Abstract
Suppression of seed germination at supraoptimal high temperature (thermoinhibiton) during summer is crucial for Arabidopsis (Arabidopsis thaliana) to establish vegetative and reproductive growth in appropriate seasons. Abscisic acid (ABA) and gibberellins (GAs) are well known to be involved in germination control, but it remains unknown how these hormone actions (metabolism and responsiveness) are altered at high temperature. Here, we show that ABA levels in imbibed seeds are elevated at high temperature and that this increase is correlated with up-regulation of the zeaxanthin epoxidase gene ABA1/ZEP and three 9-cis-epoxycarotenoid dioxygenase genes, NCED2, NCED5, and NCED9. Reverse-genetic studies show that NCED9 plays a major and NCED5 and NCED2 play relatively minor roles in high temperature-induced ABA synthesis and germination inhibition. We also show that bioactive GAs stay at low levels at high temperature, presumably through suppression of GA 20-oxidase genes, GA20ox1, GA20ox2, and GA20ox3, and GA 3-oxidase genes, GA3ox1 and GA3ox2. Thermoinhibition-tolerant germination of loss-of-function mutants of GA negative regulators, SPINDLY (SPY) and RGL2, suggests that repression of GA signaling is required for thermoinibition. Interestingly, ABA-deficient aba2-2 mutant seeds show significant expression of GA synthesis genes and repression of SPY expression even at high temperature. In addition, the thermoinhibition-resistant germination phenotype of aba2-1 seeds is suppressed by a GA biosynthesis inhibitor, paclobutrazol. We conclude that high temperature stimulates ABA synthesis and represses GA synthesis and signaling through the action of ABA in Arabidopsis seeds.
Seed germination is determined by a combination of the degree of dormancy and environmental factors such as light, oxygen, and temperature. Even after the loss of dormancy, seeds do not germinate in unfavorable conditions. Suppression of germination at supraoptimal high temperatures is called thermoinhibition (Reynolds and Thompson, 1971; Abeles, 1986; Gallardo et al., 1991). It has been shown that seed responsiveness to temperature is closely related to the level of dormancy in soil-buried seeds of winter and summer annuals (Baskin and Baskin, 1998). In the case of Arabidopsis (Arabidopsis thaliana), the maximal permissive temperature for germination rises gradually during an after-ripening period in summer, but germination is repressed by environmental temperature higher than the upper limit for germination. The seeds germinate in autumn when the temperature falls below the upper limit for germination (Baskin and Baskin, 1983). Therefore, the change in seed sensitivity to temperature plays an ecologically important role in the detection of the appropriate seasonal timing for germination in the field (Baskin and Baskin, 1998; Yoshioka et al., 1998, 2003).
Phytohormones abscisic acid (ABA) and GAs are well known to be involved in germination control. In lettuce (Lactuca sativa) seeds, reduction of ABA content is suppressed during imbibition at supraoptimal temperature, and de novo ABA biosynthesis is required for thermoinhibition (Yoshioka et al., 1998). Alleviation of thermoinhibition by exogenous GA has been reported on several plant species (Madakadze et al., 1993; Dutta et al., 1994; Carter and Stevens, 1998; Gonai et al., 2004). However, the effect of supraoptimal temperature on GA levels in the seeds and function of endogenous GA on thermoinhibition have not been clarified. We have isolated ABA-resistant germination mutants of Arabidopsis, abi3-14 and trg2, as high temperature-resistant germination mutants, and showed that the seeds of the ABA-deficient mutant aba1-1 and ABA-insensitive abi1-1 were highly tolerant to thermoinhibition (Tamura et al., 2006). These reports suggest that ABA and GA are involved in the control of seed germination by temperature, but how these hormones mediate the high-temperature signal remains unknown.
ABA is a key regulator of seed development, dormancy, germination, and adaptive responses to abiotic stresses (Zeevaart and Creelman, 1988). Endogenous ABA content is a determinant of these physiological processes, and ABA-deficient mutants exhibit reduced seed dormancy and reduced drought tolerance (McCarty, 1995). Recent genetic and genomics analyses have revealed the molecular basis of the pathway and genes/enzymes involved in ABA biosynthesis and catabolism (Nambara and Marion-Poll, 2005). In plants, ABA is synthesized from carotenoids through the indirect pathway (Zeevaart and Creelman, 1988). Zeaxanthin is converted to all-trans-violaxanthin by two-step epoxidation catalyzed by zeaxanthin epoxidase (ZEP) in plastids (Marin et al., 1996; Thompson et al., 2000a; Agrawal et al., 2001; Audran et al., 2001). 9-cis-Epoxycarotenoid dioxygenase (NCED) catalyzes oxidative cleavage of the 9-cis isomer of violaxanthin or neoxanthin and produces a C15 product xanthoxin and a C25 metabolite (Tan et al., 1997; Burbidge et al., 1999; Qin and Zeevaart, 1999; Chernys and Zeevaart, 2000; Iuchi et al., 2000, 2001). The reactions following production of xanthoxin occur in the cytosol. Xanthoxin is converted to abscisic aldehyde by short-chain dehydrogenase/reductase (SDR; Cheng et al., 2002; González-Guzmán et al., 2002), and then abscisic aldehyde is oxidized to ABA by abscisic aldehyde oxidase (Seo et al., 2000a). On the other hand, ABA is inactivated through hydroxylation and conjugation (Zeevaart and Creelman, 1988; Nambara and Marion-Poll, 2005). Among these pathways, the ABA 8′-hydroxylation pathway is shown to be the regulatory step in a variety of physiological processes. In Arabidopsis, CYP707A1 to CYP707A4 encode ABA 8′-hydroxylase (Kushiro et al., 2004; Saito et al., 2004). Gene expression and reverse-genetic analyses indicated that CYP707A2 has a major role in the rapid decrease in ABA content in the first 6 to 12 h of imbibition and suggest that CYP707A1 to CYP707A3 are involved in seed germination (Kushiro et al., 2004; Okamoto et al., 2006).
NCED was first identified by analysis of the maize (Zea mays) viviparous germination mutant vp14 (Tan et al., 1997), and is considered to be a key regulatory enzyme in ABA biosynthesis (Qin and Zeevaart, 1999, 2002; Thompson et al., 2000b; Iuchi et al., 2001). NCED is a multigene family in all plant species examined (Tan et al., 1997; Burbidge et al., 1999; Qin and Zeevaart, 1999; Chernys and Zeevaart, 2000; Iuchi et al., 2000, 2001). In Arabidopsis, phylogenetic analysis has suggested that five genes (NCED2, NCED3, NCED5, NCED6, and NCED9) are possibly involved in ABA biosynthesis, and they have been shown to catalyze NCED reaction in vitro, with the exception of NCED5 (Iuchi et al., 2001). Each member of the NCED family plays a unique regulatory role in specific environmental responses and developmental processes. Expression of the NCED3 gene is induced by drought stress, and it plays a major role in ABA synthesis in response to the stress (Iuchi et al., 2001; Ruggiero et al., 2004). NCED6 expression is regulated in a photoreversible manner via phytochrome in seeds, and it plays a role in far-red light-induced suppression of seed germination (Seo et al., 2006). NCED6 and NCED9 play a major role in the control of seed development, including dormancy (Lefebvre et al., 2006). Real-time PCR and reporter gene expression analyses indicated that NCED2 and NCED5 are expressed in specific tissues and developmental stages (Tan et al., 2003), but physiological functions of these two NCED genes remain obscure.
GA is essential for stem elongation and flowering and is also a key regulator of seed germination. Previous studies have suggested that GA responses are regulated by the modulation of GA levels and by altering the ability of cells to respond to the hormone (Richards et al., 2001). Biologically active GAs, such as GA1 and GA4, are tetracyclic diterpenoids synthesized from geranylgeranyl diphosphate, and the GA biosynthesis pathway can be divided into three stages (Hedden and Kamiya, 1997; Olszewski et al., 2002). First, geranylgeranyl diphosphate is cyclized to ent-kaurene in plastids, then ent-kaurene is oxidized to GA12 by microsomal cytochrome P450 monooxygenases, and, finally, GA12 is converted to active GA in cytosol by 2-oxoglutarate-dependent dioxygenases, GA 20-oxidase and GA 3-oxidase. Bioactive GAs are well known to be deactivated by a 2-oxoglutarate-dependent dioxygenase, GA 2-oxidase, which also catabolizes immediate precursors of active GAs. Recently, additional deactivation mechanisms by a cytochrome P450 monooxygenase and methyltransferases were reported (Zhu et al., 2006; Varbanova et al., 2007). Environmental signals regulate GA level through modulation of the late steps of GA biosynthesis and catabolism (Olszewski et al., 2002). Light signals mediated by phytochromes are critical environmental determinants for photoblastic seed germination (Borthwick et al., 1952; Shinomura et al., 1996). GA levels are regulated by phytochrome in lettuce and Arabidopsis seeds, in which genes encoding GA 3-oxidase (GA3ox) and GA 2-oxidase (GA2ox) are regulated in a photoreversible manner (Toyomasu et al., 1998; Yamaguchi et al., 1998). In Arabidopsis, GA3ox1 and GA3ox2 are up-regulated and GA2ox2 is down-regulated by red light, which promotes seed germination. In addition to light, exposure of imbibed seeds to low temperature promotes seed germination of many plant species. In Arabidopsis, expression of GA 20-oxidase genes, GA20ox1 and GA20ox2, and GA3ox1 was reported to be up-regulated by low temperature in darkness, and germination of GA3ox1 mutant (ga4-2) seeds are not stimulated by low temperature (Yamauchi et al., 2004).
