Abstract
Yeast mitogen-activated protein kinase (MAPK) signaling pathways transduce external stimuli into cellular responses very precisely. The MAPKs Slt2/Mpk1 and Hog1 regulate transcriptional responses of adaptation to cell wall and osmotic stresses, respectively. Unexpectedly, we observe that the activation of a cell wall integrity (CWI) response to the cell wall damage caused by zymolyase (β-1,3 glucanase) requires both the HOG and SLT2 pathways. Zymolyase activates both MAPKs and Slt2 activation depends on the Sho1 branch of the HOG pathway under these conditions. Moreover, adaptation to zymolyase requires essential components of the CWI pathway, namely the redundant MAPKKs Mkk1/Mkk2, the MAPKKK Bck1, and Pkc1, but it does not require upstream elements, including the sensors and the guanine nucleotide exchange factors of this pathway. In addition, the transcriptional activation of genes involved in adaptation to cell wall stress, like CRH1, depends on the transcriptional factor Rlm1 regulated by Slt2, but not on the transcription factors regulated by Hog1. Consistent with these findings, both MAPK pathways are essential for cell survival under these circumstances because mutant strains deficient in different components of both pathways are hypersensitive to zymolyase. Thus, a sequential activation of two MAPK pathways is required for cellular adaptation to cell wall damage.
INTRODUCTION
Saccharomyces cerevisiae yeast cells are exposed to rapid and extreme changes in the environment. In response to these changes, precise responses are coordinated by the cell through different mitogen-activated protein kinase (MAPK) signaling pathways. In this sense, external cues are transduced into appropriate cellular responses, allowing cells to adapt to particular environmental conditions. In budding yeast, four MAPKs, Fus3, Kss1, Hog1, and Slt2/Mpk1, control mating, filamentation/invasion, high osmolarity, and cell integrity pathways, and they are activated in response to mating pheromones, starvation, osmolarity, and cell wall damage, respectively (Qi and Elion, 2005).
Yeast cell integrity depends on a particular external envelope: the cell wall, which is necessary not only for maintaining cell morphology but also for protecting cells from extreme conditions. The components of this structure form a macromolecular complex whose mechanical strength allows cells to support turgor pressure against the plasma membrane (Levin, 2005; Lesage and Bussey, 2006). Because of the importance of the cell wall for survival, stress conditions that alter this structure lead to the activation of a cellular response that has been called the “compensatory mechanism” (Popolo et al., 2001). This response is triggered by the cell in an attempt to survive, and it is characterized by 1) an increase in β-glucan and chitin contents; 2) changes in the association between cell wall polymers; 3) an increase in the amount of several cell wall proteins (CWPs); and 4) the relocalization of important proteins from the cell wall construction machinery to the lateral cell wall.
Although several signaling pathways have been implicated in the regulation of the cell wall remodeling process, the cell wall integrity (CWI) pathway (see Levin, 2005, for a recent review) plays an essential role. In fact, the CWI is activated in response to treatment with cell wall–perturbing agents such as Congo Red, Calcofluor White, zymolyase, and pneumocandins (Ketela et al., 1999; De Nobel et al., 2000; Reinoso-Martín et al., 2003) as well as mutations that lead to a weak cell wall, such as fks1Δ or gas1Δ (de Nobel, 2000). Activation of the cell integrity pathway protects yeast cells from cell wall stress by inducing a well-characterized transcriptional program (Lagorce et al., 2003; Boorsma et al., 2004; García et al., 2004; Rodríguez-Peña et al., 2005), which includes genes related to metabolism, cell wall remodeling, and signaling pathways. In addition, other environmental conditions, including heat shock, hypo-osmotic shock, actin depolymerization, the presence of chlorpromazine, caffeine, vanadate, rapamycin, mating pheromones, and other morphological events in which cell integrity is compromised, also lead to the activation of the CWI pathway (Martín et al., 2000; Levin, 2005).
Several cell membrane proteins (Mid2, Wsc1-4, and Mtl1; Verna et al., 1997; Ketela et al., 1999; Rajavel et al., 1999) act as sensors of the CWI pathway. Among them, Mid2 and Wsc1 play the main role in sensing cell wall damage. On conditions of activation, these sensors interact with the guanine nucleotide exchange factor (GEF) Rom2, activating the small GTPase Rho1, which then interacts and activates Pkc1 (Levin, 2005). The main role of activated Pkc1 is to trigger a MAPK module. Phosphorylation of the MAPKKK Bck1 by Pkc1, activates a pair of redundant MAPKKs (Mkk1 and Mkk2), which finally phosphorylate the MAPK Slt2. The phosphorylated form of this protein acts mainly on two transcription factors: the MADS-box transcription factor Rlm1 (Watanabe et al., 1997) and SBF (Baetz et al., 2001). SBF is a heterodimeric complex of two proteins, Swi4 and Swi6, that is involved in the regulation of gene expression at the G1/S transition. Transcriptional activation of most of those genes induced in response to Congo Red and heat shock, is dependent on Rlm1 (Jung and Levin, 1999; García et al., 2004).
