, Gamarro F, Di Pietro A, Perez-Victoria JM (2005) Flavonoid structure-activity studies identify 6-prenyichrysin and tectochrysin as potent and specific inhibitors of breast cancer resistance protein ABCG2. Cancer Res 65 4852–4860 [DOI] [PubMed] [Google Scholar]
- Barron D, Ibrahim RK (1996) Isoprenylated flavonoids: a survey. Phytochemistry 43 921–982 [Google Scholar]
- Biggs DR, Welle R, Grisebach H (1990) Intracellular of prenyltransferases of isoflavonoid phytoalexin biosynthesis in bean and soybean. Planta 181 244–248 [DOI] [PubMed] [Google Scholar]
- Botta B, Vitali A, Menendez P, Misiti D, Delle Monache G (2005) Prenylated flavonoids: pharmacology and biotechnology. Curr Med Chem 12 713–739 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ (2003) Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotechnol 21 1082–1087 [DOI] [PubMed] [Google Scholar]
- Choi KB, Morishige T, Shitan N, Yazaki K, Sato F (2002) Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J Biol Chem 277 830–835 [DOI] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D (2001) Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol 127 1113–1124 [PMC free article] [PubMed] [Google Scholar]
- Cornforth RH, Popjak G (1969) Chemical synthesis of substrates of sterol biosynthesis. Methods Enzymol 15 359–390 [Google Scholar]
- De Naeyer A, Berghe WV, Pocock V, Milligan S, Haegeman G, De Keukeleire D (2004) Estrogenic and anticarcinogenic properties of kurarinone, a lavandulyl flavanone from the roots of Sophora flavescens. J Nat Prod 67 1829–1832 [DOI] [PubMed] [Google Scholar]
- Dhillon DS, Brown SA (1976) Localization, purification, and characterization of dimethylallylpyrophosphate: umbelliferone dimethylallyltransferase from Ruta graveolens. Arch Biochem Biophys 177 74–83 [DOI] [PubMed] [Google Scholar]
- Fellermeier M, Eisenreich W, Bacher A, Zenk MH (2001) Biosynthesis of cannabinoids. Incorporation experiments with 13C-labeled glucoses. Eur J Biochem 268 1596–1604 [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka Y, Yonekura-Sakakibara K, Fukuchi-Mizutani M, Nakao M, Fukui Y, Yamaguchi M, Ashikari T, Kusumi T (1998) cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J 16 421–431 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Okuhara H, Fukui Y, Nakao M, Katsumoto Y, Yonekura-Sakakibara K, Kusumi T, Hase T, Tanaka Y (2003) Biochemical and molecular characterization of a novel UDP-glucose:anthocyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol 132 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MG, Bonhoff A, Grisebach H (1985) Quantitative localization of the phytoalexin glyceollin-I in relation to fungal hyphae in soybean roots infected with Phytophthora megasperma f. sp glycinea. Plant Physiol 77 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han AR, Kang YJ, Windono T, Lee SK, Seo EK (2006) Prenylated flavonoids from the heartwood of Artocarpus communis with inhibitory activity on lipopolysaccharide-induced nitric oxide production. J Nat Prod 69 719–721 [DOI] [PubMed] [Google Scholar]
- Iwasa K, Cui W, Sugiura M, Takeuchi A, Moriyasu M, Takeda K (2005) Structural analyses of metabolites of phenolic 1-benzyltetrahydroisoquinolines in plant cell cultures by LC/NMR, LC/MS, and LC/CD. J Nat Prod 68 992–1000 [DOI] [PubMed] [Google Scholar]
- Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67 387–394 [DOI] [PubMed] [Google Scholar]
- Maitrejean M, Comte G, Barron D, El Kirat K, Conseil G, Di Pietro A (2000) The flavanolignan silybin and its hemisynthetic derivatives, a novel series of potential modulators of P-glycoprotein. Bioorg Med Chem Lett 10 157–160 [DOI] [PubMed] [Google Scholar]
- Martin D, Gershenzon J, Bohlmann J (2003) Induction of volatile terpene biosynthesis and diurnal emission by methyl jasmonate in foliage of Norway spruce. Plant Physiol 132 1586–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Tholl D, Gershenzon J, Bohlmann J (2002) Methyl jasmonate induces traumatic resin ducts, terpenoid resin biosynthesis, and terpenoid accumulation in developing xylem of Norway. Plant Physiol 129 1003–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A, Gao G, Omura M, Yano M, Ito C, Furukawa H, Takahashi D, Koshimizu K, Ohigashi H (2000) 1,1-Dimethylallylcoumarins potently suppress both lipopolysaccharide- and interferon-gamma-induced nitric oxide generation in mouse macrophage RAW 264.7 cells. Bioorg Med Chem Lett 10 59–62 [DOI] [PubMed] [Google Scholar]
