Abstract
To establish functional circuitry, retinal neurons occupy spatial domains by arborizing their processes, which requires the self-avoidance of neurites from an individual cell, and by spacing their cell bodies, which requires positioning the soma and establishing a zone within which other cells of the same type are excluded1. The mosaic patterns of distinct cell types form independently and overlap. The cues that direct these processes in the vertebrate retina are not known2,3. Here we show that some types of retinal amacrine cells from mice with a spontaneous mutation in Down syndrome cell adhesion molecule (Dscam), a gene encoding an immunoglobulin-superfamily member adhesion molecule4,5, have defects in the arborization of processes and in the spacing of cell bodies. In the mutant retina, cells that would normally express Dscam have hyperfasciculated processes, preventing them from creating an orderly arbor. Also, their cell bodies are randomly distributed or pulled into clumps rather than being regularly spaced mosaics. Our results indicate that mouse DSCAM mediates isoneuronal self-avoidance for arborization and heteroneuronal self-avoidance within specific cell types to prevent fasciculation and to preserve mosaic spacing. These functions are analogous to those of Drosophila DSCAM (ref. 6) and DSCAM2 (ref. 7). DSCAM may function similarly in other regions of the mammalian nervous system, and this role may extend to other members of the mammalian Dscam gene family.
We have identified a spontaneous mutation in mice that creates a loss-of-function allele of Dscam, the Drosophila homologues of which function in both isoneuronal self-avoidance for dendrite arborization and heteroneuronal self-avoidance for axon tiling7–12. The recessive mutation arose in the BALB/cByJ genetic background and caused an overt neurological phenotype. Mutant and wild-type mice are indistinguishable at birth, but are severely uncoordinated by postnatal day 3 (P3); as adults, the mutant mice have spontaneous seizures and kyphosis, but are fertile and long-lived (>24 months; see http://www.jax.org/research/media/wild_type.html for video). Through positional cloning (see Methods), a mutation was identified in Dscam. Sequencing of genomic and complementary DNA from affected mice revealed a 38-bp deletion in exon 17, causing a frame shift resulting in ten unique amino acids followed by a premature stop codon (Supplementary Fig. 1). This mutation truncates the protein in the second fibronectin repeat (Fig. 1a). Dscam messenger RNA levels in the brain were reduced by 70% in affected mice, consistent with nonsense-mediated decay (Fig. 1b).
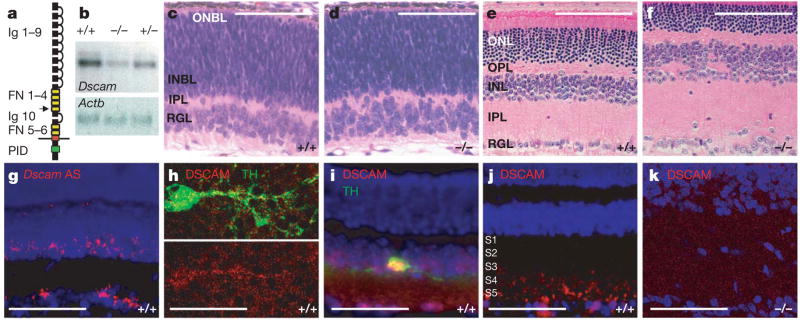
Figure 1. Identification of a mouse Dscam mutation.
a, A schematic of the DSCAM protein domain structure. The extracellular portion of DSCAM consists of ten immunoglobulin-like repeats (Ig) and six fibronectin domains (FN). The Dscam deletion truncates the coding sequence in the second fibronectin domain (arrow), which is before the transmembrane and PAK1-interacting domains (PID). b, Northern blotting of mRNA purified from whole brains of wild-type (+/+), Dscam+/− and Dscam−/− mice revealed a 70% reduction in Dscam mRNA in the mutant sample. β-actin (Actb) was used as a loading control. c–f, Haematoxylin and eosin stained sections of Dscam−/− and wild-type retinas from P0 and adult mice. The Dscam−/− retina is indistinguishable from that of the wild type during embryonic stages of development and at birth (c, d). INBL, innerneuroblast layer; ONBL, outerneuroblast layer; ONL, outernuclear layer; OPL, outerplexiform layer. e, f, In the adult Dscam−/− retina, the inner nuclear (INL), inner plexiform (IPL) and retinal ganglion (RGL) layers are disorganized compared to those of the wild-type retina, whereas other retinal layers are indistinguishable from that of the wild type. g, In Wsitu hybridization with Dscam antisense probes (Dscam AS) revealed expression in a subset of cells in the inner nuclear and ganglion cell layers. h, Whole control P15 retina stained with antibodies to DSCAM and TH. i, A section of a control adult (6–10-week-old) retina labelled using antibodies to DSCAM and TH. j, A section of wild-type P15 retina stained with DSCAM antibodies. k, A section of the Dscam−/− retina stained with an antibody to DSCAM. Scale bars: c, d, 80 μm; e, f, 106 μm; g, 160 μm; h, 45 μm; i, 120 μm; j, k, 65 μm.
Despite the overt neurological phenotype of Dscam−/− mice, examination of the central nervous system of adults and developing embryos did not reveal any gross disorganization, with the exception of a caudal folium of the cerebellum (Supplementary Fig. 2), implying a subtle phenotype or a subpopulation of affected cells.
