Abstract
Triatoma infestans is a hemiptera, vector of Chagas’ disease, that feeds exclusively on vertebrate blood in all life stages. Hematophagous insects’ salivary glands (SG) produce potent pharmacological compounds that counteract host hemostasis, including anti-clotting, anti-platelet, and vasodilatory molecules. To obtain a further insight into the salivary biochemical and pharmacological complexity of this insect, a cDNA library from its salivary glands was randomly sequenced. Also, salivary proteins were submitted to two dimentional gel (2D-gel) electrophoresis followed by MS analysis. We present the analysis of a set of 1,534 (SG) cDNA sequences, 645 of which coded for proteins of a putative secretory nature. Most salivary proteins described as lipocalins matched peptide sequences obtained from proteomic results.
Keywords: Hematophagy, Saliva, Transcriptome, Triatoma infestans, Feeding, Sialome
1. Introduction
Triatoma infestans (Hemiptera: Reduviidae) is an important vector of Trypanosoma cruzi, a protozoan parasite and etiological agent of Chagas’ disease (American trypanosomiasis) in Latin America (Dias, 1987). All instar nymphs and adults are hematophagic and need a blood meal to molt and for oviposition. The insect obtains the blood meal by injecting its maxilla into vertebrate’s skin searching for a vessel (Lavoipierre, 1965).
The (SG)s of blood-feeding arthropods show a variety of anti-hemostatic compounds that help the bug to obtain its blood meal. Like other blood-sucking arthropods that have been studied (Ribeiro and Francischetti, 2003), T. infestans is capable of counteracting host hemostatic responses triggered to prevent blood loss following tissue injury, such as vasoconstriction, blood coagulation and platelet aggregation (Ribeiro, 1995). The molecular diversity of hematophagous insect saliva represents a rich field for the discovery of novel pharmacologically active compounds and for understanding the evolutionary mechanisms leading to the insect’s adaptation to this feeding habit. Previous studies describing the sialome (set of RNA message + set of proteins found in (SG)s) of hematophagous insects and ticks (Francischetti et al., 2002; Valenzuela et al., 2002a, b) have revealed that the sialomes of these disease vectors are more complex than expected and contain many proteins to which we cannot yet ascribe a function.
Lipocalins are a large and heterogenous group of proteins that play various roles, mainly as carriers of small ligands in vertebrates and invertebrates (Flower et al., 2000). A great array of (SG) proteins belonging to the lipocalin family has generated a large number of different molecules having anti-hemostatic functions while maintaining the fundamental structure of the protein fold (Montfort et al., 2000). Lipocalins were found in the saliva of other blood-sucking insects such as Rhodnius prolixus (Ribeiro et al., 2004a) and Triatoma brasiliensis (Santos et al., 2007), and also in tick saliva (Paesen et al., 2000). In R. prolixus, three types of salivary lipocalins have been characterised: the nitrophorins consisting in a group of lipocalins working as NO carrier, and also as anti-clotting; the ADP-binding protein RPAI1, which inhibits platelet activation and aggregation (Francischetti et al., 2000); and a group of lipocalins related to T. pallidipenis thrombin inhibitor triabin (Fuentes-Prior et al., 1997). Another lipocalin from the saliva of T. pallidipenis denominated pallidipin has been ascribed as a specific inhibitor of collagen-induced platelet aggregation (Noeske-Jungblut, et al., 1994).
In this work we present the analysis of a set of 1,534 (SG) cDNA sequences, 645 of which code for proteins of a putative secretory nature. Most salivary proteins described as lipocalins – 55% of the transcripts coding for putative secreted proteins – matched peptide sequences obtained from proteomic results. We expect this work will contribute new salivary transcripts that could help the understanding of the role of salivary molecules in host/vector interactions and the discovery of novel pharmacologic agents.
2. Materials and methods
2.1. Triatomines and salivary glands cDNA library construction
Triatoma infestans were reared in an insectary room kept at 27 °C ± 1.0 °C, with a relative humidity ranging from 70 to 75% and a 16 h:8 h light:dark photoperiod. Salivary glands (SG) were dissected from Vth instar nymphs 2 days after a blood meal and transferred to RNA-Later (Ambion) solution in 1.5 mL polypropylene vials. SG were kept at −20°C for isolating polyA+ RNA.
T. infestans SG mRNA was isolated from 15 SG pairs from Vth instar nymphs, using the Micro-FastTrack mRNA isolation kit (Invitrogen). The PCR-based cDNA library was made following the instructions for the SMART (switching mechanism at 5′ end of RNA transcript) cDNA library construction kit (Clontech). This kit provides a method for producing high-quality, full-length cDNA libraries from nanogram quantities of polyA+ or total RNA. It utilises a specially designed oligonucleotide named SMART IV™ in the first-strand synthesis to generate high yields of full-length, double-stranded cDNA. T. infestans SG polyA+ RNA was used for reverse transcription to cDNA using PowerScript reverse transcriptase (Clontech), the SMART IV oligonucleotide, and the CDS III/3′ primer (Clontech). The reaction was carried out at 42 °C for 1 h. Second-strand synthesis was performed by a long-distance PCR-based protocol using the 5′ PCR primer and the CDS III/3′ primer as sense and antisense primers, respectively. These two primers also create Sfi1A and B restriction enzyme sites at the end of the cDNA. Advantage™ Taq polymerase mix (Clontech) was used to carry out the long-distance PCR reaction on a Perkin Elmer GeneAmp® PCR system 9700 (Perkin Elmer Corp.). The PCR conditions were: 95 °C for 1 min; 14 cycles of 95 °C for 10 s, 68 °C for 6 min. A small portion of the cDNA was analysed on a 1.1% agarose/EtBr (0.1 µg/mL) gel to check for the quality and range of the synthesised cDNA. Double-stranded cDNA was immediately treated with proteinase K (0.8 µg/mL) at 45 °C for 20 min and washed three times with water using Amicon filters with a 100 kDa cutoff (Millipore). The clean, double-stranded cDNA was then digested with SfiI restriction enzyme at 50 °C for 2 h followed by size fractionation on a ChromaSpin–400 drip column (Clontech). The profiles of the fractions were checked on a 1.1% agarose/EtBr (0.1 µg/mL), and fractions containing cDNA were pooled in three different groups according to their size: large, medium or small sequences. Each group was concentrated and washed three times with water using an Amicon filter with a 100 kDa cutoff. The concentrated cDNA was then ligated into a λ TriplEx2 vector (Clontech), and the resulting ligation mixture was packaged using GigaPack® Gold III Plus packaging extract (Stratagene), according to the manufacturer's instructions. The packaged library was plated by infecting log-phase XL1-Blue Escherichia coli cells (Clontech). The percentage of recombinant clones was determined by performing a blue-white selection screening on LB/MgSO4 plates containing X-gal/IPTG.
2.2. Sequencing of the T. infestans cDNA library
The T. infestans SG cDNA library was plated on LB/MgSO4 plates containing X-gal/IPTG to an average of 250 plaques per 150 mm Petri plate. Recombinant (white) plaques were randomly picked up and transferred to 96-well MICROTEST™ U-bottom plates (BD BioSciences) containing 75 µL of H2O per well. The phage suspension was either immediately used for PCR or stored at 4 °C until use.
To amplify the cDNA using a PCR reaction, 4 µL of the phage sample were used as a template. The primers were sequences from the λ TriplEx2 vector and named PT2F1 (5′-AAGTACTCTAGCAATTGTGAGC-3’) and PT2R1 (5′-CTCTTCGCTATTACGCCAGCTG-3′), positioned at the 5′ and 3′ end of the cDNA insert, respectively. The reaction was carried out in MicroAmp 96-well PCR plates (Applied Biosystems) using Platinum PCR® SuperMix (Invitrogen), on a Perkin Elmer GeneAmp® PCR system 9700 (Perkin Elmer Corp.). The PCR conditions were: 1 hold of 75 °C for 3 min, 1 hold of 94 °C for 4 min, 33 cycles of 94 °C for 1 min, 49 °C for 1 min, and 72 °C for 1 min and 20 s. The amplified products were analysed on a 1.2% agarose/EtBr gel. cDNA library clones (1800 clones) were PCR amplified, and those showing a single band were selected for sequencing. The PCR products were used as a template for a cycle-sequencing reaction using DTCS labeling kit from Beckman Coulter). The primer used for sequencing (PT2F3) is upstream from the inserted cDNA and downstream from the PT2F1 primer. The sequencing reaction was performed on a Perkin Elmer 9700 thermocycler. Conditions were 1 hold of 75 °C for 2 min, 1 hold of 94 °C for 4 min, and 30 cycles of 96 °C for 20 s, 50 °C for 20 s, and 60 °C for 4 min. After cycle-sequencing the samples, a cleaning step was performed using the multiscreen 96-well plate cleaning system (Millipore). The 96-well multiscreening plate was prepared by adding a fixed amount (manufacturer’s specification) of Sephadex-50 (Amersham Pharmacia) and 300 µL of deionised water. After partially drying the Sephadex in the multiscreen plate, the whole cycle-sequencing reaction was added to the center of each well, centrifuged at 2,500 rpm for 5 min, and the clean sample was collected on a sequencing microtiter plate (Beckman Coulter). The plate was then dried on a Speed-Vac SC110 model with a microtiter plate holder (Savant Instruments). The dried samples were immediately resuspended with 25 µL of formamide, and one drop of mineral oil was added to the top of each sample. Samples were either sequenced immediately on a CEQ 2000 DNA sequencing instrument (Beckman Coulter) or stored at −30 °C. A total of 1,534 cDNA library clones were sequenced.
2.3. Reverse phase liquid chromatography (RPLC)
Approximately 20 µL of saliva from adult insects were diluted in 200 µL buffer (50mM Tris-HCl, 120 mM NaCl pH 7.4). Proteins were loaded in a Microcon 10 (YM-10, Millipore) and centrifuged at 10,000 × g for 30 min. The flow through with proteins of low molecular weight (< 10 kDa) was kept. Microbore reversed-phase liquid chromatography (RPLC) was performed using a 0.3 mm C18 column from Phenomenex (Torrence, CA). After sample injection, the column was washed for 10 min with 95% mobile phase A (0.1% formic acid in water) at 5 µL/min and peptides were eluted using a linear gradient to 90% mobile B (100% acetonitrile and 0.1% formic acid) for 40 min. The column eluate was monitored at 220 nm. A Probot fraction collector (Dionex, Sunnylvale, CA) was used to deliver the fractions to 96-well plates containing 25 µL of water. Fractions of interest were submitted to tryptic digestion, followed by mass spectrometry as indicated below.
2.4. 2D-gel electrophoresis
2D-gel electrophoresis was performed using ZOOM IPGRunner System (Invitrogen) under manufacturer's recommended running conditions. Briefly, approximately 500 µg of sample proteins of T. infestans saliva were solubilised with 155 µL rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 20 mM DTT, 0.5% carrier ampholytes, pH 3–10). The samples were absorbed by rehydration ZOOM strips (7 cm; pH 3–10 NL) overnight at room temperature and then focused under manufacturer's recommended conditions. The focused IPG strips were reduced/alkylated/equilibrated with reducing and alkylating reagents dissolved in the sample buffer. The strips were then applied onto NuPAGE 4–12% Bis-Tris ZOOM gels (Invitrogen). The gels were run under MOPS buffer and stained with SeeBlue staining solution (Bio-Rad).
2.5. Protein identification by mass spectrometry
Protein identification of either RPLC or 2D gel-separated proteins was performed on reduced and alkylated trypsin-digested samples prepared by standard mass spectrometry protocols. Tryptic digests were analysed by coupling the Nanomate (Advion BioSciences)—an automated chip-based nano-electrospray interface source—to a quadrupole time-of-flight mass spectrometer, QStarXL MS/MS System (Applied Biosystems/Sciex). Computer-controlled, data-dependent automated switching to MS/MS provided peptide sequence information. AnalystQS software (Applied Biosystems/Sciex) was used for data acquisition. Data processing and databank searching were performed with Mascot software (Matrix Science). The NR protein database from the NCBI, National Library of Medicine, NIH, was used for the search analysis, as was a protein database generated during the course of this work.
2.6. Bioinformatic tools and procedures
Expressed sequence tags (ESTs) were trimmed of primer and vector sequences, clusterised, and compared with other databases as previously described. (Valenzuela et al., 2003). The BLAST tool (Altschul et al., 1996), CAP3 assembler (Huang et al., 1999), ClustalW (Thompson et al., 1994), and TREEview software (Page, 1996) were used to compare, assemble and align sequences, and to visualise alignments. For functional annotation of the transcripts we used the tool BlastX (Altschul et al., 1997) to compare the nucleotide sequences with the nonredundant (NR) protein database of the NCBI and to the Gene Ontology (GO) database (Ashburner et al., 2000). The tool rpsBlast (Schaffer et al., 2001) was used to search for conserved protein domains in the Pfam (Bateman et al., 2000), SMART (Letunic et al., 2002), Kog (Tatusov et al., 2003), and conserved domains (CDD) databases (Marchler-Bauer et al., 2002). We have also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI and to several organism proteomes downloaded from NCBI (yeast), Flybase (D. melanogaster), or ENSEMBL (An. gambiæ). Segments of the three-frame translations of the EST (since the libraries were unidirectional, we did not use six-frame translations) starting with a methionine in the first 100 predicted amino acids (aa)—or the predicted protein translation, in the case of complete coding sequences—were submitted to the SignalP server (Nielsen, 1997) to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc (http://www.cbs.dtu.dk/services/NetOGlyc/) (Hansen et al., 1998). Functional annotation of the transcripts was based on all the comparisons above. Following inspection of all results, transcripts were classified as either Secretory (S), Housekeeping (H), or of unknown (U) function, with further subdivisions based on function and/or protein families. Sequence alignments were done with the ClustalX software package (Thompson et al., 1997). Phylogenetic analysis and statistical neighbor-joining bootstrap tests of the phylogenies were done with the Mega package (Kumar et al., 2004). Hyperlinked Excel spreadsheets of the assembled ESTs and of the salivary protein database are supplied as Supplemental Tables 1 and 2 at the journal site.
