Abstract
Selenium (Se) is an essential element for many organisms, but excess Se is toxic. To better understand plant Se toxicity and resistance mechanisms, we compared the physiological and molecular responses of two Arabidopsis (Arabidopsis thaliana) accessions, Columbia (Col)-0 and Wassilewskija (Ws)-2, to selenite treatment. Measurement of root length Se tolerance index demonstrated a clear difference between selenite-resistant Col-0 and selenite-sensitive Ws-2. Macroarray analysis showed more pronounced selenite-induced increases in mRNA levels of ethylene- or jasmonic acid (JA)-biosynthesis and -inducible genes in Col-0 than in Ws-2. Indeed, Col-0 exhibited higher levels of ethylene and JA. The selenite-sensitive phenotype of Ws-2 was attenuated by treatment with ethylene precursor or methyl jasmonate (MeJA). Conversely, the selenite resistance of Col-0 was reduced in mutants impaired in ethylene or JA biosynthesis or signaling. Genes encoding sulfur (S) transporters and S assimilation enzymes were up-regulated by selenite in Col-0 but not Ws-2. Accordingly, Col-0 contained higher levels of total S and Se and of nonprotein thiols than Ws-2. Glutathione redox status was reduced by selenite in Ws-2 but not in Col-0. Furthermore, the generation of reactive oxygen species by selenite was higher in Col-0 than in Ws-2. Together, these results indicate that JA and ethylene play important roles in Se resistance in Arabidopsis. Reactive oxygen species may also have a signaling role, and the resistance mechanism appears to involve enhanced S uptake and reduction.
Selenium (Se) is a naturally occurring element commonly found in sedimentary rocks formed during the Carboniferous to Quaternary periods (Wilber, 1980). The accumulation of Se in surface waters or soil can become a source of toxicity for livestock, wildlife, and humans (Hamilton, 2004; Hira et al., 2004). At the same time, Se is an essential element for many organisms, including mammals, many bacteria, and some green algae (Birringer et al., 2002; Fu et al., 2002; Obata and Shiraiwa, 2005). These organisms contain selenoproteins, which require seleno-Cys (SeCys) in their active site (Stadtman, 1990, 1996). To date, there is no evidence that higher plants need Se for survival, although Se has been proposed to be a beneficial element, especially for certain Se-tolerant hyperaccumulator species that grow on Se-rich soils; these include some species of Astragalus and Stanleya that are able to accumulate Se up to 1% of their dry weight (Feist and Parker, 2001; Pickering et al., 2003; Galeas et al., 2007). High levels of Se are toxic for nonhyperaccumulator plants, as for most organisms (White et al., 2004).
Se is chemically similar to sulfur (S) and can be metabolized by S metabolic pathways (Läuchli, 1993). Due to the structural similarity between selenate (SeO42−) and sulfate, plants take up selenate, the major soluble form of Se in soil, inadvertently via sulfate transporters, and metabolize it through the sulfate assimilation pathway. This leads to the reductive assimilation of selenate to the analogs of Cys and Met, SeCys and seleno-Met, respectively (Läuchli, 1993; Terry et al., 2000; Sors et al., 2005). The incorporation of Se in Cys and Met by their selenoanalogs in proteins has been shown to diminish protein synthesis, structure, and function. For example, SeCys has been shown to inhibit the formation of S bridges (Gromer and Gross, 2002), while seleno-Met is known to affect protein synthesis (Eustice et al., 1981). Thus, when plants are exposed to high level of Se, protein synthesis is adversely affected, causing symptoms including chlorosis and stunted growth that mimic S starvation, as well as withering and drying of leaves and premature death (Terry et al., 2000).
Knowing more about factors limiting plant Se accumulation and resistance may have applications for breeding Se-fortified foods or for phytoremediation. For instance, overproduction of ATP sulfurylase, SeCys methyltransferase, or SeCys lyase in Brassica juncea was shown to lead to enhanced Se accumulation when plants were grown on Se-polluted soil (Bañuelos et al., 2005, 2007). As an alternative to generating laboratory-induced mutants or transgenic plants, another source of genetic variation can be found among naturally occurring populations of Arabidopsis (Koornneef et al., 2004). After comparison of Se resistance and genetic investigation of recombinant inbred lines between Arabidopsis (Arabidopsis thaliana) accessions Columbia (Col)-4 and Landsberg erecta-0, it was reported that the difference in selenate resistance between these accessions is controlled by multiple genes located on chromosomes 3 and 5 (Zhang et al., 2006b). In another study that compared selenate or selenite resistance and Se accumulation among 19 Arabidopsis accessions, no correlation was found between Se resistance and accumulation, either for selenate or selenite (SeO32−; Zhang et al., 2007). Although there are a few studies for natural variation of Se resistance in Arabidopsis ecotypes, the underlying mechanisms that cause the difference in Se resistance are largely unknown.
In this work, we investigated the natural difference in selenite resistance between Arabidopsis accessions Col-0 and Wassilewskija (Ws)-2. In a previous study, Zhang et al. (2006a) showed Ws was sensitive to selenite while Col was resistant. Here, the molecular mechanisms behind this physiological difference were further investigated. The new results indicate that jasmonic acid (JA) and ethylene play important roles in Se resistance in Arabidopsis. Reactive oxygen species (ROS) may also have a signaling role, and the resistance mechanism appears to involve enhanced S uptake and reduction.
RESULTS AND DISCUSSION
Ws-2 Is More Sensitive to Selenite Than Col-0
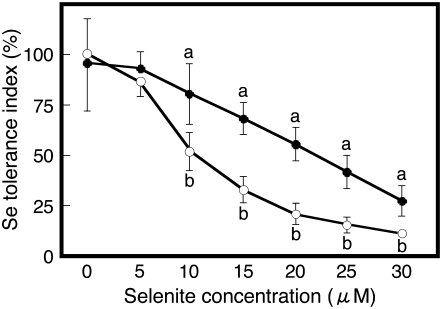
To quantify differences in selenite resistance between Col-0 and Ws-2, these accessions were grown on Murashige and Skoog medium containing different concentrations of sodium selenite for 7 d. The root growth in the absence of added Se was not different in both ecotypes (data not shown). With increasing selenite concentration in the medium, the selenite tolerance index decreased in both accessions, but the tolerance index in Ws-2 was significantly more affected than in Col-0 for the five highest selenite concentrations (Fig. 1). Thus, Ws-2 is more susceptible to selenite than Col-0. These results confirm the report by Zhang et al. (2006a) that an Arabidopsis Col accession was more resistant to selenite than Ws. The difference in tolerance index was most pronounced (2-fold) when plants were grown on medium containing 15 or 20 μm selenite, and 15 μm was chosen for all subsequent experiments.
Figure 1.
Selenite tolerance index for Col-0 (black circles) and Ws-2 (white circles). Plants were grown on control medium or on medium with various concentrations of sodium selenite for 7 d, then measured for root length. Shown are the means ± sd (n = 20). Lowercase letters indicate significant differences between Col-0 and Ws-2 for a particular selenite concentration (P < 0.05).
Sulfur Transport and Assimilation Genes Are More Induced by Selenite in Col-0 Than Ws-2
Se is chemically similar to S and known to be taken up and assimilated by plants via the same transporters and enzymes (Terry et al., 2000). Therefore, it can be expected that high levels of Se treatment prevent S uptake and assimilation, resulting in S starvation. The magnitude of S starvation induced by Se may depend on S transport and assimilation activity. To compare Col-0 and Ws-2 in this respect, macroarray analysis was used to measure the selenite-related expression of a large set of genes encoding sulfate transporters, S assimilation proteins, iron (Fe)-S cluster-related proteins, homologs of selenoproteins and Se-binding proteins, and defense-related proteins. Of the 250 genes analyzed (Supplemental Table S1), 55 genes were found to be responsive only to selenite in Col-0 but not Ws-2, using a P value of less than 0.05 and a minimal fold-change of greater than two (Table I; Supplemental Fig. S1). Among these, three encode sulfate transporters, 24 are related to S assimilation, five are Fe-S cluster related, four encode homologs of selenoprotein or Se-binding proteins, and 12 are defense-related genes (Table I). Seven genes were up-regulated only by selenite in Ws-2 but not Col-0, and 27 genes were induced in both ecotypes (Supplemental Table S1; Supplemental Fig. S1). Thus, the general trend was that genes involved in S assimilation or defense were more up-regulated by selenite treatment in Col-0 than in Ws-2.
Table I.
