SUMMARY
Insect and vertebrate eyes differ in their formation, cellular composition, neural connectivity, and visual function. Despite this diversity, Drosophila atonal and its vertebrate Ortholog in the eye, Ath5, each regulate determination of the first retinal neuron class—R8 photoreceptors and retinal ganglion cells (RGCs)—in their respective organisms. We have performed a cross-species functional comparison of these genes. In ato1 mutant Drosophila, ectopic Xenopus Ath5 (Xath5) rescues photoreceptor cell development comparably with atonal. In contrast, mouse Ath5 (Math5) induces formation of very few ommatidia, and most of these lack R8 cells. In the developing frog eye, ectopic atonal, like Xath5, promotes the differentiation RGCs. Despite strong conservation of atonal, Xath5, and Math5 structure and shared function, other factors must contribute to the species specificity of retinal neuron determination. These observations suggest that the atonal family may occupy a position in a gene hierarchy where differences in gene regulation or function can be correlated with evolutionary diversity of eye development.
INTRODUCTION
Theories of eye evolution have been debated for over a century and center around the hypothesis that the vast diversity of visual organs arose through convergent evolution. However, molecular and genetic studies (Halder et al. 1995; Loosli et al. 1998; Neumann and Nuesslein-Volhard 2000; Oliver et al. 1995; Xu et al. 1997) have challenged this idea. It has been suggested that a common Urbilaterian ancestor may have possessed an ancestral eye selector gene whose expression and function was associated with a primitive light-sensing organ (Arendt and Wittbrodt 2001; Callaerts et al. 1997; Carroll et al. 2001). In the past decade such an eye selector gene, Pax6, was identified and intensely studied (Gehring and Ikeo 1999; Kumar and Moses 2001). The ability of Pax6 to promote eye formation is strongly conserved across many metazoan phyla (Halder et al. 1995; Loosli et al. 1996; Tomarev et al. 1997; Glardon et al. 1998; Callaerts et al. 1999; Chow et al. 1999). Indeed, insect and mammalian Pax6 genes retain strongly conserved protein function (Halder et al. 1995) and cis-regulation (Kammandel et al. 1999; Xu et al. 1999). Although Pax6 specifies an eye fate, it is unlikely to directly impose morphologic, structural, or functional differences among eye types. Diversification might be explained through the downstream genes that Pax6 regulates. For example, the number and type of Pax6 “target genes” may vary in different animal species or target gene function may have diverged.
One vertebrate gene that acts as a Pax6 target gene is the basic helix-loop-helix (bHLH) transcription factor, Ath5 (Brown et al. 1998). Pax6 binding sites have recently been reported in the promoter region of mouse Ath5 (Marquardt et al. 2001), although they have not been tested functionally. In zebrafish, frogs, chickens, and mice, Ath5 regulates the formation of the first retinal neuron, retinal ganglion cells (RGCs) (Vetter and Brown 2001). Mutations in zebrafish or mouse Ath5 cause a complete loss of RGCs (Brown et al. 2001; Wang et al. 2001; Kay et al. 2001), and ectopic Ath5 expression promotes RGC fate in the chick or frog eye (Kanekar et al. 1997; Liu et al. 2001). Ath5 is the Ortholog of Drosophila atonal, which is the proneural gene for photoreceptor and chordotonal neurons (Jarman et al. 1993, 1994). atonal mutants lack the first photoreceptor neuron, the R8 cell, and ectopic expression of atonal during eye development induces extra R8 cells (Dokucu et al. 1996; Sun et al. 2000). Although Ath5 mutations cause the selective agenesis of RGCs in zebrafish and mice, loss of atonal function leads to near eyelessness in flies (Jarman et al. 1994, 1995; Brown et al. 2001; Wang et al. 2001; Kay et al. 2001). This is due to fundamental differences in the mechanisms of fly and vertebrate eye development (reiterated cell–cell induction versus renewable progenitor cells).
Previously we compared the activity of ectopic Xenopus and Mus Ath5 during frog eye development and found they do not promote the same cell types (Brown et al. 1998). Thus, the frog and mouse Ath5 genes specify RGC fate inherently but cannot substitute for one another. This further implies that amphibians and mammals have divergent aspects of retinal development. Indeed, zebrafish, frog, chick, and mammalian retinogenesis differ in their spatial patterning and temporal progression (Easter 2000; Jarman 2000). To test this idea further, we directly compared the activity of atonal, Xenopus Ath5 (Xath5), and mouse Ath5 (Math5) in the context of frog and fly eye formation. We found that atonal induces frog RGCs identically to Xath5 and Xath5 reciprocally rescues fly eye formation in an atonal mutant background nearly as well as atonal itself. In contrast, Math5 rescues atonal mutant eyes poorly and in a manner that resembles the bHLH gene, scute. We propose that Xath5 and atonal are functionally interchangeable during fly or frog retinal neuron formation but Math5 requires a mammalian-specific environment to initiate retinal neuron determination properly.
MATERIALS AND METHODS
Fly stocks and transgenic lines
The gal4-7 enhancer trap, UAS-ato, and UAS-sc constructs were previously described (Sun et al. 2000). Full-length complementary DNAs for Xenopus Ath5 (Xath5, accession number U93170), Xenopus NeuroD (accession number U28067), and mouse Ath5 (Math5, accession number AF071223) were inserted into the pUAST vector by conventional cloning and transgenic fly lines were created using standard microinjection techniques. UAS-ato, UAS-sc, UAS-Xath5, UAS-NeuroD, UAS-Math5, or gal4-7 transgenic lines were genetically recombined with the ato1 loss of function mutation (Jarman et al. 1995; Sun et al. 2000). At least three independent transgenic lines for UAS-Xath5, UAS-NeuroD, and UAS-Math5 were tested in this way. The coding regions within each construct were confirmed by DNA sequencing, and comparable activities were observed among independent lines for each construct. In all fly experiments retinal expression was induced as follows, using UAS-ato as an example: w-; gal4-7, ato1/TM6Tb X w-; UAS-ato/UAS-ato; ato1/TM6Tb. Tb+ larvae or adults were analyzed from each cross.