We did find disorganization of postnatal retinal anatomy (Supplementary Fig. 3). At birth or before, the mutant retina was indistinguishable from that of control mice (Fig. 1c, d). However, by P4, the ganglion cell layer and developing inner plexiform layer (IPL) were disorganized (not shown). This disorganization persisted into adulthood in amacrine and ganglion cells (Fig. 1e, f), and was confirmed with marker analysis (Supplementary Fig. 4). Within these cell populations, many subtypes are present13,14, and Dscam was expressed in a subset of cells in these layers (Fig. 1g). Double in situ hybridization with markers of defined amacrine subtypes indicated that Dscam was expressed in dopaminergic amacrine cells (tyrosine hydroxylase (Th)-positive) and in bNOS-positive (Nos1-expressing) cells, but not in choline acetyltransferase (Chat)-positive starburst amacrine cells (SACs) or disabled 1-positive (Dab1) AII amacrine cells (Supplementary Fig. 5). Immunofluorescence with an antibody against the extracellular domain showed that DSCAM completely overlapped dopaminergic amacrine cell staining in >400 cells examined. In whole-mount retinas at P15, anti-DSCAM staining was distributed across the soma and processes of dopaminergic amacrine cells, but redistributed to the cell body in the adult retina (Fig. 1h, i). Additional immunoreactivity was detected in retinal cross-sections in lower, more vitreal layers of the IPL, below the strata of TH-positive processes (Fig. 1j). DSCAM immunoreactivity was absent from the mutant retinas, consistent with a protein-null mutation (Fig. 1k).
Wild-type dopaminergic amacrine cells were first labelled by antiTH staining at around P6, when the neurons are still extending processes (Fig. 2a). The spacing of dopaminergic amacrine cell bodies was analysed by density recovery profiling (DRP)—a statistical measure of the cells’ spatial autocorrelation, measuring the probability of encountering a homotypic cell body at varied distances from the reference cell. Regularly spaced cells have a zone of exclusion for other homotypic cells, indicated by a below-average cell density at short distances15 (Fig. 2b). Although their spacing was normal, the Dscam−/− dopaminergic amacrine cells showed abnormal morphology at P6 (Fig. 2c, d). Examining individual cell morphologies in 20 isolated dopaminergic amacrine cells in control and Dscam−/− retinas revealed defects in arborization in the mutant retina (Fig. 2e, f). Quantification indicated no differences in total length of processes or in the number of branches per unit length, but mutant cells had a significantly larger number of processes that self-crossed (Fig. 2g–j). This phenotype was more pronounced at P10 (not shown), and eventually large fascicles of TH-positive processes from many cells form in the mutant retina (Supplementary Fig. 6). Dopaminergic processes in the wild-type adult retina form a broad, uniform plexus, and the cell bodies are evenly spaced (Fig. 2k, l). The fasciculated adult Dscam−/− dopaminergic amacrine cells had abnormal positioning of their cell bodies, which associated with the fascicles and clumped significantly (Fig. 2m, n and Supplementary Figs 6 and 7).
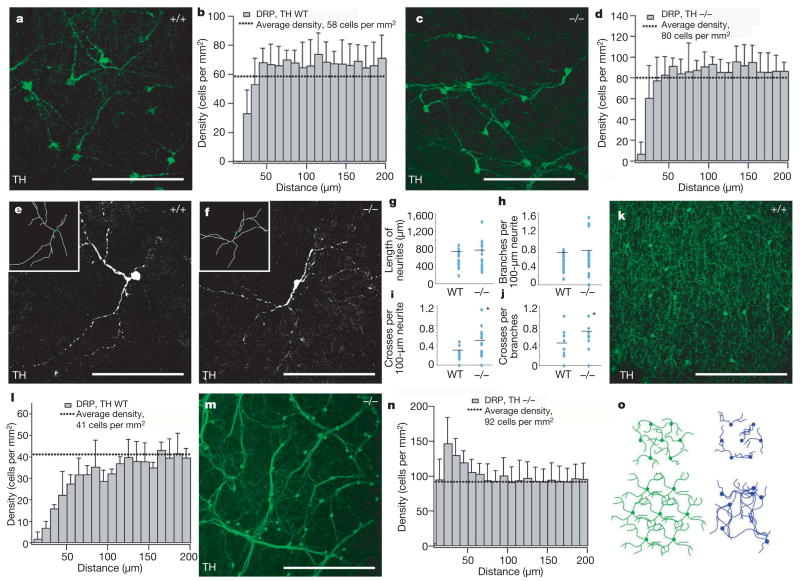
Figure 2. Arborization and mosaic patterning of dopaminergic amacrine cells in control and Dscam−/−retinas.
a, A wild-type P6 retina stained with anti-TH. Dopaminergic amacrine cell processes have little proximal contact and rarely self-cross. b, DRP analysis of cell-body spacing identified an exclusion zone surrounding dopaminergic neurons in wild-type retinas. c, A P6 Dscam−/− retina stained with anti-TH. Individual dopaminergic amacrine cells frequently self-cross. Error bars in all figures represent s.d. d, DRP analysis of Dscam−/− dopaminergic amacrine cells indicates normal soma spacing at P6. e, f, Representative isolated P6 wild-type (e) and Dscam−/− (f) dopaminergic amacrine cells are shown; insets contain traces of the neurites. g–j, Analysis of 20 isolated dopaminergic amacrine neurons from either wild-type or Dscam−/−retinas. g, No significant difference in the total neurite length was observed between wild-type and Dscam−/− neurons (t-test, P = 0.55). h, Similarly, no significant difference in neurite branches per unit length was observed (P = 0.38). i, j, A significant increase in the number of self-crossings per length of neurite (i) or per number of branches (j) was observed for the Dscam−/− neurons over controls (P = 0.003 and 0.015, respectively). k–n, Whole adult (6–8 weeks) wild-type (k, l) or Dscam−/− (m, n) retinas stained with anti-TH. TH-positive neurite fascicles run throughout the Dscam−/− retina (m) in contrast to the wild-type retina (k), in which neurites arborize evenly. l, DRP analysis indicates that dopaminergic amacrine cells in the adult wild-type retina maintain and expand their mosaic spacing and exclusion zones. n, DRP analysis of adult Dscam−/− retinas indicates aggregation of dopaminergic amacrine neurons. o, Schematic of arborization and mosaic formation in wild-type (left) and Dscam−/− (right) retinas. At P6, both wild-type and Dscam−/− dopaminergic amacrine cells are organized in a mosaic pattern; however, wild-type neurites arborize, whereas Dscam−/− neurites self-cross. By P10, wild-type dopaminergic amacrine cells have an increased exclusion zone, whereas Dscam−/− dopaminergic amacrine neurons aggregate along bundled fascicles. Scale bars: a, c, e, f, 133 μm; k, m, 388 μm.