Genes that were more induced in Col-0 than in Ws-2 by 15 μm selenite treatment
Macroarray analysis was carried out two times (experiments 1 and 2), and each macroarray membrane contained two duplicate spots per gene. Average and sd are calculated from fold-induction from the four replicate spots. Listed are genes where the fold-induction in Col-0 was >2, the fold-induction in Ws-2 was <2, and the P value between the two accessions was <0.05.
| Gene | Annotation | MIPS Code | Col-0 (Fold-Induction)
|
Ws-2 (Fold-Induction)
|
P Value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment 1
|
Experiment 2
|
Average | sd | Experiment 1
|
Experiment 2
|
Average | sd | ||||||||
| Spot 1 | Spot 2 | Spot 1 | Spot 2 | Spot 1 | Spot 2 | Spot 1 | Spot 2 | ||||||||
| Fe-S cluster-related genes | |||||||||||||||
| APO1 | Accumulation of PSI | At1g64810 | 2.86 | 1.84 | 3.15 | 1.81 | 2.42 | 0.69 | 0.79 | 1.05 | 0.89 | 1.38 | 1.03 | 0.25 | 0.011 |
| GRXS13 | Chloroplastic glutaredoxin | At1g03850 | 2.53 | 1.96 | 3.02 | 1.90 | 2.35 | 0.53 | 1.06 | 0.91 | 1.03 | 1.09 | 1.02 | 0.08 | 0.007 |
| GRX | Chloroplastic glutaredoxin | At5g13810 | 2.66 | 2.34 | 2.52 | 1.95 | 2.37 | 0.31 | 1.63 | 1.18 | 1.79 | 1.64 | 1.56 | 0.27 | 0.004 |
| GRX | Chloroplastic glutaredoxin | At5g58530 | 2.54 | 1.63 | 4.06 | 1.43 | 2.41 | 1.20 | 0.53 | 1.05 | 0.58 | 1.33 | 0.87 | 0.38 | 0.038 |
| PSAD-2 | PSI reaction center subunit II | At1g03130 | 2.69 | 1.73 | 2.67 | 2.11 | 2.30 | 0.47 | 1.25 | 1.10 | 1.16 | 1.30 | 1.20 | 0.09 | 0.008 |
| Selenoprotein | |||||||||||||||
| SFP | Selenoprotein family protein | At1g05720 | 3.25 | 1.98 | 3.50 | 2.23 | 2.74 | 1.32 | 0.94 | 1.21 | 1.17 | 1.49 | 1.20 | 0.22 | 0.008 |
| HMT3 | Homo-Cys S-methyltransferase 3 | At3G22740 | 2.32 | 2.12 | 1.88 | 2.15 | 2.12 | 0.38 | 0.96 | 1.20 | 0.86 | 1.66 | 1.17 | 0.36 | 0.018 |
| SELT | Selenoprotein related | At3g47300 | 1.99 | 2.21 | 2.51 | 2.13 | 2.21 | 0.22 | 1.28 | 1.23 | 1.58 | 1.72 | 1.45 | 0.24 | 0.002 |
| Se-binding protein | |||||||||||||||
| SBP | Putative Se-binding protein | At5g40415 | 2.54 | 1.78 | 2.62 | 2.75 | 2.42 | 0.48 | 0.89 | 1.48 | 1.03 | 2.12 | 1.38 | 0.56 | 0.026 |
| S assimilation-related genes | |||||||||||||||
| APS1 | ATP sulfurylase 1 | At3g22890 | 2.26 | 2.30 | 3.00 | 2.06 | 2.41 | 0.41 | 0.87 | 1.02 | 1.18 | 1.56 | 1.16 | 0.30 | 0.002 |
| APS2 | ATP sulfurylase 2 | At1g19920 | 2.36 | 2.92 | 2.44 | 2.72 | 2.61 | 0.26 | 0.75 | 1.16 | 0.96 | 1.93 | 1.20 | 0.52 | 0.003 |
| APS4 | ATP sulfurylase 4 | At5g43780 | 2.94 | 1.50 | 3.26 | 1.52 | 2.31 | 0.93 | 1.08 | 1.03 | 0.96 | 1.55 | 1.15 | 0.27 | 0.042 |
| APR1 | 5′-Adenylylsulfate reductase 1 | At4g04610 | 2.38 | 2.20 | 2.37 | 2.29 | 2.31 | 0.08 | 0.47 | 0.58 | 0.57 | 0.83 | 0.61 | 0.15 | 0.000 |
| APR2 | 5′-Adenylylsulfate reductase 2 | At1g62180 | 2.41 | 2.33 | 2.91 | 2.68 | 2.58 | 0.26 | 0.70 | 0.65 | 0.76 | 1.12 | 0.81 | 0.21 | 0.000 |
| APR3 | 5′-Adenylylsulfate reductase 3 | At4g21990 | 1.48 | 2.15 | 2.11 | 2.70 | 2.11 | 0.50 | 0.39 | 0.94 | 0.62 | 1.04 | 0.75 | 0.30 | 0.003 |
| SIR | Sulfite reductase | At5g04590 | 2.18 | 2.84 | 2.95 | 1.89 | 2.47 | 0.51 | 0.74 | 0.97 | 0.94 | 1.43 | 1.02 | 0.29 | 0.003 |
| SAT1 | Ser O-acetyltransferase 1 | At1g55920 | 2.12 | 1.90 | 2.50 | 2.14 | 2.16 | 0.25 | 1.31 | 1.28 | 1.49 | 1.57 | 1.41 | 0.14 | 0.002 |
| SAT3 | Ser O-acetyltransferase 3 | At3g13110 | 2.69 | 2.42 | 3.96 | 2.14 | 2.80 | 0.80 | 0.89 | 0.90 | 0.98 | 1.45 | 1.06 | 0.26 | 0.009 |
| SAT52 | Ser O-acetyltransferase 52 | At5g56760 | 4.10 | 2.15 | 4.61 | 2.46 | 3.33 | 1.21 | 1.26 | 1.08 | 1.46 | 1.91 | 1.43 | 0.36 | 0.023 |
| SAT106 | Ser O-acetyltransferase 106 | At2g17640 | 1.57 | 2.30 | 1.96 | 2.69 | 2.13 | 0.48 | 1.15 | 1.06 | 1.01 | 1.65 | 1.22 | 0.30 | 0.011 |
| CYSD1 | Cys synthase | At3g04940 | 1.99 | 2.50 | 2.87 | 2.67 | 2.51 | 0.38 | 1.36 | 1.22 | 1.28 | 1.53 | 1.35 | 0.13 | 0.003 |
| CYSD2 | Cys synthase | At5g28020 | 2.41 | 1.40 | 3.16 | 1.82 | 2.20 | 0.76 | 1.02 | 1.25 | 1.01 | 1.35 | 1.16 | 0.17 | 0.035 |
| CYSC1 | Encodes a Cys synthase isomer | At3g61440 | 1.95 | 3.07 | 3.10 | 2.41 | 2.63 | 0.56 | 1.25 | 1.31 | 1.39 | 1.55 | 1.37 | 0.13 | 0.009 |
| OAS-TL | O-acetyl-Ser (thiol) lyase | At2g43750 | 2.13 | 1.97 | 2.95 | 2.02 | 2.27 | 0.46 | 1.00 | 1.38 | 1.09 | 1.82 | 1.32 | 0.37 | 0.010 |
| GSH1 | γ-Glutamyl-Cys synthetase | At4g23100 | 2.10 | 2.64 | 3.24 | 2.54 | 2.63 | 0.47 | 0.79 | 0.96 | 0.80 | 1.21 | 0.94 | 0.20 | 0.001 |
| GSH2 | Glutathione synthetase | At5g27380 | 2.49 | 2.62 | 3.04 | 2.57 | 2.68 | 0.25 | 0.85 | 0.86 | 1.06 | 0.77 | 0.88 | 0.12 | 0.000 |
| VTC4 | 3′(2′),5′-bisphosphatenucleotidase | At5g09290 | 1.88 | 2.35 | 2.37 | 2.26 | 2.22 | 0.23 | 1.15 | 1.20 | 1.13 | 1.93 | 1.35 | 0.39 | 0.006 |
| ISU1 | Fe-S cluster assembly complex protein | At4g22220 | 2.67 | 2.36 | 3.88 | 2.28 | 2.80 | 0.74 | 1.22 | 1.16 | 1.48 | 1.30 | 1.29 | 0.14 | 0.012 |
| CPNIFS | Similar to nitrogen fixation protein | At1g08490 | 2.31 | 1.70 | 2.78 | 2.13 | 2.23 | 0.45 | 1.21 | 1.19 | 1.15 | 1.93 | 1.37 | 0.37 | 0.013 |
| NFU1 | Nitrogen fixation NifU-like family protein | At4g01940 | 2.53 | 2.25 | 2.96 | 2.13 | 2.47 | 0.37 | 1.20 | 0.93 | 1.24 | 1.49 | 1.21 | 0.23 | 0.001 |
| AtMtNlfS | Cys desulfurase | At5g65720 | 1.85 | 2.52 | 2.04 | 2.07 | 2.12 | 0.29 | 1.15 | 1.41 | 1.36 | 1.90 | 1.46 | 0.32 | 0.011 |
| NIFSL | Chloroplastic NifS-like protein | At1g18490 | 1.63 | 1.94 | 2.39 | 2.09 | 2.01 | 0.32 | 1.04 | 1.10 | 1.23 | 1.88 | 1.31 | 0.39 | 0.016 |
| ATMST1 | Mercaptopyruvate sulfurtransferase 1 | At1g79230 | 2.82 | 1.47 | 2.99 | 1.96 | 2.31 | 0.72 | 1.17 | 1.67 | 1.02 | 1.74 | 1.40 | 0.36 | 0.040 |
| Sulfur transporter | |||||||||||||||
| Sultr2;2 | Sulfate transporter | At5g10180 | 2.22 | 1.45 | 1.20 | 1.51 | 1.59 | 0.44 | 0.88 | 0.93 | 0.91 | 1.44 | 1.04 | 0.26 | 0.042 |