Scanning electron microscopy
Drosophila heads were fixed in 4% buffered glutaraldehyde for 2 h and dehydrated through a graded alcohol series. Samples were washed twice in hexamethyldisilizane (Polysciences, Warrington, PA), desiccated overnight, mounted, and sputter coated with colloidal gold. Lateral views of eyes were recorded and analyzed using an Amray 1000B (SEMTech Solutions, North Billerica, MA) scanning electron microscope. Polaroid photomicrographs were digitally scanned and processed using Photoshop 4.0 software (Adobe Systems Inc., San Jose, CA).
Histology
Sample preparation and sectioning are described in Brown et al. (1991). Sections (1 μm) were collected serially and stained with 1% toluidine blue/1% sodium borate (Sigma, St. Louis, MO). These were photographed through a Zeiss microscope (Carl Zeiss International, USA) using a 35-mm camera and Ektachrome 160T slide film (Eastman Kodak, USA). Images were digitized using a Coolscan slide scanner (Nikon, Melville, NY) and Adobe Photoshop 5.5 software.
Immunohistochemistry
Mouse anti-Scabrous (1:200) (Lee et al. 1996), rat anti-Elav (1:500) (O’Neill et al. 1994), and mouse anti-Rough (1:100) (Kimmel et al. 1990) hybridoma culture supernatants were obtained from the Developmental Studies Hybridoma Bank (University of Iowa). Mouse anti-Boss ascites (1:1000) was a gift from Helmut Kramer. Boss/Elav double or Scabrous single labels followed Cagan et al. (1992), except that imaginal discs were directly dissected in PLP fixative for Scabrous staining experiments. Rough/Elav double labels were performed as described by Kimmel et al. (1990). Directly conjugated secondary antibodies or biotinylated secondary antibodies and streptavidin-conjugated fluorochromes (Jackson ImmunoResearch Laboratory West Grove, PA) were used for antibody detection. Labeled whole-mount imaginal discs were imaged using either Biorad MRC600 or Olympus Fluoview confocal microscopes and Adobe Photoshop 5.5 or Image J software (http://rsb.info.nih.gov/ij/download/).
Xenopus in vivo lipofection
Xath5, scute, or atonal complementary DNA expression plasmids were constructed in pCS2 (Turner and Weintraub 1994). Plasmid DNA was transfected into the eye primordia of stage 17–18 frog embryos as previously described (Holt et al. 1990). The embryos were grown until stage 41 and then analyzed for effects on retinal cell fate as previously described (Kanekar et al. 1997). pEGFP-C1 plasmid DNA (BD Biosciences Clontech, Palo Alto, CA ) was included to mark the transfected cells. Images of labeled sections were digitally captured by a Xillix Microimager PMI CCD camera using Openlab software (Improvision Lexington, MA).
RESULTS
Xath5 and Math5 do not rescue the Drosophila atonal mutation identically
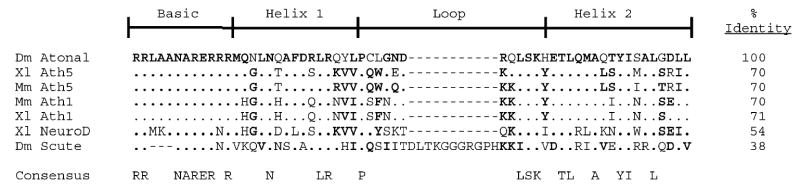
Drosophila Atonal shares significant homology in its bHLH domain with Xath5 and Math5 and with Math1 and Xath1 that are not expressed in the mammalian eye but act as an Atonal Orthologs in other regions of the vertebrate nervous system (Kim et al. 1997; Ben-Arie et al. 2000). Therefore, vertebrate Ath1 and Ath5 genes have been termed semi-Orthologs of Atonal (Brown et al. 2001, 2002; Kay et al. 2001). The amino acid homologies of these genes (Fig. 1) suggest conservation of DNA binding (basic domain) and protein interaction (HLH domain) functions. In particular, the basic domains of Atonal, Math1, Xath5, and Math5 are 100% identical (Fig. 1). We wanted to compare further the functions of the three genes that function in the eye. Atonal is normally expressed in developing eye imaginal discs within the morphogenetic furrow (MF) (Jarman et al. 1994). Sun et al. (2000) demonstrated that atonal expression within and posterior to the MF restores a sizable portion of the ato1 mutant eye. We therefore tested Xath5 and Math5 for rescue of the ato1 eye phenotype, using the same binary Gal4-UAS system (Brand and Perrimon 1993) and gal4-7 driver (Sun et al. 2000) to activate the transcription of each UAS construct during eye development. We also compared the activity of another more distantly related vertebrate bHLH gene, Xenopus NeuroD (Fig. 1), as a control. Each vertebrate gene was inserted downstream of UAS binding sites, and multiple independent transgenic fly lines were generated and tested. We compared the extent of eye rescue by scanning electron microscopy of adult flies ectopically expressing atonal, scute, Xath5, NeuroD, and Math5 (Fig. 2, A–C and G–I). These eyes were also sectioned histologically to evaluate the full complement of ommatidial photoreceptor cells (Fig. 2, D–F and J–L).
Fig. 1.
Alignment of bHLH domains for proteins analyzed in this cross-species functional comparison. ClustalW alignment of 56 (Atonal, Xath5, Math5, Math1, Xath1 NeuroD) or 64 (Scute) amino acids that comprise the basic, helix 1, loop, and helix 2 conserved domain. Consensus amino acids are indicated along the bottom, with the amino acid letter denoting perfect conservation. Dashes represent spaces introduced for optimal alignment of the basic and loop subdomains between scute and the other proteins. A “.” represents amino acids identical to Atonal, and bold type indicates those residues that are identical or similar in three or more proteins.
Fig. 2.