3. Results
General Description of the Salivary Transcriptome Database
3.1. Description of the clusterised data set / cDNA library characteristics
1,534 sequences were used to assemble a clusterised database, yielding 657 clusters of related sequences, 500 of which contained only one EST. The consensus sequence of each cluster is named either a contig (deriving from two or more sequences) or a singleton (deriving from a single sequence). In this paper, we will use the denomination contig to address sequences deriving from both consensus sequences and from singletons. The 657 contigs were compared by the program BlastX, BlastN, or rpsBlast (Altschul et al., 1997) to the nonredundant protein database of the National Center of Biological Information (NCBI), to the gene ontology database (Ashburner et al., 2000), to the conserved domains database of the NCBI (Marchler Bauer et al., 2002), and to a custom-prepared subset of the NCBI nucleotide database containing either mitochondrial or rRNA sequences. Because the libraries are unidirectional, the three frame translations of the dataset were also derived, and open reading frames (ORF) starting with methionine and longer than 40 aa residues were submitted to SignalP server (Nielsen et al., 1997) to help identify putative secreted proteins. The EST assembly, BLAST, and signal peptide results were transferred into an Excel spreadsheet for manual annotation. Five categories of expressed genes derived from the manual annotation of the contigs (Table 1). The putatively secreted (S) category contained 18% of the clusters and 42% of the sequences, with an average number of 5.5 sequences per cluster. The housekeeping (H) category had 36.4% and 35.9% of the cluster and sequences, respectively, and an average of 2.3 sequences per cluster. Forty-four percent of the clusters, containing 21% of all sequences, were classified as unknown (U) because no assignment for their function could be made; most of these consisted of singletons. Possible transposable elements originated 7 clusters, mostly singletons. We have also identified viral transcripts in our dataset. These data can be downloaded as Supplemental Table 1 for the EST data, and Supplemental File 2 for the proteome set.
Table 1.
Types of transcripts found in Triatoma infestans salivary glands.
| Types of transcripts | Clusters | Sequences | Sequences/Cluster |
|---|---|---|---|
| Secreted (S) | 118 | 645 | 5.5 |
| Housekeeping (H) | 239 | 550 | 2.3 |
| Unknown (U) | 292 | 324 | 1.1 |
| Transposable element (TE) | 7 | 11 | 1.6 |
| Viral product | 1 | 4 | 4.0 |
| Total | 657 | 1534 |
3.2. Housekeeping (H) genes
The 239 gene clusters (comprising 550 EST) attributed to H genes expressed in the (SG)s of T. infestans were further characterised into 20 groups, according to their possible function (Table 2). According to an organ specialised in secreting polypeptides and as observed in previous sialotranscriptomes (Francischetti et al., 2002; Ribeiro et al., 2004a, b; Calvo et al., 2007), the two larger sets were associated with protein synthesis machinery (298 EST in 51 clusters) and with energy metabolism (17 clusters containing 57 EST). We have also included in this category a group of 66 EST that grouped into 52 clusters and represent conserved proteins of unknown function, presumably associated with cellular metabolism. Other sequences with homology to housekeeping protein include those coding for ribosomal, cytochrome, ATP synthase subunit, and NADH-ubiquinone oxidoreductase, among other molecules.
Table 2.
Functional classification of the housekeeping genes expressed in Triatoma infestans salivary glands.
| Types of transcripts | Clusters | % | Sequences | % |
|---|---|---|---|---|
| Conserved, unknown function | 52 | 21.8 | 66 | 11.3 |
| Protein synthesis machinery | 51 | 21.3 | 298 | 54.2 |
| Signal transduction | 18 | 7.5 | 18 | 3.3 |
| Metabolism, energy | 17 | 7.1 | 57 | 10.4 |
| Transport/Storage | 14 | 5.9 | 15 | 2.7 |
| Cytoskeletal | 12 | 5.0 | 13 | 2.4 |
| Nuclear regulation | 12 | 5.0 | 13 | 2.4 |
| Protein export machinery | 10 | 4.2 | 12 | 2.2 |
| Protein modification | 10 | 4.2 | 16 | 2.9 |
| Transcription machinery | 9 | 3.8 | 10 | 1.8 |
| Proteasome machinery | 8 | 3.3 | 10 | 1.8 |
| Metabolism, carbohydrate | 6 | 2.5 | 6 | 1.1 |
| Metabolism, amino acid | 4 | 1.7 | 4 | 0.7 |
| Transcription factor | 3 | 1.3 | 3 | 0.5 |
| Metabolism, intermediate and detoxication | 3 | 1.3 | 3 | 0.5 |
| Metabolism, nucleotide | 3 | 1.3 | 3 | 0.5 |
| Lysosomal products | 2 | 0.8 | 2 | 0.4 |
| Extracellular matrix | 2 | 0.8 | 2 | 0.4 |
| Metabolism, oxidant | 2 | 0.8 | 2 | 0.4 |
| Metabolism, lipid | 1 | 0.4 | 1 | 0.2 |
| Total | 239 | 550 |
3.3. Viral product
Contig 110 produced a best BlastX match with gi|20451030|, a capsid protein precursor from a Triatoma virus (TrV), scoring with other reported capsid proteins. TrV infects triatomines and belongs to a new family of RNA viruses known as Dicistroviridae (Czibener, et al., 2000).
3.4. Transposable elements
Seven contigs on our database possibly derive from transposable elements (TE). Their translation products are similar to those of Drosophila melanogaster proteins annotated as endonuclease and reverse transcriptase. These transcripts may indicate active ongoing transposition activity in T. infestans, or with regulatory transcripts inhibiting TE genomic mobilisation.
3.5. Transcripts coding for putative secreted proteins in Triatoma infestans salivary glands
3.5.1. Lipocalins
The most abundant group of putative secreted proteins in T. infestans (SG)s is the lipocalins, corresponding to 55% of the transcripts in the S class. Lipocalins are widely distributed and heterogeneous proteins occurring in animals, plants and bacteria. Even though they have various molecular masses, the domain, determining the specific properties, is about 18–20 kDa (Flower et al., 1993). The lipocalin primary sequence shows a low percentage of similarity when comparing randomly selected members of the family. By contrast, an interesting feature of the lipocalins is their well conserved three-dimensional structure (Flower, 1995). They are typically small, extracellular proteins sharing several common molecular properties: the binding of small, principally hydrophobic molecules, binding to specific cell-surface receptors; the formation of covalent and non-covalent complexes with other soluble macromolecules. Although they have been classified mainly as transport proteins, it is now clear that members of the lipocalin family fulfill a wide variety of different functions (Flower, 2000). The aim to discover the role of salivary lipocalins in blood-sucking insects through functional genomic and proteomic studies has been able to identify anticoagulants, antiplatelets, and vasodilatory molecules (Andersen et al., 2005).
We found lipocalins similar to salivary proteins of other species of the genus Triatoma, such as pallidipin, an inhibitor of collagen-induced platelet aggregation (Noeske-Jungblut et al., 1994); and triabin, a potent and selective thrombin inhibitor, both from the bug Triatoma pallidipennis (Noeske-Jungblut et al., 1995); and procalin, a salivary allergen from T. protacta (Paddock et al., 2001). The South American T. brasiliensis was also shown to contain salivary cDNA sequences similar to these three previously described Triatoma lipocalins (Sant’Anna et al., 2002). More recently, the sialotranscriptome of T. brasiliensis revealed a high content of lipocalins in its (SG)s, comprising 93.8% of the transcripts coding for putative secreted protein (Santos et al., 2007). Lipocalins have also been found in tick saliva and in Rhodnius performing similar functions, such as histamine and serotonin binding (Ribeiro et al., 2004a; Paesen et al., 2000; Sangamnatdej et al., 2002). This scenario contrasts with mosquitoes and sandflies, where no salivary lipocalins have been described to date.
Triatomine lipocalin sequences were aligned and a neighbor-joining phylogenetic-tree constructed (Fig. 1). The bootstrap values indicated that these sequences have evolved beyond recognition of a common ancestor (not shown). These results may indicate a long evolutionary history for these proteins, or alternatively, a fast rate of evolution. It is interesting to note here that lipocalins were not found in the sialotranscriptome of Oncopeltus fasciatus, a closely related seed feeding bug, indicating that the expansion of this gene family expressed in the salivary glands of triatomines occurred as an adaptation to blood feeding (Francischetti et al., 2007).
Figure 1.
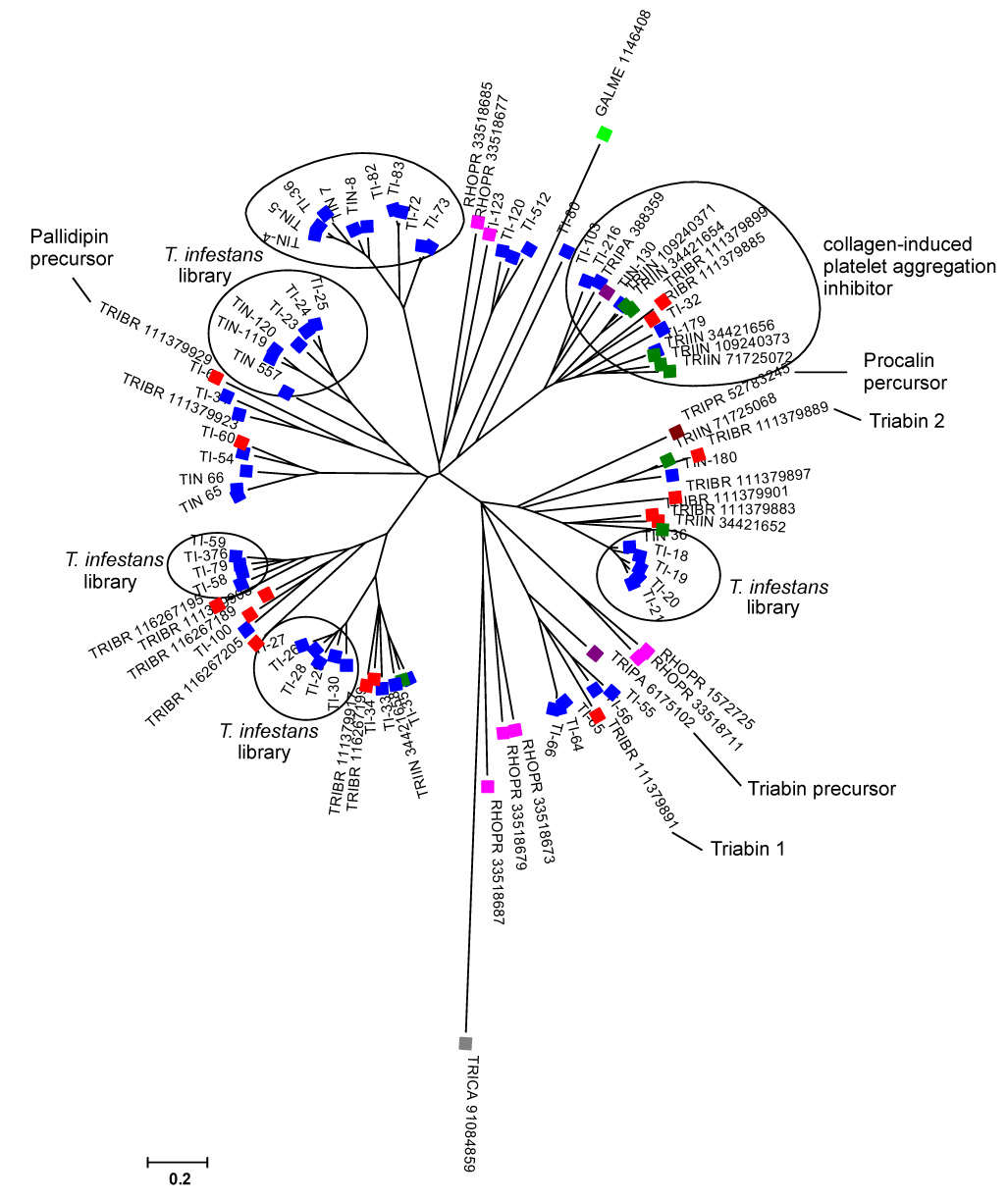
Dendrogram of the T. infestans salivary lipocalins with other insect salivary lipocalins. The sequences derived from the nonreduntant (NR) protein database of the National Center for Biotechnology Information (NCBI) are represented by five letters followed by the NCBI gi| accession number. The five letters derive from the first three letters of the genus and the first two letters from the species name. The protein sequences were aligned by the Clustal program (Thompson et al., 1997), and the dendrogram was done with the Mega package (Kumar et al., 2004) after 1,000 bootstraps with the neighbor-joining (NJ) algorithm. The bar at the bottom represents 20% amino acid substitution. The colorful squares indicate each insect species whose sequences were used: blue, T. infestans sequences from cDNA library; dark green, T. infestans proteins described previously; red, T. brasiliensis; purple, T. pallidipennis; brown, T. protacta; magenta, Rhodnius prolixus; gray, Tribolium castaneum; light green, Galleria mellonella.
3.5.1.2. Enzymes
Some transcripts coding for enzymes are identifiable. A serine protease could be involved with specific host proteolytic events that could affect clotting or the complement cascade. It could also be involved in invertebrate immunity, since prophenoloxidase-activating enzymes are serine proteinases (Söderhäll and Cerenius, 1998). A serine protease with trypsin-like activity was described in T. infestans saliva. This salivary proteolytic activity was denominated triapsin and shown to be released with ejected saliva in active form, suggesting a role in blood-feeding (Amino et al., 2001). Since some Hemiptera species utilise cysteine and aspartic proteases for proteolytic digestion (Terra et al., 1996), the presence of a serine protease could be more related to blood acquisition rather than its digestion. Or it could play a role in the innate immunity of the insect, considering the serine protease as a part of the prophenoloxidase system. This EST has similarities in the alignment with serine proteases from other insects, and phylogenetic analysis shows a strong bootstrap support for the triatomine clade (Fig. 2).