In the mutant retina, bNOS-positive amacrine cells, which normally express Dscam, also had marked differences in the cell morphology, with short bundled processes and randomly spaced cell bodies (Fig. 3a–d). Starburst and AII amacrine populations that do not express Dscam were normal in their cell-body distribution (Fig. 3e–l), maintaining an exclusion zone in the control and Dscam−/− retinas. The meshwork of processes in the IPL from star-burst cells in the inner nuclear layer was preserved in the Dscam−/− retinas, with no abnormal fasciculation seen, and the short processes of AII cells were also normal. Cross-sections of control and Dscam−/− retinas were also examined, and all cell types reached their appropriate vertical layer in the retina (Supplementary Fig. 8).
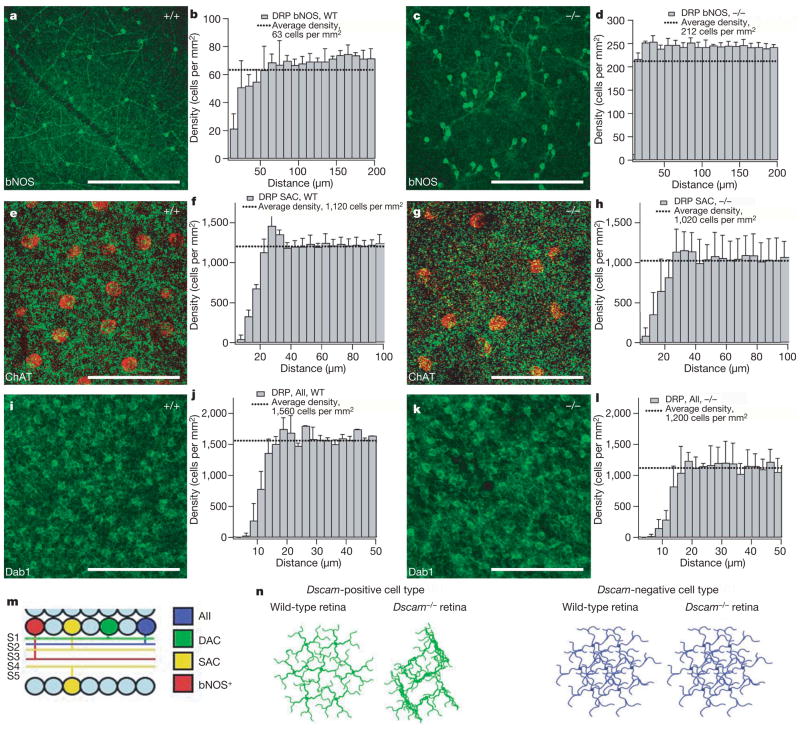
Figure 3. Morphology and cell body spacing of other amacrine cell types.
a, b, Cell bodies of bNOS-positive cells, which also express Dscam in control retinas, are spaced and arborized in a manner similar to dopaminergic amacrine neurons. c, d, The bNOS-positive amacrine cells show fasciculated processes and randomly spaced cell bodies in Dscam−/− retinas. Starburst amacrine cells have an exclusion zone around the cell body and a meshwork of arborized processes in both control (e, f) and Dscam−/− (g, h) retinas. (Cell bodies are pseudo-colored red to distinguish them from the meshwork of processes in S2 of the IPL (green).) AII amacrine cells are also evenly spaced with very short processes in control (i, j) and Dscam−/− (k, l) retinas. m, Schematic depicting the stratification of bNOS-positive, SAC, DAC and AII neurites in the inner plexiform layer. Dopaminergic amacrine neurites arborize predominantly in S1, AII amacrine neurites arborize in between S1 and S2, bNOS-positive amacrine neurites arborize in S3, and starburst amacrine neurites arborize in S2 (soma in INL) or S4 (soma in RGL). n, Schematic summarizing mosaic formation in the Dscam−/− retina. DSCAM-positive cell types arborize and maintain mosaics in the wild-type retina, but fasciculate and fail to maintain mosaics in the Dscam−/− retina. Cell types in which DSCAM expression was not detected arborize and maintain mosaics in both the wild type and the Dscam−/− retina. Scale bars: a, c, 388 μm; e, g, 62 μm; i, k, 123 μm. Different scales are used on the ordinates of DRP graphs for each cell type.
In normal retinas, the mosaic spacing of each cell type, such as AII and starburst cells, is independent, allowing dendritic overlap of functionally different cells16. If DSCAM-mediated recognition was required for this process, then cell types that express Dscam might influence the spacing of each other. We therefore examined the distribution of dopaminergic amacrine and bNOS-positive amacrine cells, relative to each other, in control and mutant retinas (Fig. 4 and Supplementary Fig. 9). These cell bodies were randomly spaced with respect to one another in both genotypes, indicating that DSCAM is necessary for maintaining mosaics in homotypic cells but is not sufficient to confer identifying signals between cell types. Also, despite the fasciculation of dopaminergic and bNOS-positive processes in the Dscam−/−retinas, no associations of processes across cell types were seen.