The high-osmolarity glycerol (HOG) pathway is mainly devoted to the adaptation of yeast cells to osmotic stress (reviewed in Hohmann, 2002; Westfall et al., 2004). This pathway has two known input branches that activate the MAPK module by different mechanisms. The first one, the Sho1 branch, is involved in response to severe hyperosmotic stress conditions and requires both Sho1 and Msb2 plasma membrane proteins. Activation of this branch involves the participation of the Rho GTPase Cdc42, Ste20, and Ste50. The target of Ste20 is the MAPKKK Ste11, which activates Pbs2 in response to hyperosmotic conditions, resulting in Hog1 activation (de Nadal et al., 2002; Hohmann, 2002). The second branch, leading to the activation of Pbs2, involves a “two-component” phospho-relay signaling system with the participation of the transmembrane protein, Sln1 and the response regulators proteins Ypd1 and Ssk1 (Posas and Saito, 1998). In this branch, two redundant MAPKKKs (Ssk2/Ssk22) are responsible for the phosphorylation of Pbs2, the MAPKK common for both branches of the HOG pathway that is responsible for the final activation of Hog1. Once Hog1 is activated, it coordinates the transcriptional program necessary for cellular adaptation to osmotic stress. Additionally to the osmotic stress response, the Hog1 pathway has also been recently implicated in the regulation of the response to different kind of stresses, including adaptation to citric acid stress (Lawrence et al., 2004), presence of methylglyoxal (Aguilera et al., 2005), temperature downshift (Panadero et al., 2006), and tolerance to arsenite (Thorsen et al., 2006).
Although the cell integrity pathway has been shown to be the main pathway responsible for cell wall construction, other pathways like the STE vegetative growth pathway (SVG) have also been reported to be involved in maintaining cell integrity (Lee and Elion, 1999). Furthermore, in silico analysis of the promoters of the genes that are induced transcriptionally in response to cell wall damage suggests that the calcineurin signaling pathway, the HOG pathway, and the regulatory machinery from the general stress response could also act as regulators of the response to cell wall damage (Lagorce et al., 2003; Boorsma et al., 2004; García et al., 2004).
In an attempt to characterize the molecular mechanisms that govern the transcriptional induction of CRH1 (a gene encoding a glycosylphosphatidylinositol [GPI]-cell wall protein that is part of the compensatory response) under cell wall damage growth conditions, we identified in this work a new connection between the HOG and the cell integrity (SLT2) pathways. Therefore, both pathways seem to be essential for the adaptation of yeast to zymolyase-mediated cell wall stress. Evidence of a new role for the HOG pathway in yeast cell wall biogenesis and a cooperative role for both pathways controlling cell integrity is presented.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
The yeast strains used in this work are listed in Table 1. To generate the CR001, RG001, and RG002 double mutant strains, the SLT2, MSN2, and SKO1 genes were replaced by URA3, HIS3, and KanMX4 markers, respectively, in the corresponding single-deletant strains (hog1Δ, msn4Δ, and TM233) using a PCR-based method described by Wach et al. (1997).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf, Germany |
| slt2Δ | BY4741 isogenic, slt2::KanMX4 | Euroscarf, Germany |
| hog1Δ | BY4741 isogenic, hog1::KanMX4 | Euroscarf, Germany |
| fus3Δ | BY4741 isogenic, fus3::KanMX4 | Euroscarf, Germany |
| kss1Δ | BY4741 isogenic, kss1::KanMX4 | Euroscarf, Germany |
| rlm1Δ | BY4741 isogenic, rlm1::KanMX4 | Euroscarf, Germany |
| swi4Δ | BY4741 isogenic, swi4::KanMX4 | Euroscarf, Germany |
| swi6Δ | BY4741 isogenic, swi6::KanMX4 | Euroscarf, Germany |
| wsc1Δ | BY4741 isogenic, wsc1::KanMX4 | Euroscarf, Germany |
| wsc2Δ | BY4741 isogenic, wsc2::KanMX4 | Euroscarf, Germany |
| wsc3Δ | BY4741 isogenic, wsc3::KanMX4 | Euroscarf, Germany |
| wsc4Δ | BY4741 isogenic, wsc4::KanMX4 | Euroscarf, Germany |
| mid2Δ | BY4741 isogenic, mid2::KanMX4 | Euroscarf, Germany |
| bck1Δ | BY4741 isogenic, bck1::KanMX4 | Euroscarf, Germany |
| rom2Δ | BY4741 isogenic, rom2::KanMX4 | Euroscarf, Germany |
| sho1Δ | BY4741 isogenic, sho1::KanMX4 | Euroscarf, Germany |
| sko1Δ | BY4741 isogenic, sko1::KanMX4 | Euroscarf, Germany |