Figure 2.
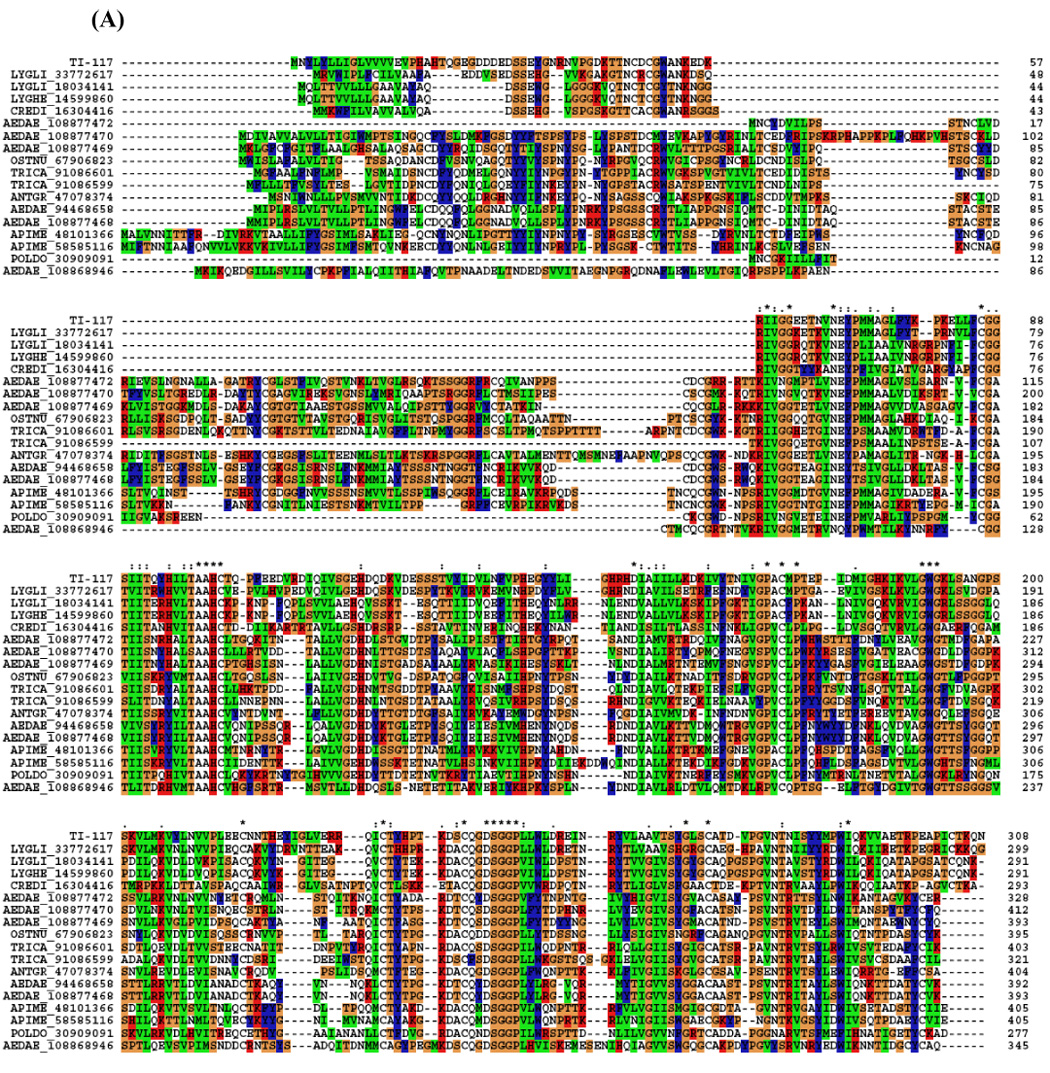

(A) ClustalW alignment of members of the serine protease family deriving from salivary glands of T. infestans and from other insects. (B) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 10% amino acid divergence in the sequences.
3.5.1.3 Apyrase
Partial coding sequences (truncated in the 5′ region) for the enzyme apyrase, a member of the 5′ nucleotidase family, were found. This enzyme was described before in T. infestans (SG)s (Faudry et al., 2004), where it helps the acquisition of blood meals by the degradation of adenosine diphosphate (ADP), a mediator of platelet aggregation and inflammation (Ribeiro and Francischetti, 2003).
3.5.1.4 Inositol phosphatase family
Inositol phosphates are involved in many cellular processes related to signal transduction, secretion and cytoskeletal structure. Numerous enzymes are involved in the metabolism of inositol phosphates and phosphoinositides, including kinases, phosphatases, and phospholipases (Erneux et al., 1998; Guo et al., 1999). Inositol polyphosphate 5-phosphatases (IPPs) share a 5-phosphatase domain and consist of a large family of enzymes acting on different substrates. They regulate the pool of 5-phosphorylated inositol phosphates and phosphoinositides, thereby influencing cellular processes, for which second messenger functions have been reported: Ins(1,4,5)P3, Ins(1,3,4,5)P4, and the lipids PtdIns(4,5)P2 and PtdIns(3,4,5)P3 (Majerus, et al., 1999); (Toker et al., 1997).
Alignment of two contigs from the cDNA library with other IPP sequences from vertebrates and insects showed they share similarities and conserved (aa) sequence (Fig. 3A). The phylogram shows a clade with Triatoma and Rhodnius sequences but apart from other insects, and even the Strongylocentrotus purpuratus sequence is more conserved with vertebrates than with insects (Fig. 3B).
Figure 3.
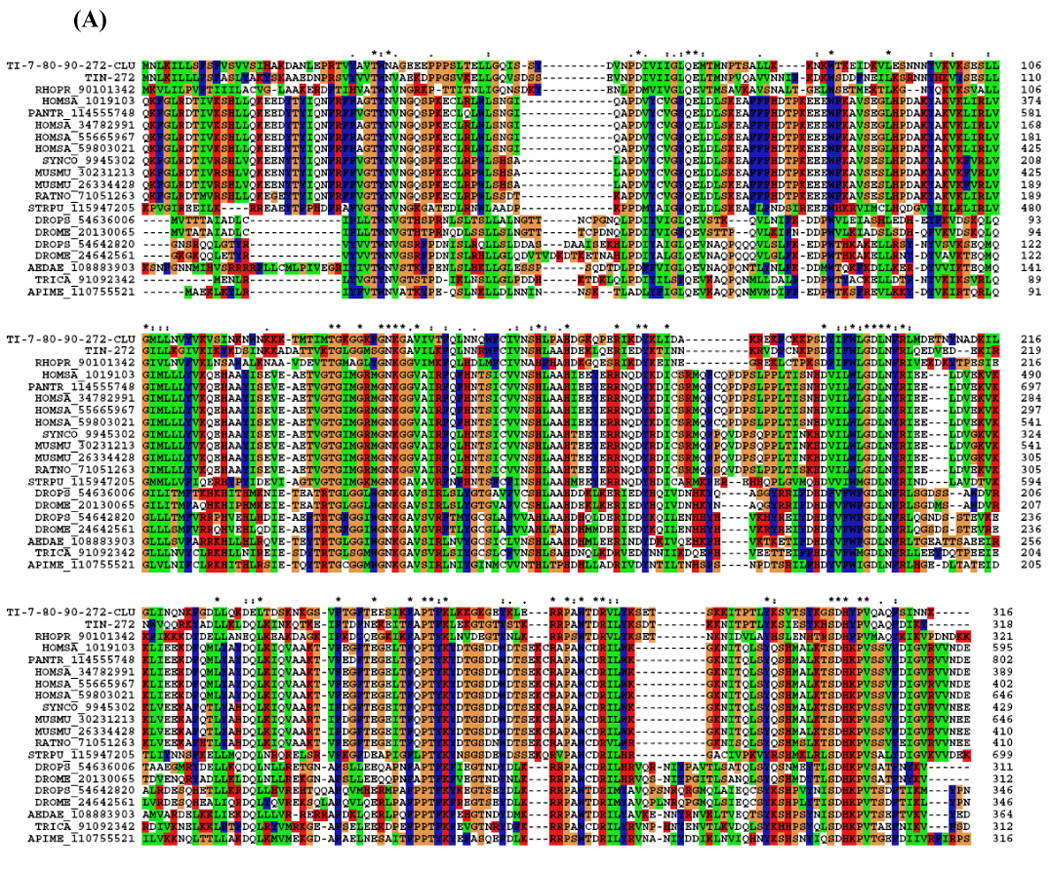
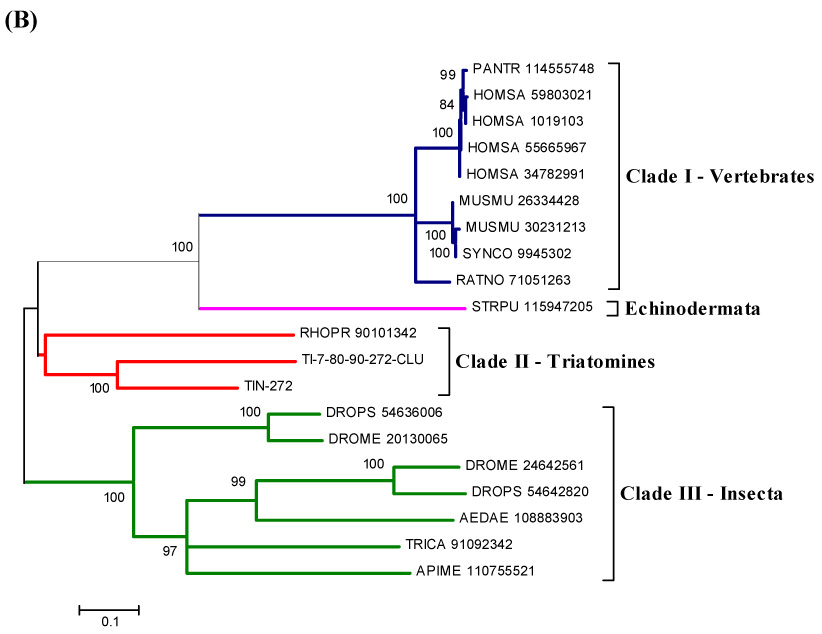
(A) ClustalW alignment of the T. infestans inositol polyphosphate 5-phosphatases (IPP) with IPP from other organisms. (B) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 10% amino acid divergence in the sequences.
Rhodnius prolixus also secretes into its saliva an inositol polyphosphate 5-phosphatase during blood feeding (Ribeiro et al., 2004a). It seems to reduce the concentration of some phospholipids in the plasma membrane of cells and platelets; this could have effects such as elimination of substrates for phospholipase C and PI3-kinase, changes in cytoskeletal architecture, and changes in membrane trafficking and vesicle secretion (Andersen et al., 2006). The presence of this phosphatase in Triatoma saliva can also be related to antithemostatic response, since changes in platelets membrane could interfere with their aggregation and facilitate the acquisition of blood meal. This protein was also identified in the 2D gel (Fig. 12).
Figure 12.
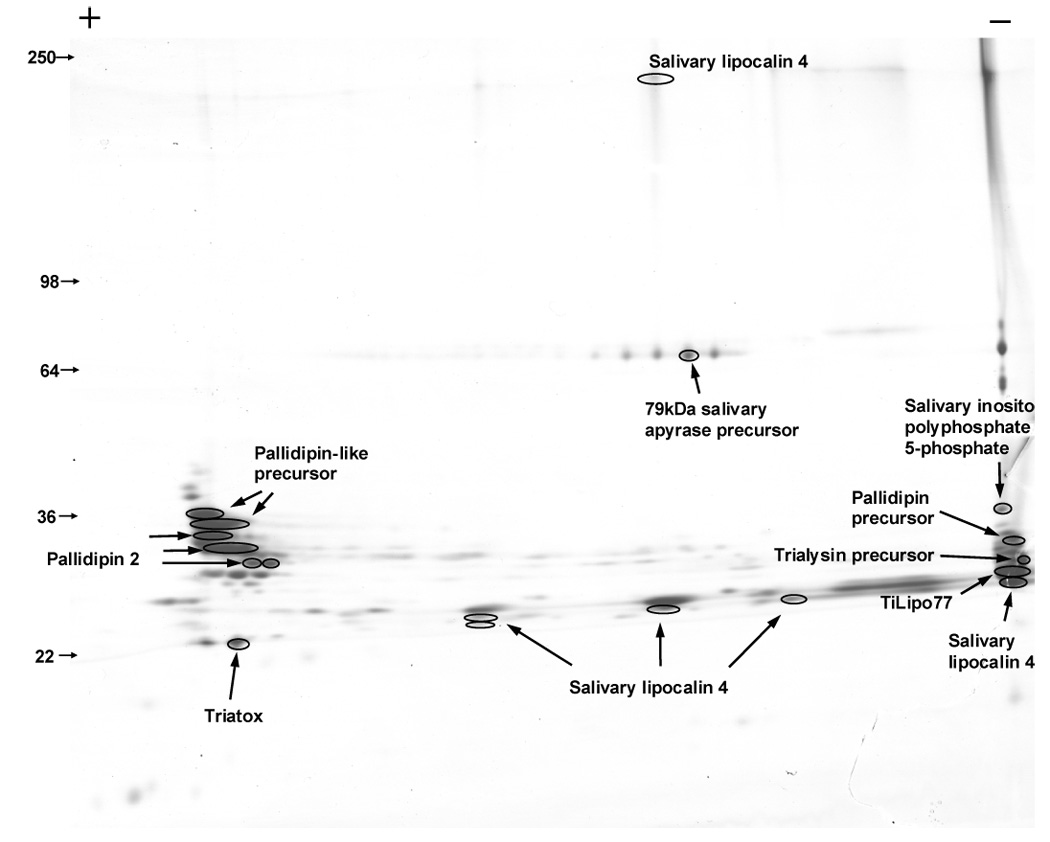
Two-dimensional (2D)-gel electrophoresis of Triatoma infestans salivary proteins. Numbers on the left indicate molecular weight marker positions in the gel. The + and − signs indicate the anode or cathode side of the isoelectrophocusing dimension, which ranged from pH 3–10. Gel bands that were identified to a protein (following tryptic digestion and mass spectrometry) are shown in the gel. In some cases, more than one band accounted for the same protein, possibly due to trailing or multiple isoforms. For experimental details, see Materials and methods.