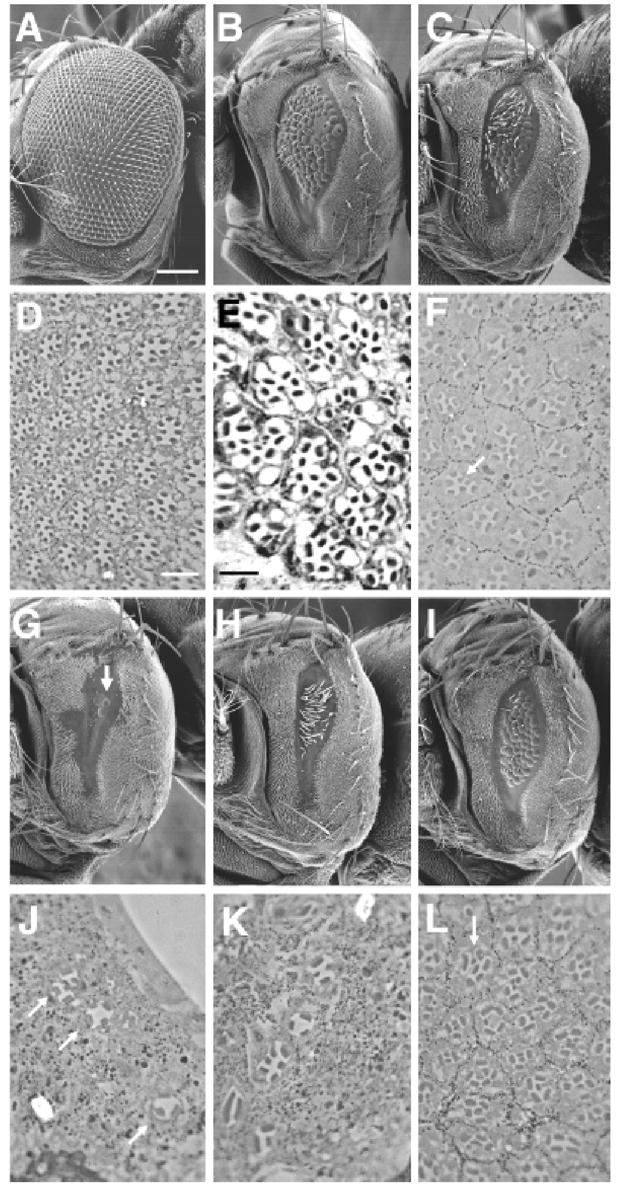
Xath5 and Math5 rescue fly photoreceptor development differently. Scanning electron micrographs (A–C, G–I) and histologic sections (D–F, J–L) of adult Drosophila compound eyes. All histologic sections are at the proximal level of the retina where R8 photoreceptor cells are found. (A and D) w-; gal4-7, ato1/TM6Tb eyes are normal in size (A) and photoreceptor cell composition (D). (B and E) atonal-rescued eye with extra R8 cells in many ommatidia (E and Sun et al. 2000). (C and F) Xath5-rescued eye is slightly smaller size than atonal rescue. Cells that morphologically resemble R8s are present in these ommatidia (arrow in F). (G and J) Math5-rescued eye with two ommatidia (arrow in G). The total number of ommatidia observed varied from 2 to 20 (n≥20 eyes). Histologic sections of these rare ommatidia contain fewer than eight photoreceptor cells per ommatidium (arrows in J) with no clearly identifiable R8 cells. (H and K) Scute-rescued eye. Scute induces extra interommatidial bristles (H) and small numbers of ommatidia, mostly lacking R8 cells (K and Sun et al. 2000) (I and L) NeuroD-rescued eye is the same size as Xath5- rescued eye (compare I with C) and has R8-containing ommatidia (arrow in L). Scale bars: A, 100 μm; D, 10 μm; E, 3 μm.
In these experiments Xath5 restored fly eye development nearly as well as atonal (compare Fig. 2, C with B and F with E), including cells that morphologically resemble R8 photoreceptors in the proximal retina (arrow in Fig. 2F). However, Math5 rescued only a small number of ommatidia (<25 per eye) (Fig. 2, G and J), and none of these contained photoreceptor cells with clear R8 cell morphology. All Math5-rescued ommatidia examined contained fewer than the normal complement of eight photoreceptor cells. Among the three UAS-Math5 fly lines tested two displayed no rescue, whereas a third induced small numbers of ommatidia (Fig. 2, G and J). This phenotype is similar to that of gal4-7 driven scute expression during fly eye development (Fig. 2, H and K) (Sun et al. 2000). Although scute is not normally expressed during photoreceptor neuron formation and is predicted to have different DNA-binding properties (Chien et al. 1996), it can rescue a small number of ommatidia when expressed by eye disc cells within and posterior to the MF (Sun et al. 2000). Consistent with its known role in interommatidial bristle formation (Brown et al. 1991), scute-rescued eyes contain numerous bristles (Fig. 2H). However, Math5 was unable to induce interommatidial bristles (Fig. 2G). Xenopus NeuroD, a relatively divergent member of the Atonal family (Fig. 1) (Brown et al. 1998; Hassan and Bellen 2000), rescued the ato1 eye phenotype equivalently to atonal or Xath5 (Fig. 2, I and L).
Xath5 and NeuroD induce early stages of ommatidial formation but Math5 does not
In the Drosophila eye, R8 is the founding photoreceptor neuron in each ommatidium and atonal is a key regulator of R8 determination (Jarman et al. 1994). To test whether Xath5 or Math5 can induce R8 cells, we examined eye imaginal discs for Bride-of-sevenless (Boss) protein expression. Boss is an early marker for terminally differentiated R8 cells (green punctate staining in Fig. 3). We colabeled eye discs with the pan-differentiation marker, Elav, to delineate photoreceptor neurons within forming ommatidia (cells in red in Fig. 3). UAS-atonal was previously shown to induce Boss expression in ato1 eye discs, whereas UAS-scute similarly expressed within and posterior to the MF rarely does (Sun et al. 2000). We compared the ability of UAS-Xath5, UAS-Math5, and UAS-NeuroD to induce Boss expression. Both Xath5-rescued (Fig. 3C) and NeuroD-rescued eyes (Fig. 3F) expressed Boss immediately posterior to the MF (Fig. 3B). However, Math5 induced only sparse Boss expression, usually in 1–3 ommatidia per eye disc (vertical arrows in Fig. 3D). This phenotype is identical to that of UAS-scute (Fig. 3E), although UAS-scute discs contained more ommatidia.