Genetic studies have identified several GA-signaling components in Arabidopsis, some of which play a role in GA-induced seed germination, as shown by their loss-of-function mutant phenotypes (Olszewski et al., 2002). The DELLA subfamily of GRAS proteins and SPINDLY (SPY), a Ser/Thr O-linked N-acetylglucosamine transferase, acts as negative regulators of GA-dependent seed germination (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Lee et al., 2002; Wen and Chang, 2002). SPY is thought to increase the activity of DELLA proteins by N-acetylglucosamine modification (Silverstone et al., 2007). SPY also acts as a positive regulator of cytokinin signaling (Greenboim-Wainberg et al., 2005). In rice (Oryza sativa), the SPY ortholog OsSPY works as a negative regulator of GA signaling as in Arabidopsis and may also function as a negative regulator of brassinosteroid biosynthesis (Shimada et al., 2006). One of the five DELLA protein genes in Arabidopsis, RGL2, plays a major role on GA-dependent germination of the seeds (Lee et al., 2002; Tyler et al., 2004). The germination phenotypes of multiple DELLA protein mutants in the ga1-3 background indicated that GAI, RGA, and RGL1 enhance the function of RGL2 (Cao et al., 2005).
Seed germination is considered to be determined by the balance of the negative and positive effects of ABA and GA, respectively. Interaction between ABA and GA signaling has been well studied in cereal aleurone cells as a model system and recent reports shed light on the interaction between ABA and GA in their metabolism. Gonai et al. (2004) suggested that exogenously applied GA3 alleviates thermoinhibition of lettuce seeds by enhancing the catabolism of ABA. Seo et al. (2006) indicated that endogenous ABA suppresses GA biosynthesis in developing seeds and in far-red light-treated mature seeds during imbibition through suppression of GA20ox and GA3ox genes in Arabidopsis. Zentella et al. (2007) showed that the transcript levels of GA20ox1 in Arabidopsis plants are significantly reduced by exogenous ABA.
In this study, we show that both enhancement of ABA synthesis and suppression of GA synthesis and signaling during imbibition are required for thermoinhibition of Arabidopsis seeds. Our data suggest that high temperature enhances ABA synthesis by stimulating expression of ZEP and three of the five NCED genes, NCED2, NCED5, and NCED9, and suppresses GA synthesis through suppression of GA20ox and GA3ox genes. In addition, high temperature may suppress GA signaling by enhancing SPY gene expression and maintaining RGL2 gene expression. Our data also suggest that GA synthesis and signaling are suppressed through the action of ABA in the seeds at high temperature.
RESULTS
ABA Synthesis in Imbibed Seeds Is Responsible for Thermoinhibition
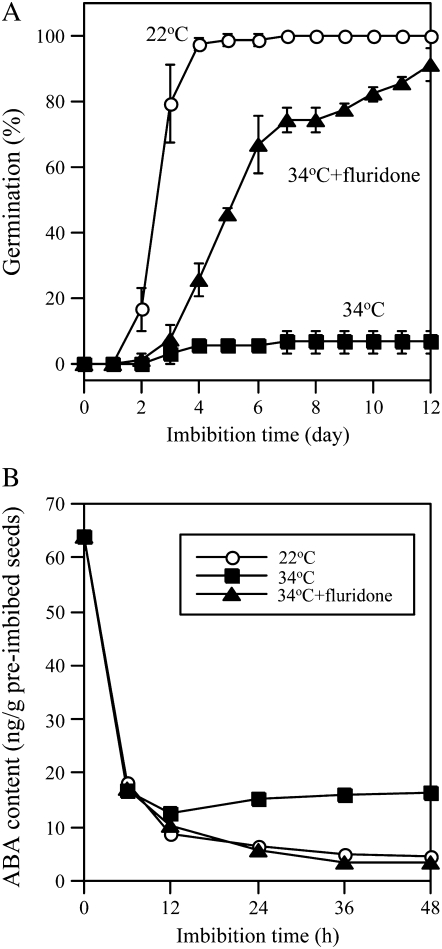
To examine whether ABA synthesis is responsible for thermoinhibition of Arabidopsis seeds, we first applied an ABA biosynthesis inhibitor to Columbia (Col) accession seeds during imbibition at supraoptimal temperatures. We used fluridone, which inhibits phytoene desaturase in the carotenoid biosynthesis pathway, as a potent inhibitor of ABA biosynthesis. As shown in Figure 1A, 10 μm fluridone rescued germination of the seeds at supraoptimal high temperature (34°C). This suggests that thermoinhibition of Arabidopsis seeds requires ABA biosynthesis de novo after the onset of imbibition as observed in lettuce and several winter annual species (Yoshioka et al., 1998). To examine whether the ABA level is altered by high temperature, we quantitated ABA in the imbibed seeds by gas chromatography (GC)-mass spectrometry (MS; Fig. 1B). At optimal temperature for germination (22°C), ABA levels decreased remarkably by 6 h of imbibition and continued to decrease until 48 h as reported previously (Kushiro et al., 2004; Millar et al., 2006; Okamoto et al., 2006). At 36 h after the start of imbibition, when the radicle starts to emerge, the ABA level decreased to one-tenth of the initial content (Fig. 1B). The decrease within the first 6 h also occurred at 34°C, but the reduction speed slowed down after 6 h and the level turned to increase gently after 12 h. The application of 10 μm fluridone suppressed the increase of ABA completely at 34°C, and the levels were comparable to those at 22°C. These results suggest that high temperature stimulates synthesis of ABA in Arabidopsis seeds.
Figure 1.
ABA biosynthesis in imbibed seeds is required for thermoinhibition. After-ripened Col seeds were imbibed at 22°C (white circles), 34°C (black squares), and at 34°C in the presence of 10 μm fluridone (black triangles) under continuous illumination. A, Effect of ABA biosynthesis inhibitor fluridone on thermoinhibition. Error bars show sd (n = 3). B, Effects of high temperature and ABA biosynthesis inhibitor fluridone on endogenous ABA levels in imbibed seeds. ABA levels were quantified by GC-MS using two independent seed batches and similar results were obtained.
High Temperature Up-Regulated ABA Biosynthesis Genes, ABA1/ZEP and NCED, and Down-Regulated the ABA Catabolism Gene CYP707A
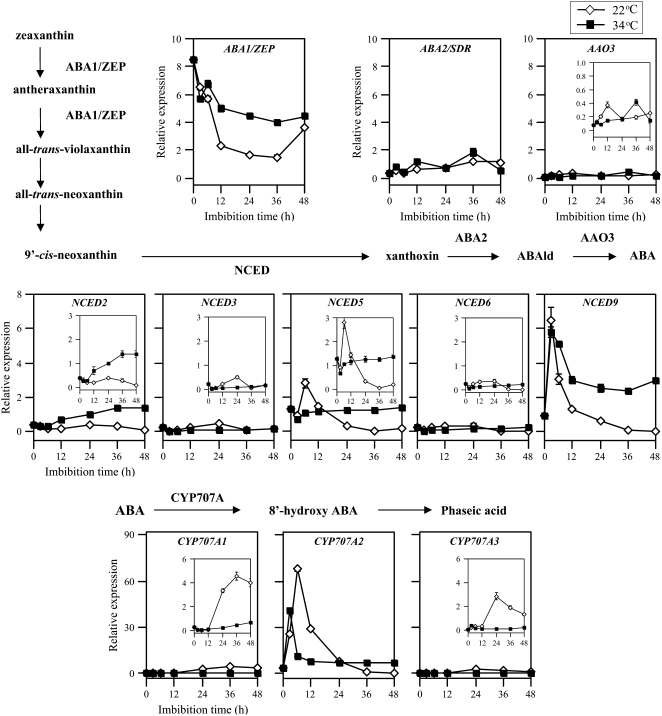
To investigate the regulation step of ABA synthesis by high temperature, we analyzed the expression of genes that code for ABA biosynthesis enzymes by the quantitative reverse transcription (RT)-PCR method. ABA biosynthesis enzymes, zeaxanthin epoxidase (ABA1/ZEP), short-chain dehydrogenase/reductase (ABA2/SDR), and abscisic aldehyde oxidase (AAO3) have been reported to be coded by single genes in Arabidopsis (Marin et al., 1996; Seo et al., 2000b; Cheng et al., 2002). Dry seeds contained a considerable level of ABA1/ZEP transcript, and the transcript decreased significantly in the first 12 h of imbibition at 22°C and continued to decrease until 36 h (Fig. 2). At 34°C, the transcript also decreased, but reduced the rate of decrease after 3 h and kept considerably higher level than at 22°C. ABA1/ZEP transcript levels were approximately 2 times higher at 34°C than at 22°C at 12 h when the reduction rate of the ABA level slowed down (Fig. 1B). ABA2/SDR transcript levels slightly increased during imbibition both at 22°C and 34°C, and the levels were not remarkably different between the two temperature conditions. The levels of AAO3 transcript were low throughout the imbibition period, and there was no significant difference between the two temperatures. These results suggest that ABA2/SDR and AAO3 genes have no regulatory function on modulation of ABA level in response to high temperature in seeds.
Figure 2.
Effect of high temperature on expression of ABA biosynthesis and catabolism genes in imbibed seeds. Total RNA was prepared from after-ripened Col seeds imbibed at 22°C (white diamonds) or at 34°C (black squares) under continuous illumination. Transcript levels were quantified by quantitative RT-PCR as described in “Materials and Methods.” Experiments were repeated twice with different seed batches and similar results were obtained. Data from one of the replicates are shown. Values are means with sds from three measurements.
In Arabidopsis, five NCED genes are reported to be involved in ABA biosynthesis, and NCED2, NCED3, NCED6, and NCED9 proteins have been reported to have the NCED activity to produce xanthoxin in vitro (Iuchi et al., 2001). To identify the enzyme activity of NCED5, the NCED5 gene was expressed in Escherichia coli to obtain glutathione S-transferase (GST)-fused recombinant protein, and the proteins were incubated with 9′-cis-neoxanthin. HPLC analysis of the reaction products indicated that NCED5 catalyzes the cleavage of 9′-cis-neoxanthin to produce a C25 product (Supplemental Fig. S1A). The enzyme activity of the GST-NCED5 fusion protein was further confirmed by GC-MS analysis in which cis-xanthoxin was identified as another product from 9′-cis-neoxanthin (Supplemental Fig. S1B). In both experiments, GST-NCED3 recombinant protein was used as a control to give similar results (Supplemental Fig. S1, C and D). These results confirm that NCED5 encodes an active NCED enzyme, which is probably engaged in ABA biosynthesis.