3.5.1.5 Serpins
Serpins constitute a family of proteins, most of which function as serine proteinase inhibitors. They are important regulators of serine proteinases involved in inflammation, blood coagulation, fibrinolysis, and complement activation. Other roles for serpins could be protection against microbial proteinases, regulation of both endogenous proteinases and phenoloxidase activation (Kanost, 1999). Among arthropods, serpins have been described from lepidopteran insects, crayfish, ticks, and others (Jiang, et al., 1996; Liang et al., 1995; Prevot, et al, 2006). One of the contigs from the cDNA library matched for a serpin but the sequence was truncated (Table 3).
Table 3.
Classification of transcripts coding for putative secreted proteins in Triatoma infestans salivary glands.
| Types of transcripts | Clusters | Sequences | % S type sequence | |
|---|---|---|---|---|
| Lipocalins | 69 | 355 | 55.0 | |
| Trialysin | 6 | 74 | 11.5 | |
| Peptide with trialysin signal peptide | 7 | 64 | 9.9 | |
| Hemolysin-domain protein | 7 | 32 | 5.0 | |
| Defensin and immunity related peptides | 6 | 32 | 5.0 | |
| Enzymes | 7 | 20 | 3.1 | |
| Kazal domain-containing peptides | 3 | 17 | 2.6 | |
| Antigen 5 protein family | 2 | 13 | 2.0 | |
| Other peptides | 5 | 9 | 1.4 | |
| Odorant binding protein family | 2 | 9 | 1.4 | |
| Triatox | 1 | 8 | 1.2 | |
| Similar to Culicoides salivary protein | 1 | 6 | 0.9 | |
| Serpin | 1 | 4 | 0.6 | |
| Thrombospondin-like | 1 | 2 | 0.3 | |
| Total | 118 | 645 |
3.5.1.6 Kazal domain-containing peptides
Two contigs with transcripts encoding a polypeptide with a mature molecular mass of 8.9 kDa and containing a single Kazal domain are predicted. Although this domain is associated with serine protease inhibitors, it is also found in other extracellular proteins without this function and containing antimicrobial activity (Fogaça et al., 2005). Kazal-type serine protease inhibitors are single- or more commonly multi-domain proteins, can be classical or non-classical Kazal-type inhibitors, and share a conserved sequence motif and molecular conformation (one central α-helix and three small antiparallel β- sheets) (Stubbs et al., 1997). Several Kazal-type family members have been previously described to be present in vertebrate and invertebrate animals. The role of Kazal-type inhibitors is not always known. Some invertebrate Kazal-type inhibitors, such as rhodniin, bdellin B-3 and infestin 1–2 (Friedrich et al., 1993; Fink, et al., 1986; Sommerhoff et al., 1994; Campos, et al., 2002) are identified as non-classical Kazal-type inhibitors. Thrombin inhibitors of the Kazal-type family have been found in triatomines. Infestin is the longest Kazal-type proteinase inhibitor precursor described in triatomines. It seems to be similar to rhodniin and dipetalogastin precursors but has more Kazal-type domains (Lovato et al., 2006). Rhodniin, isolated from the blood-sucking insect R. prolixus, is such a Kazal-type inhibitor composed of two domains, and strongly inhibits thrombin, the key enzyme of the blood coagulation cascade (van de Locht et al., 1995).
Alignment with other sequences containing a Kazal domain shows the presence of five conserved cysteines (Fig. 4A). These sequences also showed similarity to the vasodilator named vasotab from the horse fly Hybomitra bimaculata. Vasotab is a member of the Kazal-type protease inhibitor family (Takác et al., 2006). The phylogram analysis indicates a strong bootstrap support for the triatomine clade (Fig. 4B). A transcript encoding this polypeptide was also found in the mosquitoes Ae. aegypti (Ribeiro et al., 2007) and Ae. albopictus (Arcà et al., 2007). And recently, the sialotranscriptome of the triatomine Triatoma brasiliensis demonstrated the presence of nine clusters coding for Kazal-type peptides (Santos, et al., 2007).
Figure 4.
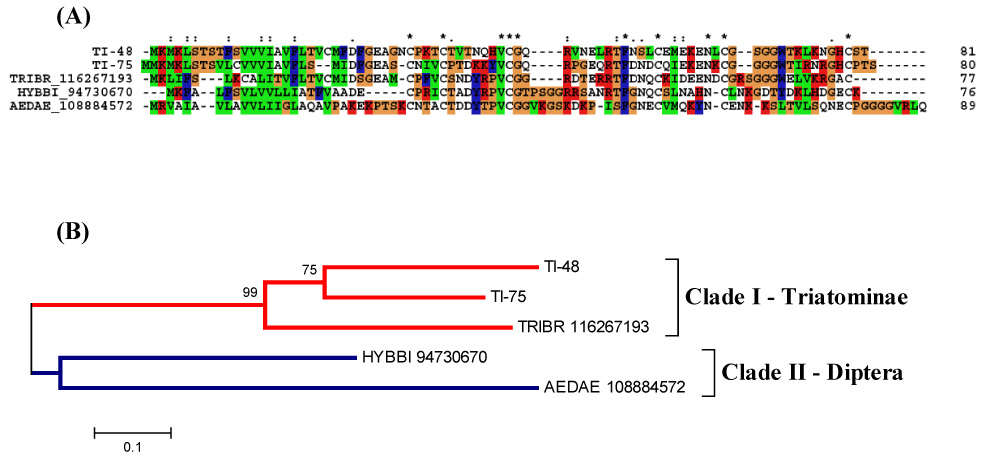
The salivary Kazal-domain containing peptides of T. infestans. (A) ClustalW alignment. Six conserved cysteines are shown in black with white background and marked with an asterisk (*). (B) Phylogenetic tree showing the sequence distance relationships between members of the family. The bar shows 10% divergence at the amino acid level. The numbers indicate the bootstrap value from 1000 trials.
3.5.1.7 – Antigen-5 protein family
This is a family of secreted proteins that belong to the CAP family (cysteine-rich secretory proteins; AG5 proteins of insects; pathogenesis-related protein 1 of plants) (Megraw et al., 1998). The CAP family is related to venom allergens in social wasps and ants (Hoffman, 1993; King and Spangfort, 2000) and to antifungal proteins in plants (Stintzi et al., 1993; Szyperski et al., 1998). Members of this protein family are found in the (SG)s of many blood-sucking insects (Francischetti et al., 2002; Li et al., 2001; Valenzuela et al., 2002b; Arcà et al., 2005; Calvo et al., 2007). The function of any AG5 protein in the saliva of any blood-sucking arthropod is still unknown. Other animals have shown some insight into their activity: snake venom proteins of the same family have been shown to contain smooth muscle-relaxing activity (Yamazaki et al., 2002; Yamazaki and Morita, 2004), and the salivary neurotoxin of the venomous lizard Heloderma horridum is also a member of this protein family (Nobile et al., 1996).
We here report a salivary member of the antigen-5 family found in T. infestans (SG)s. Alignment and phylogenetic analysis of insect members of this family indicates that T. infestans salivary antigen-5 protein clusters with other reported antigen-5 proteins from R. prolixus, vespids and other insects. The hymenoptera proteins, from vespids, have an additional domain after the signal peptide (Fig. 5), indicating that probably their evolution diverged at some point from the others. Both Rhodnius and Triatoma proteins have a basic tail. This content of a poly-Lys tail in salivary proteins may direct these proteins to the surface of activated platelets, where they could bind and/or release biologically active ligands or interact with other proteins on the platelet surface. They could also block the interaction of coagulation factors with the membrane surface or direct the inhibitor to negatively charged membranes critical for productive blood coagulation complex assembly (Broze Jr., 1995). Demonstration that proteins rich in positively charged residues effectively block the coagulation cascade comes from studies performed with a recombinant R. prolixus salivary lipocalin (nitrophorin-7, NP-7). NP-7 contains a cluster of positively charged residues in the N-terminus and specifically binds to anionic phospholipids, preventing thrombin formation by the prothrombinase complex (Andersen et al., 2004).
Figure 5.

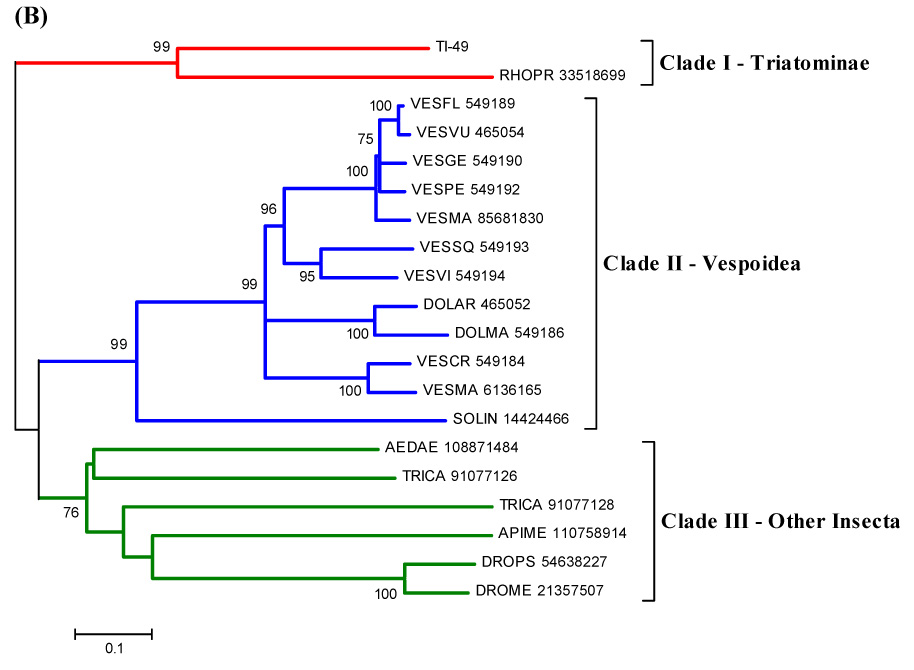
(A) ClustalW alignment of T. infestans salivary antigen-5 with members of the same family. (B) Phylogenetic tree of selected members of the antigen-5 family of proteins from Triatominae, Vespoidea and other Insecta, obtained by the NJ method. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 10% amino acid divergence in sequences. For details, see text.
3.5.1.8 Odorant-binding protein family
Many airborne molecules, such as hydrophobic odorants and pheromones, must be recognised by a specialised class of proteins that facilitate their delivery to the olfactory receptors (OR) (Forêt and Maleszka, 2006). In both insects and vertebrates this function is provided by odorant-binding proteins (OBPs) (Pelosi et al., 1996; Krieger and Breer, 1999; Deyu and Leal, 2002). They are generally thought to solubilise hydrophobic odorants and carry them to the respective receptors (Vogt et al., 1991; Pelosi et al., 1994; Prestwich et al., 1995). Besides contributing to the recognition of odorants in insects, they may also function as carriers in other developmental and physiological processes.
Insect OBPs are small, water soluble molecules expressed in both olfactory and gustatory sensilla, as well as in other specialised tissues (Pelosi et al., 2005). OBPs have also been found in non-olfactory tissue, suggesting that their roles may be related to general carrier capabilities with broad specificity for lipophilic compounds (Forêt and Maleszka, 2006). The heme-binding protein of R. prolixus (Paiva-Silva et al., 2002) is one of these OBPs implicated in non-olfactory functions. In fact, there is increasing evidence that OBPs do play an active role in odorant recognition rather than merely serving as passive odorant carriers. One line of evidence is the large number of OBPs present within a variety of insect species. Several proteins of the odorant-binding protein (OBP) family have been described in D. melanogaster, Apis mellifera, and the hemiptera R. prolixus, where a putative OBP with a signal peptide indicative of secretion was identified (Hekmat-Scafe et al., 2000; Forêt and Maleszka, 2006; Ribeiro et al., 2004a). We found transcripts coding for members of the OBP family in T. infestans. They share similarities with OBPs from the mosquitoes An. gambiae and Ae. aegypti. The OBP-like sequences have four conserved cysteines and good bootstrap support for the triatomine clade (Fig. 6). Sequences in Diptera clade do not have a strong bootstrap support, except for some members that probably originate from genetic duplication and/or function conservation. It has been proposed that a moderate number of OBPs could act in a combinatorial manner with a moderate number of OR to greatly increase the discriminating power of an insect’s olfactory system (Hekmat-Scafe, et al., 2002).
Figure 6.