Figure 4. Co-existence of dopaminergic and bNOS-positive amacrine cells.
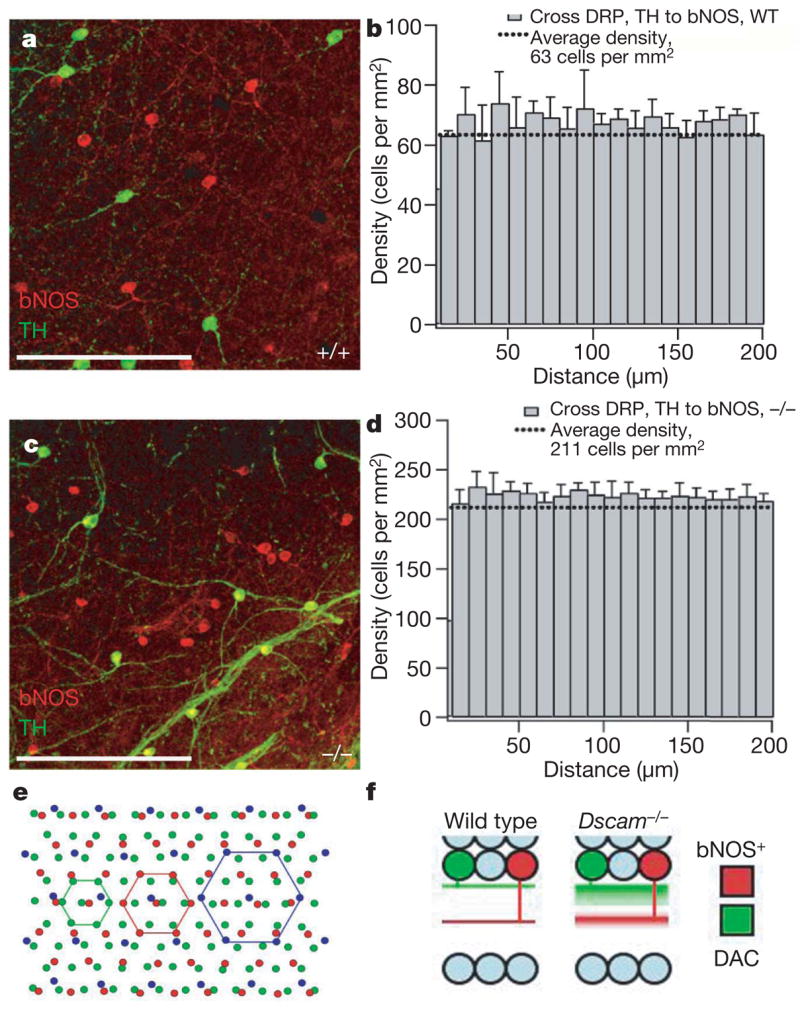
a, In wild-type retinas double-labelled for two Dscam-expressing cell populations (dopaminergic amacrine and bNOS-positive), the cell bodies and processes appear independent. b, This was confirmed by cross-DRP analyses comparing dopaminergic to bNOS-positive cells, which indicated no difference from average density (random spacing). c, In Dscam−/− mutant retinas, the fasciculation and cell-body-spacing defects are evident, but the spacing of cells remains independent and they do not aggregate, as indicated by cross-DRP analyses (d). In addition, the fascicles of processes also remain independent. e, Schematic depicting the independence of retinal neuron mosaic formation. Retinal neurons form organized mosaics that are independent of the spacing of other cell types. f, Schematic depicting dopaminergic amacrine and bNOS-positive neuron arborization in the wild-type and Dscam−/− retina. Wild-type dopaminergic amacrine neurons arborize in S1, whereas bNOS-positive neurons arborize in S3. In the Dscam−/− retina, the arborization of dopaminergic amacrine and bNOS-positive neurites becomes diffuse but, although neurites of both cell types fasciculate with homotypic neurites (dopaminergic amacrine with dopaminergic amacrine or bNOS-positive with bNOS-positive), they do not fasciculate with heterotypic neurites (dopaminergic amacrine with bNOS-positive). Scale bars in a and c represent 240 μm.
Mammalian DSCAM is therefore required for isoneuronal self-avoidance and for heteroneuronal recognition within a cell type. In the absence of DSCAM, the processes of an individual dopaminergic or bNOS-positive amacrine cell fail to form proper arbors, and instead fasciculate with processes of other cells of the same type, secondarily pulling the cell bodies out of their mosaic position. However, different cell types expressing Dscam do not influence each other’s spacing. A neurodevelopmental role for vertebrate DSCAM was not previously established beyond an early, non-neuron-specific, developmental phenotype seen in zebrafish17.
Our results are consistent with previous studies showing that retinal mosaics form very early, before neurons have extensive arbors, and that arborization and mosaic formation are largely independent18,19. The normal spacing of cells in the P6 Dscam−/− retina indicates that the early stages of mosaic formation are intact. However, DSCAM-mediated heteroneuronal self-avoidance is needed to prevent fasciculation of homotypic cells, which destroys their mosaic pattern.