| Sultr3;1 | Sulfate transporter | At3g51895 | 2.06 | 1.78 | 2.64 | 1.96 | 2.11 | 0.37 | 0.60 | 0.74 | 0.58 | 1.03 | 0.74 | 0.21 | 0.001 |
| Sultr3;5 | Sulfate transporter | AT5g19600 | 2.23 | 1.39 | 2.55 | 2.00 | 2.04 | 0.49 | 1.11 | 1.60 | 1.31 | 1.14 | 1.29 | 0.23 | 0.023 |
| Defense and hormone biosynthesis/signaling genes | |||||||||||||||
| ACS6 | 1-Aminocyclopropane-1-carboxylate synthase 6 | At4g11280 | 5.41 | 4.40 | 5.14 | 2.24 | 4.30 | 1.43 | 1.63 | 1.49 | 1.56 | 1.05 | 1.43 | 0.26 | 0.013 |
| SAM1 | S-adenosyl-Met synthetase 1 | At1g02500 | 4.45 | 4.23 | 5.37 | 2.62 | 4.17 | 1.14 | 0.92 | 2.46 | 0.86 | 2.60 | 1.71 | 0.95 | 0.009 |
| ERF1 | Ethylene response factor 1 | At3g23240 | 2.87 | 2.70 | 3.31 | 1.81 | 2.67 | 0.63 | 1.53 | 1.83 | 1.56 | 1.85 | 1.69 | 0.17 | 0.024 |
| PR4 | Pathogenesis-related protein 4 | At3g04720 | 3.05 | 2.67 | 4.18 | 2.15 | 3.01 | 1.37 | 0.89 | 1.24 | 0.94 | 1.42 | 1.17 | 0.72 | 0.009 |
| PDF1.2 | Plant defensin 1.2 | At5g44420 | 3.33 | 4.54 | 3.43 | 2.74 | 3.51 | 0.75 | 0.94 | 1.52 | 1.06 | 1.67 | 1.30 | 0.35 | 0.002 |
| LOX2 | Lipooxigenase 2 | At3g45140 | 2.84 | 2.70 | 3.14 | 2.30 | 2.24 | 0.89 | 0.72 | 1.36 | 0.56 | 2.31 | 1.17 | 1.33 | 0.012 |
| AOS | Allene oxide synthase | At5g42650 | 3.45 | 2.25 | 3.14 | 1.55 | 2.60 | 0.86 | 0.86 | 1.75 | 1.18 | 0.81 | 1.15 | 0.43 | 0.018 |
| VSP1 | Vegetative storage protein 1 | At5g24780 | 3.94 | 2.29 | 2.96 | 1.26 | 2.61 | 0.63 | 1.04 | 1.28 | 1.08 | 1.27 | 1.17 | 0.12 | 0.041 |
| PIN2 | Proteinase inhibitor 2 | At2g02100 | 2.32 | 2.35 | 2.16 | 1.96 | 2.20 | 0.18 | 1.16 | 1.32 | 1.20 | 1.35 | 1.25 | 0.09 | 0.000 |
| JR | JA-responsive gene | At3g16470 | 2.79 | 2.00 | 3.31 | 1.40 | 2.37 | 0.85 | 0.71 | 1.86 | 0.80 | 1.73 | 1.28 | 0.61 | 0.042 |
| BAH1 | Benzoic acid hypersensitive 1 | At1g02860 | 3.00 | 3.94 | 3.98 | 3.25 | 3.54 | 0.49 | 0.94 | 1.44 | 1.35 | 1.59 | 1.33 | 0.27 | 0.000 |
| MPK6 | MAP kinase 6 | At2g43790 | 2.44 | 2.37 | 4.43 | 3.14 | 3.10 | 0.96 | 0.79 | 1.24 | 0.51 | 1.24 | 0.94 | 0.36 | 0.007 |
| Others | |||||||||||||||
| CA1 | Carbonic anhydrase 1 | At3g01500 | 2.40 | 1.22 | 3.07 | 1.37 | 2.01 | 0.88 | 0.80 | 0.84 | 0.81 | 1.08 | 0.88 | 0.13 | 0.040 |
| PE3 | Pectinesterase family protein | At5g04960 | 2.34 | 1.71 | 3.50 | 2.31 | 2.46 | 0.75 | 1.02 | 1.45 | 1.13 | 1.59 | 1.30 | 0.27 | 0.023 |
| SAUR | Auxin-responsive family protein | At2g46690 | 2.41 | 2.13 | 2.61 | 1.89 | 2.26 | 0.32 | 0.98 | 0.76 | 1.00 | 1.45 | 1.05 | 0.29 | 0.001 |
| APX1 | Cytosolic ascorbate peroxidase | At1g07890 | 2.72 | 3.41 | 2.87 | 3.03 | 3.01 | 0.30 | 0.73 | 0.90 | 0.86 | 1.16 | 0.91 | 0.18 | 0.000 |
| APX4 | Ascorbate peroxidase, thylakoid-bound (tAPX) | At1g77490 | 3.78 | 2.09 | 3.08 | 1.74 | 2.67 | 0.93 | 1.23 | 1.80 | 1.10 | 1.98 | 1.53 | 0.43 | 0.043 |
| cytDHAR | Dehydroascorbate reductase, cytosol | At1g75270 | 2.10 | 2.68 | 2.99 | 1.52 | 2.32 | 0.65 | 1.04 | 1.46 | 0.92 | 1.38 | 1.20 | 0.26 | 0.017 |
| MSD1 | Mitochondrial, similar to MnSOD | At3g10920 | 1.64 | 2.83 | 1.62 | 2.28 | 2.09 | 0.58 | 0.78 | 1.69 | 1.10 | 1.75 | 1.33 | 0.47 | 0.045 |
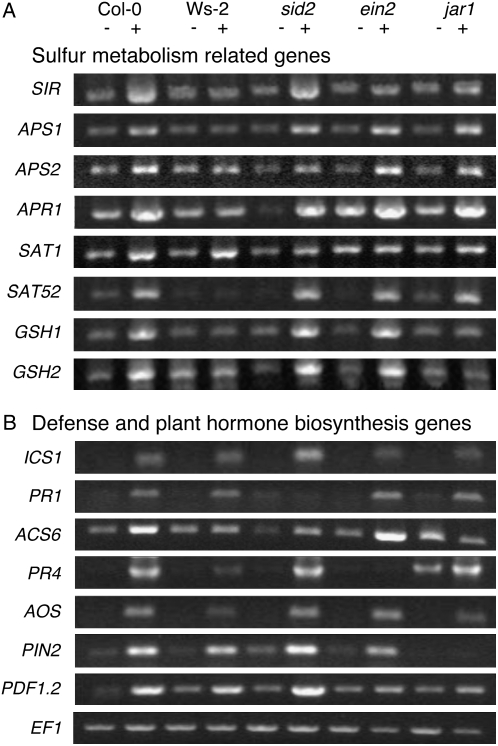
Genes specifically up-regulated in the selenite-resistant Col-0 accession but not in selenite-sensitive Ws-2 included three sulfate transporters (Sultr2;2, Sultr3;1, and Sultr3;5), three ATP sulfurylases (APS1, APS2, and APS4), three 5′-adenylylsulfate reductases (APR1, APR2, and APR3), a sulfite reductase (SIR), four Ser O-acetyltransferases (SAT1, SAT3, SAT52, and SAT106), three Cys synthases (CYSD1, CYSD2, and CYSC1), all involved in sulfate to Cys assimilation, and two glutathione biosynthesis genes encoding γ-glutamyl-Cys synthetase (GSH1) and glutathione synthetase (GSH2; Table I). The specific induction by selenite of several of these genes in Col-0 plants but not Ws-2 was further investigated by determining their expression profiles using semiquantitative reverse transcription (RT)-PCR. The mRNA levels of SIR, APS1, APR1, SAT52, GSH1, and GSH2 again increased only with selenite treatment in Col-0 but not in Ws-2 (Fig. 2A). Induction of APR2 and SAT1 with selenite in Ws-2 was lower than in Col-0 (Fig. 2A). Therefore, the results from the macroarray and semiquantitative RT-PCR approaches both indicate that the expression of several key genes involved in S uptake and assimilation are more enhanced upon selenite treatment in Col-0 than in Ws-2. In this context, it is interesting to note that both APS1 and a SAT gene (SAT1) are located in the chromosome 3 quantitative trait loci region that was shown earlier to be associated with selenate tolerance in Col (Zhang et al., 2006b). Coordinated induction of S uptake and assimilation genes, including Sultr2;1, APR2, APR3, and GSH1, has also been observed in S-starved plants (for review, see Hirai and Saito, 2004), suggesting that part of the selenite responses observed in this experiment could be induced by S deficiency. However, the selenite-induced genes encoding homologs of selenoproteins (At1g05720, At3G22740, and At3g47300) and a Se-binding protein (At5g40415) were not induced by S deficiency. Thus, the induction of some genes by selenite treatment may occur via S starvation, while others may be induced in a different, selenite-specific response.
Figure 2.
Semiquantitative RT-PCR confirms the gene expression patterns identified in macroarray analysis. A, Induction of sulfur metabolism-related genes in Col-0, Ws-2, sid2, ein2, and jar1 grown without (−) and with (+) 15 μm selenite. B, Induction of defense and plant hormone biosynthesis-related genes in 7-d-old Col-0, Ws-2, sid2, ein2, and jar1 plants grown without (−) and with (+) 15 μm selenite. EF1 is used as control for RT-PCR.