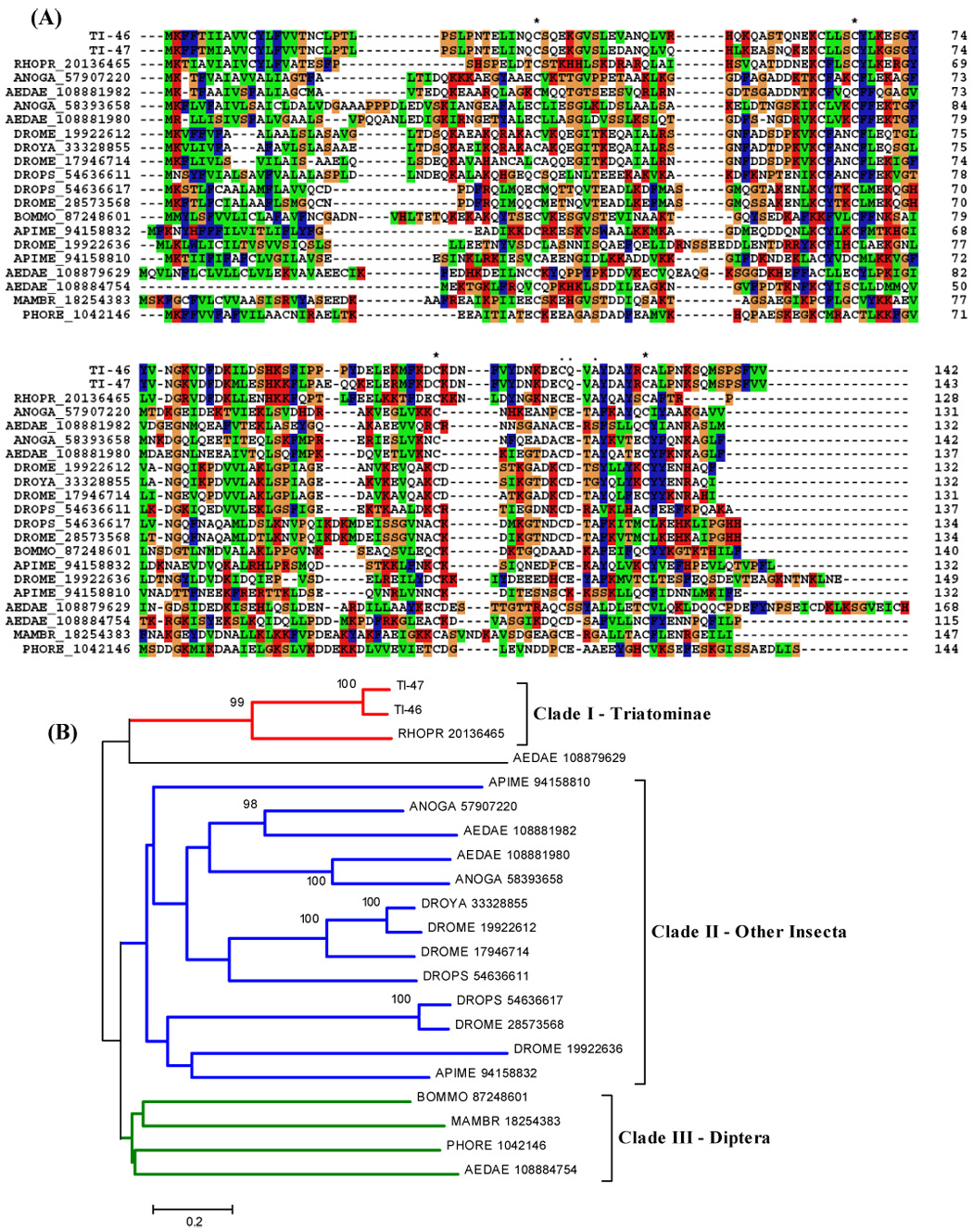
(A) ClustalW alignment of the OBP family of salivary peptides. Four conserved cysteines are shown in black with white background and marked with an asterisk (*). (B) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 20% amino acid divergence in sequences.
It is possible to speculate whether these salivary OBP’s could be acting as partners in gustatory perception in Triatoma, as they are related to gustatory perception in Drosophila (Galindo and Smith, 2001). If this is the case, saliva would be important not only for its anti-hemostatic function, but also in the chemoreception of phagostimulants (Friend and Smith, 1977). Against this hypothesis is the previous observation that Rhodnius prolixus surgically deprived of salivary glands were able to feed normally on an artificial feeder (Ribeiro and Garcia, 1981).
3.5.1.9 Similar to Culicoides salivary protein
This EST sequence found in T. infestans shows similarity with Culicoides and Fungi sequences (Fig. 7). Culicoides are competent vectors of a wide range of economically important pathogens that affect both domestic and wild animals (Mellor et al., 2000). A broad range of genes encoding salivary proteins were characterised from a (SG) cDNA library of C. sonorensis and revealed many proteins involved in antihemostasis, immunomodulation, and digestion (Campbell et al., 2005). In addition to acting as a vector, Culicoides are the primary cause of an extremely pruritic allergic dermatitis, known colloquially as “summer eczema”, or “insect bite hypersensitivity” in horses (Anderson, et al., 1993; Kurotaki, et al., 1994). The saliva of kissing bugs (Triatominae) contains several allergens responsible for severe allergic reactions such as urticaria, dyspnea, and anaphylaxis in humans (Moffitt et al., 2003). This transcript, similar to one found in Culicoides, could be related to hypersensitivity or allergic responses by the human host.
Figure 7.
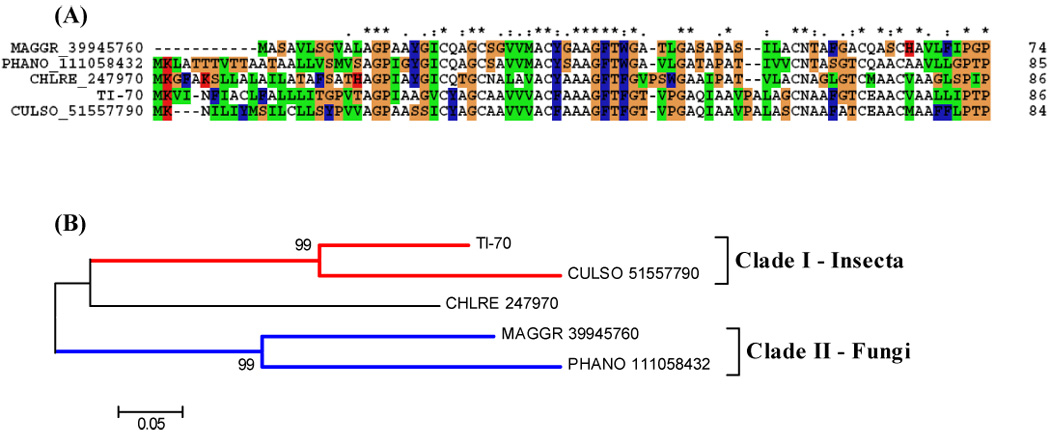
(A) ClustalW alignment of a predicted sequence from a contig that showed similarity with a sequence from Culicoides. (B) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 5% amino acid divergence in sequences.
3.5.1.10 Defensin and immunity-related peptides
Antimicrobial peptides (AMPs) are widely distributed throughout the animal and plant kingdoms. Despite sharing some common features, such as a small size (often below 10 kDa) and a cationic character, most AMPs differ in their aa sequence and mode of action (Fogaca et al., 2004). Within this AMP family, defensins constitute one of the major families that have been characterised (Bulet et al., 2004). Defensins are cationic peptides with molecular weights of about 4 kDa, three disulphide bridges, formed by six cysteines residues and three characteristic domains, an amino terminal flexible loop, followed by an α-helix and a carboxy-terminal anti-parallel β-sheet (Bonmantin et al., 1992; Bulet et al., 1999). This peptide can be found in different organisms, such as plants, fungi, mollusks, scorpions, insects, and birds, and also in different cells of various mammals. In humans, insects, and plants, defensins contribute significantly to the host defense against invasion by microorganisms (Raj et al., 2002). In addition to the diversity of structure and mode of action, the site and the regulation of the synthesis of AMPs also differs among arthropod groups. In insects, AMPs are mainly synthesised in the fat body and their gene transcription is strongly induced after an injury and/or infection (Bulet et al., 2002). Defensins have been isolated and characterised as part of the innate immune response from the hemolymph of all insect species so far investigated, like Odonata, Diptera, Coleoptera, Lepidoptera, Acari and Hemiptera (Bartholomay et al., 2004; Bulet et al., 1992; Ceraul et al., 2003; Ishibashi et al., 1999; Johns et al., 2001; Lamberty et al., 1999; Lopez et al., 2003).
Tick defensins have also been found to exist in (SG)s, an important organ for blood feeding and pathogen transmission (Valenzuela et al., 2002). A defensin-like molecule from Ixodes ricinus was recently described and found to be induced following microbial challenge (Rudenko et al., 2005). In triatomines, defensins have been investigated in R. prolixus, being isolated, purified and sequenced (Lopez et al., 2003).
From the alignment of the defensin sequence found in T. infestans (SG)s with other defensins, we notice the presence of the six conserved cysteines, characteristic in this family and important for the disulphide bridges conformation (Fig. 8A). The phylogram reveals a triatomine clade sharing sequences from T. brasiliensis and R. prolixus, and showing a bootstrap support (Fig. 8B).
Figure 8.
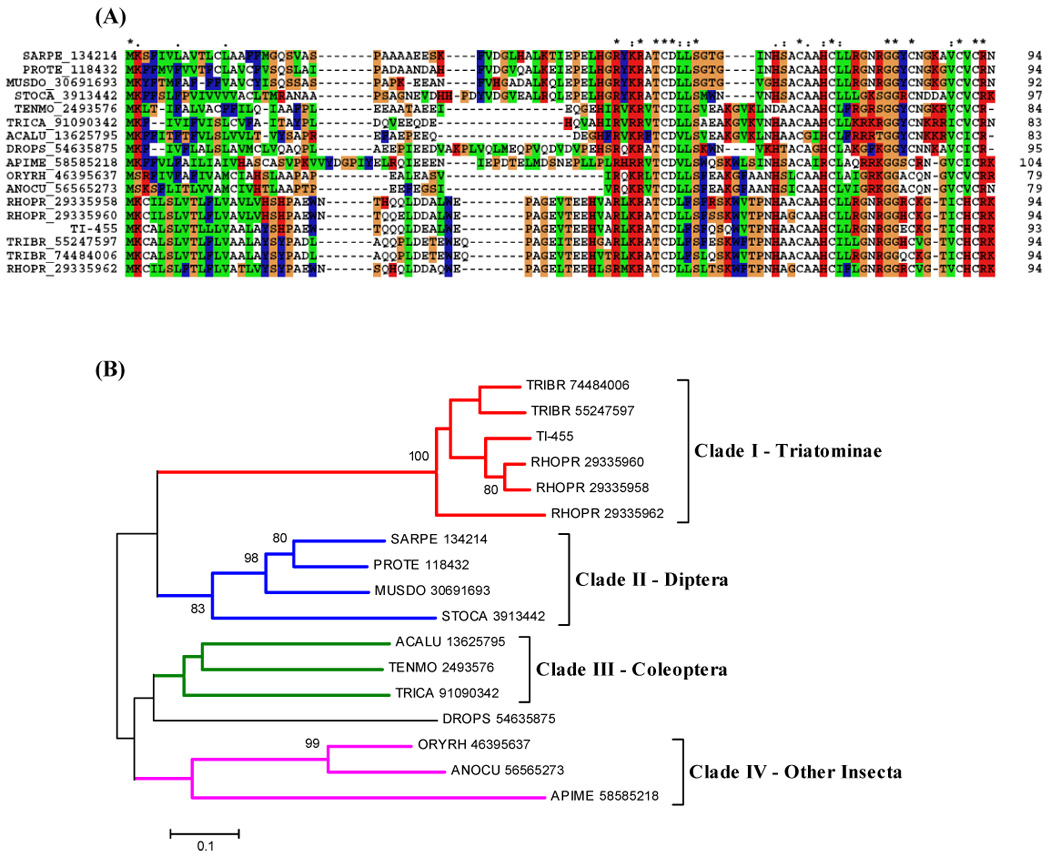
(A) ClustalW alignment of the defensin family of salivary peptides. Six conserved cysteines are shown in black with white background and marked with an asterisk (*). (B) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 10% amino acid divergence in sequences.
3.5.1.11 Similarity to bacterially induced peptide Hdd1 (from Hyphantria cunea)
In a previous work, 11 inducible genes were isolated after bacterial challenge in H. cunea, one of the most serious insect pests in Korea. Hdd1 (Hyphantria differentially displayed clones) was one of this immune-related cDNA but it didn’t produce any significant homology with any known protein (Shin et al., 1998). One contig from the T. infestans cDNA library matched this peptide but the function remains unknown.
3.5.2 Coding for proteins found only in triatomines
3.5.2.1 Trialysin
We identified two contigs that matched the protein trialysin, a pore-forming molecule present in the (SG)s of T. infestans (Amino et al., 2002; Martins et al., 2006) (Fig. 9). It is a 22-kDa protein synthesised as a precursor and processed by limited proteolysis. It exerts its potent cytolytic activity on a large variety of cell types, from bacteria to mammalian cells, and it has been hypothesised that it favors the maintenance of the salivary fluid free of microorganisms and parasites. Trialysin possesses a basic amphipathic lytic motif in the N-terminal region containing 27 aa residues, similar to antimicrobial lytic peptides, and a protein portion that increases the lytic specificity toward eukaryotic cells. Studies in which synthetic peptides containing portions of the N-terminus of trialysin have been tested for their lytic properties against the infective stage of T. cruzi, Escherichia coli and human erythrocytes revealed differences in specificity and activity for the tested targets. The cell membrane is the main target of antimicrobial peptides, thus the diverse lipid composition and distribution or the presence of other components in the cell membrane of the various organisms play an important role in the modulation of peptide/membrane interaction, leading to the differentiation of their action (Amino et al., 2002; Martins et al., 2006).
Figure 9.