Decreased cell death may also indirectly contribute to the disruption of mosaics, based on the increase in dopaminergic and bNOS cell densities in the mutant retina. The absolute density of cells that would normally express Dscam was increased by ~250% for dopaminergic amacrine cells, and by ~300% for bNOS-positive cells in the mutant retina (DRPs in Figs 2 and 3). In contrast, the densities of the ChAT-positive (starburst) and DAB1-positive (AII) cells were slightly decreased. Dopaminergic amacrine cells rely in part on cell death to sculpt their mosaic pattern20. We saw decreased TdT-mediated dUTP nick end labelling (TUNEL)-positive cells in the developing Dscam−/− retina, but saw no change in the number of proliferating cells (Supplementary Fig. 10); we also identified that clumped dopaminergic amacrine cells were not clonal, indicating that they are not the result of overproliferation from a progenitor, but consistent with lateral migration of cell bodies (Supplementary Fig. 11). DSCAM-mediated self-recognition may therefore be upstream of cell death in the wild-type retina, whereas non-Dscam-expressing cells may use other mechanisms for spacing21.
The fact that DSCAM mediates both isoneuronal and heteroneuronal self-avoidance in given amacrine cell types is interesting given recent work in Drosophila. In flies, Dscam is alternatively spliced to generate tens of thousands of protein isoforms6. The homophilic interaction of DSCAM is highly isoform-specific, and this specificity is determined by the variable immunoglobulin domains8,22,23. Therefore, Drosophila neurons use DSCAM for isoneuronal self-avoidance and arborization because each cell recognizes only its own processes, which express the same set of isoforms8,9,24. Reducing or eliminating the diversity of DSCAM isoforms perturbs Drosophila circuit organization25,26. Dscam2 in Drosophila is not extensively alternatively spliced and mediates heteroneuronal cell avoidance in tiling the axons of L1 neurons in the medulla of the Drosophila visual system7. Mouse DSCAM functions in a highly analogous way, promoting self-avoidance for both arborization (isoneuronal) and the prevention of fasciculation (heteroneuronal). Although the vertebrate genes are not extensively alternatively spliced, they do undergo homophilic and paralogue-specific binding5,27,28. The coexistence of Dscam-expressing amacrine cell populations suggests that stratification in different IPL layers may allow reuse of DSCAM as a recognition signal. Alternatively, additional co-signals may distinguish distinct cell populations, consistent with the continued lack of interaction between bNOS and dopaminergic cells in the Dscam−/− retina. Additional complexity for self-recognition could also be introduced by other Dscam gene family members, such as Dscam-like 1 (Dscaml1), which is expressed in a similar but discrete population of retinal neurons27,28 (P.G.F. and R.W.B., unpublished) or the highly homologous Sidekick proteins29.
METHODS SUMMARY
Genetics and Dscam analysis
The Dscam mutation was mapped by breeding to C57BL6/J and establishing linkage in F2 mice with polymorphic markers, and the mutation was identified by positional cloning approaches. Mice were genotyped by PCR from genomic DNA spanning the 38-bp deletion (Dscam forward, CTTTGCGCGTTATGATCCT; Dscam reverse, GTGGTGTCGATACTGATG). Control mice were littermates of the Dscam−/− animals to control for genetic background effects and age.
Immunocytochemistry
Whole-mount dissected retinas or cryostat sections were stained using standard immunofluorescence protocols. Antibody sources are listed in the Methods. Fluorescence images were collected using a Leica SP5 confocal microscope or a Nikon epifluorescence microscope with a digital camera.
In situ hybridization
In situ hybridization was performed using digoxygenin-and fluorescein isothiocyanate (FITC)-labelled riboprobes, detected by HRP-conjugated secondary antibodies and TSA-Plus fluorescent substrates (Perkin Elmer).
Analysis of mosaics
The spacing of cell bodies was analysed using DRP as described previously, with the modification that the closest bin in the analysis was corrected to account for the diameter of the cell body15,16.
METHODS
Animal husbandry
Mice were used in accordance with protocols approved by the Animal Care and Use Committee at the Jackson Laboratory. The Jackson Laboratory is accredited by the Association for the Assessment and Accreditation or Laboratory Animal Care (AAALAC). Animals were housed in PIV caging and given food and water ad libitum.
Genetic mapping
The Dscamdel17 mutation arose spontaneously in the Balb/cJ mapping partner of an unrelated F1 mapping cross. The mutation was initially localized to the distal region of chromosome 16 by typing 140 strain-specific markers, spaced approximately every 10 Mb, on a total of 10 mutant and 10 wild-type mice, the phenotypes of which were confirmed by retinal histology. The mutation was fine-mapped against DBA/2J, C57BL/6J and CAST/EiJ strains to maximize the number of polymorphic markers available. In total, more than 1,800 meioses were typed at the flanking markers D16Mit106 and D16Mit94. The interval was narrowed to 1.71 Mb (0.2 cM), between D16Mit219 and D16Mit205. This interval contained 14 annotated genes including mouse Dscam. For analysis of the Dscam gene, mRNA from affected and control brain was prepared using Trizol (Invitrogen), reverse transcribed with SuperscriptIII (Invitrogen) for PCR and sequencing, or used for northern blot analysis.
Mouse strains
The official strain designation of the Dscam mutation is Dscamdel17, reflecting the molecular lesion. It is referred to in this paper as Dscam−. In segregating genetic backgrounds containing BALB/c or 129 loci, these mice survive to adulthood and will breed as homozygotes. However, as the background is made increasingly C57BL/6, the affected mice die neonatally. The experiments described were therefore carried out on a mixed, but predominantly BALB/cByJ, background. Littermates of experimental mutants were used as controls whenever possible to account for age and genetic background. The yellow fluorescent protein (YFP) transgenic strain used in Supplementary Fig. 6 is the MitoZ strain.