Levels of Selenite-Induced Expression in Ethylene- and JA-Modulated Genes Are Higher in Col-0 Than in Ws-2
Earlier studies have shown that S deficiency induces the expression of 12-oxophytodienoate reductase, involved in JA biosynthesis (Hirai et al., 2003; Nikiforova et al., 2003). JA is known to be a plant hormone whose production is induced by various environmental stresses (Pieterse and van Loon, 1999). In addition to JA, salicylic acid (SA) and ethylene are also known as stress-inducible phytohormones (Nürnberger and Scheel, 2001). To study the involvement of JA, SA, and ethylene in selenite resistance, the selenite-related expression of genes either involved in the biosynthesis of these phytohormones or known to be responsive to these hormones was analyzed in the two accessions using macroarrays. The selenite-induced expression of all SA-related genes tested, i.e. the biosynthetically involved isochorismate synthase 1 (ICS1) and Phe ammonia-lyase 2 (PAL2), and the SA-responsive pathogenesis-related protein 1 (PR1), PR2, PR5, enhanced disease susceptibility 1 (EDS1), and EDS5 were all similar in Col-0 and Ws-2 (Supplemental Table S1). In contrast, the induction levels of genes known to be involved in ethylene or JA biosynthesis, i.e. 1-aminocyclopropane-1-carboxylate synthase 6 (ACS6), S-adenosyl-Met synthetase 1 (SAM1), lipooxigenase 2 (LOX2), and allene oxide synthase (AOS), were remarkably higher in Col-0 than in Ws-2 (Table I). Moreover, induction of ethylene response factor 1 (ERF1), PR4, plant defensin 1.2 (PDF1.2), vegetative storage protein 1 (VSP1), proteinase inhibitor 2 (PIN2), and JA-responsive gene (JR), which are responsive to ethylene and/or JA, was also significantly more pronounced in Col-0 (Table I). To confirm the selenite-induced gene expression observed from macroarray experiments with an independent experimental approach and biological replicate, semiquantitative RT-PCR was performed for selected genes, which were indeed significantly more induced in Col-0 than in Ws-2 by the selenite treatment. Consistent with the macroarray data, the expression of genes ICS1 and PR1 was increased in both accessions, and the expression of genes ACS6, PR4, AOS, PIN2, and PDF1.2 was more induced by selenite in Col-0 than in Ws-2 (Fig. 2B). The expression of PR4 is known to be induced by ethylene (Lawton et al., 1994) and widely used as a good marker for ethylene signaling. Induction of the PIN2 gene is JA dependent (Farmer et al., 1992), and concomitant triggering of the ethylene and JA pathways is required for PDF1.2 induction (Penninckx et al., 1998). ACS is the rate-limiting enzyme and governs the major regulatory step in stress-induced ethylene production (Yang and Hoffman, 1984; Bleecker and Kende, 2000), and AOS is also a key enzyme in JA synthesis (Stenzel et al., 2004). Together, these results show that ethylene and JA biosynthesis and/or signaling are induced by selenite in Col-0 but much less in Ws-2.
Induction of S Assimilation Genes by Selenite Is Repressed in Ethylene- and JA-Signaling Mutants
To further analyze the importance of JA, SA, and ethylene in selenite-induced gene expression, we investigated whether genes up-regulated by selenite in Col-0 but not Ws-2 were still up-regulated by selenite in Col-0 mutants impaired in the biosynthesis or signaling of these plant hormones: sid2 (lacking SA due to the mutation of the biosynthetic ICS1 gene; Wildermuth et al., 2001), ein2 (completely lacking ethylene signaling; Guzmán and Ecker, 1990), and jar1 (deficient for JA signaling; Staswick et al., 1992). To verify the relevance of these mutants, we first checked the induction of genes that are induced by the phytohormones in question. As expected, the expression of PR1, induced by SA, was completely absent in the sid2 mutant (Fig. 2B). Furthermore, expression of PR4, the marker for ethylene signaling, was not identified in the ein2 mutant, and the JA-inducible PIN2 transcript was not observed in the jar1 mutant. Expression of PDF1.2 is regulated by ethylene and JA synergistically, and induction of this gene was moderately suppressed in ein2 and jar1 mutants. These results indicate that SA biosynthesis and ethylene and JA signaling are indeed impaired in sid2, ein2, and jar1 mutants, respectively.
These same mutants were tested for the induction of selected S-related genes by selenite using semiquantitative RT-PCR. The induction of gene expression of SIR and SAT1 by selenite was suppressed in both ein2 and jar1 mutants but not in sid2 mutants (Fig. 2A). Selenite induction of GSH1 and GSH2 expression was also inhibited in the jar1 mutant and not in ein2 or sid2 mutants. The selenite-related APS, APR, and SAT52 induction was not affected in the mutants. Together, these observations suggest that ethylene and JA act as signal molecules in the selenite-mediated induction of gene expression of some, but not all, S assimilation genes; SA does not appear to act as a signal.
Ethylene and JA Are Important Factors in the Difference in Selenite Resistance between Col-0 and Ws-2
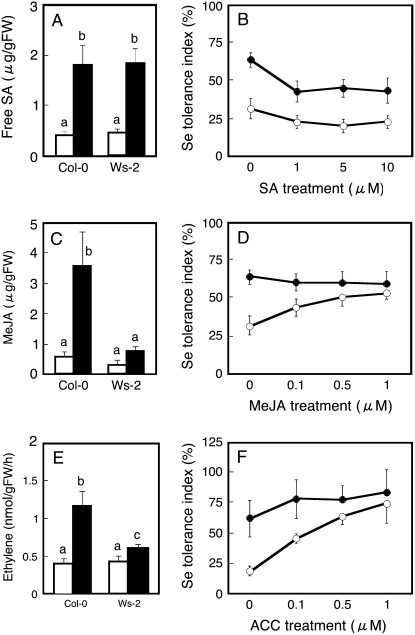
The observation that the expression of known ethylene- and JA-inducible genes was more extensively enhanced by selenite in Col-0 than in Ws-2 may suggest that Col-0 generates higher levels of these phytohormones and/or is more sensitive to these hormones than Ws-2 under selenite treatment. To test these hypotheses, first the levels of these plant hormones were measured in Col-0 and Ws-2 plants treated with or without selenite. Free SA accumulation was similar in both accessions and increased 4-fold with selenite treatment in both (Fig. 3A). Interestingly, the shoot concentration of methyl jasmonate (MeJA) was approximately 2-fold higher in Col-0 than in Ws-2 (Fig. 3C) and also increased more with selenite treatment in Col-0 (7-fold) than in Ws-2 (2-fold). Ethylene generation from seedlings grown without selenite was not different in both accessions but increased more with selenite treatment in Col-0 (3-fold) than in Ws-2 (1.4-fold; Fig. 3E). Thus, the observed higher level of production of ethylene and JA in selenite-treated Col-0 in comparison with Ws-2 is in agreement with the observed higher level of induction of genes that contribute to ethylene (ACS6 and SAM1) and JA biosynthesis (AOS and LOX2; Table I; Fig. 2B). Also, the observed increase in SA levels upon selenite treatment, to a similar extent in Col-0 and Ws-2, corresponds well with the observed ICS1 and PAL2 expression levels.
Figure 3.
Effect of selenite supply on endogenous SA (A), MeJA (C), and ethylene (E) content in Col-0 and Ws-2, and effect on selenite tolerance index of exogenous supply with SA (B), MeJA (D), and ACC (F). The plants shown in A, C, and E were grown for 7 d without (white bars) or with (black bars) treatment with 15 μm selenite. Values are means ± sd from three replicate samples, each consisting of several seedlings. For the results shown in B, D, and F, Col-0 (black circles) and Ws-2 (white circles) plants were grown on control medium or on medium with 15 μm selenite that also contained various concentrations of SA (B), MeJA (D), or ACC (F). After 7 d of growth, root length was measured and selenite resistance index was calculated. Values are means ± sd (n = 20). Different lowercase letters above each bar denote significant differences (P < 0.05).
The above results indicate that increased ethylene and JA signaling, caused by production of ethylene and JA by selenite treatment, correlates with enhanced selenite resistance in Col-0. Alternatively, Col-0 may be more sensitive to ethylene and/or JA than Ws-2. To compare phytohormone sensitivity between the two accessions, we examined the SA-, ethylene-, and JA-induced inhibition of root growth in seedlings of Col-0 and Ws-2. Note that rather than ethylene gas, its precursor, 1-aminocyclopropane-1-carboxylate (ACC), was supplied. As shown in Supplemental Figure S2, the degree of inhibition was not statistically different between Col-0 and Ws-2 (approximately 20% and 45% inhibition at 1 and 10 μm SA; approximately 20% and 32% inhibition at 1 and 0.1 μm MeJA; approximately 15% and 42% inhibition at 0.1 and 1 μm ACC, respectively). This suggests that the enhanced ethylene and JA signaling in Col-0 was not due to higher ethylene or JA sensitivity.
To assess whether the selenite resistance of the sensitive accession Ws-2 is limited by its lower ethylene or JA concentration, we next tested whether selenite sensitivity could be mitigated via external supply with MeJA or ACC. Indeed, the selenite tolerance index of Ws-2 increased with increasing MeJA content in the medium (Fig. 3D). As a result, no significant difference in selenite tolerance index was observed any more between Col-0 and Ws-2 when grown on 0.5 or 1 μm MeJA (Fig. 3D). Treatment with ACC resulted in a similar trend. Accession Ws-2 showed lower resistance to selenite than Col-0 without ACC, but its selenite tolerance index increased with increasing ACC content in the media, and it became the same as Col-0 when the plants were grown at or above 0.5 μm ACC (Fig. 3F). In contrast, growing the plants on a series of different concentrations of SA had no positive effect on the selenite tolerance index of Ws-2 (Fig. 3B); Col-0 was even significantly inhibited by external SA (Fig. 3B).