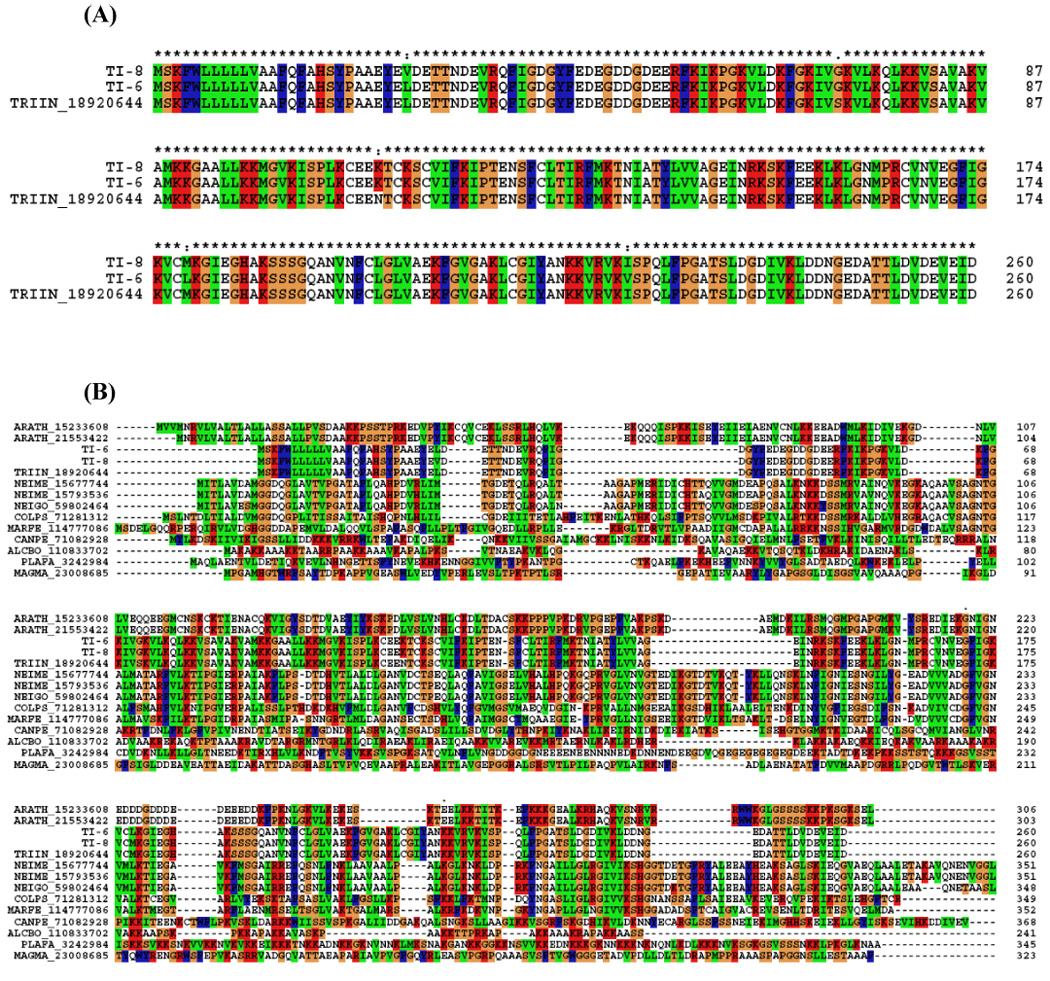
(A) ClustalW alignment of the unique family of trialysin salivary peptides. The sequences shown are from the cDNA library (TI-6, TI-8) and trialysin from NCBI database (|gi18920644|). (B) ClustalW alignment of the trialysin sequences with other proteins that showed similarity to trialysin. (C) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 20% amino acid divergence in sequences.
Further analysis of the near relatives of trialysin found in the nonredundant database indicates related bacterial genes, a proteobacteria clade being strongly associated with the T. infestans sequences (Fig 9b,c).
3.5.2.2 Peptides with trialysin signal peptide
This cDNA library also revealed a group of short proteins sharing the signal peptide from trialysin protein. They do not share similarities with other proteins and their function is unknown.
3.5.2.3. Hemolysin-domain protein
Hemolysins are proteins considered to permeate target cells, forming pores in cytoplasmic membranes of erythrocytes, leukocytes, and other cells, causing the modification of cellular functions and/or lysis of host cells. Hemolysins are more often described in microorganisms, such as Escherichia coli, Staphylococcus aureus, Clostridium septicum, and others (Gouaux, 1998; Melton et al., 2004; Wassenaar, T.M., 2005). The family of hemolysins consists of secreted, water-soluble proteins that form transmembrane, polymeric channels. They are considered a virulence factor from these microorganisms and also a member of the RTX (repeats in toxin) protein family (Welch, 2001). The presence of amphipathic and hydrophilic domains confers to the protein an overall amphiphilic character, which explains its tendency both to aggregate and to interact with membranes (Hyland et al., 2001; Schindel et al., 2001; Soloaga et al., 1999).
We found contigs with proteins containing a hemolysin domain (Fig. 10). These proteins could act as cytolytic proteins, causing erythrocyte lysis in saliva and helping the early steps of the digestion process. Also, it could be related to defense, participating in lysis of microorganisms in insect saliva.
Figure 10.
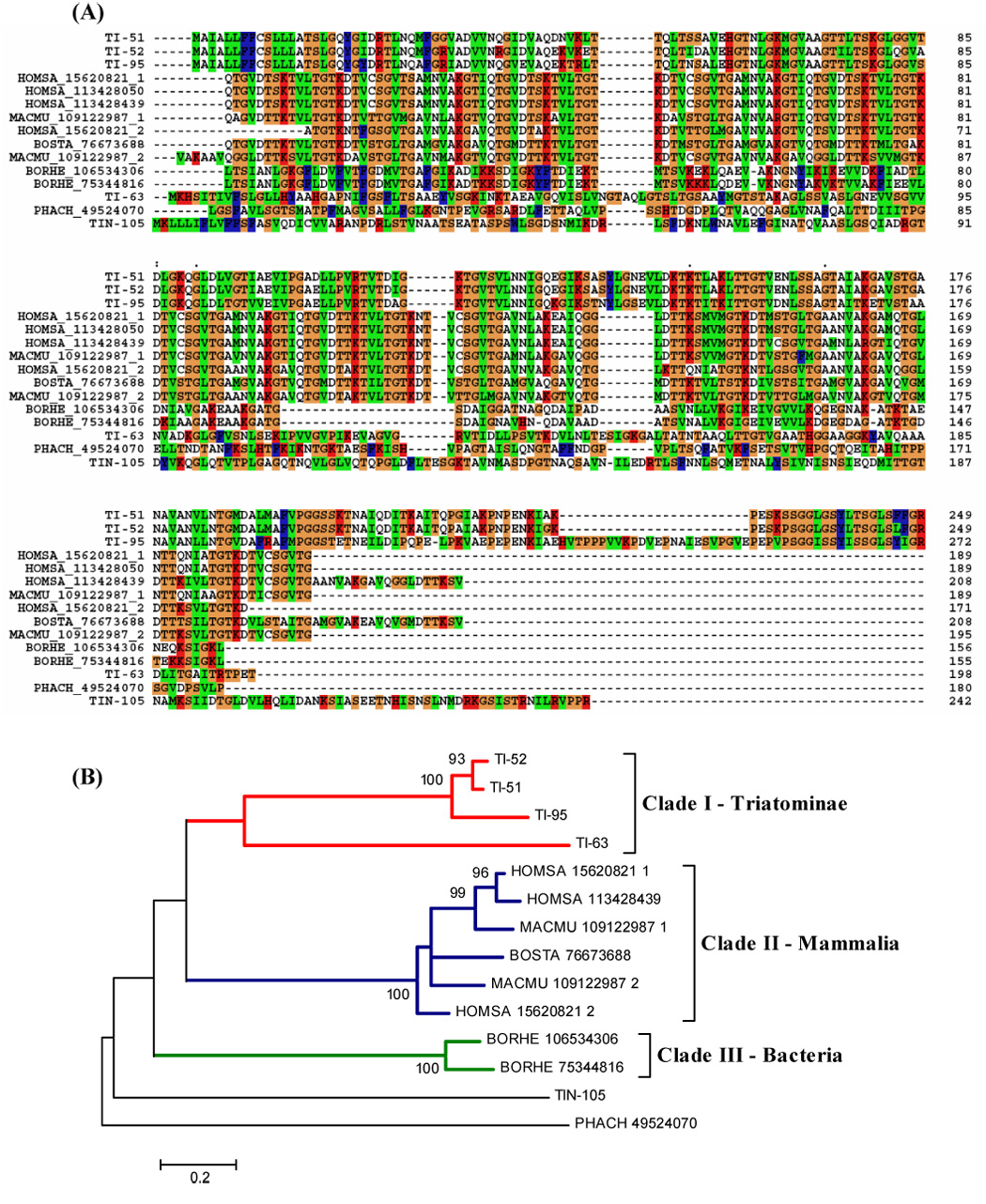
(A) ClustalW alignment of the mature forms of T. infestans hemolysin-domain containing proteins with other proteins with hemolysin-domain. The signal peptide region is not shown. (B) Phylogram. Numbers on branches indicate bootstrap value for 1000 trials. Bar indicates 20% distance in amino acid sequence.
3.5.2.4 Triatox
One of the contigs matched a toxin previously described in T. infestans saliva as triatox (|gi71725070|) (Fig. 11A). We also found this protein in the 2D gel (Fig. 12). By secondary structure prediction, this protein shows to be an amphipathic α-helix (Fig. 11B). α-helical peptides are linear molecules that exist in aqueous media and become amphipathic helices upon interaction with hydrophobic membranes, like cecropin (Christensen et al., 1988), magainins (Zasloff, 1987), and melittins (Andreu et al., 1992). The amphipathic character of antimicrobial peptides makes them surface-active products, as their biological activity occurs at lipid membrane interfaces (Maget-Dana, 1999). Their amphipathy allows them to be both soluble in an aqueous medium, such as the extracellular medium, and to diffuse towards polar/apolar interfaces such as the extracellular medium/cell membrane interface. Linear peptides that can assume an active, amphipathic, α-helical structure are among the most abundant in nature. They have evolved to act against several microbial targets and appear to represent an important role in innate defense (Giangaspero et al., 2001). The structure of triatox suggests it can function as an antimicrobial peptide and be related to the innate defense of the insect in the (SG)s.
Figure 11.
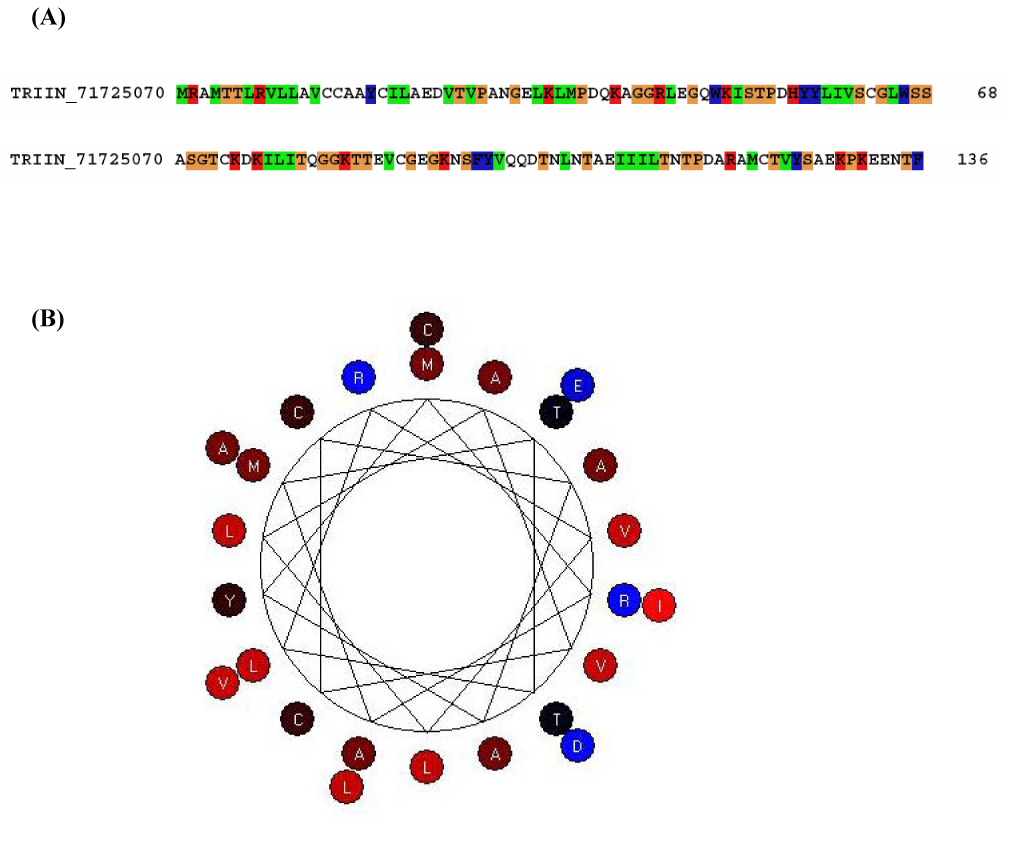
(A) triatox sequence gi71725070. (B) Prediction of triatox (TI-57) secondary structure using BioEdit program.
3.5.2.5 Thrombospondin-like
This protein shows weak similarity with larger proteins of Plasmodium annotated as “thrombospondin-related adhesive protein.” Thrombospondin domains usually bind to sulphated glycoconjugates found on cell surfaces, allowing for host-cell specificity of binding (Dubremetz, et al., 1998). The function is unknown in other genera.
3.6. 2D-Gel electrophoresis/Mass spectrometry proteomic investigation
The (SG)s of T. infestans were shown to contain proteins such as apyrase and the pore-forming protein trialysin (Amino et al., 2002; Faudry et al., 2004). To obtain further information on the salivary proteins of T. infestans, electrophoresis of saliva was performed by two-dimensional (2D) SDS-PAGE followed by mass spectrometry. Figure 12 shows the pattern of separation of T. infestans salivary proteins by 2D-gel. We were able to identify several salivary lipocalins, and the proteins named “triatox” (|gi71725070|), salivary apyrase precursor (|gi34481604|), salivary inositol polyphosphate 5-phosphate (approximately 37-kDa apparent salivary precursor |gi34481604 “salivary inositol polyphosphate 5-phophate (approximately 37-kDa apparent molecular mass) similar to pallidipin precursor, putative trialysin precursor, similar to lipocalin-like TiLipo77, and similar to pallidipin-2. Supplemental tables S1 and S2 have columns indicating the peptide sequences found in the proteomic experiment.
3.7 Salivary peptide identification by RP-HPLC/Mass spectrometry
Because the gel used for the proteomic experiment above does not well identify polypeptides below 10 kDa, we obtained a 10-kDa filtrate from 20 µL of T. infestans saliva, submitted it to RP-HPLC and performed tryptic digestion of the fractions to obtain peptide sequences by MS/MS. This allowed identification of four members of the short trialysin-like family, which have predicted mature molecular weights of 6.3 kDa. Several fragments matching the hemolysin-like proteins were also identified, indicating this protein family, whose members have more than 20 kDa, may be proteolytically processed in saliva, possibly by triapsin, the molecularly uncharacterised salivary serine protease of T. infestans (Amino et al., 2001). Supplemental tables S1 and S2 have columns indicating the peptide sequences found in this proteomic experiment.