Fig. 3.
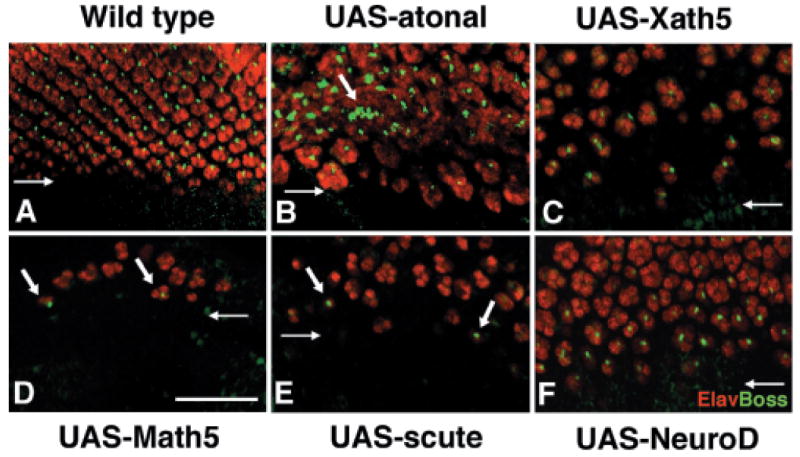
Xath5 induces fly R8 formation but Math5 does so rarely. Drosophila imaginal discs double labeled with anti-Boss (green) to mark R8 photoreceptor cells and anti-Elav (red) to mark differentiated photoreceptor neurons. In each panel, a horizontal arrow marks the position of morphogenetic furrow (MF) and anterior is down. (A) w-; gal4-7, ato1/TM6Tb control. Elav expression highlights normal ommatidial patterning. One R8 cell expressing Boss (green punctate staining) is present within each ommatidium posterior to the MF. (B) As reported by Sun et al. (2000), atonal induces extra R8 cells (vertical arrows) and ommatidial patterning is slightly disorganized. (C) Xath5 induces R8 cell formation. Ommatidial patterning is similar to atonal rescues (compare B and C). (D) Math5 induces very few ommatidia, and these are always located within a small posterior patch of the imaginal disc. Vertical arrows point to two ommatidia with Boss expression (in green). (E) Infrequent induction of R8-containing ommatidia (vertical arrows) by UAS-scute. (F) UAS-NeuroD rescued eye discs with R8 cells, as in UAS-Xath5 (compare F and C). Scale bar, 25 μm.
It is possible that Math5 may activate very early steps of fly R8 or ommatidial development that cannot be maintained into later stages of retinal formation. To test this hypothesis, we examined eye discs for expression of Rough. This homeodomain transcription factor is normally expressed by MF cells that abut but do not overlap those expressing Atonal (Dokucu et al. 1996). At different times during R8 determination and differentiation, Rough represses Atonal expression (Kimmel et al. 1990; Dokucu et al. 1996). Rough-positive cells in the MF are at the “precluster” stage of development (green nuclear staining in Fig. 4A), which precedes R8 cell selection and high levels of Atonal expression. Precluster expression of Rough also precedes Elav within terminally differentiated photoreceptor cells (red cells in Fig. 4). Posterior to the MF, Rough is coexpressed with Elav in R2, R3, R4, and R5 cells (Kimmel et al. 1990) (Fig. 4A). In atonal-rescued eye discs, both precluster (green nuclei) and R cell expression (red cells with green nuclei) of Rough can be discerned (Fig. 4B). In UAS-Xath5 (Fig. 4C) and UAS-NeuroD (Fig. 4F) eye discs, Rough is mainly expressed within precluster cells, but in older larvae we observed Rough coexpression with Elav posterior to the MF (not shown). In UAS-Math5 (Fig. 4D) and UAS-scute (Fig. 4E) discs, however, precluster Rough expression was not observed, although Rough is expressed by more mature R cells that coexpress Elav. Thus, atonal, Xath5, and NeuroD rescue the early precluster cell development that precedes R8 selection, whereas Math5 and Scute do not. In contrast, all bHLH genes tested facilitate late Rough expression in differentiating R cells.
Fig. 4.
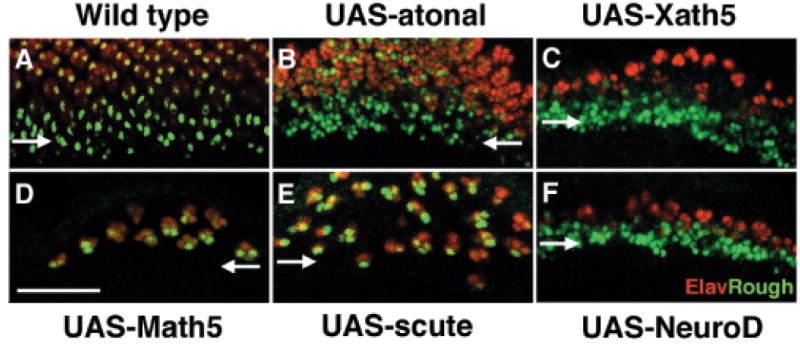
Math5 does not induce ommatidial precluster formation. Confocal micrographs of Rough (green) and Elav (red) expression in developing Drosophila eye discs. In all panels, horizontal arrows mark the position of the morphogenetic furrow (MF) and anterior is down. (A) w-; gal4-7, ato1/TM6Tb control. In the MF, Rough is expressed within R8, R2, and R5 cells of ommatidial preclusters (green nuclei). After differentiation these cells coexpress Rough and Elav (cells with green nuclei and red cytoplasm). (B) atonal-rescued eyes initiate precluster formation within the MF. Some ommatidia have extra Rough-positive nuclei. (C) Xath5-rescued discs initiate precluster formation, marked by Rough expression (green nuclei alone) in the MF. (D) In UAS-Math5 eye discs, cells expressing only Rough were never observed. However, many differentiated photoreceptors (labeled in red) also express Rough. (E) Scute-rescued eye disc also fail to initiate Rough precluster expression. Similar to Math5, many differentiated photoreceptors coexpress Rough and Elav. (F) Rough MF precluster expression in NeuroD-rescued eyes. Scale bar, 25 μm.