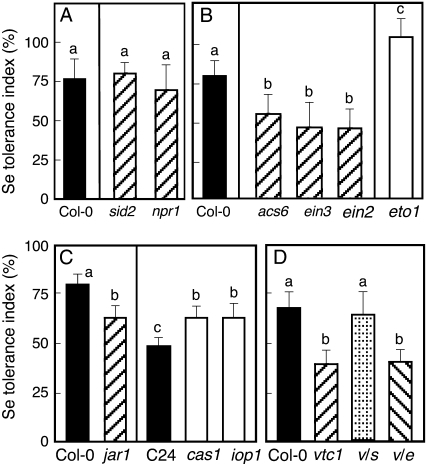
The importance of SA, ethylene, and JA for acquisition of selenite resistance in Arabidopsis was also investigated using mutants with defects in phytohormone biosynthesis or signaling. Mutants incapable of SA production (sid2) or signaling (npr1; Cao et al., 1997) showed no statistical difference in selenite tolerance index in comparison to their wild type, Col-0 (Fig. 4A). In contrast, selenite resistance in mutants defective in ethylene production (acs6) or signaling (ein3 and ein2) was less than that in wild-type Col-0 (Fig. 4B). Furthermore, an ethylene-overproducing mutant in the Col-0 background, eto1, showed higher resistance than wild-type Col-0 (Fig. 4B). Selenite resistance in a mutant deficient for JA signaling, jar1, was significantly lower than in Col-0 (Fig. 4C). Conversely, JA-hypersensitive mutant iop1 (Penninckx et al., 2003) and the constitutive JA-producing mutant cas1 (Kubigsteltig and Weiler, 2003) were more tolerant to selenite than their wild type, C24 (Fig. 4C). Taken together, these results indicate that the selenite susceptibility of Ws-2 is highly dependent on ethylene and JA biosynthesis, while there is no evidence of SA involvement.
Figure 4.
Selenite tolerance index for SA-, ethylene-, and JA-signaling and -biosynthesis mutants in comparison with their respective wild-type controls. A, SA-biosynthesis (sid2) and SA-signaling (npr1) mutants in Col-0 background. B, Ethylene-biosynthesis (acs6), ethylene-signaling (ein3 and ein2), and ethylene-overproducing (eto1) mutants in Col-0 background. C, JA-signaling (jar1) mutant in Col-0 background, and JA-overproducer (cas1) and hypersensitive for JA (iop1) mutants in C24 background. D, AsA-biosynthesis (vtc1) mutant and double mutants v/s (vtc1 and sid2) and v/e (vtc1 and ein2) in Col-0 background. Shown values represent mean of tolerance index ± sd (n = 20). Different lowercase letters above each bar denote significant differences (P < 0.05).
In view of the finding that both ethylene and JA appear to play important roles in selenite resistance in Arabidopsis, it is interesting to note that in earlier studies JA was implicated to be involved in the regulation of S metabolism because S starvation induced genes involved in JA synthesis, as well as JA-responsive genes (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003). The above-mentioned studies showed no indication that ethylene plays a role in S deficiency responses. However, a recent comprehensive gene expression analysis showed that transcripts regulating ethylene synthesis (ACS6) and signaling (ERF) were up-regulated by selenate treatment, and plants overexpressing ERF1 exhibited an increase in selenate resistance (Van Hoewyk et al., 2008). These results indicate that Se resistance achieved through ethylene signaling is not mediated by S starvation resulting from the Se treatment but is a Se-specific response.
Sulfur Assimilation Is More Up-Regulated by Selenite Treatment in Col-0 Than Ws-2 Affecting Levels of S Metabolites
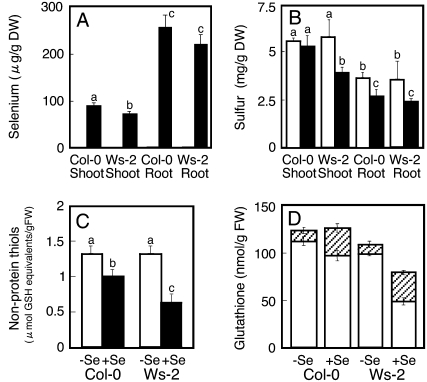
As described above, several S transport and assimilation genes were up-regulated only by selenite in Col-0 and not in Ws-2. To investigate whether this affected Se and S uptake differently, total Se and S concentrations were measured in both accessions. When grown on 15 μm selenite, the shoot and root Se levels were approximately 10% higher in Col-0 than Ws-2 (P < 0.05 for shoot only; Fig. 5A). Shoot and root total S levels were the same in Col-0 and Ws-2 under control conditions (Fig. 5B). When treated with selenite, Ws-2 showed reduced shoot and root S levels compared to control conditions (Fig. 5B). In Col-0 roots, total S levels were also lower in selenite-treated plants than under control conditions, but Col-0 shoot S levels were not affected by selenite (Fig. 5B). As a consequence, on selenite the shoot S levels were approximately 50% higher in Col-0 than in Ws-2 plants (Fig. 5B).
Figure 5.
Se, S, nonprotein thiols, and glutathione content in Arabidopsis. Col-0 and Ws-2 plants were grown on media without (white bars) and with (black bars) 15 μm selenite. For results shown in A and B, seedlings were treated with Se for 1 month and Se (A) and S (B) were analyzed in shoot and root tissues. For results shown in C and D, seedlings were treated with Se for 7 d, and shoot tissues were analyzed for nonprotein thiols (C) and reduced (white bars) and oxidized (hatched bars) glutathione (D). Values are means ± sd from three replicate samples, each consisting of several seedlings. Different lowercase letters above each bar denote significant differences (P < 0.05).
It is known that reduced organic S metabolites compose a large fraction of the S pool (Nikiforova et al., 2006). To determine whether levels of reduced organic S metabolites are affected by selenite treatment, the levels of nonprotein thiols were measured in shoot tissue. Under control conditions, both accessions contained the same nonprotein thiol level (Fig. 5C). When grown with 15 μm selenite, the nonprotein thiol levels were reduced in both accessions compared to control conditions (Fig. 5C). However, the magnitude of this reduction in nonprotein thiol level was significantly higher in Ws-2 than in Col-0. As a result, selenite-treated Col-0 plants contained approximately 65% higher nonprotein thiol levels than Ws-2 plants on the same medium.
A large fraction of the nonprotein thiol pool consists of glutathione, which is involved in regulating the redox state of the cell as well as the storage and transport of reduced S in plants (Nikiforova et al., 2006). While the level of reduced glutathione (GSH) was not affected by selenite treatment in Col-0, it was nearly 2-fold lower in selenite-treated Ws-2 compared to control conditions and to selenite-treated Col-0 plants (Fig. 5D, white bars). The level of oxidized glutathione (GSSG) doubled with selenite treatment in both accessions (Fig. 5D, hatched bars); no significant difference in GSSG content was found between Col-0 and Ws-2 grown either without or with selenite. Therefore, it appears that S uptake is inhibited by selenite and that Col-0 is better able to maintain S status in the presence of selenite than Ws-2. Col-0 appears to also be better able to maintain a sufficiently large pool of reduced S compounds, especially glutathione, when treated with selenite, which may in part be responsible for its resistance. Incidentally, a reduction in GSH content was also observed in S-starved Arabidopsis (Hirai et al., 2003), indicating that selenite treatment induced S starvation in Ws-2 plants.
Optimal Levels of ROS Are Necessary for Selenite Resistance
From the experiments described above, it appears that ethylene and JA play important roles in selenite resistance in Arabidopsis. Ethylene and JA are known to be required for plant defense responses such as pathogen attack, drought stress, and resistance to air pollutants (Dong, 1998; Zhu, 2002; Overmyer et al., 2003). During defense responses to these environmental stresses, generation of ROS often precedes ethylene and JA production (Dong, 1998; Overmyer et al., 2003). Optimal levels of ROS act as signal molecules that activate defense responses (Overmyer et al., 2003). To obtain further insight into the biochemical mechanisms underlying the observed difference in selenite resistance between Col-0 and Ws-2, we compared ROS generation in these accessions.
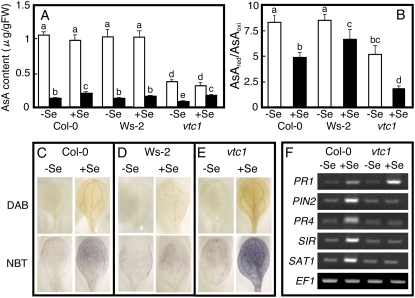
Before ROS detection, we measured reduced (AsA) and oxidized (DHA) ascorbic acid content in the plants because AsA is the major antioxidant molecule in plants (Smirnoff et al., 2001). The AsA content was similar in the two accessions and not affected by selenite treatment (Fig. 6A). The level of DHA was also similar in both accessions under control conditions but increased significantly in Col-0 when plants were treated with selenite, in contrast to Ws-2 (Fig. 6A). The vtc1 mutant (Conklin et al., 1996) that is impaired in AsA production was also used in this experiment. As expected, the vtc1 mutant contained significantly lower levels of AsA compared to the wild type (Col-0), which were not affected by selenite treatment; the DHA level was also reduced under control conditions but increased with selenite treatment, similar to the Col-0 wild type (Fig. 6A). The AsA redox state was lower (i.e. more oxidized) in the presence of selenite in all plant types (Fig. 6B). Under control conditions, the AsA redox state was similar in Col-0 and Ws-2, but due to the higher DHA level in selenite-treated Col-0, the AsA redox state was significantly lower in Col-0 than in Ws-2 on selenite medium (Fig. 6B). The AsA redox state in the vtc1 mutant was significantly lower than in Col-0 and Ws-2, both with and without selenite.
Figure 6.
Effect of selenite treatment on the AsA content, redox state, and generation of ROS. A, Shoot AsA content. White bars, Reduced form of AsA; black bars, oxidized form of AsA. Values are means ± sd from three replications. B, AsA redox state, calculated as reduced/oxidized AsA. Col-0 (C), Ws-2 (D), and vtc1 (E) plants were grown for 7 d on media with or without 15 μm selenite. In situ detection of the ROS hydrogen peroxide and superoxide in leaves was carried out with DAB and NBT staining, respectively. The presence of the brown precipitate and the purple formazan precipitate indicates the location of hydrogen peroxide and superoxide, respectively. F, Induction of selected genes in 7-d-old Col-0 and vtc1 plants grown without and with 15 μm selenite. Values are means ± sd from three replicate samples, each consisting of several seedlings. Different lowercase letters above each bar denote significant differences (P < 0.05).