4. Concluding remarks
In an attempt to improve our understanding of the variety of proteins and transcripts expressed in T. infestans (SG)s, we have performed a cDNA library and a 2D-gel using, respectively, mRNA and proteins from this same tissue. We described the set of cDNA present in the (SG)s of Triatoma infestans. Expression and bioassay of the novel proteins will ultimately characterise the salivary pharmacologic complexity from T. infestans evolution to blood feeding.
We believe this cDNA library should help our continuing effort to understand the evolution of blood sucking in vector arthropods and the discovery of novel pharmacologically active compounds.
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases. TCFA was a recipient of a CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Ministry of Education, Brazil) fellowship. We are grateful to the NIAID Research Technologies Branch, directed by Dr. Robert Hohman, for support in the proteomic studies, to Nancy Shulman for editorial assistance to Chuong Huynh from NCBI for help with posting the data, and Drs. Robert Gwadz and Katherine Zoon for encouragement and support.
Abbreviations
- aa
amino acid
- AMP
antimicrobial peptides
- EST
expressed sequence tags
- OBP
odorant-binding protein
- H
putative housekeeping transcripts
- S
putative secreted transcripts
- U
unknown function transcripts
- SG
salivary glands
- Ti
Triatoma infestans
- 2D
two dimensional
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amino R, Martins RM, Procopio J, Hirata IY, Juliano MA, Schenkman S. Trialysin, a Novel Pore-forming Protein from Saliva of Hematophagous Insects Activated by Limited Proteolysis. J. Biol. Chem. 2002;277:6207–6213. doi: 10.1074/jbc.M109874200. [DOI] [PubMed] [Google Scholar]
- Amino R, Tanaka AS, Schenkman S. Triapsin, an unusual activatable serine protease from the saliva of the hematophagous vector of Chagas’ disease Triatoma infestans (Hemiptera: Reduviidae) Insect Biochem. Mol. Biol. 2001;31:465–472. doi: 10.1016/s0965-1748(00)00151-x. [DOI] [PubMed] [Google Scholar]
- Andersen JF, Gudderra NP, Francischetti IMB, Ribeiro JMC. The Role of Salivary Lipocalins in Blood Feeding by Rhodnius prolixus. Arch. Insect Biochem. Physiol. 2005;58:97–105. doi: 10.1002/arch.20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Guderra NP, Francischetti IM, Valenzuela JG, Ribeiro JM. Recognition of anionic phospholipid membranes by an antihemostatic protein from a blood-feeding insect. Biochemistry. 2004;43:6987–6994. doi: 10.1021/bi049655t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Ribeiro JMC. A Secreted Salivary Inositol Polyphosphate 5–Phosphatase from a Blood-Feeding Insect: Allosteric Activation by Soluble Phosphoinositides and Phosphatidylserine. Biochemistry. 2006;45:5450–5457. doi: 10.1021/bi052444j. [DOI] [PubMed] [Google Scholar]
- Anderson GS, Belton P, Kleider N. Hypersensitivity of horses in British Columbia to extracts of native and exotic species of Culicoides (Diptera: Ceratopogonidae) J. Med. Entomol. 1993;30:657–663. doi: 10.1093/jmedent/30.4.657. [DOI] [PubMed] [Google Scholar]
- Andreu D, Ubach J, Boman A, Wahlin B, Wade D, Merrifield RB, Boman HG. Shortened cecropin A-mellitin hybrids. Significant size reduction retains potent antibiotic activity. FEBS Lett. 1992;296:190–194. doi: 10.1016/0014-5793(92)80377-s. [DOI] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Francischetti IMB, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JMC. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochemisty and Molecular Biology. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JMC. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J. Exp. Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomay LC, Fuchs JF, Cheng L-L, Beck ET, Vizioli J, Lowenberger C, Christensen BM. Reassessing the role of defensin in the innate immune response of the mosquito, Aedes aegypti. Insect Mol. Biol. 2004;13:125–132. doi: 10.1111/j.0962-1075.2004.00467.x. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonmantin JM, Bonnat JL, Gallet X, Vovelle F, Ptak M, Reichhart J-M, Hoffmann JA, Keppi E, Legrain M, Achstetter T. Two-dimensional 1H NMR study of recombinant insect defensin A in water: resonance assignments, secondary structure and global folding. J. Biomol. NMR. 1992;2:235–256. doi: 10.1007/BF01875319. [DOI] [PubMed] [Google Scholar]
- Broze GJ., Jr. Tissue factor pathway inhibitor and the current concept of blood coagulation. Blood Coagul. Fibrinolysis. 1995;6:S7–S13. doi: 10.1097/00001721-199506001-00002. [DOI] [PubMed] [Google Scholar]
- Bulet P, Charlet M, Hetru C. Antimicrobial peptides in insect immunity. In: Ezekowitz AB, Hoffmann JA, editors. Infectious disease: Innate immunity. Totowa, NJ: Humana Press; 2002. pp. 89–107. [Google Scholar]
- Bulet P, Cociancich S, Reuland M, Sauber F, Bischoff R, Hegy G, Van Dorsselaer A, Hetru C, Hoffmann JA. A novel insect defesin mediates the inducible antibacterial activity in larvae of the dragonfly Aeschna cyanea (Paleoptera, Odonata) Eur. J. Biochem. 1992;209:977–984. doi: 10.1111/j.1432-1033.1992.tb17371.x. [DOI] [PubMed] [Google Scholar]
- Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects; structure and functions. Dev. Comp. Immunol. 1999;23:329–344. doi: 10.1016/s0145-305x(99)00015-4. [DOI] [PubMed] [Google Scholar]
- Bulet P, Stocklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- Calvo E, Dao A, Pham VM, Ribeiro JMC. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem. Mol. Biol. 2007;37:164–175. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Mol. Biol. 2005;14:121–136. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- Campos IT, Amino R, Sampaio CA, Auerswald EA, Friedrich T, Lemaire HG, Schenkman S, Tanaka AS. Infestin, a thrombin inhibitor presents in Triatoma infestans midgut, a Chagas’ disease vector: gene cloning, expression and characterization of the inhibitor. Insect Biochem. Mol. Biol. 2002;32:991–997. doi: 10.1016/s0965-1748(02)00035-8. [DOI] [PubMed] [Google Scholar]
- Ceraul SM, Sonenshine DE, Ratzlaff RE, Hynes WL. An arthropod defensin expressed by the hemocytes of the American dog tick, Dermacentor variabilis (Acari:Ixodidae) Insect Biochem. Mol. Biol. 2003;33:1099–1103. doi: 10.1016/s0965-1748(03)00122-x. [DOI] [PubMed] [Google Scholar]
- Christensen B, Fink J, Merrifield RB, Mauzerall D. Channel-forming properties of cecropins and related model compounds incorporated into planar lipid membranes. Proc. Natl. Acad. Sci. USA. 1988;85:5072–5076. doi: 10.1073/pnas.85.14.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czibener C, La Torre JL, Muscio O, Ugalde RA, Scodeller EA. Nucleotide sequence analysis of Triatoma virus shows that it is a member of a novel group of insect RNA viruses. J. General Virol. 2000;81:1149–1154. doi: 10.1099/0022-1317-81-4-1149. [DOI] [PubMed] [Google Scholar]
- Deyu Z, Leal W. Conformational isomers of insect odorant-binding proteins. Arch. Biochem. Biophys. 2002;397:99–105. doi: 10.1006/abbi.2001.2660. [DOI] [PubMed] [Google Scholar]
- Dias JC. Control of Chagas disease in Brazil. Parasitology Today. 1987;3(11):336–441. doi: 10.1016/0169-4758(87)90117-7. [DOI] [PubMed] [Google Scholar]
- Dubremetz JF, Garcia-Réguet N, Conseil V, Fourmaux MN. Apical organelles and host-cell invasion by Apicomplexa. Int. J. Parasitol. 1998;28:1007–1013. doi: 10.1016/s0020-7519(98)00076-9. [DOI] [PubMed] [Google Scholar]
- Erneux C, Govaerts C, Communi D, Pesesse X. The diversity and possible functions of the inositol polyphosphate 5-phosphatases. Biochim. Biophys. Acta. 1998;1436:185–199. doi: 10.1016/s0005-2760(98)00132-5. [DOI] [PubMed] [Google Scholar]
- Faudry E, Lozzi SP, Santana JM, D’Souza-Ault M, Kieffer S, Felix CR, Ricart CA, Sousa MV, Vernet T, Teixeira AR. Triatoma infestans apyrases belong to the 5’-nucleotidase family. J. Biol. Chem. 2004;279(19):19607–19613. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- Fink E, Rehm H, Gippner C, Bode W, Eulitz M, Machleidt W, Fritz H. The primary structure of bdellin B-3 from the leech Hirudo medicinalis. Bdellin B-3 is a compact proteinase inhibitor of a “non-classical” Kazal type. It is present in the leech in a high molecular mass form. Biol. Chem. Hoppe Seyler. 1986;367:1235–1242. doi: 10.1515/bchm3.1986.367.2.1235. [DOI] [PubMed] [Google Scholar]
- Flower DR. Multiple molecular recognition properties of the lipocalin protein family. J. Mol. Recogn. 1995;8:185–195. doi: 10.1002/jmr.300080304. [DOI] [PubMed] [Google Scholar]
- Flower DR, North AC, Attwood TK. Structure and sequence relationships in the lipocalins and related proteins. Protein Sci. 1993;2:753–761. doi: 10.1002/pro.5560020507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim. Biophys. Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Fogaça AC, Lorenzini DM, Kaku LM, Esteves E, Bulet P, Daffre S. Cysteine-rich antimicrobial peptides of the cattle tick Boophilus microplus: isolation, structural characterization and tissue expression profile. Dev. Comp. Immunol. 2004;28:191–200. doi: 10.1016/j.dci.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Forêt S, Maleszka R. Function and evolution of a gene family encoding odorant binding-like protein in a social insect, the honey bee (Apis mellifera) Genome Res. 2006;16:1404–1413. doi: 10.1101/gr.5075706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Ribeiro JM, Champagne D, Andersen J. Purification, cloning, expression, and mechanism of action of a novel platelet aggregation inhibitor from the salivary gland of the blood-sucking bug, Rhodnius prolixus. J. Biol. Chem. 2000;275(17):12639–12650. doi: 10.1074/jbc.275.17.12639. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J. Exp. Biol. 2002;205:2429–2451. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Lopes AH, Dias FA, Pham VM, Ribeiro JM. An insight into the sialotranscriptome of the seed-feeding bug, Oncopeltus fasciatus. Insect Biochem Mol Biol. 2007;37:903–910. doi: 10.1016/j.ibmb.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T, Kroger B, Bialojan S, Lemaire HG, Hoffken HW, Reuschenbach P, Otte M, Dodt J. A Kazal-type inhibitor with thrombin specificity from Rhodnius prolixus. J. Biol. Chem. 1993;268:16216–16222. [PubMed] [Google Scholar]
- Friend WG, Smith JJB. Factors affecting feeding by bloodsucking insects. Ann. Rev. Entomol. 1977;22:309–331. doi: 10.1146/annurev.en.22.010177.001521. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Noeske-Jungblut C, Donner P, Schleuning W-D, Huber R, Bode W. Structure of thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc. Natl. Acad. Sci. USA. 1997;94:11845–11850. doi: 10.1073/pnas.94.22.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangaspero A, Sandri L, Tossi A. Amphipathic α helical antimicrobial peptides. A systematic study of the effects of structural and physical properties on biological activity. Eur. J. Biochem. 2001;268:5589–5600. doi: 10.1046/j.1432-1033.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- Gouaux E. α-Hemolysin from Staphylococcus aureus: An Archetype of β-Barrel, Channel-Forming Toxins. J. Structural Biol. 1998;121:110–122. doi: 10.1006/jsbi.1998.3959. [DOI] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconjugate J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Dorit RL, Carlson JR. Molecular Evolution of Odorant-Binding Protein Genes OS-E and OS-F in Drosophila. Genetics. 2000;155:117–127. doi: 10.1093/genetics/155.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-Wide Analysis of the Odorant-Binding Protein Gene Family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DR. Allergens of Hymenoptera venom. XXV: The amino acid sequences of antigen 5 molecule sand the structural basis of antigenic cross-reactivity. J. Allergy Clin. Immunol. 1993;92:707–716. doi: 10.1016/0091-6749(93)90014-7. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland C, Vuillard L, Hughes C, Koronakis V. Membrane interaction of Escherichia coli hemolysin: flotation and insertion-dependent labeling by phospholipids vesicles. J. Bacteriol. 2001;183:5364–5370. doi: 10.1128/JB.183.18.5364-5370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi J, Saido-Sakanaka H, Yang J, Sagisaka A, Yamakawa M. Purification, cDNA cloning and modification of a defensin from the coconut rhinoceros beetle, Oryctes rhinoceros. Eur. J. Biochem. 1999;266:616–623. doi: 10.1046/j.1432-1327.1999.00906.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Huang Y, Mulnix AB, Kadel J, Cole K, Kanost MR. Organization of serpin gene-1 from Manduca sexta: evolution of a family of alternate exons encoding the reactive site loop. J. Biol. Chem. 1996;271:28017–28023. doi: 10.1074/jbc.271.45.28017. [DOI] [PubMed] [Google Scholar]
- Johns R, Sonenshine DE, Hynes WL. Identification of a defensin from the hemolymph of the American dog tick, Dermacentor variabilis. Insect Biochem. Mol. Biol. 2001;31:857–865. doi: 10.1016/s0965-1748(01)00031-5. [DOI] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Develop. Comp. Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- King TP, Spangfort MD. Structure and biology of stinging insect venom allergens. Int. Arch. Allergy Immunol. 2000;123(2):99–106. doi: 10.1159/000024440. [DOI] [PubMed] [Google Scholar]