| pbs2Δ | BY4741 isogenic, pbs2::KanMX4 | Euroscarf, Germany |
| opy2Δ | BY4741 isogenic, opy2::KanMX4 | Euroscarf, Germany |
| rom1Δ | BY4741 isogenic, rom1::KanMX4 | Euroscarf, Germany |
| tus1Δ | BY4741 isogenic, tus1::KanMX4 | Euroscarf, Germany |
| mtl1Δ | BY4741 isogenic, mtl1::KanMX4 | Euroscarf, Germany |
| ssk1Δ | BY4741 isogenic, ssk1::KanMX4 | Euroscarf, Germany |
| ste11Δ | BY4741 isogenic, ste11::KanMX4 | Euroscarf, Germany |
| mkk1Δ mkk2Δ | BY4741 isogenic, mkk2::KanMX4; mkk1::SpHIS5 | Jiménez-Sánchez et al. (2007) |
| CR001 | BY4741 isogenic, hog1::KanMX4; slt2::URA3 | This study |
| RG001 | BY4741 isogenic, msn4::KanMX4; msn2::HIS3 | This study |
| TM141 | MATa; his3Δ0; leu2Δ0; trp1Δ0; ura3Δ0 | Posas et al. (1998) |
| FP66 | TM141 isogenic, ste50::HIS3 | Posas et al. (1998) |
| YEN44 | TM141 isogenic, smp1::HIS3 | de Nadal et al. (2003) |
| TM233 | TM141 isogenic, hog1::TRP1 | de Nadal et al. (2003) |
| FP53 | TM141 isogenic, ssk2::LEU2; ssk22::LEU2; ste20::HIS3 | F. Posas lab |
| RG002 | TM141 isogenic, hog1::TRP1; sko1::KanMX4 | This study |
| TM257 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 | Posas et al. (1998) |
| FP67 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ste50::HIS3 | Posas et al. (1998) |
| KT063 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 hkr1::natMX4 msb2::kanMX6 | Tatebayashi et al. (2007) |
| W303-1A | MATa; ade2-1; ura3-1; leu2-3,112; trp1-1; his3-11,15; can1-100 | Rep et al. (1999) |
| YEN29 | W303-1A isogenic, hot1::KanMX4 | Rep et al. (1999) |
| CML128 | MATa leu2-3, 112 ura3-52 trp1 his4 can1r | de la Torre et al. (2002) |
| NML344 | CML128 isogenic, pkc1::LEU2 | de la Torre-Ruiz et al. (2002) |
| HAS17-3B | MATα; ura3-52; leu2-3,112; his3-11,15 | Straede and Heinisch (2007) |
| HAS17-3D | MATa; ura3-52; leu2-3,112; his3-11,15; wsc1:: SpHIS5; mid2:: KlLEU2 | Straede and Heinisch (2007) |
| MCH-7B | MATα; ura3-52; leu2-3,112; his3-11,15Δtrp1::loxP MAL SUC GAL | Lorberg et al. (2001) |
| MALY7-7B | MATα; Δrom2::Sphis5+ura3-52; leu2-3,112; his3-11,15 Δtrp1::loxP MAL SUC GAL | Lorberg et al. (2001) |
Yeast cells were grown, depending on the experimental approaches, on YPD (2% glucose, 2% peptone, 1% yeast extract) or SD medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose) supplemented with the required amino acids. Routinely, cells were grown overnight in liquid media (YPD or SD in the case of cells bearing plasmids) at 220 rpm and 24°C to an optical density of 0.8–1 (OD600). This culture was diluted to 0.2 (OD600) in fresh YPD and were grown at 24°C for an additional 3 h. Then, the culture was divided into two parts. One part was allowed to continue growing under the same conditions (the untreated culture), and the other one was supplemented with Congo Red (30 μg/ml; Merck, Darmstadt, Germany) or zymolyase 100T from Arthrobacter luteus (0.4 U/ml; MP Biomedicals, Aurora, OH). Cells were collected at the indicated times and processed depending on the experimental approach, as described below.
Plasmids
To obtain the plasmid pCRH1-LACZ, a 1.1-kb (EcoRI/BamHI) and a 0.5-kb (XbaI/HindIII) fragment upstream and downstream from CRH1 obtained from genomic DNA by PCR amplification were cloned into the episomal shuttle vector YEp352. Finally, the lacZ gene from the YEp356R plasmid was inserted between the fragments described above to drive its expression under the control of the CRH1 promoter. The pJV89GL plasmid, containing a functional CRH1-green fluorescent protein (GFP) fusion, has recently been described in Cabib et al., 2007. The plasmid pCYC-R2 consists of a derivative of the plasmid pLG669-Z (Guarente and Ptashne, 1981), in which a SmaI/SmaI fragment of the CYC1 promoter localized upstream from the UAS was deleted and the Rlm1-binding box 2 (fragment −365 to −344 with respect to the start codon) from the CRH1 promoter was sequentially cloned into the unique XhoI site. In sum, in this construction the expression of the lacZ gene is under the control of the CYC1 promoter driven by a functional Rlm1-binding site from the CRH1 promoter (our unpublished results). Plasmids containing HOG1-GFP, hog1(KM)-GFP (consisting of a catalytically inactive protein), and hog1(TAYA)-GFP (a nonphosphorylatable derivative) have been described previously (Ferrigno et al., 1998). These plasmids were constructed using the centromeric vector pRS416.