To assay cellular hydrogen peroxide and superoxide accumulation, we performed in situ ROS detection as shown in Figure 6, C to E. The top row shows the accumulation of hydrogen peroxide in Col-0 (C), Ws-2 (D), and vtc1 (E). Hydrogen peroxide is visualized in situ as a reddish-brown precipitate, as 3,3′-diaminobenzidine (DAB) polymerizes on contact with hydrogen peroxide in a reaction requiring peroxidase (Torres et al., 2002). Brown precipitates were observed in selenite-treated Col-0, Ws-2, and vtc1 leaves, but almost no stain was detected in untreated plants. The density of brown precipitates in selenite-treated Col-0 appears higher than that in Ws-2 (Fig. 6, A and B). The leaves of the vtc1 mutant were stained more than Col-0 and Ws-2, likely due to its lack of AsA and low ROS-scavenging ability (Fig. 6B). The bottom row shows the accumulation of superoxide in Col-0 (C), Ws-2 (D), and vtc1 (E), monitored in situ via the precipitation of purple formazan from the reaction of nitro blue tetrazolium (NBT) with superoxide. Similar to the DAB staining, selenite-treated plants showed more superoxide than control plants, and Col-0 accumulated more superoxide than Ws-2; as expected, a high level of superoxide was also detected in the selenite-treated vtc1 mutant. Thus, selenite treatment resulted in the formation of hydrogen peroxide and superoxide, and this response was more pronounced in Col-0 than Ws-2.
The observation that the selenite-resistant accession Col-0 generates more ROS than selenite-sensitive Ws-2 when plants are treated with selenite may suggest that the generation of ROS is important for acquisition of selenite resistance. If this is the case, the vtc1 mutant that generates higher levels of ROS than Col-0 may also show enhanced selenite resistance. However, the vtc1 mutant actually showed lower selenite resistance than Col-0 (Fig. 4D). It has been reported that extreme levels of ROS can trigger the production of SA that in turn leads to more ROS via a self-amplifying loop, activating the oxidative cell death cycle (Overmyer et al., 2003; Kangasjärvi et al., 2005). Indeed, a previous study showed that the vtc1 mutant produces high levels of SA after inoculation with a pathogen (Barth et al., 2004). To assess whether a high level of SA was also present in the selenite-treated vtc1 mutant, we carried out semiquantitative RT-PCR to evaluate the expression of the PR1 gene, which encodes the SA-responsive PR1, which is frequently used as a marker for SA signaling (Cao et al., 1997). Selenite treatment enhanced PR1 expression in Col-0 and vtc1, but induction of PR1 in vtc1 was higher than in Col-0 (Fig. 6F), suggesting that selenite induces SA production and that this effect is more pronounced in the vtc1 mutant, perhaps because of its higher ROS levels. To further investigate whether the reduced selenite resistance of the vtc1 mutant is due to higher levels of selenite-induced SA production, we tested the selenite resistance of two double mutants: a vtc1/ein2 mutant that lacks AsA biosynthesis and ethylene signaling, and a vtc1/sid2 mutant lacking AsA and SA biosynthesis. The selenite resistance in the vtc1/ein2 mutant was not different from the vtc1 mutant, but the selenite resistance in the vtc1/sid2 mutant was restored to the level of Col-0 (Fig. 4D). The finding that the vtc1 mutant, which produces more SA when treated with selenite, is more selenite sensitive but if SA production is knocked out in vtc1/sid2 selenite then resistance is restored, suggests that SA inhibits the acquisition of selenite resistance. Indeed, we show that treatment of SA in Col-0 inhibited selenite resistance (see Fig. 3B). The next intriguing question is how the SA inhibits selenite resistance in plants. Our results indicate SA may inhibit JA and/or ethylene signaling because we observed that selenite induction of PR4 (a marker for ethylene signaling) and PIN2 (a marker for JA signaling) in Col-0 was completely repressed in the vtc1 mutant (Fig. 6F). Inhibition of JA and/or ethylene signaling by SA was also observed earlier in plants suffering biotic and abiotic stresses (Pena-Cortes et al., 1993; Doares et al., 1995; Berrocal-Lobo et al., 2002). Therefore, it appears that in the selenite-treated vtc1 mutant, an increase in SA production inhibited JA and ethylene signaling, leading to impaired Se resistance. Indeed, the vtc1 mutant showed no selenite-related induction of SIR and SAT1, whose expression is regulated by JA and/or ethylene (see Figs. 2A and 6F).
Taken together, our results suggest that an excess level of ROS production such as in the vtc1 mutant leads to a high level of SA, which inhibits JA and ethylene signaling, thereby impeding S assimilation and selenite resistance. However, a low ROS response such as in Ws-2 appears to be associated with a low Se resistance as well. Thus, an optimal level of ROS may be needed to acquire selenite resistance. Recently, ROS induction upon selenite treatment was also observed in a cell suspension of coffee (Gomes et al., 2007). The cellular mechanisms regulating ROS production in response to Se in plants are not clear at this point and will require further study. In this context, it is interesting to note that Zhang et al. (2006a) showed that a molecular marker on chromosome 4 (ciw7) appeared to be linked to selenite resistance in Col. Although no ethylene- or JA-related gene is found around the marker, a potential defense-related gene, LSD1-like 2 (LOL2; At4g21610), is located close to the marker. LOL2 encodes a member of a small family of LSD1 proteins that contain three highly related zinc fingers, and may function as either a transcriptional regulator or a scaffold protein (Dietrich et al., 1997). Eppel et al. (2003) showed LOL and LSD1 may function as antagonistic transcriptional regulators or scaffolds that control attenuation of cell death through regulation of ROS and/or SA production level. In future studies, it will be interesting to test the involvement of LOL genes in selenite tolerance in Arabidopsis.
CONCLUSION
The results presented here indicate that the higher selenite resistance of accession Col-0 compared to Ws-2 is dependent on its higher level of selenite-induced JA and ethylene synthesis. Selenite-related ROS production was also higher in Col-0 than in Ws-2. This may indicate that JA and ethylene production require an optimal level of ROS production to lead to Se resistance in plants. The resistance mechanism may involve JA- and ethylene-enhanced S uptake and assimilation, as observed in Col-0. The higher levels of organic S compounds observed in Col-0 may enable it to more efficiently prevent Se analogs from replacing S in proteins and other S compounds. However, Se levels were also higher in Col-0 plants compared to Ws-2. It is intriguing to speculate that the Se-binding protein homolog that was induced by selenite in Col-0 but not in Ws-2 may play an additional role in alleviating Se toxicity. This would be in agreement with the study by Agalou et al. (2005), where overexpression of Se-binding protein resulted in enhanced Se resistance. Another possibility for increasing Se resistance in Col-0 is caused by its higher antioxidant levels, because higher Se causes the extra oxidative stress. JA might also be involved in this process because GSH and AsA biosynthetic pathways were enhanced after MeJA treatment (Sasaki-Sekimoto et al., 2005).
Se is an essential element for animals, including humans (Rayman, 2000). The recommended dietary allowance is 40 to 70 μg/d. However, human diets in several countries lack sufficient Se, which leads to enhance susceptibility to cancer, viral infections, and heart problems (Rayman, 2000). On the other hand, soils containing >0.5 mg Se kg−1 are considered seleniferous, and forage produced on such soils often contains more than the maximum permissible level for animal consumption. Soils with elevated levels of Se are found in many countries, including Australia, China, India, and the United States (Dhillon and Dhillon, 2003). Se accumulation by plants may help alleviate both Se deficiency and toxicity. A better understanding of the mechanisms and rate-limiting factors controlling plant Se uptake and assimilation will be vital for the optimal use of plants to alleviate dietary Se deficiency or for cleanup of Se-polluted areas. Several transgenic plants with enhanced Se accumulation and resistance have already been developed (Pilon-Smits et al., 1999; Agalou et al., 2005; Bañuelos et al., 2005, 2007; Van Hoewyk et al., 2005) and may be useful for Se phytoremediation (for review, see Pilon-Smits and Freeman, 2006). The new knowledge obtained here of the genes and processes that impact Se accumulation and resistance in Arabidopsis may lead to further development of plants with increased Se content.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Selenite Resistance Assays
Seeds of Arabidopsis (Arabidopsis thaliana) Col-0, Ws-2, npr1, acs6, ein3, ein2, eto1, jar1, and vtc1 were obtained from the Arabidopsis Biological Resource Center (ABRC; Columbus, OH). The sid2 mutant and iop1 mutant were obtained from Christiane Nawrath (University of Fribourg) and Bart P.H.J. Thomma (Wageningen University), respectively. C24 and cas1 mutant were provided from Ines Kubigsteltig, Ruhr-Universität Bochum. The double mutants vtc1/sid2 and vtc1/ein2 were created by selecting F2 individuals from the cross between vtc1 and sid2 or vtc1 and ein2. For selecting vtc1/sid2 double mutants, F2 plants that showed low levels of AsA were identified as described by Conklin et al. (2000). F3 lines lacking ozone-inducible SA accumulation were selected, then the point mutation of the ICS1 gene in the sid2 mutant was identified with DNA sequencing. For selecting vtc1/ein2 double mutants, F2 plants lacking AsA biosynthesis were identified as described above. Ethylene-insensitive F3 lines were selected on plates containing 20 μm ACC by screening for the lack of the triple response (Guzmán and Ecker, 1990).