Expression of Scabrous is absent in Math5-rescued eye discs
In the anterior MF ubiquitous expression of Atonal protein is rapidly refined to groups of cells from which R8s emerge (Jarman et al. 1994, 1995; Dokucu et al. 1996). Simultaneously, restricted groups of Atonal-expressing cells and nascent R8 cells secrete Scabrous, which acts to inhibit excess R8 specification (Frankfort and Mardon 2002; Hsiung and Moses 2002). Scabrous precedes Boss and is the earliest known marker of forming R8 cells (Baker et al. 1990). We tested the ability of atonal, Xath5, Math5, NeuroD, and scute to induce Scabrous expression in ato1 rescued eye discs (Fig. 5). Figure 5A shows the normal pattern of Scabrous expression within secretory vesicles of MF cells. Initially, this expression is found in more precluster cells than will become R8 cells. However, once R8 is selected, Scabrous is rapidly restricted to the forming R8 neuron. In UAS-atonal rescued eyes, Scabrous is expressed by cells immediately posterior to the MF (Fig. 5B). This delay is due to the gal4-7 driver, which activates UAS constructs several rows posterior to the normal up-regulation of Atonal or Scabrous protein in the anterior MF (Sun et al. 2000). Both UAS-Xath5 and UAS-NeuroD induce Scabrous expression similarly to UAS-atonal (compare Fig. 5, B, C, and F). In UAS-Math5 discs, Scabrous expression was virtually absent. Only one Scabrous-expressing cell (arrowhead in Fig. 5D) was found in 50 eye discs tested. More cells expressing Scabrous were observed in scute-rescued eye discs (Fig. 5E), but these are randomly spaced throughout the posterior disc. Thus, Math5 and scute differ in their ability to activate Scabrous in ato mutant eyes.
Fig. 5.
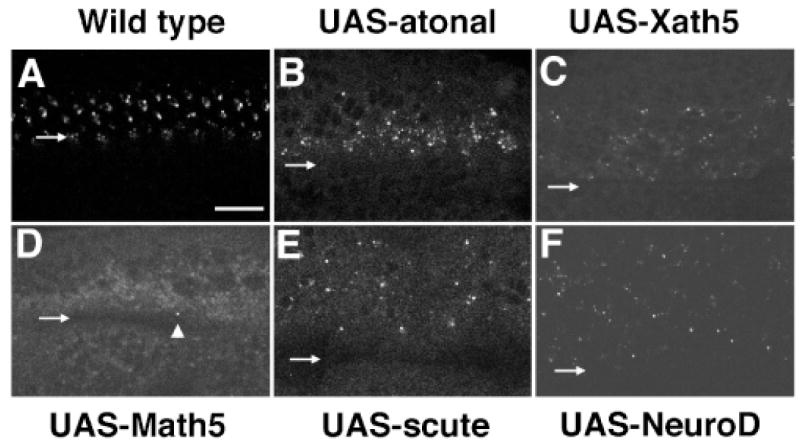
Differing expression of Scabrous in ato1 imaginal discs when atonal, Xath5, NeuroD, Math5, or scute are ectopically expressed. Confocal micrographs of Scabrous expression in the MF. (A) w-;gal4-7, ato1/TM6Tb control. Normal expression highlights ommatidial precluster formation within the morphogenetic furrow (MF). (B) In atonal-rescued eyes, Scabrous expression initiates immediately posterior to the MF in a broad band of cells. (C) Xath5-rescued eyes also initiate Scabrous expression immediately posterior to the MF similarly to atonal. (D) Math5 fails to activate Scabrous expression. The arrowhead points to the lone Scabrous-positive cell observed throughout these experiments. (E) scute induces randomly spaced Scabrous-expressing cells posterior to the MF. (F) The expression of Scabrous in NeuroD rescued eye discs was nearly identical to that of Xath5-rescues. Scale bar, 25 μm.
atonal induces vertebrate RGCs in the developing frog eye
To determine whether atonal can function equivalently to Xath5 or Math5 in the context of a vertebrate eye, we overexpressed it in Xenopus retinal progenitors by in vivo lipofection (Holt et al. 1990). In these experiments, lipofection of Xath5 DNA caused an increase in the representation of RGCs (Fig. 6, B and D) compared with green fluorescent protein (GFP) control lipofections (Fig. 6, A and D) as was previously shown (Kanekar et al. 1997). In the same context, however, lipofection of Math5 DNA paradoxically promotes a bipolar cell fate rather than an RGC fate (Brown et al. 1998).
Fig. 6.