Quantitative RT-PCR Assays
Total RNA was isolated from cells (5 × 107) with the mechanical disruption protocol using the RNeasy MINI kit (QIAGEN, Hilden, Germany), following the manufacturer's instructions. RNA concentrations were determined by measuring absorbance at 260 nm. RNA purity and integrity were assessed using RNA Nano Labchips in an Agilent 2100B Bioanalyzer (Agilent Technologies, Palo Alto, CA). Real-time quantitative RT-PCR (Q-RT-PCR) assays were performed as previously described (García et al., 2004), using an ABI 7700 instrument (Applied Biosystems, Foster City, CA). For quantification, the abundance of each gene at different time intervals was determined relative to the standard transcript of ACT1 and to the t = 0-h sample (1× sample) following the 2−ΔΔCt method, as described in Livak and Schmittgen (2001). Finally, the ratio of the treated/nontreated condition for each data point was calculated. The following forward and reverse primers, respectively, were used: ACT1, 5′-ATCACCGCTTTGGCTCCAT-3′ and 5′-CCAATCCAGACGGAGTACTTTCTT-3′; and CRH1, 5′-ACTACCCAGATATCAAGCAAATACACA-3′ and 5′-TCAGCACCGTTAGAAATTTGAACA-3′.
β-Galactosidase Assays
Yeast cell extracts were prepared by harvesting cells by centrifugation from 5 ml of an exponentially growing culture. Then, cells were resuspended in 250 μl of breaking buffer (100 mM Tris-HCl, pH 8, 1 mM dithiothreitol, 20% glycerol) and glass beads (Glasperlen ca. 1 mm, Sartorius AG, Goettingen, Germany) were added to break cells in a Fast-Prep machine. Finally, extracts were clarified by centrifugation, and protein concentrations were measured using the Bradford method. β-Galactosidase assays were performed using the crude extracts obtained as described previously (Amberg et al., 2005), scaling the protocol to a 96-well microtiter plate format. Ten microliters of cell extract was mixed with 90 μl of Z buffer plus β-mercaptoethanol (0.03%) and 20 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG; 4 mg/ml in Z buffer). The absorbance of the enzymatic reaction was measured at 415 nm on a microplate reader (Model 680, Bio-Rad Laboratories, Hercules, CA) after at least 10 min of incubation at 30°C and after the addition of 50 μl of 1 M Na2CO3 to stop the reaction. β-Galactosidase activity was expressed as nanomoles of ONPG converted per minute per milligram protein. These experiments were performed at least in triplicate from independent yeast transformants.
Western Blotting Assays
Immunoblot analyses, including cell collection, lysis, fractionation of proteins by SDS-PAGE, and transfer to nitrocellulose membranes, were carried out as previously described (Martín et al., 2000). The detection of phosphorylated Slt2 and Hog1 was accomplished using anti-phospho-p44/p42 MAPK (thr202/tyr204; Cell Signalling Technology, Beverly, MA), and anti-phospho-p38 MAPK (thr180/tyr182; Santa Cruz Biotechnology, Santa Cruz, CA) polyclonal antibodies, respectively. The detection of GFP was performed using the anti-GFP JL-8 mAb (Clontech, Palo Alto, CA). Hog1 was detected with anti-p38 MAPK (thr180/tyr182) polyclonal antibody (NEB). To monitor protein loading, actin levels were determined using the mouse anti-actin mAb C4 (ICN Biomedicals, Aurora, OH). For quantification of the bands from autoradiography films, densitometric analysis was performed using the QuantityOne package (Bio-Rad Laboratories). Act1 data were used for sample normalization.
Phenotypic Analysis
To determine the sensitivity of the different strains to Congo Red, yeast cells were grown as indicated above, and 5 μl of log-phase cultures (∼15 × 103 cells) plus three 1:5 serial dilutions were spotted onto YPD solid media containing different concentrations of Congo Red. Growth was monitored on the plates after incubation over 2 d at 30°C. Zymolyase sensitivity was performed in 96-well microtiter plates, preparing twofold serial dilutions of zymolyase 20T (ImmunO, MP Biomedicals) to give concentrations ranging from 125 to 0.25 U/ml in a final volume of 150 μl of YPD. Each well was inoculated with ∼104 cells from an exponentially growing culture. Plates were incubated for 24–48 h at 24°C, and cell growth was determined by measuring absorbance at 550 nm on an ELISA microplate reader.