Relatively low levels of NCED3 and NCED6 transcripts were found in dry seeds, and their levels were not changed significantly by imbibition at 22°C and at 34°C (Fig. 2). The level of NCED2 transcript was also low in dry seeds and did not increase during imbibition at 22°C. In contrast, a marked increase in the transcript level was detected at 12 h at 34°C, and the expression continued to increase until 36 h. In dry seeds, transcript levels of NCED5 and NCED9 were 2.5 to 6 times more abundant than those of other NCEDs. They increased transiently and decreased to low levels during imbibition at 22°C. At 34°C, NCED5 transcript level did not change significantly and the transcript was 4 to 8 times more abundant than at 22°C at 24 h and later time points. NCED9 expression peaked at 3 h at 34°C, and its level was similar to that at 22°C at this point. However, the rate of decrease was significantly slower at 34°C than at 22°C and it ceased to decrease after 12 h. The NCED9 transcript was the most abundant among five NCEDs throughout the imbibition period at 34°C. NCED2 and NCED9 transcript levels were significantly higher at 34°C than at 22°C at 12 h after the start of imbibition when the decrease rate of ABA slowed down at 34°C (Fig. 1B). These observations suggest that NCED2, NCED5, and NCED9 contribute to enhanced ABA biosynthesis ability in the seeds at supraoptimal temperature.
We analyzed transcript levels of ABA 8′-hydroxylase genes, CYP707A1, CYP707A2, and CYP707A3, and found that the expression of all three genes was significantly reduced at 34°C (Fig. 2). Expression of CYP707A2 was most abundant among the three ABA 8′-hydroxylase genes and was transiently increased during imbibition even at 34°C. CYP707A2 transcript in dry and imbibed seeds might be translated in the first 6 h and contribute to ABA catabolism in the thermoinhibited seeds as well as in germinating seeds because the reduction of ABA levels in the initial 6 h was commonly observed at 22°C and at 34°C, and the decrease rate of ABA at 34°C in the presence of fluridone was comparable to that at 22°C (Fig. 1B). ABA catabolism by the 8′-hydroxylation pathway may also work in thermoinhibited lettuce seeds because dihydrophaseic acid, which is an ABA catabolite in the 8′-hydroxylation pathway, is accumulated in the seeds imbibed at supraoptimal temperature (Chiwocha et al., 2003). ABA content in thermoinhibited seeds showed only a slight increase after 12 h of imbibition (Fig. 1B). This fine tuning of the ABA level may be achieved by the balance between synthesis and catabolism since CYP707A2 transcript became more abundant at 34°C than at 22°C after 36 h of imbibition (Fig. 2).
NCED2, NCED5, and NCED9 Have Unequally Redundant Functions on Thermoinhibition and ABA Level of the Seeds
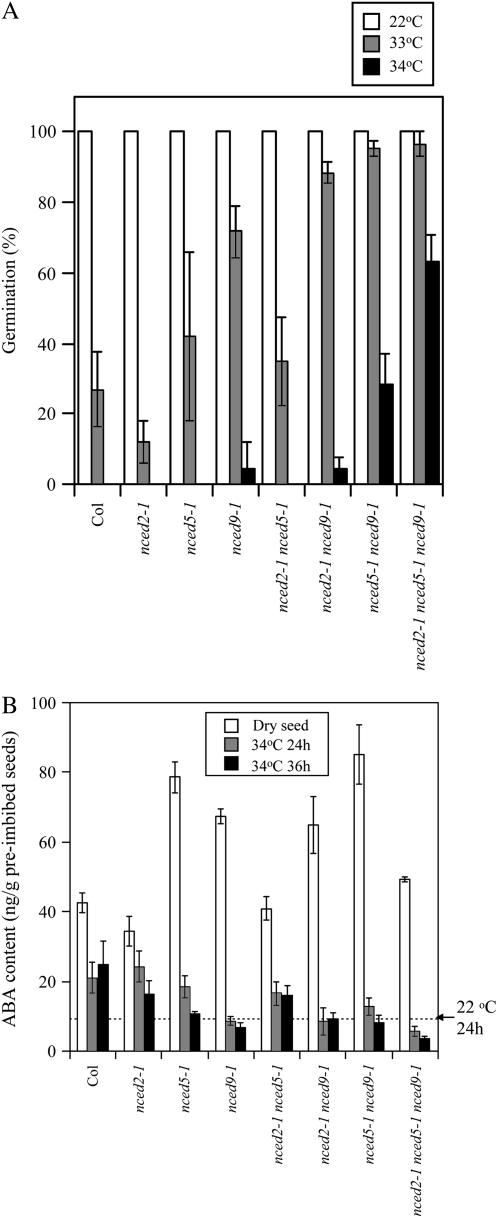
To evaluate the physiological function of ABA biosynthesis genes on thermoinhibition, we analyzed germination phenotypes of their loss-of-function mutants. The seeds of aba1-1, aba2-1, and aao3-4 showed apparent thermoinhibition resistance at 34°C (Supplemental Fig. S2B). ABA1/ZEP, ABA2/SDR, and AAO3, which are involved in the indirect pathway, should be responsible for ABA biosynthesis in Arabidopsis seeds also at supraoptimal temperature conditions. T-DNA insertion lines for NCED2, NCED5, and NCED9 were obtained from the Arabidopsis Biological Resource Center (ABRC; Alonso et al., 2003), and we analyzed the corresponding T-DNA insertion homozygotes (Supplemental Fig. S2A). The seeds of nced9 mutant alleles showed considerable tolerance to thermoinhibition, but those of nced2 and nced5 showed no apparent phenotypes (Fig. 3A; Supplemental Fig. S2B). This suggests that NCED9 has a major role in thermoinhibition. To further evaluate the contribution of each NCED gene to thermoinhibition, we generated double and triple mutants of NCED2, NCED5, and NCED9, and investigated their germination phenotypes at supraoptimal temperature conditions. The seeds of the nced5-1 nced9-1 double mutant showed higher thermoinhibition tolerance than nced9-1, but the nced2-1 nced5-1 double mutant showed similar germination to its parents and the wild type (Fig. 3A). Seeds of the nced2-1 nced5-1 nced9-1 triple mutant showed higher thermoinhibition tolerance than the double mutants. These results suggest that NCED5 and NCED2 genes have relatively minor, but unequally redundant (Briggs et al., 2006), function with NCED9, and their up-regulation by high temperature is critical for thermoinhibition of Arabidopsis seeds.
Figure 3.
NCED2, NCED5, and NCED9 are unequally redundant on thermoinhibition and ABA synthesis in the seeds. A, Thermoinhibition resistance of single and multiple mutants of nced2, nced5, and nced9. Seeds were imbibed at 22°C (white bars), 33°C (gray bars), and 34°C (black bars) under continuous illumination for 5 d. Error bars show sd (n = 3). Germination assays were confirmed in three independent batches. B, ABA content in seeds of nced single and multiple mutants. ABA was extracted from dry seeds (white bars) and the seeds imbibed at 34°C for 24 h (gray bars) and 36 h (black bars) under continuous illumination. ABA was quantified by liquid chromatography-tandem mass spectrometry. Dotted line indicates ABA content in wild-type seeds imbibed at 22°C for 24 h. Error bars show sd (n = 3).
To evaluate the contribution of each NCED gene on de novo ABA biosynthesis during thermoinhibition, we analyzed the endogenous ABA level in nced mutants. The levels of ABA in dry seeds of single and multiple nced mutants were not lower, but rather higher in some cases, than those in wild-type seeds (Fig. 3B). It is possible that the mutations are compensated or overcompensated by other NCED genes during seed development. When seeds were imbibed at 34°C for 24 h, the nced9-1 single mutant, double mutants except for nced2-1 nced5-1, and the triple mutant reduced ABA content considerably; ABA levels were comparable to those of wild-type seeds imbibed at 22°C for 24 h. The ABA content in the triple mutant was lower than that of the double mutants. These results indicate that NCED2, NCED5, and NCED9 are responsible for ABA biosynthesis in the seeds imbibed at supraoptimal temperature conditions and support the idea that their up-regulation is a key regulatory point in ABA biosynthesis and germination inhibition by high temperature.
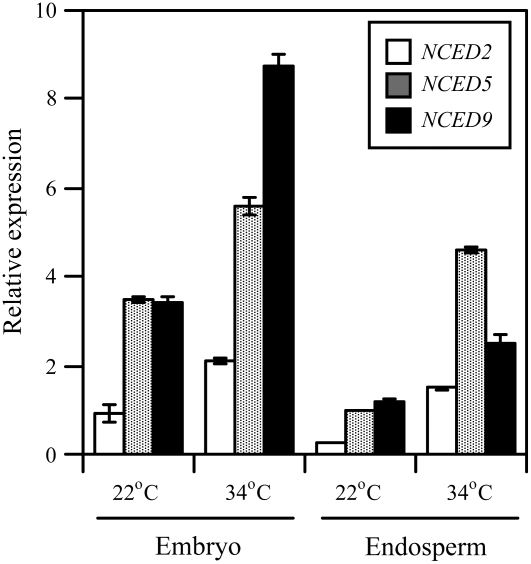
To investigate contributions of ABA biosynthesis in the embryo and the endosperm to thermoinhibition, we divided the 24-h imbibed Col seeds into the embryo and the remaining part, including endosperm and testa, and RNA was extracted from each tissue for quantitative RT-PCR. Quantification of an embryo-specific marker gene, GA3ox2 (Yamaguchi et al., 2001), and an endosperm-specific marker gene, EPR1 (Dubreucq et al., 2000; Penfield et al., 2004), indicated that there was no cross-contamination between the two fractions (Supplemental Fig. S3). In both the embryo and the endosperm, all three NCED transcripts were detected, and they were more abundant at 34°C than at 22°C (Fig. 4). Expression of NCED9 was predominant over the other two NCED genes in the embryos from the seeds imbibed at 34°C. In the endosperm, on the other hand, NCED5 transcript was most abundant among the three NCED genes. ABA synthesized in embryo may have a major contribution to germination inhibition at supraoptimal temperatures because seeds of the nced9-1 single mutant showed thermoinhibition tolerance, but those of the nced5-1 single mutant and the nced2 nced5 double mutant were intolerant (Fig. 3A; Supplemental Fig. S2B).