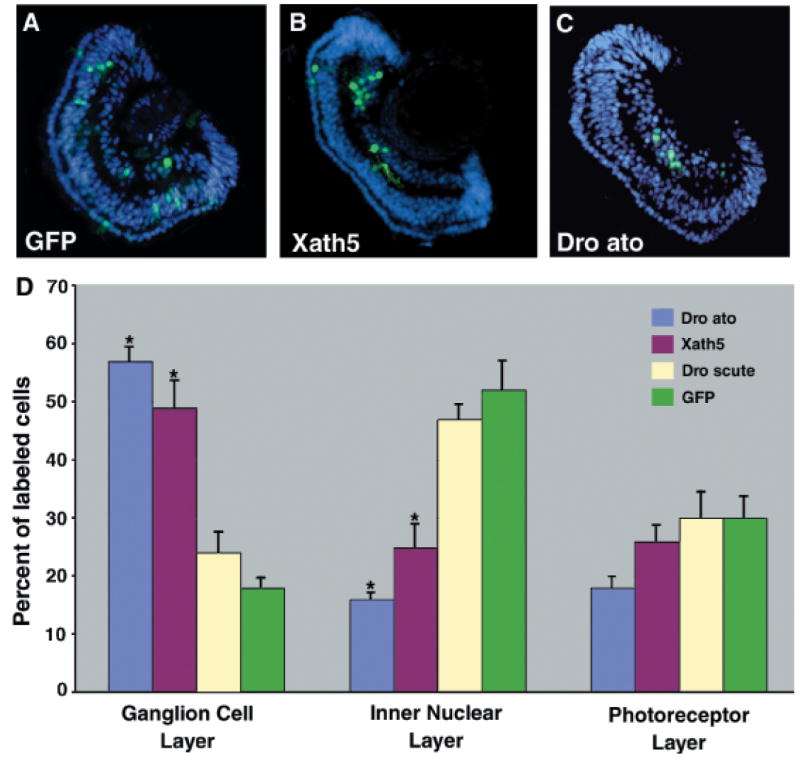
Drosophila atonal mimics Xath5 over expression by promoting vertebrate retinal ganglion cell fate. (A–C) Cryostat sections through the retina of stage 41 embryos lipofected at stage 17 with green fluorescent protein DNA alone or in combination with either Xath5 or Drosophila atonal (Dro ato) DNA as indicated. Nuclei were stained with Hoechst dye (blue) to visualize the retinal cell layers. Overexpression of Xath5 (B) or Dro ato (C) caused an increase in the representation of GFP-labeled cells in the ganglion cell layer. (D) Overexpression of either Xath5 or Dro ato promoted an increase in retinal ganglion cell differentiation compared with control GFP lipofections, whereas overexpression of Drosophila scute did not. n = 472 cells from five embryos for GFP, 939 cells from six embryos for Xath5, 634 cells from five embryos for scute, and 950 cells from seven embryos for Dro ato. Error represents SEM; the asterisks represent significant difference as compared with GFP controls, P<0.01 by Student’s t-test.
Drosophila atonal behaved similarly to Xath5 upon DNA lipofection, causing a dramatic increase in the representation of labeled cells in the ganglion cell layer (Fig. 6, C and D). Overexpression of Drosophila scute, a nonretinal bHLH gene chosen for comparison, did not have this effect (Fig. 6D). Instead, scute caused an increase in bipolar cell differentiation (data not shown; Moore et al. 2002). Both of these Drosophila bHLH genes can promote neural development in cleavage-stage frog embryos and ectopic neurogenesis in neural plate stage embryos (data not shown) similar to Xath5 or Math5 (Kanekar et al. 1997; Brown et al. 1998).
Our findings together with previous functional comparisons of Xath5 and Math5 (Brown et al. 1998) suggest that atonal and Xath5 act analogously in both the fly or vertebrate frog retina. However, Math5 does not function like atonal or Xath5 when tested outside the context of the mammalian retina. In the fly and frog eye, Math5 instead behaves like scute, another bHLH gene that does not normally function during fly retinal development.
DISCUSSION
Current debates regarding eye evolution focus on the emergence of a growing number of conserved developmental genes and pathways. Genes from different species are termed Orthologs when they share coding sequence homology, expression patterns, and endogenous function (Abouheif 1997). Beyond these criteria, however, few Orthologs have been reciprocally tested for conserved function in both invertebrate and vertebrate eyes. Such assessments are needed to identify divergent functions that correlate with different eye types. Because early (Pax6) and late (photoreceptor opsins and lens crystallins) acting proteins appear to have invariant roles in highly polymorphous bilaterian eyes (Arendt and Wittbrodt 2001 and references therein), it is plausible that gene networks required for early eye specification and visual function have significantly diverged. Here we compared the cross-species functions of Drosophila atonal, Xenopus Xath5, and MusMath5 during insect and amphibian eye development.
Drosophila proneural genes induce vertebrate retinal neurons
Ectopic expression of atonal in the developing Xenopus eye biases retinal progenitors toward an RGC fate. atonal thus functions as well as Xath5 in this vertebrate eye assay. By comparison, Drosophila scute promotes bipolar cell fate in DNA lipofection and 16-cell RNA injection studies. This result is consistent with the behavior of its vertebrate Orthologs, Xenopus Xash1 and mouse Mash1 (Kanekar et al. 1997; Brown et al. 1998; Moore et al. 2002) and the differing roles of atonal and scute in Drosophila. During fly neurogenesis, atonal and scute exhibit mutually exclusive expression patterns and promote chordotonal and photoreceptor versus external sensory neuron fates, respectively. Thus, specification of neuronal subtype might be controlled through the spatiotemporal regulation of these genes. However, we expressed atonal and scute equivalently in time and space within the frog retina. This is similar to vertebrate neurogenesis where multiple bHLH genes function simultaneously, sometimes within the same cell. How do atonal and scute promote different vertebrate retinal fates? As in Drosophila, they may have different intrinsic properties and so activate different target genes (Chien et al. 1996; Sun et al. 2000) or may respond differently to regulatory mechanisms.
Recently, Moore et al. (2002) demonstrated that phosphorylation of some Xenopus bHLH proteins modulates the timing of their function and, subsequently, the neuronal fates they promote. In the frog retina, GSK3β kinase phosphorylates NeuroD and Xash1 and so delays their action, but it does not phosphorylate Xath5. Drosophila Scute can be similarly phosphorylated in Xenopus by GSK3β kinase, and this modulates its activity to induce bipolar neurons upon its ectopic expression (Moore et al. 2002). However, when coexpressed with a dominant negative (dn) form of GSK3β, Scute instead induces RGCs. Thus, posttranslational regulation of Scute by GSK3β controls the timing of Scute function in the frog eye just as it does for its Ortholog, Xash1 (Moore et al. 2002). In principle, posttranslational modification of Drosophila bHLHs may also help regulate the timing of fly neurogenesis. However, not all differences between bHLH factors can be accounted for by protein phosphorylation. During Xenopus retinal development, Math5 overexpression promotes bipolar differentiation (Brown et al. 1998), but this activity is not altered by coexpression with dnGSK3β (Moore et al. 2002). The different effects of ectopic Math5 and Xath5 in the Xenopus retina thus cannot be attributed to GSK3β phosphorylation. This suggests that frog and mouse retinal environments are not equivalent.