Seeds were surface sterilized with 15% bleach and germinated on agar plates containing 0.5× Murashige and Skoog medium and 1% Suc with or without added sodium selenite at the indicated concentrations. Seedlings were grown on plates in a growth chamber at 24°C under a photosynthetic photon flux density of 150 μmol m−2 s−1 at a 16-h-light/8-h-dark cycle for 7 d.
For analysis of selenite resistance, plants were vertically grown for 7 d, and seedling root length was measured from digital photographs of the seedlings with the Image J program (http://rsb.info.nih.gov/ij/). The Se tolerance index was calculated as root length in the presence of selenite divided by mean of root length on control medium × 100%.
Macroarray Analysis
The expression of 250 genes was studied by custom-made cDNA macroarray using cDNA clones from the ABRC and RIKEN BioResource Center (Ibaraki, Japan). These cDNA clones were resequenced for confirmation before use. PCR-amplified sample was blotted onto Hybond N+ nylon membranes with MultiPin Blotter 96 (Atto). Each gene was spotted on a membrane in duplicate. λDNA was used as negative control. The constitutively expressed gene EF1α (At5G12110) was included as internal standard. For the macroarray studies, Col-0 and Ws-2 plants were grown on agar plates with or without 15 μm sodium selenite for 7 d. Then shoots were separated from roots and frozen with liquid nitrogen for total RNA extraction. Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen). Hybridization, probe labeling, and signal detection were carried out according to Tamaoki et al. (2003b). The signal intensity of each spot was obtained as described previously (Tamaoki et al., 2003b). In brief, we subtracted the value of the signal intensity of the negative control (λDNA) from the signal intensity of each spot, then normalized the signal intensities against the intensity of EF1α, the expression of which had been confirmed to be unchanged with or without 15 μm sodium selenite (see Fig. 2).
Expression Analysis via Semiquantitative RT-PCR
Plants were grown for 7 d on 0.5× Murashige and Skoog medium with or without 15 μm sodium selenite. Total RNA was isolated from shoots as described above. Five micrograms of DNase-treated total RNA was reverse transcribed using the First Strand cDNA synthesis kit (Fermentas International) following the manufacturer's instructions. PCR reactions were carried out as described previously (Schiavon et al., 2007). A list of primers used in these experiments is presented in Supplemental Table S2.
In Vitro Treatments and in Situ ROS Detection
The effect of exogenous SA, ACC, or MeJA on plant selenite resistance was analyzed by sowing sterilized Arabidopsis seeds on plates containing 0.5× Murashige and Skoog with a range of concentrations of SA, ACC, or MeJA with or without 15 μm sodium selenite. Seedlings were grown on vertical plates for 7 d. Seminal root length was measured from digital photographs of the seedlings with the ImageJ program (http://rsb.info.nih.gov/ij/). The Se tolerance index was calculated as described above.
Se-induced in situ accumulation of superoxide was detected with NBT (Boehringer Mannheim) as described by Jabs et al. (1996). To visualize in situ accumulation of hydrogen peroxide, DAB staining was performed as described by Torres et al. (2002).
Measurement of SA, MeJA, and JA
MeJA, salicylate, and jasmonate levels in shoot tissue were determined in plants grown as described above with or without 15 μm sodium selenite for 7 d. For measurement of SA and MeJA, the extracts were prepared as described by Wilbert et al. (1998). The extracts were analyzed by liquid chromatography-mass spectrometry (LC-MS) using a Hewlett-Packard Agilent 1100 series HPLC and a Finnigan LcQDuo thermoquest MS system equipped with Xcalibur software. Through 30-mL injections, these extracts were separated at 40°C using a Phenomenex Hypersil 5-mm C18 (ODS) column (250 × 2 mm, 5 mm) at a flow rate of 0.32 mL/min using two eluents: A, water + 0.1% formic acid; and B, 100% methanol + 0.1% formic acid. The following gradient program was used during the 23-min run: 0 to 7 min, 50% A and 50% B; 7 to 9 min, 30% A and 70% B; 9 to 12 min, 100% B; 12 to 13 min, 50% A and 50% B, with a 10-min postrun, column wash 50% A and 50% B. Standard curves were established using chemicals purchased from Sigma Chemical; MeJA (catalog no. 392707) had a retention time of 2.5 min, SA (catalog no. A–6262) had a retention time of 4.45 min, and jasmonate (catalog no. J2500) had a retention time of 6.85 min. Through MS, the different metabolites were measured at their appropriate masses and retention times observed for each of the standards. The MS detector settings were 1 to 3.5 min in positive ion mode using parameters generated with the MeJA standard and the automated tune program, 3.5 to 5.5 min in negative ion mode using parameters generated with the SA standard and the automated tune program, and 5.5 to 13 min in negative ion mode using parameters generated with the JA standard and the automated tune program. Samples were kept at room temperature (25°C) in the autosampler. The previously published (Wilbert et al., 1998) and observed precursor and product ions for these standards were exactly the same. These masses and those from the dimmers for MeJA and JA were used to quantify these compounds.
Measurement of Ethylene, Nonprotein Thiols, Ascorbic Acid, and Glutathione
For measurement of ethylene, 10 seedlings were enclosed in a 60-mL vial and incubated for 12 h with illumination in a growth chamber. A 25-mL gaseous phase of the vial was subjected into a Fisions 8000 gas chromatograph equipped with a flame ionization detector. A 2-m Altec Hayesep N 80/100 column was used with isothermic oven temperature at 70°C and flame ionization detector at 200°C. The program was 2 min in length with the ethylene peak running from 1.180 to 1.633 min. Ethylene peak area was determined by the PeakSimple program (ver. 3.39, 6 channel; SRI Instruments). The amount of ethylene generated from the seedlings was estimated from the peak area compared to that of ethylene standard.
Measurement of nonprotein thiol levels was performed using Ellman's reagent as described (Zhu et al., 1999). To measure total AsA, DHA, GSH, and GSSG levels, 100 mg of fresh plants was homogenized in 2 mL of cold 5% (w/v) metaphosphoric acid with sea sand. The AsA and DHA contents were quantified as described previously (Tamaoki et al., 2003a). GSH and GSSG contents were measured as described by Yoshida et al. (2006). All experiments were carried out in three replicates, each consisting of 20 to 30 pooled plants.
Quantification of Se and S Accumulation
Col-0 and Ws-2 plants were grown on 0.5× Murashige and Skoog agar medium with or without 15 μm selenite in a growth chamber for 3 weeks. Root and shoot materials were harvested separately, rinsed with distilled water, and dried at 37°C for a week. Three replicates consisting of 30 to 50 seedlings were acid-digested and analyzed for Se and S by inductively coupled plasma atomic emission spectrometry as described by Pilon-Smits et al. (1999).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identified up-regulated selenite-responsive genes (P < 0.05, fold change >2) that are categorized with biological function.
Supplemental Figure 2. Sensitivity to SA, ACC, and MeJA in seedlings of Col-0 and Ws-2.
Supplemental Table S1. Macroarray study showing induction of genes in Col-0 and Ws-2 by 15 μm selenite treatment.
Supplemental Table S2. Primers used in semiquantitative RT-PCR.
Supplementary Material
Acknowledgments
We are grateful to Dr. Christiane Nawrath and Dr. Silvia Heck (University of Fribourg, Fribourg, Switzerland) for the gift of the sid2 mutant and to Dr. Bart P.H.J. Thomma (Wageningen University, Wageningen, Netherlands) for providing the iop1 mutant. We also thank Dr. Ines Kubigsteltig (Ruhr-Universität Bochum, Bochum, Germany) for providing C24 and the cas1 mutant.