Figure 4.
Expression of NCED2, NCED5, and NCED9 in embryo and endosperm of thermoinhibited seeds. Total RNA was prepared from embryo and endosperm/testa separated from Col seeds imbibed at 22°C or at 34°C under continuous illumination for 24 h. Transcript levels of NCED2 (white bars), NCED5 (gray bars), and NCED9 (black bars) were quantified by quantitative RT-PCR with a set of primers and a Taq-Man probe specific to each gene. To compare transcript levels across different NCED genes, standard curves were generated using a series of known concentration of target sequences (genomic DNA). Results for each tissue were normalized to the amplification of the 18S rRNA control and the amount of total RNA extracted from each tissue of one seed. Experiments were repeated three times with different seed batches and similar results were obtained. Data from one of the replicates are shown. Values are means with sds from three measurements.
Alleviation of Thermoinhibition by Exogenous GA and Suppression of Endogenous GA Level at High Temperature
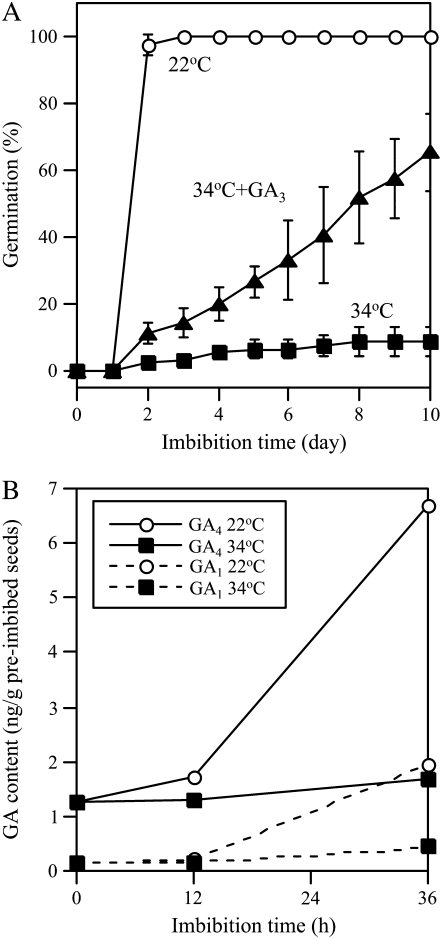
GA is known to be essential for germination of Arabidopsis seeds (Koornneef and van der Veen, 1980). To investigate the effect of GA on thermoinhibition of Arabidopsis seeds, seeds were imbibed at 34°C in the presence of GA3. Thermoinhibition of Col seeds was alleviated by 50 μm GA3 (Fig. 5A). This result suggests that enhancement only of ABA biosynthesis in seeds is not sufficient, but suppression of an increase in bioactive GA level is also required for thermoinhibition of Arabidopsis seeds. To evaluate this possibility, we determined endogenous GA levels during imbibition at optimal and supraoptimal temperature conditions. In Arabidopsis, GA4 is a main active GA and GA1 is a minor component during seed germination. At 22°C, both active GAs increased in abundance at 12 h after the start of imbibition, and the level of GA4 at 36 h was 5 to 6 times higher than the initial level (Fig. 5B). At 34°C, on the other hand, no increase in the levels of GA4 and GA1 was observed. Germination may be inhibited not only by ABA but also by the absence of active GAs at supraoptimal temperature conditions.
Figure 5.
Alleviation of thermoinhibition by exogenous GA and suppression of bioactive GA levels by high temperature. A, Effect of exogenous GA on thermoinhibition. After-ripened Col seeds were imbibed at 22°C (white circles), 34°C (black squares), and 34°C with 50 μm GA3 (black triangles) under continuous illumination. Error bars show sd (n = 3). B, Effect of temperature on bioactive GA levels in seeds. GA levels were quantified by GC-MS using two independent seed batches and similar results were obtained.
Suppression of GA Biosynthesis Gene Expression at High Temperature
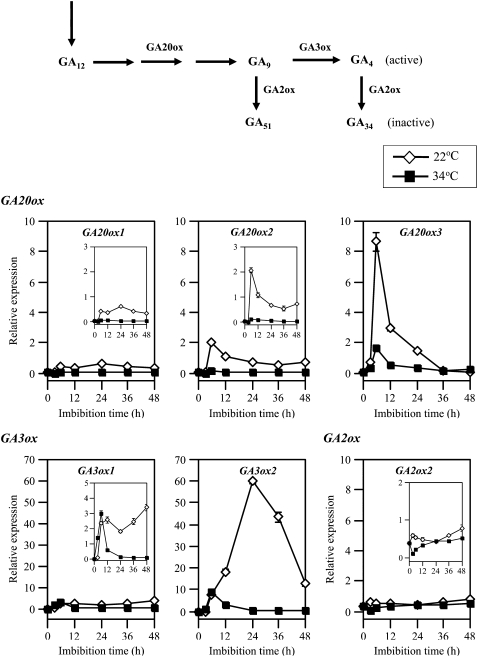
To investigate the regulation mechanism of GA biosynthesis by high temperature in seeds, we analyzed the expression of GA 20-oxidase (GA20ox1, GA20ox2, and GA20ox3) and GA 3-oxidase (GA3ox1 and GA3ox2) genes that are involved in the late steps of GA biosynthesis in Arabidopsis seeds (Ogawa et al., 2003). Expression of the three GA20ox genes was induced significantly within 6 h of imbibition at 22°C (Fig. 6). The transcript level of GA20ox1 was maintained from 6 to 48 h after the start of imbibition. GA20ox2 and GA20ox3 transcript levels peaked at 6 h and decreased thereafter. GA20ox3 showed the highest expression among the three GA20ox genes at 6 h, but its expression decreased to very low level at 36 h. At 34°C, on the other hand, expression of all three GA20ox genes was greatly suppressed (Fig. 6). Expression of the two GA3ox genes was also up-regulated within the first 6 h of imbibition at 22°C. GA3ox1 mRNA level was maintained from 6 h to 48 h of imbibition, but GA3ox2 expression showed a peak at 24 h (Fig. 6). At 34°C, expression of the two GA3ox genes was greatly suppressed, as observed for GA20ox genes. These results suggest that high temperature repressed bioactive GA synthesis through suppression of GA20ox and GA3ox gene expression in Arabidopsis seeds. Suppression of GA3ox1 and GA3ox2 expression by high temperature may be critical for thermoinhibition of Arabidopsis seeds.
Figure 6.
Expression of GA biosynthesis and deactivation genes was suppressed at high temperature. Total RNA was prepared from after-ripened Col seeds imbibed at 22°C (white diamonds) or at 34°C (black squares) under continuous illumination. Transcript levels were quantified by quantitative RT-PCR as described in “Materials and Methods.” Experiments were repeated twice with similar results. Data from one of the replicates are shown. Values are means with sds from three measurements.
To evaluate the contribution of GA deactivation to thermoinhibition, we analyzed transcript levels of GA2ox genes in germinating and thermoinhibited seeds. Quantitative RT-PCR experiments indicated that expression of GA2ox2, whose expression is photoreversibly regulated and most highly expressed in seeds among the eight GA2ox genes (Oh et al., 2006; Seo et al., 2006), stayed at low level during imbibition even at 34°C (Fig. 6). Other GA2ox genes (GA2ox1, GA2ox3–GA2ox8) also showed low expression levels, and there was no difference in expression levels between germinating and thermoinhibited seeds (data not shown). These results suggest that active GA levels are dependent on their synthesis in thermoinhibited seeds in the light.
GA Negative Regulators, SPY and RGL2, Are Involved in Thermoinhibition of Arabidopsis Seeds
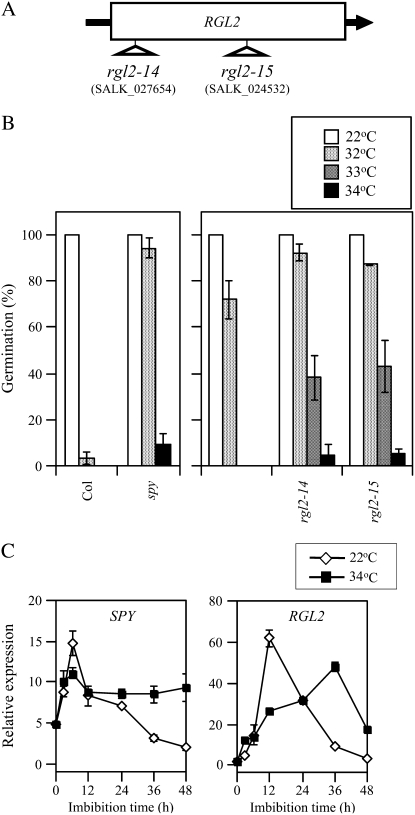
GA response is repressed by negative regulators, DELLA proteins and SPY, and GA signal is transduced through degradation of DELLA protein (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Sun and Gubler, 2004). To evaluate the role of GA negative regulators in thermoinhibition of Arabidopsis seeds, we investigated thermoinhibition resistance of the loss-of-function mutants of them. Seeds of spy-4 showed clear resistance to thermoinhibition (Fig. 7B). T-DNA insertion lines for all five DELLA protein genes were available at the ABRC, and we analyzed the homozygous T-DNA insertion mutants. Seeds of gai, rga, rgl1, and rgl3 knockout lines showed no clear resistance to thermoinhibition (data not shown). Seeds of rgl2 alleles showed slightly, but significantly, higher germination percentages at supraoptimal temperature conditions (Fig. 7B). These germination studies suggest that SPY and RGL2 are involved in the thermoinhibition mechanism of Arabidopsis seeds.
Figure 7.