Genotyping
The primers Dscam17F and Dscam17R (CTTTGCGCGTT-ATGATCCT and GTGGTGTCGATACTGATG, respectively), which flank the Dscamdel17 deletion, were used to genotype mice. Tail biopsies were digested overnight in 100 μl of 1 mg ml−1 proteinase-K in buffer containing 100 mM Tris, pH 8.5, 10 mM EDTA and 200 mM NaCl. Samples were then boiled for 15 min to denature the DNA and to inactivate the proteinase-K. PCR was performed using Eppendorf master taq with the following program: 94 °C for 1 min, followed by 35 cycles of 94 °C for 20 sec, 53 °C for 30 sec and 72 °C for 30 sec, followed by a single 72 °C extension for 2 min. PCR of DNA from homozygous wild-type mice amplifies a single 170 bp band, whereas PCR of DNA from homozygous mutant mice have a single 133 bp band, with both bands apparent in PCR samples run using DNA from heterozygous mice.
Analysis of mRNA
For RNA isolation, total retinal or brain RNA was isolated by dissecting the retina or brain of wild-type and Dscam−/− mice, which were then homogenized in 500 μl or 2 ml Trizol reagent, respectively, according to the manufacturer’s protocol. RNA samples were resuspended in diethyl pyrocarbonate (DEPC)-treated H2O. Reverse transcription was performed using SuperScript First-Strand Synthesis kit according to manufacturer’s instructions using Superscript III reverse transcriptase and a mix of random hexamer and oligo-dT primers (Invitrogen). For northern blot analysis, a probe to the amino-terminal region of the Dscam transcript was generated by PCR with reverse transcription (RT–PCR) with primers Dscam1 forward and Dscam1 reverse (TCCTTCAAGCTTCAGCAC and CATTTCCTGTCACACTGC, respectively). This fragment was labelled with 32P using the Amersham Rediprime II Random labelling kit according to the manufacturer’s instructions (GE Healthcare). Total RNA (25 μg) from each sample was heated at 75 °C for 5 min and loaded onto a 2% formaldehyde gel. Electrophoresis of samples was conducted for 5 h at 50 volts (V). RNA was transferred to a charged nylon membrane for 16–18 hours using the standard sandwich technique. RNA was crosslinked to the membrane with ultraviolet light and prehybridized at 50 °C in 10 ml hybridization buffer for 1–2 hours (5× SSC, 1× Denhardt’s solution, 1% salmon sperm DNA (100 μg ml−1), 50% formamide and 0.5% SDS). Labelled probe (10 μCi) was boiled in TE (10 mM Tris, pH 7.5, and 1 mM EDTA) for 5 min to denature and was added to 5 ml of hybridization buffer. Blots were hybridized at 65 °C for 16–18 hours. Blots were washed two times for 15 min in 2× SSC and 0.5% SDS followed by two 15-min washes in 0.2× SSC and 0.1% SDS at 65 °C. Transcript levels were compared in three Dscam−/− mice, three Dscam+− mice and three Dscam+/+ mice at ten days of age.
RNA in situ hybridization
Probe templates were prepared by PCR from first-strand brain cDNA. The primers contained T7 (anti-sense) or SP6 (sense) transcription initiation sequences at the 3′ and 5′ ends, respectively, of the PCR products. RNA probes labelled with digoxygenin or fluorescein dUTP were prepared using the following reaction mixture: 1 μg DNA, 2 μl 10× T7 or SP6 buffer (Roche), 1.5 μl T7 or SP6 RNA polymerase (Roche), 0.5 μl RNase out (Invitrogen), 2 μl digoxyigenin or fluorescein dNTP mixture (Roche), and H2O to 20 μl. Samples were incubated at 37 °C for 2 h, after which 1 μl of RNase-free DNase I (Roche) was added. Samples were incubated for an additional 15 min, after which 1 μl 0.5 M EDTA was added to stop the reaction. The RNA was precipitated by adding 2.5 μl of 4 M NH4Cl and 75 μl of chilled (−20 °C) ethanol, and resuspended in 40 μl of DEPC-treated H2O. Typical yields were 300–400 ng μl−1 of labelled riboprobe.
For hybridization, eyes were enucleated and fixed in 4% paraformaldehyde at 4 °C for 4 h and were frozen in cyro-embedding media. Sections were cut at 15–20 μm with a cryostat and incubated at 65 °C for 10 min. Sections were again fixed with 4% paraformaldehyde for 10 min at room temperature (20 °C), followed by three washes with DEPC-treated PBS. Sections were digested with proteinase-K for 5 min at room temperature (1 μg ml−1 proteinase-K in 50 mM Tris, pH 7.5, and 5 mM EDTA). After digestion, samples were fixed again in 4% paraformaldehyde for 5 min at room temperature. Sections were washed three times in DEPC-treated PBS. Sections were then acetylated for 10 min at room temperature (59 ml H2O, 0.8 ml triethanolamine, 0.105 ml HCl and 0.15 ml acetic anhydride). After acetylation, sections were permeabilized in 1% Triton X-100/PBS for 30 min at room temperature, followed by three 10-min rinses in DEPC-treated PBS. Slides were prehybridized for one hour at room temperature in hybridization solution (50% formamide, 5× SSC, 5× Denhart’s solution, 250 μg ml−1 yeast tRNA, 500 μg salmon sperm DNA and 50 μg ml−1 heparin). While sections were prehybridizing, probes were prepared at a concentration of 200 ng ml−1 in hybridization solution. Probe solution (50 μl) was prepared per slide and incubated at 80 °C for 5–10 min to denature. Prehybridization solution was replaced with hybridization solution, and the slides were coverslipped with 50-mm glass coverslips and incubated in a humidified chamber (50% formamide 5×SSC) at 65–70 °C overnight.