- Okada K, Ohara K, Yazaki K, Nozaki K, Uchida N, Kawamukai M, Nojiri H, Yamane H (2004) The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol Biol 55 567–577 [DOI] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) Ntr1 encodes a high-affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370 264–268 [DOI] [PubMed] [Google Scholar]
- Sadre R, Gruber J, Frentzen M (2006) Characterization of homogentisate prenyltransferases involved in plastoquinone-9 and tocochromanol biosynthesis. FEBS Lett 580 5357–5362 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Ohara K, Yazaki K (2005) Gene expression and characterization of isoprene synthase from Populus alba. FEBS Lett 579 2514–2518 [DOI] [PubMed] [Google Scholar]
- Savidge B, Weiss JD, Wong YHH, Lassner MW, Mitsky TA, Shewmaker CK, Post-Beittenmiller D, Valentin HE (2002) Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol 129 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn HY, Son KH, Kwon CS, Kwon GS, Kang SS (2004) Antimicrobial and cytotoxic activity of 18 prenylated flavonoids isolated from medicinal plants: Morus alba L., Morus mongolica Schneider, Broussnetia papyrifera (L.) Vent, Sophora flavescens Ait and Echinosophora koreensis Nakai. Phytomedicine 11 666–672 [DOI] [PubMed] [Google Scholar]
- Stevens JF, Page JE (2004) Xanthohumol and related prenylflavonoids from hops and beer: to your good health! Phytochemistry 65 1317–1330 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Nakayama T, Yonekura-Sakakibara K, Fukui Y, Nakamura N, Yamaguchi MA, Tanaka Y, Kusumi T, Nishino T (2002) cDNA cloning, heterologous expressions, and functional characterization of malonyl-coenzyme A: anthocyanidin 3-O-glucoside-6″-O-malonyltransferase from dahlia flowers. Plant Physiol 130 2142–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara S, Ibrahim RK (1995) Prenylated isoflavonoids: an update. Phytochemistry 38 1073–1094 [Google Scholar]
- Tahara S, Katagiri Y, Ingham JL, Mizutani J (1994) Prenylated flavonoids in the roots of yellow lupin. Phytochemistry 36 1261–1271 [DOI] [PubMed] [Google Scholar]
- Venkatesh TV, Karunanandaa B, Free DL, Rottnek JM, Baszis SR, Valentin HE (2006) Identification and characterization of an Arabidopsis homogentisate phytyltransferase paralog. Planta 223 1134–1144 [DOI] [PubMed] [Google Scholar]
- Wang BH, Ternai B, Polya G (1997) Specific inhibition of cyclic AMP-dependent protein kinase by warangalone and robustic acid. Phytochemistry 44 787–796 [DOI] [PubMed] [Google Scholar]
- Wiseman H, Casey K, Clarke DB, Barnes KA, Bowey E (2002) Isoflavone aglycon and glucoconjugate content of high- and low-soy U.K. foods used in nutritional studies. J Agric Food Chem 50 1404–1410 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Ichimura M, Ishikawa N, Tanaka T, Iinuma M, Mizuno M (1992) Localization of prenylated flavonoids in Sophora flavescens var. angustifolia plants. Z Naturforsch 47c 535–539 [Google Scholar]
- Yamamoto H, Kawai S, Mayumi J, Tanaka T, Iinuma M, Mizuno M (1991) Prenylated flavanone production in callus cultures of Sophora flavescens var. angustifolia. Z Naturforsch 46c 172–176 [Google Scholar]
- Yamamoto H, Senda M, Inoue K (2000) Flavanone 8-dimethylallyltransferase in Sophora flavescens cell suspension cultures. Phytochemistry 54 649–655 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Yatou A, Inoue K (2001) 8-Dimethylallylnaringenin 2′-hydroxylase, the crucial cytochrome P450 mono-oxygenase for lavandulylated flavanone formation in Sophora flavescens cultured cells. Phytochemistry 58 671–676 [DOI] [PubMed] [Google Scholar]
- Yazaki K, Kunihisa M, Fujisaki T, Sato F (2002) Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and characterization of a key enzyme in shikonin biosynthesis. J Biol Chem 277 6240–6246 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Smith DA (1983) Concurrent metabolism of the phytoalexins phaseollin, kievitone and phaseollinisoflavan by Fusarium solani f. sp. phaseoli. Physiol Plant Pathol 23 89–100 [Google Scholar]
- Zhao P, Inoue K, Kouno I, Yamamoto H (2003) Characterization of leachianone G 2″-dimethylallyltransferase, a novel prenyl side-chain elongation enzyme for the formation of the lavandulyl group of sophoraflavanone G in Sophora flavescens Ait. cell suspension cultures. Plant Physiol 133 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.