Thermoinhibition-resistant germination of spy-4 and rgl2 seeds and expression of the GA negative regulator genes at high temperature. A, T-DNA insertion sites in the RGL2 gene. Arrow indicates 5′ to 3′ direction of the gene. B, Effect of imbibition temperature on germination of spy and rgl2 mutant seeds. Seeds were imbibed at 22°C (white bars), 32°C (light gray bars), 33°C (dark gray bars), and 34°C (black bars) under continuous illumination for 5 d. Error bars show sd (n = 3). C, Effect of temperature on expression of SPY and RGL2 genes. Total RNA was prepared from after-ripened Col seeds imbibed at 22°C (white diamonds) or at 34°C (black squares) under continuous illumination. Transcript levels were quantified by quantitative RT-PCR as described in “Materials and Methods.” Experiments were repeated twice with similar results. Data from one of the replicates are shown. Values are means with sds from three measurements.
To investigate whether the expressions of GA negative regulators are regulated by high temperature or not, we quantified the transcripts in imbibed seeds by quantitative RT-PCR. SPY gene expression was induced rapidly and transiently during imbibition at 22°C and reduced to a low level at 48 h (Fig. 7C). Expression of SPY was also rapidly induced by imbibition at 34°C and maintained at high level by 48 h. RGL2 showed significant induction by imbibition at 22°C and its expression peaked at 12 h. At 34°C, induction was delayed and a peak was observed at 36 h. These gene expression studies suggest that SPY and RGL2 work as negative regulators of GA signaling in thermoinhibited seeds and that high temperature enhances suppressive function of SPY by maintenance of its transcript level.
High Temperature Suppresses GA Action through ABA
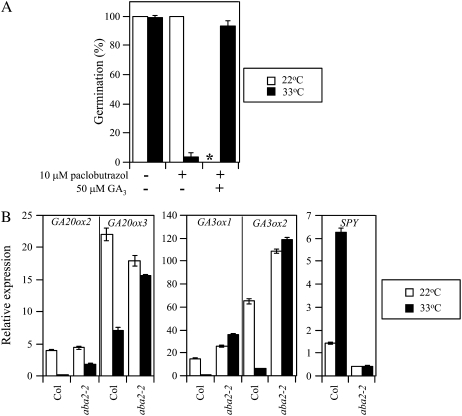
Recent studies have suggested that ABA suppresses GA levels in dark imbibed seeds treated with a far-red light pulse and in seedlings of Arabidopsis (Seo et al., 2006; Zentella et al., 2007). We therefore hypothesized that GA action might be suppressed by high temperature through the action of ABA in thermoinhibited seeds. To examine this possibility, we first analyzed germination of ABA-deficient mutant seeds imbibed at supraoptimal temperature in the presence of a GA biosynthesis inhibitor, paclobutrazol (PAC). The high temperature-resistant germination phenotype of aba2-1 mutant seeds was almost completely suppressed by PAC and the suppression was recovered by GA3 (Fig. 8A), demonstrating that GA biosynthesis is required for germination of aba2-1 seeds at supraoptimal temperature. These observations suggest that GA action is derepressed in the absence of ABA at supraoptimal temperature conditions. In other words, high temperature may regulate GA action through the action of ABA. To evaluate this possibility, expression of GA synthesis and signaling genes in aba2-2 mutant seeds was analyzed at supraoptimal temperature conditions. In this experiment, germination of wild-type (Col) seeds was almost completely suppressed (5.2% germination after 7 d of imbibition), and the seeds of aba2-2 showed thermoinhibition tolerance at 33°C (98.0% germination after 7 d of imbibition). Quantitative RT-PCR analysis indicated that the transcript levels of GA3ox1 and GA3ox2 in 24-h imbibed aba2-2 seeds were similar between 22°C and 33°C (Fig. 8B). Transcript levels of GA20ox2 and GA20ox3 in aba2-2 seeds imbibed at 33°C for 12 h were less abundant than those at 22°C, but more abundant than those of wild-type seeds imbibed at 33°C. These results suggest that expression of GA3ox1 and GA3ox2 genes is suppressed by ABA, and expression of GA20ox2 and GA20ox3 genes is suppressed by both ABA and other factors affected by temperature. These observations suggest that bioactive GA content in Arabidopsis seeds may be suppressed mainly through the action of ABA, which is enhanced by high temperature in the light (Fig. 9). We also found that the transcript level of SPY in aba2-2 seeds was not enhanced by high temperature (Fig. 8B). These results suggest that expression of SPY genes is up-regulated by ABA (Fig. 9).
Figure 8.
Modulation of GA synthesis and signaling by ABA in thermoinhibited seeds. A, Effect of GA biosynthesis inhibitor PAC on thermoinhibition-tolerant germination of ABA-deficient seeds. The seeds of aba2-1 were imbibed at 22°C and at 33°C under continuous illumination for 6 d in the presence or absence of PAC and GA3. All the imbibition solutions contained 10 μm fluridone to ensure ABA deficiency. Asterisk denotes that the germination test at 22°C in the presence of both PAC and GA3 was not done. Error bars show sd (n = 3). B, Expression of GA synthesis and signaling genes in ABA-deficient mutant seeds. Seeds of wild type (Col) and aba2-2 were imbibed at 22°C (white bars) and 33°C (black bars) under continuous illumination. Transcript levels were quantified by quantitative RT-PCR at 12 h (GA20ox) or at 24 h (GA3ox and SPY) after the start of imbibition. Experiments were repeated three times with different seed batches and similar results were obtained. Data from one of the replicates are shown. Values are means with sds from three measurements.
Figure 9.
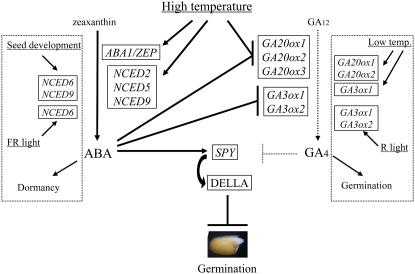
Regulation of ABA synthesis, GA synthesis, and GA signaling by high temperature in Arabidopsis seeds. High temperature enhances expression of specific ABA biosynthesis genes (ABA1/ZEP, NCED2, NCED5, and NCED9) under continuous illumination. High temperature also enhances expression of a GA negative regulator gene (SPY) and suppresses expression of GA biosynthesis genes (GA20ox1, GA20ox2, GA20ox3, GA3ox1, and GA3ox2) not directly, but through the action of ABA. Expression of GA20ox2 and GA20ox3 may be regulated by both ABA and high temperature. Transcripts of DELLA protein genes are not increased by high temperature, but their protein activity may be enhanced by SPY. A different set of NCED genes (NCED6 and NCED9) works during seed development and contributes to dormancy development. Expression of NCED6 is induced by far-red (FR) light, and those of GA3ox1 and GA3ox2 are induced by red (R) light. Expression of a set of GA biosynthesis genes (GA20ox1, GA20ox2, and GA3ox1) is induced by low temperature and suppressed by high temperature and may contribute to germination control in a wide range of temperatures. [See online article for color version of this figure.]
DISCUSSION
Regulation of ABA Biosynthesis by High Temperature in Seeds
Pharmacological studies have suggested that ABA biosynthesis is required for thermoinhibition of the seeds of lettuce, chickweed, and several winter annual species when imbibed in darkness (Yoshioka et al., 1998, 2003). In this study, we confirmed that ABA synthesis de novo after the start of imbibition is essential for germination inhibition of Arabidopsis seeds at supraoptimal temperature in the light (Figs. 1 and 9). We have reported genetic evidence that shows that ABA has a critical function in thermoinhibition of Arabidopsis seeds and that not all, but a subset of ABA signaling components, including ABI1, ABI3, and possibly ABI2, are required for germination inhibition at high temperature (Tamura et al., 2006). These observations suggest that seeds have a specific mechanism to modulate ABA content in response to high temperature and use a specific ABA-signaling pathway for germination inhibition.
Our aim in this study was to elucidate how ABA and GA action (metabolism and responsiveness) is controlled by high temperature. We observed the up-regulation of ABA1/ZEP expression before the increase of ABA level in thermoinhibited seeds (Figs. 1 and 2), suggesting that enhanced expression of ABA1/ZEP contributes to the elevated ABA production at high temperature. In Nicotiana plumbaginifolia, overexpression of NpZEP by the cauliflower mosaic virus 35S promoter causes an increase in endogenous ABA content in the seed and delays germination (Frey et al., 1999). They recently demonstrated, however, that ABA content does not correlate with the level of ABA1/ZEP expression in transgenic N. plumbaginifolia seeds in the NpZEP loss-of-function mutant background (Frey et al., 2006). These results indicate that altered ABA1/ZEP expression alone is not sufficient to affect the level of ABA accumulation. However, it would still be possible for altered ABA1/ZEP expression to contribute to the change in ABA level if a downstream limiting step is coregulated. Analysis of endogenous epoxycarotenoids and tissue- and developmental stage-specific suppression/overexpression studies are required to evaluate the impact of ABA1/ZEP expression on the level of ABA during thermoinhibition of Arabidopsis seed germination.
NCED is considered to be a key regulatory enzyme in ABA biosynthesis in drought-stressed plants (Qin and Zeevaart, 1999, 2002; Iuchi et al., 2001). In this study, we identified catalytic activity of recombinant NCED5 protein in vitro (Supplemental Fig. S1), and found that expression of NCED2, NCED5, and NCED9 was up-regulated by high temperature in seeds (Fig. 2). We confirmed the physiological relevance of NCED2, NCED5, and NCED9 genes to thermoinhibition by analyzing thermoinhibition-resistant germination phenotypes of the nced9 single mutant and multiple nced mutants. Interestingly, the degree of reductions in ABA levels in single and multiple nced mutant seeds imbibed at 34°C were roughly correlated with the sum of the expression levels of each NCED gene during the imbibition period in wild-type seeds (Figs. 2 and 3B). In addition, the increase in ABA level was preceded by the increase in NCED2 and NCED9 transcript levels in thermoinhibited seeds. These results suggest that enhancement of ABA synthesis by high temperature is regulated at the transcript level of NCED as in drought-stressed plants. Transcript levels of NCED2 and NCED5 were about one-half those of NCED9 in thermoinhibited wild-type seeds, but seeds of the nced2 nced5 double mutant had a higher level of ABA than the nced9 single mutant (Figs. 2 and 3B). The relatively high expression level of NCED9 after 3 to 12 h of imbibition at 34°C may contribute to the above difference. It is also possible that the spatial distribution of carotenoid precursors and expression of ABA biosynthesis genes, including NCED itself, in a seed affect the contribution of each NCED to ABA biosynthesis.