For probe detection, after hybridization, sections were washed twice for 5 min with TBS at room temperature. Peroxidase-conjugated antibodies to digoxygenin or fluorescein (Roche) were diluted 1:2,000 in TNB (TSA plus system) and incubated on sections at 4 °C overnight. Sections were washed in TBST (0.05% Tween) three times for 10 min at room temperature. Probes were detected using the TSA plus system (Perkin-Elmer). For double-labelling experiments, POD-antibodies against FITC and digoxygenin (DIG) were applied sequentially, and the peroxidase conjugate of the first detection antibody was inactivated by H2O2 treatment before applying the second antibody. Different tyramide conjugates were used when developing the signals to achieve different colours, and detection reactions were allowed to proceed for 30 min. Dscam and amacrine-marker expression was examined at P12. At least four independent retinas were examined. Sense-strand controls were used in all experiments to confirm specificity.
Histology and immunocytochemistry
Tissues were fixed in Bouin’s fixative overnight. Sections (4 μm) were cut with a microtome and stained using a standard haemotoxylin and eosin stain. For immunocytochemistry, embryos or enucleated eyes were fixed in 4% paraformaldehyde overnight. Tissue was equilibrated in 30% sucrose/PBS and frozen in OCT media. Thicker sections (8 μm) were cut using a cryostat and used immediately or stored for up to two weeks at −20 °C. Sections were blocked in 3% normal horse serum in 1× PBS and 0.1% Triton X-100. Primary antibodies (indicated in the Methods Summary) were incubated overnight at 4 °C and subsequently washed twice for 10 min in PBS. Sections were incubated at room temperature with secondary antibodies for one hour, and then washed four times in PBS for 10 min. The last wash contained 2 μl of 1 mg ml−1 DAPI per 40 ml PBS.
Whole-mounted retinas
Mice were perfused transcardially with PBS and 4% paraformaldehyde. Eyes were enucleated and placed in a dish containing PBS. A small hole was made at the junction between the cilliary body and retina with a 30-gauge needle. The eye was hemisected and the retina was gently teased away from the sclera. Retinas were fixed a second time in 4% paraformaldehyde for one hour, followed by a rinse in PBS. Retinas were incubated with primary antibodies in PBS, supplemented with 3% normal horse serum and 0.5% Triton X-100, for 3–5 days at 4 °C with gentle rocking. Primary antibodies were washed off overnight in PBS, and the retinas were incubated with secondary antibodies in PBS, 3% normal horse serum and 0.5% Triton. Retinas were washed for 2–4 h in PBS and were flat-mounted. Whole-mount images were collected from mice between six and ten weeks of age, and at least six mutant and control animals were included in each analysis.
Antibodies used
The following primary antibodies were used in standard immunofluorescence and western blotting protocols. Goat anti-human DSCAM (R&D Systems), rabbit anti-tyrosine hydroxylase (Novus Biologicals), goat anti-choline acetyltransferase (Chemicon), rabbit anti-Disabled1 (gift from B. Howell), anti-CHX10 (Santa Cruz Biotechnology), anti-BRN3b (Santa Cruz Biotechnology), mouse anti-neurofilament (2H3, Developmental Studies Hybridoma Bank), mouse anti-PAX6 (Developmental Studies Hybridoma Bank), rabbit anti-bNOS (Sigma-Aldrich), mouse anti-TH (Novocastro), and mouse anti-Ki67 (Dako).
Western blot analysis
A P0 retina was dissected and homogenized in hypertonic sucrose lysis buffer (0.32 M sucrose, 1 mM NaHCO3, 1 mM MgCl2 and 0.5 mM CaCl2) containing protease inhibitors with a glass dounce homogenizer. Total protein (40 μg) was loaded onto 6% acrylamide gels, run and transferred. DSCAM antibodies were used at a concentration of 1:500. The specificity of the DSCAM antibody in western blotting was established using COS-7 cells transfected with c-Myc-tagged Dscam cDNA. A single band of approximately 220 kD was observed when using an antibody to either c-Myc or DSCAM in the transfected sample only. Western blots from tissue were performed using the DSCAM antibody after pre-absorption against mutant tissue to reduce background staining. As a result, immunoreactivity to truncated protein products specific to the mutant could have been missed. The serum used in Fig. 1 for immunocytochemistry was not pre-absorbed.
Cell morphologies
P6 retinas were collected and stained with anti-TH as described previously. TH-positive amacrine cells that were sufficiently isolated from neighbouring cells to allow their processes to be definitively identified were imaged using a ×63 objective lens on a Leica SP5 confocal microscope. The total length of 20 wild-type and 20 Dscam−/− dendrites was traced, and measured using ImageJ.
DRP and nearest neighbour analysis analysis
DRP and nearest neighbour analysis (NNA) were performed as described previously23,24. The entire retinas were imaged and reconstructed for analysis of TH and bNOS amacrine cells. Four 775-μm fields for each wild type and Dscam−/− retina were imaged and used to analyse starburst and AII amacrine cells. Cholinergic amacrine cells were imaged in the inner nuclear layer for analysis. Independent samples from at least three animals, ages P6 and six to ten weeks, were included in each analysis.