This work was supported by the Ministry of Education, Science, Sports and Culture of Japan (grant no. 18780006 to M.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Masanori Tamaoki (mtamaoki@nies.go.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agalou A, Roussis A, Spaink HP (2005) The Arabidopsis selenium-binding protein confers resistance to toxic levels of selenium. Funct Plant Biol 31 881–890 [DOI] [PubMed] [Google Scholar]
- Bañuelos G, LeDuc DL, Pilon-Smits EAH, Terry N (2007) Transgenic Indian mustard overexpressing selenocysteine lyase or selenocysteine methyltransferase exhibit enhanced potential for selenium phytoremediation under field conditions. Environ Sci Technol 41 599–605 [DOI] [PubMed] [Google Scholar]
- Bañuelos G, Terry N, LeDuc DL, Pilon-Smits EAH, Mackey B (2005) Field trial of transgenic Indian mustard plants shows enhanced phytoremediation of selenium-contaminated sediment. Environ Sci Technol 39 1771–1777 [DOI] [PubMed] [Google Scholar]
- Barth C, Moeder W, Klessig DF, Conklin PL (2004) The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiol 134 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29 23–32 [DOI] [PubMed] [Google Scholar]
- Birringer M, Pilawa S, Flohe L (2002) Trends in selenium biochemistry. Nat Prod Rep 19 693–718 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16 1–18 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Saracco SA, Susan R, Norris SR, Last RL (2000) Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Williams EH, Last RL (1996) Environmental stress sensitivity of an ascorbic acid deficient Arabidopsis mutant. Proc Natl Acad Sci USA 93 9970–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doares SH, Narvaez-Vasquez J, Conconi A, Ryan CA (1995) Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol 108 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon KS, Dhillon SK (2003) Distribution and management of seleniferous soils. Adv Agron 79 119–184 [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88 685–694 [DOI] [PubMed] [Google Scholar]
- Dong X (1998) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1 316–323 [DOI] [PubMed] [Google Scholar]
- Eppel P, Mack AA, Morris VRF, Dangl JL (2003) Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc Natl Acad Sci USA 100 6831–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice DC, Kull FJ, Shrift A (1981) Selenium toxicity: aminoacylation and peptide bond formation with selenomethionine. Plant Physiol 67 1054–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Johnson RR, Ryan CA (1992) Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol 98 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist LJ, Parker DR (2001) Ecotypic variation in selenium accumulation among populations of Stanleya pinnata. New Phytol 149 61–69 [DOI] [PubMed] [Google Scholar]
- Fu LH, Wang XF, Eyal Y, She YM, Donald LJ, Standing KG, Ben-Hayyim G (2002) A selenoprotein in the plant kingdom: mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii glutathione peroxidase. J Biol Inorg Chem 277 25983–25991 [DOI] [PubMed] [Google Scholar]
- Galeas ML, Zhang LH, Freeman JL, Wegner M, Pilon-Smits EAH (2007) Seasonal fluctuations of selenium and sulfur accumulation in selenium hyperaccumulators and related non-accumulators. New Phytol 173 517–525 [DOI] [PubMed] [Google Scholar]
- Gomes RA Jr, Gratão PL, Gaziola SA, Mazzafera PM, Lea PJ, Azevedo RA (2007) Selenium-induced oxidative stress in coffee cell suspension cultures. Funct Plant Biol 34 449–456 [DOI] [PubMed] [Google Scholar]
- Gromer S, Gross JH (2002) Methylseleninate is a substrate rather than an inhibitor of mammalian thioredoxin reductase. Implications for the antitumor effects of selenium. J Biol Chem 277 9701–9706 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SJ (2004) Review of selenium toxicity in the aquatic food chain. Sci Total Environ 326 1–36 [DOI] [PubMed] [Google Scholar]
- Hira CK, Partal K, Dhillon K (2004) Dietary selenium intake by men and women in high and low selenium areas of Punjab. Public Health Nutr 7 39–43 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Saito K (2004) Post-genomic approaches for the elucidation of plant adaptive mechanism to sulfur deficiency. J Exp Bot 55 1871–1879 [DOI] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular super oxide. Science 273 1853–1856 [DOI] [PubMed] [Google Scholar]
- Kangasjärvi J, Jaspers P, Kollist H (2005) Signalling and cell death in ozone-exposed plants. Plant Cell Environ 28 1021–1036 [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vregdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55 141–172 [DOI] [PubMed] [Google Scholar]
- Kubigsteltig II, Weiler EW (2003) Arabidopsis mutants affected in the transcriptional control of allene oxide synthase, the enzyme catalyzing the entrance step in octadecanoid biosynthesis. Planta 217 748–757 [DOI] [PubMed] [Google Scholar]
- Läuchli A (1993) Selenium in plants: uptake, functions, and environmental toxicity. Bot Acta 106 455–468 [Google Scholar]
- Lawton KA, Potter SL, Uknes S, Ryals J (1994) Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 6 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling of sulphur responsive genes in Arabidopsis reveals global effects of sulphur nutrition on multiple metabolic pathways. Plant Physiol 132 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulphur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Bielecka M, Gakière B, Krueger S, Rinder J, Kempa S, Morcuende R, Scheible WR, Hesse H, Hoefgen R (2006) Effect of sulfur availability on the integrity of amino acid biosynthesis in plants. Amino Acids 30 173–183 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Scheel D (2001) Signal transmission in the plant immune response. Trends Plant Sci 6 372–379 [DOI] [PubMed] [Google Scholar]
- Obata T, Shiraiwa Y (2005) A novel eukaryotic selenoprotein in the Haptophyte alga Emiliania huxleyi. J Biol Chem 280 18462–18468 [DOI] [PubMed] [Google Scholar]
- Overmyer K, Broschė M, Kangasjärvi J (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8 335–342 [DOI] [PubMed] [Google Scholar]
- Pena-Cortes H, Albrecht T, Prat S, Weiler EW, Willmitzer L (1993) Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 191 123–128 [Google Scholar]
- Penninckx IAMA, Eggermont K, Schenk PM, Ackerveken GVD, Cammue BPA, Thomma BPHJ (2003) The Arabidopsis mutant iop1 exhibits induced over-expression of the plant defensin gene PDF1.2 and enhanced pathogen resistance. Mol Plant Pathol 4 479–486 [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchla A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathway is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering IJ, Wright C, Bubner B, Ellis D, Persans MW, Yu EY, George GN, Prince RC, Salt DE (2003) Chemical form and distribution of selenium and sulfur in the selenium hyperaccumulator Astragalus bisulcatus. Plant Physiol 131 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, van Loon LC (1999) Salicylic acid-independent plant defence pathway. Trends Plant Sci 4 52–58 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Freeman LJ (2006) Environmental cleanup using plants: biotechnological advances and ecological considerations. Front Ecol Environ 4 203–210 [Google Scholar]
- Pilon-Smits EAH, Hwang S, Lytle CM, Zhu YL, Tai JC, Bravo RC, Chen Y, Leustek T, Terry N (1999) Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiol 119 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman MP (2000) The importance of selenium to human health. Lancet 356 233–241 [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Yokota-Hirai M, Noji M, Saito K, et al (2005) Coordinated activation of metabolic pathway for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44 653–668 [DOI] [PubMed] [Google Scholar]
- Schiavon M, Zhang L-H, Abdel-Ghany SE, Pilon M, Malagoli M, Pilon-Smits EAH (2007) Variation in copper tolerance in Arabidopsis thaliana accessions Columbia, Landsberg erects and Wassilewskija. Physiol Plant 129 342–350 [Google Scholar]
- Smirnoff N, Conklin PL, Loewus FA (2001) Biosynthesis of ascorbic acid in plants: a renaissance. Annu Rev Plant Physiol Plant Mol Biol 52 437–67 [DOI] [PubMed] [Google Scholar]
- Sors TG, Ellis DR, Salt DE (2005) Selenium uptake, translocation, assimilation and metabolic fate in plants. Photosynth Res 86 373–389 [DOI] [PubMed] [Google Scholar]
- Stadtman TC (1990) Selenium biochemistry. Annu Rev Biochem 59 111–127 [DOI] [PubMed] [Google Scholar]
- Stadtman TC (1996) Selenocysteine. Annu Rev Biochem 65 83–100 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Mauzher H, Weichert H, Ziegler J, Feussner I, Wasternack C (2004) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51 895–911 [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Mukai F, Asai N, Nakajima N, Kubo A, Aono M, Saji H (2003. a) Light-controlled expression of a gene encoding L-galactono-γ-lactone dehydrogenase which affects ascorbate pool size in Arabidopsis thaliana. Plant Sci 164 1111–1117 [Google Scholar]
- Tamaoki M, Nakajima N, Kubo A, Aono M, Matsuyama T, Saji H (2003. b) Transcriptome analysis of O3-exposed Arabidopsis reveals that multiple signal pathways act mutually antagonistically to induce gene expression. Plant Mol Biol 53 443–456 [DOI] [PubMed] [Google Scholar]
- Terry N, Zayed AM, de Souza MP, Tarun AS (2000) Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol 51 401–432 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Garifullina GF, Ackley AR, Abdel-Ghany SE, Marcus MA, Fakra S, Ishiyama K, Inoue E, Pilon M, Takahashi H (2005) Overexpression of AtCpNifS enhances selenium tolerance and accumulation in Arabidopsis. Plant Physiol 139 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Takahashi H, Hess A, Tamaoki M, Pilon-Smits EAH (2008) Transcriptome and biochemical analyses give insights into selenium-stress responses and selenium tolerance mechanisms in Arabidopsis. Physiol Plant 132 236–253 [DOI] [PubMed] [Google Scholar]
- White PJ, Bowen HC, Parmagure P, Fritz M, Spracklen WP, Spiby RE, Meacham MC, Mead A, Harriman M, Trueman LJ, et al (2004) Interaction between selenium and sulfur nutrition in Arabidopsis thaliana. J Exp Bot 55 1927–1937 [DOI] [PubMed] [Google Scholar]
- Wilber CG (1980) Toxicology of selenium: a review. Clin Toxicol 17 171–230 [DOI] [PubMed] [Google Scholar]
- Wilbert SM, Ericsson LH, Gordon MP (1998) Quantification of jasmonic acid, methyl jasmonate, and salicylic acid in plants by capillary liquid chromatography electrospray tandem mass spectrometry. Anal Biochem 257 186–194 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 414 562–565 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35 155–189 [Google Scholar]
- Yoshida S, Tamaoki M, Shikano T, Nakajima N, Ogawa D, Ioki M, Aono M, Kubo A, Kamada H, Inoue Y, et al (2006) Cytosolic dehydroascorbate reductase is important for ozone tolerance in Arabidopsis thaliana. Plant Cell Physiol 47 304–308 [DOI] [PubMed] [Google Scholar]
- Zhang LH, Abdel-Ghany SE, Freeman JL, Ackley AR, Schiavon M, Pilon-Smits EAH (2006. a) Investigation of selenium tolerance mechanism in Arabidopsis thaliana. Physiol Plant 128 212–223 [Google Scholar]
- Zhang LH, Ackley AR, Pilon-Smits EAH (2007) Variation in selenium tolerance and accumulation among 19 Arabidopsis thaliana accessions. J Plant Physiol 164 327–36 [DOI] [PubMed] [Google Scholar]
- Zhang LH, Byrne PF, Pilon-Smits EAH (2006. b) Mapping quantitative trait loci associated with selenate tolerance in Arabidopsis thaliana. New Phytol 170 33–42 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Pilon-Smits EAH, Jouanin L, Terry N (1999) Overexpression of glutathione synthetase in Brassica juncea enhances cadmium tolerance and accumulation. Plant Physiol 119 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.