Flow Cytometry and Fluorescence Microscopy
For Crh1-GFP analysis, cells were collected, washed twice with PBS, and then analyzed by flow cytometry and fluorescence microscopy. In this case, cells were visualized with a Nikon TE2000 fluorescence inverted microscope equipped with CCD (Melville, NY). Digital images were acquired with an Orca C4742-95-12ER camera (Hamamatsu, Bridgewater, NJ) and processed with the Aquacosmos Imaging Systems software. A CyAn MLE flow cytometer (Dako, Glostrup, Denmark) was used for flow cytometry analysis, acquiring green fluorescence through a 530/30 BP filter (BFP). The marker was set using unstained yeasts as controls. Dead cells were discriminated from the analysis using propidium iodide.
RESULTS
CRH1 Expression Is Induced in Response to Cell Wall Damage
CRH1 encodes a cell wall transglycosidase responsible for the cross-linking between chitin and β-1,6 glucan (Rodríguez-Peña et al., 2000; Cabib et al., 2007). Previous genome-wide analyses revealed that the expression of this gene is induced in response to several types of cell wall stress (Agarwal et al., 2003; Lagorce et al., 2003; García et al., 2004). We studied the kinetics of CRH1 transcriptional activation in response to the transient cell wall damage caused by Congo Red, a compound that binds to chitin, interfering with proper cell wall construction (Roncero and Duran, 1985), and zymolyase, which affects cell wall integrity as a result of the presence of a main β-1,3 glucanase and a residual protease activity (Zlotnik et al., 1984). At the concentrations assayed (30 μg/ml Congo Red and 0.4 U/ml zymolyase), both compounds elicited a similar response, with about a threefold increase in CRH1 expression attained after 1 h of exposure, with a sustained response for the following 3 h and a substantial fall at 6 h of treatment (Figure 1A). CRH1 induction correlated with an increase in the amount of Crh1 as deduced from Crh1-GFP monitoring of zymolyase-treated and nontreated cells by flow cytometry (Figure 1B). This increase was also clearly observed with fluorescence microscopy analysis. In the absence of stress, Crh1-GFP mainly localized to polarized cell growth sites, in particular to the bud neck (Figure 1C), as previously described (Rodríguez-Peña et al., 2000). Interestingly, although the protein still localized to the bud neck in cells challenged with zymolyase, much of the overinduced protein localized to the lateral cell wall, especially in large daughter cells, suggesting that Crh1 plays an important role in the remodeling of the daughter cell lateral wall under stress.
Figure 1.
CRH1 is induced under cell wall damage conditions. (A) The expression of CRH1 was examined after Congo Red (30 μg/ml) and zymolyase (0.4 U/ml) treatment in the WT strain (BY4741) carrying the pCRH1-LACZ plasmid. Cells were collected at different time points, and both CRH1 mRNA levels and β-galactosidase activity were determined, as described in Materials and Methods. Congo Red treatment corresponds to black bars (β-galactosidase activity) and squares (CRH1 mRNA). Zymolyase is represented by white bars (β-galactosidase activity) and squares (CRH1 mRNA). In both cases, the results are expressed as the ratio of treated versus untreated cells. Data represent means and SDs of three independent experiments. (B) BY4741 WT cells expressing Crh1-GFP were analyzed by flow cytometry after treatment with 0.4 U/ml zymolyase (gray line) or nontreated (black line) at the different times indicated. Dotted line corresponds to control BY4741 cells not expressing Crh1-GFP. (C) Localization of Crh1-GFP in WT cells (BY4741) growing exponentially in YPD and treated for 3 h with zymolyase (0.4 U/ml; right) or not treated (left).
Adaptation to Zymolyase-mediated Cell Wall Damage Depends on the Slt2-MAPK Pathway But Does Not Require the Wsc and Mid2 CWI Sensors
Current knowledge supports the notion that the regulation of transcriptional responses to cell wall damage mainly depends on the MAPK Slt2. In agreement with this, treatment with zymolyase caused a rapid increase in the levels of phosphorylated Slt2 15 min upon stress (Figure 2A), as measured by Western blotting using an antibody against the phosphorylated form of this MAPK. The kinetics of Slt2 phosphorylation revealed a peak after 1–2 h of treatment, although phosphorylation was still detected even after 6 h (Figure 2A). The activation of Slt2 was abrogated in the bck1Δ and mkk1Δ mkk2Δ mutants (Figure 2B). The protein kinase Pkc1 was also required. In this case, pkc1Δ cells and its isogenic wild-type (WT) CML128 were maintained in 1 M sorbitol to prevent cell lysis. As shown in Figure 2B, a functional Pkc1 was mandatory for Slt2 phosphorylation by zymolyase. In agreement with the phosphorylation data, elements of the cell integrity MAPK module, including the MAPKKK Bck1 and the MAPK Slt2, were essential for transcriptional activation of CRH1 by zymolyase (Figure 2C). We then checked the role of transcription factors regulated by Slt2. Although the basal levels of CRH1 seemed to be regulated by Swi4, CRH1 expression was induced by zymolyase in swi4Δ and swi6Δ mutants (Figure 2C). In contrast, the transcription factor Rlm1 was essential for CRH1 induction (Figure 2C).