Is Math5 a functional Ortholog of atonal and Xath5?
Our further functional comparison of mammalian Math5 within the developing Drosophila eye demonstrates that it cannot functionally substitute for atonal or Xath5. Nevertheless, within the mouse eye Math5 is the proneural gene for RGCs (Brown et al. 2001; Wang et al. 2001), demonstrating that it functions equivalently to other known vertebrate Ath5 genes (Kanekar et al. 1997; Liu et al. 2001; Kay et al. 2001). For example, zebrafish Ath5 mutants exhibit a loss of RGCs (Kay et al. 2001), whereas ectopic expression Xenopus or chick Ath5 promotes excess RGC formation (Kanekar et al. 1997; Liu et al. 2001). Instead, within both the frog and fly retina, mouse Math5 acts like Drosophila scute (this study) or Mash1 (Brown et al. 1998; Moore et al. 2002). Several mechanisms can explain this conundrum. First, the tempo of eye development differs widely among mice, frogs, and fruit flies. Xenopus retinogenesis is completed in 1 day and Drosophila retinogenesis in 3 days. This contrasts with the rodent eye where retinal neurogenesis occurs during a 3-week period. Neither atonal nor Xath5 have been tested in the rodent eye, so is it unclear whether they can induce RGCs during a protracted period of retinal development. Interestingly, ectopic Math5 can induce RGCs in the chick retina (Liu et al. 2001), which develops on a time scale similar to rodents. Cath5 activity has not yet been tested in the Xenopus or Drosophila assays.
Second, Math5 may be unable to form functional heterodimers with daughterless, the Drosophila bHLH partner for atonal. Perhaps amino acid differences between Xath5 and Math5 or a putative transactivation domain in the C-terminus of Xath5, but not Math5, allow Xath5 to function with Daughterless in the fly eye. We do not favor this explanation because Scute and Daughterless make functional heterodimers elsewhere in the fly nervous system and the phenotypes of Math5 and Scute are nearly identical in both the fly and frog retina. Instead, Math5 may interact differently with other components of the transcriptional complex. This suggests that Math5 protein has low activity outside of a mammalian cell. Math5 may require a mammalian-specific modification or cofactor. Alternatively, a conserved fly/frog component may prevent Math5 from inducing either fly R8 or frog RGC neurons.
Finally, atonal, Xath5, and Math5 may activate different target genes within their respective eye types. Atonal transcriptionally regulates scabrous by binding to its promoter in the fly eye (data not shown), whereas other bHLH proneural genes regulate scabrous outside the eye (Mlodzik et al. 1990). Here we show that Xath5, NeuroD, and scute (but not Math5) activate scabrous in the developing fly eye. Scabrous expression is more widely spaced in discs rescued by Xath5 and NeuroD compared with atonal. However, Scabrous expression is extremely dispersed in UAS-scute eyes and essentially absent in UAS-Math5 eyes. Because Math5 and Atonal share 100% amino acid homology in their DNA-binding domains, our results cannot be simply explained by divergent basic domains binding DNA differently. Because no vertebrate eye homologue of scabrous has been described, it may represent a downstream pathway component that is not shared by all three eye types. Downstream transcriptional targets have been identified for each of the bHLH genes tested here, but none has been shown to function in fly, amphibian, and mouse retinal development.
Although Math5 does not act analogous to atonal or Xath5 in the fly and frog retina, it is a semi-Ortholog of atonal (Sharman 1999). This designation is based on the intrinsic function of vertebrate Ath5 genes in zebrafish, frog, chick, and mouse retinal development; the partitioning of atonal function between vertebrate Ath5 and Ath1 genes; and the failure of loss or gain of function Ath1 experiments to provoke eye phenotypes. The expression patterns and functions of Math1 and Math5 in mice are minimally overlapping but together encompass those of atonal in fruit flies. This implies that atonal was duplicated during evolution and tissue enhancers partitioned between the two Ath genes. After divergence, Math1 acquired a new function within intestinal secretory cells that is not paralleled in Drosophila (Yang et al. 2001). Recently, Wang and colleagues (2002) performed a comprehensive functional comparison of atonal and Math1. In fruit flies, ectopic Math1 rescued all atonal mutant phenotypes, including R8 photoreceptor formation (Wang et al. 2002). In mice, homologous recombination of atonal into the Math1 locus fully compensated for loss of Math1, even within developing intestinal secretory cells (Wang et al. 2002).
The ability of Math1 activity to rescue Drosophila photoreceptor development allows us to speculate that Math5 retinal function was modified relatively recently. The properties of Math5 are thus relatively derived in comparison with Math1, which appears to retain more basal characteristics. Phylogenetic and amino acid analyses support this idea (Brown et al. 2002). In particular, 10 bHLH amino acid residues that resolve Ath5 or Ath1 from Atonal show pattern differences between these clades (Brown et al. 2002). Interestingly, all affected residues are predicted to affect protein–protein interactions and not DNA binding. These studies point to the importance of testing the functional equivalence of Math1 and Math5 in mice by reciprocal substitution. If mutant phenotypes fail to rescue, this would suggest that differences in (unconserved) protein structure and/or tissue-specific factors are responsible for the divergence of Math1 and Math5 function.
We propose that the atonal gene family represents one position, within a network of early eye development genes, where gene regulation and/or protein interaction have diverged during eye evolution. To understand whether functional divergence within the atonal gene family is correlated with eye types, it will be necessary to identify and study these genes in more animal taxa.