Environmental, Developmental, and Spatial Regulation of NCED Gene Family Members Is Involved in Regulation of ABA Level and Germination
Among the five NCED genes in Arabidopsis, expression of NCED3 is induced by drought stress in plants (Iuchi et al., 2001; Ruggiero et al., 2004). In this study, we found that expression of NCED2, NCED5, and NCED9 was up-regulated by high temperature in after-ripened seeds under continuous illumination. NCED6 and NCED9 are responsible for ABA synthesis during seed development and dormancy (Lefebvre et al., 2006). In contrast to NCED9, NCED6 showed weak expression during imbibition, and T-DNA insertion mutants of NCED6 showed no resistance to thermoinhibition (Supplemental Fig. S2). Recently, by using a strongly dormant Arabidopsis accession, Cape Verde Islands (Cvi), Cadman et al. (2006) reported that expression of NCED6 is up-regulated in primary dormant and secondary dormant seeds imbibed in the dark. Seo et al. (2006) reported that NCED6 expression in seeds is induced during dark imbibition after far-red light treatment and suppressed by subsequent red-light treatment. Therefore, NCED6 may work for ABA production and contribute to acquire dormancy during seed development and to maintain the resting/dormant state of mature imbibed seeds in the dark condition. These results indicate that a subset of NCED paralogs responds to specific environmental and developmental cues, and they encode enzymes that catalyze ABA production for proper response of the plant.
The endosperm is thought to be a barrier for radicle protrusion and ABA has been shown to inhibit the endosperm rupture of Arabidopsis and cress (Lepidium sativum) seeds (Müller et al., 2006). They also found that the endosperm weakens prior to its rupture and that ABA delays the onset and decreases the rate of this weakening process. In lettuce seeds, thermoinhibition is alleviated by removing or puncturing the endosperm (Dutta and Bradford, 1994). From spatial expression analysis of NCED genes during development of Arabidopsis seeds, Lefebvre et al. (2006) suggested that ABA synthesized in the endosperm contributes to the induction of seed dormancy. We found that expression of NCED9 was predominant over other NCED genes in the embryo, whereas NCED5 expression was predominant in the endosperm of thermoinhibited seeds (Fig. 4). Thermoinhibition tolerance of nced9 and the thermoinhibition-sensitive phenotype of the nced5 single mutant and the nced2 nced5 double mutant may suggest that ABA synthesis in the embryo is critical for suppression of germination at supraoptimal temperature. Expression of the three NCED genes in the embryo and the endosperm and enhancement of their expression at high temperature in both tissues may suggest that ABA synthesized in the endosperm is also involved in thermoinhibition of the seeds. It is also possible that compensation by NCED9 in the absence of NCED2 and NCED5 expression in the endosperm enhances the barrier function of the endosperm. Whatever the case, both tissues may cooperatively regulate seed germination by sensing and responding to temperature.
Suppression of GA Synthesis Is Critical for Thermoinhibition
Alleviation of thermoinhibition by exogenous GA3 in lettuce and Arabidopsis seeds suggests that suppression of active GA synthesis is required for thermoinhibition (Gonai et al., 2004; Fig. 5A). Lettuce seeds do not increase bioactive GA (GA1) content by 12 h of imbibition at supraoptimal temperature (33°C) in the dark (Gonai et al., 2004). Because bioactive GA synthesis is regulated by light during seed germination (Yamaguchi and Kamiya, 2002), it is not clear whether low GA content is due to the absence of light or to high temperature in thermoinhibited lettuce seeds. In this study, we showed that thermoinhibited Arabidopsis seeds did not increase bioactive GA content even in the light condition. This indicates that the bioactive GA level is controlled by high temperature regardless of the light condition. Gonai et al. (2004) reported that thermoinhibition of lettuce seeds at 33°C is not rescued by fluridone or GA3 alone, but by combined application of fluridone and GA3. They also showed that exogenous GA3 lowers ABA content by enhancing catabolism of ABA presumably through the 8′-hydroxylation pathway. In Arabidopsis seeds, application of fluridone or GA3 alleviated thermoinhibition, but the seeds did not show 100% germination at 34°C (Figs. 1 and 5A). Combined application of fluridone and GA3 allowed almost 100% germination at 34°C (data not shown). We also examined germination at 36°C and found that fluridone or GA3 alone rescued germination only slightly, but the combined application allowed 60% to 70% of the seeds to germinate (data not shown), as observed in lettuce seeds (Gonai et al., 2004). Thermoinhibition might be achieved by maintaining ABA content above the threshold level of germination not only through its biosynthesis, but also through repression of its catabolism mediated by GA also in Arabidopsis seeds. We found that expression of three ABA 8′-hydroxylase genes, CYP707A1, CYP707A2, and CYP707A3, was suppressed at 34°C, but the ABA content showed a similar decreased rate in the presence of fluridone at 34°C and in the absence of fluridone at 22°C (Figs. 1 and 2). This suggests that ABA catabolism has little contribution to ABA content at 34°C, but it is possible that suppression of ABA catabolism contributes to up-regulation of ABA levels at higher temperature conditions. It is interesting to know whether expression of CYP707A genes is modulated by the action of GA o r not.
During germination of Arabidopsis seeds, GA20ox genes, GA20ox1, GA20ox2, and GA20ox3, and GA3ox genes, GA3ox1 and GA3ox2, are involved in GA synthesis (Ogawa et al., 2003). Suppression of all these GA20ox and GA3ox genes at supraoptimal temperature (Fig. 6) may be responsible for suppression of GA synthesis. Mitchum et al. (2006) reported that seeds of the ga3ox1 ga3ox2 double mutant do not germinate in the light at 22°C and their germination is rescued by application of bioactive GA. Therefore, suppression of GA3ox1 and GA3ox2 genes at high temperature should be critical for thermoinhibition. Interestingly, expression of GA20ox1, GA20ox2, and GA3ox1 has been reported to be up-regulated by low temperature (Yamauchi et al., 2004). Thus, GA synthesis is likely to be regulated by a wide range of temperature through modulation of GA20ox and GA3ox gene expression (Fig. 9). Expression of GA3ox genes in lettuce and Arabidopsis is well known to be photoreversibly regulated by light through phytochrome (Toyomasu et al., 1998; Yamaguchi et al., 1998). Understanding the cross talk between light and temperature signals on the regulation of GA3ox gene expression may give us insight into how plants transduce multiple signals and how plants respond to complex environmental stimuli.
The GA2ox2 gene showed little expression during imbibition even at supraoptimal temperature conditions (Fig. 6). Expression of GA2ox2 is regulated via phytochrome in a photoreversible manner as observed for NCED6 (Seo et al., 2006). Therefore, this gene might be suppressed because of continuous illumination conditions adopted in this study. In fact, expression of GA2ox2 is up-regulated in primary dormant and secondary dormant Cvi seeds imbibed in the dark (Cadman et al., 2006). Thus, it will be informative to examine the effect of high temperature on GA2ox2 expression in imbibed seeds in darkness. In the dark condition, for example, when the seeds are buried in the soil in summer, GA deactivation may contribute to thermoinhibition of the seeds.
Function of GA Negative Regulators on Thermoinhibition
Loss-of-function mutations in GA negative regulators, SPY and DELLA proteins, allow GA response in the absence of active GA (Jacobsen and Olszewski, 1993; Peng and Harberd, 1997; Silverstone et al., 1997, 1998; Dill and Sun, 2001; King et al., 2001; Lee et al., 2002). Overexpression of SPY in the wild-type and spy-3 backgrounds reduces GA response and enhances sensitivity to ABA during seed germination (Swain et al., 2001). Overexpression lines also show slow germination under normal conditions. Our results indicated that the seeds of loss-of-function mutant spy-4 were highly resistant to thermoinhibition (Fig. 7B). SPY transcripts decreased to low level at 22°C, but maintained relatively high level at 34°C (Fig. 7C). In addition to its role as a GA negative regulator, SPY also acts as a positive regulator of cytokinin signaling (Greenboim-Wainberg et al., 2005). If SPY exerts its function on thermoinhibition through cytokinin signaling, cytokinin should inhibit germination of seeds. However, application of cytokinin alleviates thermoinhibition of lettuce seeds (Odegbaro and Smith, 1969). These observations support the idea that up-regulation of SPY expression by high temperature is involved in the thermoinhibition mechanism of Arabidopsis seeds through regulation of the GA response (Fig. 9). In rice, the SPY ortholog (OsSPY) is suggested to work as a negative regulator of brassinosteroid biosynthesis (Shimada et al., 2006). Application of brassinosteroids to the seeds of GA-deficient and GA-insensitive mutants of Arabidopsis partially rescues germination phenotypes (Steber and McCourt, 2001). It is therefore also possible that SPY suppresses germination by repressing brassinosteroid biosynthesis.
High temperature-resistant germination of rgl2 seeds suggests that RGL2 works as a repressor of Arabidopsis seed germination at supraoptimal temperature conditions. The thermoinhibition-resistant germination phenotype of rgl2, however, was mild when compared with the germination of spy-4 and ABA-deficient mutant seeds (Figs. 3 and 7B; Supplemental Fig. S2). It is possible that SPY acts as a positive regulator of ABA action in addition to its role as a GA negative regulator, as reported in barley (Hordeum vulgare) aleurone (Robertson et al., 1998). A germination study of multiple DELLA protein mutants suggested that RGL2 is the predominant repressor of Arabidopsis seed germination in the light and GAI, RGA, and RGL1 enhance the function of RGL2 (Cao et al., 2005). Recently, RGA and GAI have been shown to play a role in inhibiting germination of dark-imbibed seeds in the absence of active phytochromes (Oh et al., 2007). DELLA proteins other than RGL2 may enhance RGL2 function also in supraoptimal temperature conditions. It is also possible that high temperature stabilizes DELLA protein through an increase in ABA levels because GFP-fused RGA protein was reported to be stabilized by exogenous ABA in root cells (Achard et al., 2006). However, a recent report showed that ABA has no effect on the stability of endogenous RGA protein in seedlings of the GA-deficient mutant, ga1-3, in the absence or presence of exogenous GA (Zentella et al., 2007). Further analysis on the levels of DELLA proteins in seeds will increase our knowledge on the effect of ABA on GA signaling and germination inhibition.