Figure 2.
Adaptation to zymolyase requires different elements of the Slt2 MAPK pathway but not the sensors or the GEFs of this pathway. (A) Time course of Slt2 activation in WT (BY4741) cells growing at 24°C to midlog phase and exposed to zymolyase (0.4 U/ml) at the indicated times. The protein load was monitored using a mouse anti-actin mAb. (B) Phosphorylation of Slt2 after 2 h of zymolyase treatment in slt2Δ, mkk1Δ mkk2Δ, bck1Δ, and pkc1Δ mutants. (C and D) Expression of CRH1-lacZ was determined in WT, slt2Δ, bck1Δ, rlm1Δ, swi4Δ, and swi6Δ strains (C) and WT, mid2Δ, wsc1Δ, wsc2Δ, wsc3Δ, wsc4Δ, and mtl1Δ strains (D), in the absence (□) and presence (3 h) of zymolyase 100T (0.4 U/ml; ■). (E) Slt2-phosphorylation by zymolyase in mid2Δ, wsc1Δ, and mid2Δ wsc1Δ double mutant (strain HAS17-3D) compared with corresponding WT strains, BY4741 and HAS17-3B. (F) Slt2 activation in the rom2Δ mutant compared with the WT (MCH-7B) in untreated cells or cells growing in the presence of Congo Red (30 μg/ml) or zymolyase (0.4 U/ml) for 2 h. (G) Phosphorylation of Slt2 in rom1Δ, rom2Δ, and tus1Δ mutants (BY4741 background) after 2 h of zymolyase treatment. Graphics in F and G represent quantification, by densitometric analysis, of the Phospho-Slt2 bands from Western blots shown above, normalized with respect to the actin bands.
A family of cell surface sensors has been implicated in the detection and signaling of cell wall damage, leading to activation of the cell integrity pathway. As shown in Figure 2D, the induction of CRH1 by zymolyase was not abrogated in the mutants deleted in any of these sensors (wsc1Δ, wsc2Δ, wsc3Δ, wsc4Δ, mtl1Δ, and mid2Δ), although slight decreases were detected for wsc1Δ and wsc3Δ. Correspondingly, Slt2 phosphorylation, induced by zymolyase, was not abrogated in single wsc1Δ and mid2Δ mutants (Figure 2E). To rule out the possibility that in the absence of one sensor the other could be functioning, we checked Slt2 phosporylation by zymolyase in a double wsc1Δ mid2Δ strain. As shown in Figure 2E, Slt2 was clearly activated by zymolyase in this mutant, suggesting that the main sensors of the cell integrity pathway are not important for sensing zymolyase-mediated cell wall damage. The same result was obtained in a wsc1Δ mid2Δ strain with a different genetic background (data not shown). In contrast, induction of CRH1 by Congo Red clearly decreased in a mid2Δ strain (Supplementary Figure S1).
Under conditions of many other types of cell integrity pathway activation (Calcofluor White, pneumocandins, etc.), these sensors interact with the GEF Rom2, which in turn activates Rho1, leading to the activation of the Slt2 MAPK cascade. However, as shown in Figure 2F, although the Slt2 activation caused by Congo Red demanded the participation of Rom2, the levels of phosphorylated Slt2 due to the presence of zymolyase were clearly increased in a rom2Δ, indicating that activation caused by this stress does not require this GEF. Additionally, none of the other two GEFs of this pathway described already, Rom1 or Tus1, were required for Slt2 activation in the presence of zymolyase (Figure 2G). It should be noted that basal levels of Slt2-P were higher in the rom2Δ and tus1Δ mutants, but that zymolyase still increased phosphorylation of Slt2 in these mutants. The possibility that in the absence of one GEF another one might substitute it cannot be ruled out, but all these data are certainly in agreement with a model in which the activation of the Slt2 MAPK by zymolyase, in contrast to other stimuli activating this pathway, does not require the upstream elements of the cell integrity pathway, i.e., neither the sensors nor the GEFs Rom1, Rom2, and Tus1.
Adaptation to Zymolyase-mediated Cell Wall Damage Not Only Requires Slt2 But Also the Hog1 MAPK
To study the possible participation of other pathways in the adaptation to zymolyase, we analyzed the induction of CRH1 in strains deficient in other MAPKs. CRH1-lacZ expression was analyzed in the slt2Δ, hog1Δ, fus3Δ, and kss1Δ strains challenged with zymolyase or Congo Red. Although the induction of CRH1 by Congo Red was only dependent on the MAPK Slt2 (Figure 3A), transcriptional activation of this gene by zymolyase required both the Slt2 and Hog1 MAPKs (Figure 3A). These results were also confirmed by measuring Crh1 protein levels in cells expressing Crh1-GFP. As shown in Figure 3B, overproduction of Crh1 by Congo Red was dependent on Slt2 but not on Hog1, whereas the increase in Crh1 mediated by zymolyase was dependent on both MAPKs.