Acknowledgments
We thank Carol Sattler of the McArdle Cancer Laboratory at the University of Wisconsin for histology, Bruce Donohoe of the University of Michigan Microscopy & Image Analysis Laboratory for scanning electron microscopy, Bob Holmgren for the use of his microscope, Helmut Kramer for anti-Boss antibody, and the Developmental Studies Hybridoma Bank, under the auspices of the NICHD and maintained by The University of Iowa. We are indebted to Bill Goossens for technical advice, Jim Lauderdale for valuable discussions, and David Blackburn and Ross Cagan for critical reading of the manuscript. This work was supported by NIH grants EY12274 (to M. L. V.), EY14259 (to T. G.), and EY13612 (to N. L. B.); the Pew Scholars Program in the Biomedical Scholars Program, sponsored by the Pew Charitable Trust (to M. L. V.); and the Howard Hughes Medical Institute (to Y.-N. J.).
References
- Abouheif E. Developmental genetics and homology: a hierarchical approach. Trends Ecol Evol. 1997;12:405–408. doi: 10.1016/s0169-5347(97)01125-7. [DOI] [PubMed] [Google Scholar]
- Arendt D, Wittbrodt J. Reconstructing the eyes of Urbilateria. Philos Trans R Soc Lond B Biol Sci. 2001;356:1545–1563. doi: 10.1098/rstb.2001.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science. 1990;250:1370–1377. doi: 10.1126/science.2175046. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brown NL, Dagenais S, Chen CM, Glaser T. Molecular characterization and mapping of ATOH7, a human atonal homolog with a predicted role in retinal ganglion cell development. Mammal Gen. 2002;13:95–101. doi: 10.1007/s00335-001-2101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski JA, Glaser T. Math5 is required for retinal ganglion cell and optic nerve development. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Markey DR, Carroll SB. hairy gene function in the Drosophila eye: normal expression is dispensable but ectopic expression alters cell fates. Development. 1991;113:1245–1256. doi: 10.1242/dev.113.4.1245. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Kramer H, Hart AC, Zipursky SL. The bride of sevenless and sevenless interaction: internalization of a transmembrane ligand. Cell. 1992;69:393–399. doi: 10.1016/0092-8674(92)90442-f. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Munoz-Marmol AM, Glardon S, Castillo E, Sun H, Li WH, Gehring WJ, Salo E. Isolation and expression of a Pax-6 gene in the regenerating and intact. Planarian Dugesia(G)tigrina. Proc Natl Acad Sci USA. 1999;96:558–563. doi: 10.1073/pnas.96.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Blackwell Science; Malden, MA: 2001. [Google Scholar]
- Chien CT, Hsiao CD, Jan LY, Jan YN. Neuronal type information encoded in the basic helix-loop-helix domain of proneural genes. Proc Natl Acad Sci USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–4147. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Easter SS., Jr Let there be sight. Neuron. 2000;27:193–195. doi: 10.1016/s0896-6273(00)00026-x. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development. 2002;129:1295–1306. doi: 10.1242/dev.129.6.1295. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Glardon S, Holland LZ, Gehring WJ, Holland ND. Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): insights into eye and photoreceptor evolution. Development. 1998;125:2701–2710. doi: 10.1242/dev.125.14.2701. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hassan BA, Bellen HJ. Doing the MATH: is the mouse a good model for fly development? Genes Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- Holt CE, Garlick N, Cornel E. Lipofection of cDNAs in the embryonic vertebrate central nervous system. Neuron. 1990;4:203–214. doi: 10.1016/0896-6273(90)90095-w. [DOI] [PubMed] [Google Scholar]
- Hsiung F, Moses K. Retinal development in Drosophila: specifying the first neuron. Hum Mol Genet. 2002;11:1207–1214. doi: 10.1093/hmg/11.10.1207. [DOI] [PubMed] [Google Scholar]
- Jarman AP. Developmental genetics: vertebrates and insects see eye to eye. Curr Biol. 2000;10:R857–R859. doi: 10.1016/s0960-9822(00)00821-6. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous sytem. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, Jan YN, Vetter ML. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger KC, Roeser T, Staub W, Baier H. Retinal ganglion cell determination requires lakritz, a zebrafish homolog of Drosophila atonal. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Kim P, Helms AW, Johnson JE. XATH-1, a vertebrate homolog of Drosophila atonal, induces a neuronal differentiation within ectodermal precursors. Dev Biol. 1997;187:1–12. doi: 10.1006/dbio.1997.8572. [DOI] [PubMed] [Google Scholar]
- Kimmel BE, Heberlein U, Rubin GM. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. Eye specification in Drosophila: perspectives and implications. Semin Cell Dev Biol. 2001;12:469–474. doi: 10.1006/scdb.2001.0270. [DOI] [PubMed] [Google Scholar]
- Lee EC, Hu X, Yu SY, Baker NE. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16:1179–1188. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci USA. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, Kmita-Cunisse M, Gehring WJ. Isolation of a Pax6 homolog from the ribbonworm Lineus sanguineus. Proc Natl Acad Sci USA. 1996;93:2658–2663. doi: 10.1073/pnas.93.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, Koster RW, Carl M, Krone A, Wittbrodt J. Six3, a medaka homologue of the Drosophila homeobox gene sine oculis is expressed in the anterior embryonic shield and the developing eye. Mech Dev. 1998;74:159–164. doi: 10.1016/s0925-4773(98)00055-0. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- Moore KB, Schneider ML, Vetter ML. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/s0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- Sharman AC. Some new terms for duplicated genes. Semin Cell Dev Biol. 1999;10:561–563. doi: 10.1006/scdb.1999.0338. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Ectopic scute induces Drosophila ommatidia development without R8 founder photoreceptors. Proc Natl Acad Sci USA. 2000;97:6815–6819. doi: 10.1073/pnas.110154497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomarev SI, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J. Squid Pax-6 and eye development. Proc Natl Acad Sci USA. 1997;94:2421–2426. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12:491–498. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Hassan BA, Bellen HJ, Zaghbi HY. Drosophila atonal fully rescues the phenotypes of Math1 null mice: new functions evolve in new cellular contexts. Curr Biol. 2002;12:1611–1616. doi: 10.1016/s0960-9822(02)01144-2. [DOI] [PubMed] [Google Scholar]
- Xu PX, Woo I, Her H, Beier DR, Maas RL. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, Maas RL. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]