TUNEL staining and analysis
TUNEL cell death detection kit was used to perform TUNEL analysis according to the manufacturer’s instructions for difficult tissues (Roche). Eyes were enucleated and immediately frozen and cut in 10-μm sections using a cryostat. After finishing the TUNEL reaction, the slides were washed in PBS for 5 min, and immunocytochemistry was performed using a PAX6 antibody as detailed previously. TUNEL-positive cells were counted according to their location: retinal ganglion layer (including the inner plexiform layer), the inner neuroblast layer and the outer neuroblast layer. The inner and outer neuroblast layers were distinguished by PAX6 immunoreactivity. At least three slides with eight sections on each slide for each of three mice were recorded at each time point for each genotype.
Mitotic index
The mitotic index in the wild-type and Dscam−/− retina was determined at three time points, E14, E17 and P1. BrDU injections were administered at a dosage of 10 mg kg−1 20 min before tissue collections. Embryo heads or enucleated eyes were fixed overnight in 4% paraformaldehyde and were imbedded in paraffin blocks. Sections were cut at 5 μm and stained with antibodies to BrDU and Ki67 after boiling in citrate buffer for 10 min. The number of BrDU-positive/Ki67-positive cells (S-phase) were divided by the number of Ki67-positive cells (G2/M phase). Cells that did not label for either marker were also counted and no significant difference was observed in their number. At least three slides with eight sections on each slide for each of three mice were recorded at each time point for each genotype.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank G. Cox and S. Ackerman for comments on the manuscript, and M. Yamagata and J. Sanes for sharing unpublished results. The authors are supported by the NEI, the Knights Templar Eye Foundation (P.G.F.) and Research to Prevent Blindness (R.H.M.). We also thank the scientific services at The Jackson Laboratory (supported by NCI Cancer Center).
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Wassle H, Boycott BB. Functional architecture of the mammalian retina. Physiol Rev. 1991;71:447–480. doi: 10.1152/physrev.1991.71.2.447. [DOI] [PubMed] [Google Scholar]
- 2.Novelli E, Resta V, Galli-Resta L. Mechanisms controlling the formation of retinal mosaics. Prog Brain Res. 2005;147:141–153. doi: 10.1016/S0079-6123(04)47011-3. [DOI] [PubMed] [Google Scholar]
- 3.Galli-Resta L. Local, possibly contact-mediated signalling restricted to homotypic neurons controls the regular spacing of cells within the cholinergic arrays in the developing rodent retina. Development. 2000;127:1509–1516. doi: 10.1242/dev.127.7.1509. [DOI] [PubMed] [Google Scholar]
- 4.Yamakawa K, et al. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet. 1998;7:227–237. doi: 10.1093/hmg/7.2.227. [DOI] [PubMed] [Google Scholar]
- 5.Agarwala KL, Nakamura S, Tsutsumi Y, Yamakawa K. Down syndrome cell adhesion molecule DSCAM mediates homophilic intercellular adhesion. Brain Res Mol Brain Res. 2000;79:118–126. doi: 10.1016/s0169-328x(00)00108-x. [DOI] [PubMed] [Google Scholar]
- 6.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 7.Millard SS, Flanagan JJ, Pappu KS, Wu W, Zipursky SL. Dscam2 mediates axonal tiling in the Drosophila visual system. Nature. 2007;447:720–724. doi: 10.1038/nature05855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan XL, et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 10.Soba P, et al. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews BJ, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Hughes ME, et al. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong JH, Fish DR, Rockhill RL, Masland RH. Diversity of ganglion cells in the mouse retina: unsupervised morphological classification and its limits. J Comp Neurol. 2005;489:293–310. doi: 10.1002/cne.20631. [DOI] [PubMed] [Google Scholar]
- 15.Rodieck RW. The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis Neurosci. 1991;6:95–111. doi: 10.1017/s095252380001049x. [DOI] [PubMed] [Google Scholar]
- 16.Rockhill RL, Euler T, Masland RH. Spatial order within but not between types of retinal neurons. Proc Natl Acad Sci USA. 2000;97:2303–2307. doi: 10.1073/pnas.030413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yimlamai D, Konnikova L, Moss LG, Jay DG. The zebrafish Down syndrome cell adhesion molecule is involved in cell movement during embryogenesis. Dev Biol. 2005;279:44–57. doi: 10.1016/j.ydbio.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Galli-Resta L, Resta G, Tan SS, Reese BE. Mosaics of islet-1-expressing amacrine cells assembled by short-range cellular interactions. J Neurosci. 1997;17:7831–7838. doi: 10.1523/JNEUROSCI.17-20-07831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin B, Wang SW, Masland RH. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron. 2004;43:475–485. doi: 10.1016/j.neuron.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Raven MA, Eglen SJ, Ohab JJ, Reese BE. Determinants of the exclusion zone in dopaminergic amacrine cell mosaics. J Comp Neurol. 2003;461:123–136. doi: 10.1002/cne.10693. [DOI] [PubMed] [Google Scholar]
- 21.Resta V, Novelli E, Di Virgilio F, Galli-Resta L. Neuronal death induced by endogenous extracellular ATP in retinal cholinergic neuron density control. Development. 2005;132:2873–2882. doi: 10.1242/dev.01855. [DOI] [PubMed] [Google Scholar]
- 22.Wojtowicz WM, et al. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meijers R, et al. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- 24.Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- 25.Hattori D, et al. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BE, et al. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 27.Agarwala KL, et al. Cloning and functional characterization of DSCAML1, a novel DSCAM-like cell adhesion molecule that mediates homophilic intercellular adhesion. Biochem Biophys Res Commun. 2001;285:760–772. doi: 10.1006/bbrc.2001.5214. [DOI] [PubMed] [Google Scholar]
- 28.Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. doi: 10.1038/nature06469 (this issue). [DOI] [PubMed] [Google Scholar]
- 29.Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.