Figure 3.
CRH1 expression is regulated by the MAPK Slt2 or by Hog1 and Slt2 MAPKs simultaneously, depending on the nature of the cell wall stress. (A) Expression of CRH1-LacZ was studied in WT (BY4741) and slt2Δ, hog1Δ, kss1Δ, and fus3Δ mutant cells growing in presence of Congo Red (30 μg/ml; ■) or zymolyase (0.4 U/ml; □). β-Galactosidase activities are expressed as the ratio of Congo Red or zymolyase-treated cells versus untreated cells. Mean and SDs derived from three independent experiments. (B) Levels of Crh1-GFP were examined by Western blot in WT BY4741, slt2Δ, and hog1Δ untreated and treated (Congo Red or zymolyase) cells transformed with plasmid pJV89GL. The protein load was determined using an anti-actin antibody.
The induction of CRH1 by zymolyase was recovered in hog1Δ cells transformed with a plasmid carrying a WT HOG1 gene (data not shown). Therefore, our data suggest that the cellular response to different kinds of cell wall damage seems to be regulated by different mechanisms. Second, in addition to the cell integrity pathway, the HOG pathway is also required for adaptation of the cell to the cell wall damage caused by zymolyase.
Activation of the Hog1 MAPK by phosphorylation has mainly been described to occur in response to osmotic stress. On the basis of the results reported above, we were prompted to test whether Hog1 was activated by zymolyase using antibodies that recognize the phosphorylated form of Hog1. As shown in Figure 4A, zymolyase activated Hog1 and this activation depended on the presence of Pbs2. However, Hog1 overphosphorylation was still present in a slt2Δ strain under the same conditions (Figure 4B). It is worth to note that levels of phosphorylation of Hog1 by zymolyase treatment were much lower than in osmotic stress conditions like 0.4 M NaCl (Figure 4A). To check if these conditions affected the localization pattern of Hog1, Hog1-GFP was followed at different times after the addition of zymolyase (Figure 4C). As shown in Figure 4C, the Hog1-GFP fusion protein did not translocated from the cytoplasm to the nucleus in response to zymolyase stress, in contrast to what happens under osmotic stress conditions (Ferrigno et al., 1998; Figure 4C). Moreover, as expected, Hog1 was not overphosphorylated in cells growing in presence of Congo Red (Figure 4D). Induction of CRH1 was then assayed in hog1Δ cells transformed with either plasmids expressing the hog1 (TA/YA) allele, which abolish the phosphorylation of Hog1 by Pbs2, or the hog1 (KM) mutant allele, which results in a catalytically inactive protein. As shown in Figure 4E, there was no induction of CRH1 in the absence of HOG1 (plasmid pRS416) or in cells containing KM or TA/YA hog1 mutant alleles, indicating that not only the presence of Hog1 but also that its catalytic activity are essential for the transcriptional response to cell wall damage induced by zymolyase.
Figure 4.
Hog1 is slightly activated by zymolyase and the presence of an active form of this MAPK is necessary for the induction of CRH1. (A) Effect of zymolyase in Hog1 activation. Exponentially growing WT cells were collected before and after different intervals of zymolyase treatment, and Hog1 activation was examined by immunoblotting total extracts with an anti-phospho-p38 antibody. The levels of phosphorylation of Hog1 by 0.4 M NaCl are also shown. The phosphorylation of Hog1 by zymolyase was dependent on the presence of the MAPKK Pbs2. (B) Hog1 is activated by zymolyase in a slt2Δ strain. Exponentially growing WT and slt2Δ cells were collected before and after different intervals of zymolyase treatment and Hog1 activation was examined. (C) Hog1 is not translocated to the nucleus after zymolyase-induced stress. A hog1Δ strain was transformed with a GFP-tagged Hog1 (pRS-HOG1-GFP). Cells were grown exponentially, exposed (Zym +) or not (Zym −) to 0.4 U/ml zymolyase 100T at the indicated times and visualized by fluorescence microscopy. (D) Congo Red does not activate the MAPK Hog1 in a WT strain. Exponentially growing WT cells were collected before and after different intervals of Congo Red treatment at 30 μg/ml, and Hog1 activation was examined. (E) CRH1 mRNA levels (represented as the ratio of treated vs. untreated cells) were analyzed by Q-RT-PCR in hog1Δ cells transformed with pRS416 (empty vector), the WT HOG1 allele (pRS416-HOG1), or inactive mutant alleles of hog1 (hog1 TA-174/YA-176 and hog1 KM-78) after 2 h of zymolyase treatment.