Regulation of GA Action by ABA
Suppression of the thermoinhibition-resistant germination phenotype of ABA-deficient mutant seeds by a GA biosynthesis inhibitor, PAC, indicates that GA is necessary for germination in the absence of ABA at supraoptimal temperature conditions. This suggests the possibility that GA biosynthesis is suppressed at high temperature through the action of ABA (Figs. 8 and 9). The significant expression of GA3ox1, GA3ox2, GA20ox2, and GA20ox3 genes in ABA-deficient mutant seeds at high temperature strongly suggests that these GA biosynthesis genes are regulated by ABA. The partial derepression of GA20ox2 and GA20ox3 expression at the ABA-deficient conditions may indicate that these two genes are regulated by both ABA and other factors induced by high temperature. Expression of GA20ox1 is reported to be significantly reduced by exogenous ABA in seedlings (Zentella et al., 2007). Seo et al. (2006) suggested that ABA down-regulated GA levels through suppression of GA3ox1 and GA3ox2 genes in far-red light-irradiated and dark-imbibed seeds, but ABA plays a relatively minor role in regulating GA levels compared with the role played by light. In our illuminated conditions, expression of GA3ox1 and GA3ox2 was severely repressed at high temperature in wild-type seeds, but their expression was completely derepressed in ABA-deficient mutant seeds at high temperature (Fig. 8). Our present data suggest that ABA plays a major role in the suppression of GA levels at high temperature in the light. The suppression of SPY gene expression in ABA-deficient mutant seeds imbibed at high temperature suggests that expression of SPY is up-regulated through the action of ABA. ABA may inhibit GA action by suppressing GA biosynthesis and also by suppressing GA signaling (Fig. 9).
A Common Germination Control Mechanism in Dormant and After-Ripened Seeds Changes Sensitivity to Temperature during After-Ripening?
Fluridone and GA treatments induce germination of dormant seeds of N. plumbaginifolia and Arabidopsis (Grappin et al., 2000; Jullien et al., 2000). De novo ABA synthesis and suppression of GA synthesis may be required for maintaining dormancy as well as for thermoinhibition of the after-ripened seeds. Expression of ABA1/ZEP, NCED2, and NCED9 is up-regulated and GA3ox2 is down-regulated in primary and secondary dormant Cvi accession seeds as in thermoinhibited Col seeds (Fig. 2; Cadman et al., 2006). Up-regulation of NCED6 and GA2ox2 in the dormant seeds imbibed in darkness, which was not observed in the thermoinhibited seeds imbibed in the light, may be the result of the difference in the light condition because the expression of these two genes is regulated by phytochrome (Seo et al., 2006). The significant decrease in ABA level, which was observed in the thermoinhibited seeds in the first several hours of imbibition, was also reported for dormant seeds of Arabidopsis C24 and Cvi accessions imbibed at room temperature (Ali-Rachedi et al., 2004; Millar et al., 2006). CYP707A2 expression in dormant C24 seeds also increases transiently during the first 3 h of imbibition, but the peak level is about one-half of after-ripened seeds, which is similar to what we observed in thermoinhibited seeds (Millar et al., 2006; Fig. 2). These results suggest that a common genetic mechanism is involved in ABA and GA regulation in dormant seeds imbibed at room temperature and in after-ripened seeds imbibed at high temperature. Seed responsiveness to temperature is closely related to the level of dormancy, and the maximal temperature for germination rises gradually during an after-ripening period in field-grown soil-buried seeds of winter annual species and in laboratory-grown Arabidopsis seeds (Baskin and Baskin, 1998; Tamura et al., 2006). Tamura et al. (2006) isolated high temperature-resistant germination mutants of Arabidopsis from after-ripened seed batches and showed that they have reduced seed dormancy. They also indicated that the seeds of Arabidopsis reduced-dormancy mutants rdo1, rdo2, rdo3, and rdo4 are thermoinhibition tolerant. Among five ABA-insensitive mutants of Arabidopsis, the seeds of abi1-1, abi2-1, and abi3 alleles show reduced dormancy and thermoinhibition tolerance and the seeds of abi4 and abi5 alleles show normal levels of dormancy and normal thermoinhibition sensitivity. Dormant and after-ripened seeds may utilize a common genetic mechanism to modulate germination and the change in sensitivity to temperature may occur during after-ripening on the modulation mechanism of ABA and GA levels in the seeds.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
We used the Col-0 accession of Arabidopsis (Arabidopsis thaliana) for germination, hormone, and gene expression analyses. Seeds of spy-4 were kindly provided by Stephen M. Swain. Seeds of aba1-1 (CS21); T-DNA insertion lines of SALK (Alonso et al., 2003), nced2-1 (SALK_026541), nced2-2 (SALK_052555), nced2-3 (SALK_090937), nced2-4 (SALK_004231), nced9-1 (SALK_033388), nced9-2 (SALK_123975), rgl2-14 (SALK_027654), and rgl2-15 (SALK_024532); and Wisconsin, nced5-1 (CS853457, WiscDsLox377–380M20) and nced6-3 (CS852600, WiscDsLox356H02), were obtained from the ABRC (The Ohio State University). Plants were grown as described previously (Tamura et al., 2006).
Isolation of T-DNA Insertion Homozygotes and nced Multiple Mutants
Plants homozygous for T-DNA insertion in each gene were identified by the method described in SIGnAL (http://signal.salk.edu) of the Salk Institute Genomic Analysis Laboratory with T-DNA left-border primers LB_6313R for SALK lines and p745 (http://www.hort.wisc.edu/krysan/2010/default.htm) for WiscDsLox lines and the gene-specific primers listed in Supplemental Table S1. T-DNA insertion points were identified by sequencing the amplified left-border flanking regions.
To generate multiple nced mutants, we first crossed nced2-1 and nced9-1, and genotypes of F2 plants were identified by PCR with the left-border primers and the gene-specific primers. Then we crossed the nced2 nced9 double mutant with nced5-1, and the triple and nced2 nced5 and nced5 nced9 double mutants were isolated from the segregants by PCR as described above.
Germination Tests
Seeds were harvested at physiological maturity when about one-half of the fruits on a plant turned to yellow and were stored in a desiccator for after-ripening for about 2 months at room temperature. Thirty seeds were imbibed with 300 μL of water in a well of a 24-well plate. Seeds were imbibed at constant temperature under continuous fluorescent light (18 μmol m−2 s−1). Fluridone (recrystallized; Daw Elanco) and GA3 (Sigma) were first dissolved in dimethylsulfoxide (DMSO) and then diluted to the final concentrations with water. The final concentration of DMSO was usually 0.01%, and the maximal concentration was restricted to 0.1% because DMSO affects germination of Arabidopsis seeds at higher than 0.1%. Germination was scored as radicle protrusion. All germination tests were done at least in two independent seed batches with three replicates in each. A typical result was presented because there were some differences in germination percentages from batch to batch, but the results were almost parallel.
GA and ABA Analysis
One gram and 0.4 g of dry seeds were used for GA and ABA measurement, respectively. Quantitative analysis of GA and ABA by GC-MS was performed using 2H-labeled GAs and 13C-labeled ABA as internal standards, as described previously (Gawronska et al., 1995; Cheng et al., 2002). GA and ABA levels were determined twice using independent seed batches. In the case of nced single and multiple mutants, ABA was extracted from 5 (nonimbibed) or 10 mg (imbibed) seeds with deuterium-labeled d6-ABA purchased from ICON SERVICES as an internal standard, and quantified by liquid chromatography-tandem mass spectrometry as described previously (Saika et al., 2007).
Quantitative Real-Time PCR
Total RNA was isolated using an RNAqueous kit with plant RNA isolation aid (Ambion). Total RNA (2 μg) was treated with RNase-free DNase (Promega) to eliminate genomic DNA contamination. First-strand cDNA was synthesized with random hexamers using a SuperScript first-strand synthesis system according to the manufacturer's instructions (Invitrogen). Quantitative RT-PCR with Taq-Man technology (Holland et al., 1991) was performed using first-strand cDNA as a template on a sequence detector system (model 7700; Applied Biosystems). For quantitative RT-PCR, 12.5 μL of QuantiTect Probe PCR kit (Qiagen), 5 μL of cDNA, 0.2 μm of Taq-Man probe, and 0.8 μm of each primer were used in triplicate 25-μL reactions and subjected to the following cycling conditions: 50°C for 2 min, followed by 95°C for 15 min, then 45 cycles of 95°C for 15 s, and 60°C for 1 min. The sequence of primers and Taq-Man probes for ZEP, NCEDs, ABA2, AAO3, CYP707As, GA20oxs, GA3oxs, and GA2ox2 were as described previously (Ogawa et al., 2003; Kushiro et al., 2004; Seo et al., 2004; Yamauchi et al., 2004). Primers and probes for SPY and RGL2 are listed in Supplemental Table S2. Data were analyzed using ABI Prism 7700 SDS software (Applied Biosystems). To compare mRNA levels across different genes, the absolute amount of each target transcript was determined by generating standard curves using a series of known concentrations of target sequences. Genomic DNA (ABA1/ZEP, ABA2, AAO3, NCED2, NCED3, NCED5, NCED6, NCED9; Figs. 2 and 4) and cDNA (GA20ox1, GA20ox2, GA20ox3, GA3ox1, GA3ox2, GA2ox2, SPY, RGL2; Figs. 6, 7C, and 8B) were used as the target sequences. Then the data were normalized to the amplification of 18S rRNA internal control. For each sample, the mean value from triplicate real-time PCRs was adapted to calculate the transcript abundance, and the mean values were plotted with the sds. To confirm biological reproducibility, experiments were performed at least twice using different seed batches and we obtained similar results. We also obtained similar results by two-color microarray analysis (Arabidopsis II 22k array; Agilent) with RNA samples from another two different seed batches (A. Imamura, S. Toh, and N. Kawakami, unpublished data).