- Krieger J, Breer H. Olfactory reception in invertebrates. Science. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kurotaki T, Narayama K, Oyamada T, Yoshikawa H, Yoshikawa T. Immunopathological study on equine insect hypersensitivity (“kasen”) in Japan. J. Comp. Pathol. 1994;110:145–152. doi: 10.1016/s0021-9975(08)80186-7. [DOI] [PubMed] [Google Scholar]
- Lamberty M, Ades S, Uttenweiler-Joseph S, Brookharts G, Bushey D, Hoffmann JA, Bulet P. Insect immunity: isolation from the lepidopteran Heliothis virescens of a novel insect defensin with potent antifungal activity. J. Biol. Chem. 1999;274:9320–9326. doi: 10.1074/jbc.274.14.9320. [DOI] [PubMed] [Google Scholar]
- Lavoipierre MM. Feeding mechanism of blood-sucking arthropods. Nature (London) 1965;208:302–303. doi: 10.1038/208302a0. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Kwon J, Aksoy S. Characterization of genes expressed in the salivary glands of the tsetse fly, Glossina morsitans morsitans. Insect Mol. Biol. 2001;10(1):69–76. doi: 10.1046/j.1365-2583.2001.00240.x. [DOI] [PubMed] [Google Scholar]
- Liang Z, Söderhäll K. Isolation of cDNA encoding a novel serpin of crayfish hemocytes. Comp. Biochem. Physiol. 1995;112B:385–391. doi: 10.1016/0305-0491(95)00105-0. [DOI] [PubMed] [Google Scholar]
- Lopez L, Morales G, Ursic R, Wolff M, Lowenberger C. Isolation and characterization of a novel insect defensin from Rhodnius prolixus, a vector of Chagas disease. Insect Biochem. Mol. Biol. 2003;33:439–447. doi: 10.1016/s0965-1748(03)00008-0. [DOI] [PubMed] [Google Scholar]
- Lovato DV, Campos ITN, Amino R, Tanaka AS. The full-length cDNA of anticoagulant protein infestin revealed a novel releasable Kazal domain, a neutrophil elastase inhibitor lacking anticoagulant activity. Biochimie. 2006;88:673–681. doi: 10.1016/j.biochi.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Maget-Dana R. The monolayer technique: a potent tool for studying the interfacial properties of antimicrobial and membrane-lytic peptides and their interactions with lipid membranes. Biochim. Biophys. Acta. 1999;1462:109–140. doi: 10.1016/s0005-2736(99)00203-5. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marejus PW, Kisseleva MV, Norris FA. The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 1999;275:20110–20116. doi: 10.1074/jbc.274.16.10669. [DOI] [PubMed] [Google Scholar]
- Martins RM, Sforça ML, Amino R, Juliano MA, Oyama S, Jr., Juliano L, Pertinhez TA, Spisni A, Schenkman S. Lytic Activity and Structural Differences of Amphipathic Peptides Derived from Trialysin. Biochemistry. 2006;45:1765–1774. doi: 10.1021/bi0514515. [DOI] [PubMed] [Google Scholar]
- Megraw T, Kaufman TC, Kovalick GE. Sequence and expression of Drosophila Antigen 5-related 2, a new member of the CAP gene family. Gene. 1998;222(2):297–304. doi: 10.1016/s0378-1119(98)00489-2. [DOI] [PubMed] [Google Scholar]
- Mellor PS, Boorman J, Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu. Rev. Entomol. 2000;45:307–340. doi: 10.1146/annurev.ento.45.1.307. [DOI] [PubMed] [Google Scholar]
- Melton JA, Parker MW, Rossjohn J, Buckley JT, Tweten RK. The Identification and Structure of the Membrane-spanning Domain of the Clostridium septicum Alpha Toxin. J. Biol. Chem. 2004;279:14315–14322. doi: 10.1074/jbc.M313758200. [DOI] [PubMed] [Google Scholar]
- Moffitt JE, Venarske D, Goddard J, Yates AB, de Shazo RD. Allergic reactions to Triatoma bites. Ann. Allergy Asthma Immunol. 2003;91:122–128. doi: 10.1016/S1081-1206(10)62165-5. [DOI] [PubMed] [Google Scholar]
- Montfort WR, Weichsel A, Andersen JF. Nitrophorins and related antihemostatic lipocalins from Rhodnius prolixus and other blood-sucking arthropods. Biochim. Biophys. Acta. 2000;1482(1–2):110–118. doi: 10.1016/s0167-4838(00)00165-5. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nobile M, Noceti F, Prestipino G, Possani LD. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp. Brain Res. 1996;110:15–20. doi: 10.1007/BF00241369. [DOI] [PubMed] [Google Scholar]
- Noeske-Jungblut C, Haendler B, Donner P, Alagon A, Possani L, Scheleuning W-D. Triabin, a highly potent exosite inhibitor of thrombin. J. Biol. Chem. 1995;270:28626–28634. doi: 10.1074/jbc.270.48.28629. [DOI] [PubMed] [Google Scholar]
- Noeske-Jungblut C, Krätzschmar J, Haendler B, Alagon A, Possani L, Verhallen P, Donner P, Scheleuning W-D. An inhibitor of collagen-induced platelet aggregation from the saliva of Triatoma pallidipennis. J. Biol. Chem. 1994;269:5050–5053. [PubMed] [Google Scholar]
- Paddock CD, McKerrow JH, Hansell E, Foreman KW, Hsieh I, Marshall N. Identification, cloning, and recombinant expression of procalin, a major triatomine allergen. J. Immunol. 2001;167:2694–2699. doi: 10.4049/jimmunol.167.5.2694. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Adams PL, Nuttall PA, Stuart DL. Tick histamine-binding proteins: lipocalins with a second binding cavity. Biochim. Biophys. Acta. 2000;1482:92–101. doi: 10.1016/s0167-4838(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Paiva-Silva GO, Sorginea MHF, Benedetti CE, Meneghini R, Almeida IC, Machado EA, Dansa-Petretski M, Yepiz-Plascencia G, Law JH, Oliveira PL, Masuda H. On the biosynthesis of Rhodnius prolixus heme-binding protein. Insect Biochem. Mol. Biol. 2002;32:1533–1541. doi: 10.1016/s0965-1748(02)00074-7. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins. Crit. Rev. Biochem. Mol. Biol. 1994;29:199–228. doi: 10.3109/10409239409086801. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Perireceptor events in olfaction. J. Neurobiol. 1996;30:3–19. doi: 10.1002/(SICI)1097-4695(199605)30:1<3::AID-NEU2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Pelosi P, Calvello M, Ban L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses. 2005;30:i291–i292. doi: 10.1093/chemse/bjh229. [DOI] [PubMed] [Google Scholar]
- Prestwich GD, Du G, LaForest S. How is pheromone specificity encoded in proteins? Chem. Senses. 1995;20:461–469. doi: 10.1093/chemse/20.4.461. [DOI] [PubMed] [Google Scholar]
- Prevot P-P, Adam B, Boudjeltia KZ, Brossard M, Lins, Cauchie P, Brasseur R, Vanhaeverbeek M, Vanhamme L, Godfroid E. Anti-hemostatic Effects of a Serpin from the Saliva of the Tick Ixodes ricinus. J. Biol. Chem. 2006;281:26361–26369. doi: 10.1074/jbc.M604197200. [DOI] [PubMed] [Google Scholar]
- Raj PA, Dentino AR. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol. Lett. 2002;206:9–18. doi: 10.1111/j.1574-6968.2002.tb10979.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Garcia ES. The role of saliva in feeding in Rhodnius prolixus. J. Exp. Biol. 1981;94:219–230. [Google Scholar]
- Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: Sialome and post-sialome perspectives. Annu. Rev. Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem. Mol. Biol. 2004b;34:543–563. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC. Blood-feeding arthropods: Live syringes or invertebrate pharmacologists? Infect. Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JMC, Andersen J, Silva-Neto MAC, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem. Mol. Biol. 2004a;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Arcà B, Lombardo F, Calvo E, Pham VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Edwards MJ, Grubhoffer L. Differential expression of Ixodes ricinus tick genes induced by blood feeding or Borrelia burgdorferi infection. J. Med. Entomol. 2005;42:36–41. doi: 10.1093/jmedent/42.1.36. [DOI] [PubMed] [Google Scholar]
- Sangamnatdej S, Paesen GC, Slovak M, Nuttall PA. A high affinity serotonin- and histamine-binding lipocalin from tick saliva. Insect Mol. Biol. 2002;11:79–86. doi: 10.1046/j.0962-1075.2001.00311.x. [DOI] [PubMed] [Google Scholar]
- Sant’ Anna MR, Araújo JG, Pereira MH, Pesquero JL, Diotaiuti L, Lehane SM, Lehane MJ. Molecular cloning and sequencing of salivary gland-specific cDNAs of the blood-sucking bug Triatoma brasiliensis (Hemiptera: Reduviidae) Insect Mol. Biol. 2002;11:585–593. doi: 10.1046/j.1365-2583.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- Santos A, Ribeiro JM, Lehane MJ, Gontijo NF, Veloso AB, Sant’anna MR, Nascimento Araújo R, Grisard EC, Pereira MH. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae) Insect Biochem. Mol. Biol. 2007;37(7):702–712. doi: 10.1016/j.ibmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindel C, Zitzer A, Schulte B, Gerhards A, Stanley P, Hughes C, Koronakis V, Bhakdi S, Palmer M. Interaction of Escherichia coli hemolysin with biological membranes. A study using cysteine scanning mutagenesis. Eur. J. Biochem. 2001;268:800–808. doi: 10.1046/j.1432-1327.2001.01937.x. [DOI] [PubMed] [Google Scholar]
- Shin SW, Park S-S, Park D-S, Kim MG, Kim SC, Brey PT, Park H-Y. Isolation and characterization of immune-related genes from the fall webworm, Hyphantria cunea, using PCR-based differential display and subtractive cloning. Insect Biochem. Mol. Biol. 1998;28:827–837. doi: 10.1016/s0965-1748(98)00077-0. [DOI] [PubMed] [Google Scholar]
- Söderhäll K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 1998;10:23–28. doi: 10.1016/s0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- Soloaga A, Veiga MP, Garcia-Segura LM, Ostolaza H, Brasseur R, Goni FM. Insertion of Escherichia coli alpha-haemolysin in lipid bilayers as a non-transmembrane integral protein: prediction and experiment. Mol. Microbiol. 1999;31:1013–1024. doi: 10.1046/j.1365-2958.1999.01225.x. [DOI] [PubMed] [Google Scholar]
- Sommerhoff CP, Sollner C, Mentele R, Piechottka GP, Auerswald EA, Fritz H. A Kazal-type inhibitor of human mast cell tryptase: isolation from the medical leech Hirudo medicinalis, characterization, and sequence analysis. Biol. Chem. Hoppe Seyler. 1994;375:685–694. doi: 10.1515/bchm3.1994.375.10.685. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Heitz T, Prasard V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- Stubbs MT, Morenweiser R, Sturzebecher J, Bauer M, Bode W, Huber R, Piechottka GP, Matschiner G, Sommerhoff CP, Fritz H, Auerswald EA. The three-dimensional structure of recombinant leech-derived tryptase inhibitor in complex with trypsin. Implications for the structure of human mast cell tryptase and its inhibition. J. Biol. Chem. 1997;272:19931–19937. doi: 10.1074/jbc.272.32.19931. [DOI] [PubMed] [Google Scholar]
- Szyperski T, Fernandez C, Mumenthaler C, Wuthrich K. Structure comparison of human glioma pathogenesis-related protein GliPR and the plant pathogenesis-related protein P14a indicates a functional link between the human immune system and a plant defense system. Proc. Nat. Acad. Sci. USA. 1998;95:2262–2266. doi: 10.1073/pnas.95.5.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takác P, Nunn MA, Mészáros J, Pechánová O, Vrbjar N, Vlasáková P, Kozánek M, Kazimírová M, Hart G, Nuttall PA, Labuda M. Vasotab, a vasoactive peptide from horse fly Hybomitra bimaculata (Diptera, Tabanidae) salivary glands. J. Exp. Biol. 2006;209:343–352. doi: 10.1242/jeb.02003. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BioMed Central Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra WR, Ferreira C, Jordão BP, Dillon RJ. Biology of the Insect Midgut. London: Chapman & Hall; 1996. p. 400. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A, Cantley LC. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol. 2003;33:717–732. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Mather TN, Ribeiro JMC. Exploring the sialome of the tick, Ixodes scapularis. J. Exp. Biol. 2002a;205:2843–2864. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JMC. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2002b;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Van de Locht A, Lamba D, Bauer M, Huber R, Friedrich T, Kröger B, Höffken W, Bode W. Two heads are better than one: crystal structure of the insect derived double domain Kazal inhibitor rhodniin in complex with thrombin. EMBO J. 1995;14:5149–5157. doi: 10.1002/j.1460-2075.1995.tb00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG, Prestwich GD, Lerner MR. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J. Neurobiol. 1991;22:74–84. doi: 10.1002/neu.480220108. [DOI] [PubMed] [Google Scholar]
- Wassenaar TM. Use of antimicrobial agents in veterinary medicine and implications for human health. Crit. Rev. Microbiol. 2005;31:155–169. doi: 10.1080/10408410591005110. [DOI] [PubMed] [Google Scholar]
- Welch RA. RTX toxin structure and function: a story of numerous anomalies and few analogies in toxin biology. Curr. Top. Microbiol. Immunol. 2001;257:85–111. doi: 10.1007/978-3-642-56508-3_5. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Koike H, Sugiyama Y, Motoyoshi K, Wada T, Hishinuma S, Mita M, Morita T. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 2002;269:2708–2715. doi: 10.1046/j.1432-1033.2002.02940.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Morita T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 2004;44:227–231. doi: 10.1016/j.toxicon.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