Assay of NCED Activity
The procedure used for the assay of NCED enzyme activity was as described previously (Iuchi et al., 2000). Recombinant NCED5 or NCED3 protein was incubated with 9′-cis-neoxanthin under 37 mm BisTris, pH 6.7, 0.05% (v/v) Triton X-100, 1 mm ascorbate, and 500 μm FeSO4 at room temperature for 1 h (total volume 100 μL). After the addition of 1 mL of water, the reaction mixture was extracted twice with 1 mL of ethyl acetate. The ethyl acetate fractions were combined, concentrated, and analyzed by HPLC on a column of Senshu Pak ODS H 3151 (150 mm length, 8 mm i.d.; Senshu Scientific). The column was eluted with a linear gradient between solvent A (85:15 [v/v], methanol:water) and solvent B (1:1 [v/v], chloroform:methanol) at a flow rate of 1.5 mL min−1. The concentration of solvent B was increased from 10% to 50% in 25 min and kept at 50% for 5 min. The elution from the column was monitored with a UV/visible detector at 440-nm wavelength to detect substrate carotenoids and C25 products.
To identify cis-xanthoxin, the ethyl acetate extract of the reaction mixture was submitted to HPLC purification using the column described above. The column was eluted with 50% (v/v) aqueous methanol at a flow rate of 1.5 mL min−1, and the elution from the column was monitored with a UV detector at 260-nm wavelength. The predicted cis-xanthoxin fraction was collected and submitted to GC-MS analysis.
Separation of Embryo and Endosperm with Testa
Col seeds were imbibed at 22°C and 34°C for 24 h and endosperm/testa and embryo were separated under a microscope. Approximately 850 embryos and endosperms/testas were pooled for each RNA isolation. RNA extraction, cDNA synthesis, and quantitative RT-PCR were done as described above, but cDNA was made from 1 μg of total RNA and quantitative RT-PCR was done with 10-μL reactions by the following cycling conditions: 95°C for 20 s, 45 cycles of 95°C for 3 s, and 60°C for 30 s. Results from embryo and endosperm/testa were normalized to the amplification of the 18S rRNA control and the amount of total RNA extracted from each tissue of one seed.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. NCED5 has 9-cis-epoxicarotenoid dioxygenase activity in vitro.
Supplemental Figure S2. Thermoinhibition-resistant germination of seeds from ABA-deficient mutants and lines with T-DNA insertion in NCED genes.
Supplemental Figure S3. Evaluation of embryo and endosperm separation by quantitative RT-PCR of EPR1 and GA3ox2 transcripts.
Supplemental Table S1. Gene-specific PCR primer sequences for T-DNA insertion analysis.
Supplemental Table S2. Sequences of gene-specific primers and Taq-Man probes for quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Mitsunori Seo, Mikihiro Ogawa, and Yukika Yamauchi for critical discussion. We also thank Stephen M. Swain and Hiroyuki Hirano for providing spy-4 seeds and fluridone, respectively. T-DNA insertion lines of SALK and Wisconsin were obtained from the ABRC.
This work was supported by the Japan Society for the Promotion of Science for Young Scientists (research fellowship to S.T.).
This article is dedicated to the memory of Akira Watanabe.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Naoto Kawakami (kawakami@isc.meiji.ac.jp).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abeles FB (1986) Role of ethylene in Lactuca sativa cv. ‘Grand Rapids’ seed germination. Plant Physiol 81 780–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94 [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H (2001) Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol 125 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, Jullien M (2004) Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219 479–488 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Audran C, Liotenberg S, Gonneau M, North H, Frey A, Tap-Waksman K, Vartanian N, Marion-Poll A (2001) Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Aust J Plant Physiol 28 1161–1173 [Google Scholar]
- Baskin CC, Baskin JM (1998) Germination ecology of seeds with nondeep physiological dormancy. In Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination. Academic Press, San Diego, pp 49–85
- Baskin JM, Baskin CC (1983) Seasonal changes in the germination responses of buried seeds of Arabidopsis thaliana and ecological interpretation. Bot Gaz 144 540–543 [Google Scholar]
- Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK (1952) A reversible photoreaction controlling seed germination. Proc Natl Acad Sci USA 38 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS (2006) Unequal genetic redundancies in Arabidopsis—a neglected phenomenon? Trends Plant Sci 11 492–498 [DOI] [PubMed] [Google Scholar]
- Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB (1999) Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J 17 427–431 [DOI] [PubMed] [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46 805–822 [DOI] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113 [DOI] [PubMed] [Google Scholar]
- Carter AK, Stevens R (1998) Using ethephon and GA3 to overcome thermoinhibition in “Jalapeno M” pepper seed. HortScience 33 1026–1027 [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JA (2000) Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol 124 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, Kermode AR (2003) A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J 35 405–417 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreucq B, Berger N, Vincent E, Boisson M, Pelletier G, Caboche M, Lepiniec L (2000) The Arabidopsis AtEPR1 extensin-like gene is specifically expressed in endosperm during seed germination. Plant J 23 643–652 [DOI] [PubMed] [Google Scholar]
- Dutta S, Bradford KJ (1994) Water relations of lettuce seed thermoinhibition. II. Ethylene and endosperm effects on base water potential. Seed Sci Res 4 11–18 [Google Scholar]
- Dutta S, Bradford KJ, Nevins DJ (1994) Wall autohydrolysis in isolated endosperms of lettuce (Lactuca sativa L.). Plant Physiol 104 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A, Audran C, Marin E, Sotta B, Marion-Poll A (1999) Engineering seed dormancy by the modification of zeaxanthin epoxidase gene expression. Plant Mol Biol 39 1267–1274 [DOI] [PubMed] [Google Scholar]
- Frey A, Boutin JP, Sotta B, Mercier R, Marion-Poll A (2006) Regulation of carotenoid and ABA accumulation during the development and germination of Nicotiana plumbaginifolia seeds. Planta 224 622–632 [DOI] [PubMed] [Google Scholar]
- Gallardo M, Delgado MD, Sanchez-Calle IM, Matilla AJ (1991) Ethylene production and 1-aminocyclopropane-1-carboxylic acid conjugation in thermoinhibited Cicer arietinum L. seeds. Plant Physiol 97 122–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronska H, Yang Y-Y, Furukawa K, Kendrick R, Takahashi N, Kamiya Y (1995) Effects of low irradiance stress on gibberellin levels in pea seedlings. Plant Cell Physiol 36 1361–1367 [Google Scholar]
- Gonai T, Kawahara S, Tougou M, Satoh S, Hashiba T, Hirai N, Kawaide H, Kamiya Y, Yoshioka T (2004) Abscisic acid in the thermoinhibition of lettuce seed germination and enhancement of its catabolism by gibberellin. J Exp Bot 55 111–118 [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M (2000) Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 210 279–285 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48 431–460 [DOI] [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH (1991) Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 88 7276–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27 325–333 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2000) A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol 123 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien M, Bouinot D, Ali-Rachedi S, Sotta B, Grappin P (2000) Abscisic acid control of seed dormancy expression in Nicotiana plumbaginifolia and Arabidopsis thaliana. In J-D Viemont, J Crabbe, eds, Dormancy in Plants. CABI Publishing, Wallingford, UK, pp 195–210
- King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen J (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58 257–263 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45 309–319 [DOI] [PubMed] [Google Scholar]
- Madakadze R, Chirco EM, Khan AA (1993) Seed germination of three flower species following matriconditioning under various environments. J Am Soc Hortic Sci 118 330–334 [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- McCarty DR (1995) Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol 46 71–93 [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J 45 942–954 [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45 804–818 [DOI] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47 864–877 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56 165–185 [DOI] [PubMed] [Google Scholar]
- Odegbaro OA, Smith OE (1969) Effects of kinetin, salt concentration, and temperature on germination and early seedling growth of Lactuca sativa L. J Am Soc Hortic Sci 94 167–170 [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006) Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47 124–139 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14 S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113 1051–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T, Thompson P (1971) Characteristics of the high temperature inhibition of germination of lettuce (Lactuca sativa). Physiol Plant 24 544–547 [Google Scholar]
- Richards DE, King KE, Ait-Ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52 67–88 [DOI] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE (1998) Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero B, Koiwa H, Manabe Y, Quist TM, Inan G, Saccardo F, Joly RJ, Hasegawa PM, Bressan RA, Maggio A (2004) Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol 136 3134–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, et al (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48 287–298 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45 1694–1703 [DOI] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, et al (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48 354–366 [DOI] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T (2000. a) Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J 23 481–488 [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000. b) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48 390–402 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996) Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PY, Martinez EC, Sun TP (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, McCourt P (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55 197–223 [DOI] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Olszewski NE (2001) Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol 126 1174–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N, Yoshida T, Tanaka A, Sasaki R, Bando A, Toh S, Lepiniec L, Kawakami N (2006) Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol 47 1081–1094 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis-epoxycarotenoid dioxygenase gene family. Plant J 35 44–56 [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000. a) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42 833–845 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Symonds RC, Mulholland BJ, Dadswell AR, Blake PS, Burbidge A, Taylor IB (2000. b) Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J 23 363–374 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y (1998) Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol 118 1517–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, Yu F, Jikumaru Y, Ross J, Cortes D, et al (2007) Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y (2002) Gibberellin and light-stimulated seed germination. J Plant Growth Regul 20 369–376 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun TP (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28 443–453 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun TP (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Endo T, Satoh S (1998) Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol 39 307–312 [Google Scholar]
- Yoshioka T, Gonai T, Kawahara S, Satoh S, Hashiba T (2003) The regulation of the thermoinhibition of seed germination in winter annual plants by abscisic acid. In G Nicolás, KJ Bradford, D Côme, HW Pritchard, eds, The Biology of Seeds: Recent Research Advances. CABI Publishing, Oxon, pp 217–223
- Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39 439–473 [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, et al (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.