Adaptation to Zymolyase Depends on the Sho1 Branch of the HOG Pathway
We next investigated the components of the HOG pathway that were necessary for zymolyase-mediated CRH1 induction. The HOG1 pathway has two known input branches that activate the MAPK module by different mechanisms. The Sho1 branch requires both Sho1 and Msb2 plasma membrane proteins. The second branch is activated through the transmembrane protein Sln1. In contrast to what we had observed in the case of the sensors of the cell integrity pathway, the induction of CRH1 by zymolyase was completely lost in a sho1Δ mutant (Figure 5). Additionally, the presence of the MAPKK Pbs2, the MAPKKKs Ste11, but not Ssk2/22, was essential for CRH1 induction (Figure 5). Moreover, mutations in other elements of the Sln1 branch (ssk1Δ) did not affect CRH1 induction, but mutations in elements of the Sho1 branch, such as Ste50 and Ste20, completely abolished CRH1 transcriptional activation (Figure 5). All these results indicate that in addition to the cell integrity pathway the induction of CRH1 expression in response to zymolyase is regulated through the Sho1 branch of the HOG pathway.
Figure 5.
CRH1 induction by zymolyase requires the Sho1 branch of the HOG pathway. Expression of CRH1-LacZ was studied in the WT BY4741 and the corresponding single mutants sho1Δ, ssk1Δ, pbs2Δ, and ste11Δ and in the WT TM141 and its mutant-derived strains ste50Δ, ssk2Δ ssk22Δ, and ssk2Δ ssk22Δ ste20Δ growing exponentially in the absence (□) and in the presence of 0.4 U/ml zymolyase for 3 h (■). Results are represented as means with SDs derived from three independent experiments.
Cells Deficient on the HOG and SLT2 Signaling Pathways are Hypersensitive to Zymolyase
Because the functional HOG1 and SLT2 pathways were required for adaptation to zymolyase, we were prompted to study whether the lack of induction of the transcriptional compensatory response might affect the ability of cells mutated in these pathways to survive under cell wall stress. The phenotype of cells lacking different elements of the high osmolarity and cell integrity pathways in the presence of zymolyase was characterized in liquid media containing increasing amounts of zymolyase. As shown in Figure 6A, in contrast to the extreme hypersensitivity of slt2Δ, mkk1Δ mkk2Δ, and bck1Δ strains to zymolyase, the growth of strains deleted in either of the two sensors of this pathway—mid2Δ and wsc1Δ—or the Rho1 GEF Rom2 (rom2Δ) in the presence of zymolyase was similar to the WT strain (Figure 6A). Additionally, lack of any of the other two GEFs, Rom1 and Tus1, did not render zymolyase-sensitive strains (data not shown). On the other hand, strains deleted in elements of the Sho1 branch of the HOG1 pathway including sho1Δ, ste50Δ, ste11Δ, pbs2Δ, and hog1Δ (Figure 6B) were also highly hypersensitive to zymolyase. As expected, the growth of strains deleted in elements of the Sln1 branch, such as the ssk1Δ (Figure 6B) and ssk2Δ ssk22Δ (not shown) strains, was not affected by the presence of this stress. In contrast to zymolyase, the sho1Δ, ste50Δ, ste11Δ, pbs2Δ, and hog1Δ strains were not sensitive to 50 μg/ml Congo Red (Supplementary Figure S2). Therefore, the sensitivity of these strains was consistent with the lack of CRH1 transcriptional induction in the mutants, indicating that the activation of both the HOG1 pathway through the Sho1 branch and the cell integrity pathway is essential for cell survival under zymolyase-mediated cell wall stress.
Figure 6.
Mutant strains belonging to the CWI pathway, and the Sho1 branch of the HOG pathway are hypersensitive to zymolyase. (A) Sensitivity to zymolyase of WT BY4741 and mutant strains mid2Δ, wsc1Δ, rom2Δ, bck1Δ, mkk1Δ mkk2Δ, and slt2Δ. (B) Sensitivity to zymolyase of WT BY4741 and the mutant strain derivatives ssk1Δ, ste11Δ, sho1Δ, ste50Δ, pbs2Δ, and hog1Δ. Sensitivity was measured as described in Materials and Methods.
The Effect of Both SLT2 and HOG Pathways on the Transcriptional Activation of CRH1 Is Exerted through Rlm1
The induction of CRH1 by zymolyase requires the coordination of the two signaling pathways that converge at the promoter of CRH1. As shown above, the transcription factor Rlm1, regulated by the MAPK Slt2, plays an essential role in the induction of CRH1. We therefore examined whether the transcriptional factors regulated by Hog1 might also be involved. Because the induction of CRH1 was dependent on the presence of both Slt2 and Hog1, a possible mechanism could be a simultaneous transcriptional regulation of CRH1 by Rlm1, regulated by the MAPK Slt2 and transcriptional modulators regulated by Hog1. We first tested if zymolyase was able to induce CRH1 transcription in a double sko1Δ hog1Δ mutant strain. Sko1 could be phosphorylated under cell wall stress by the MAPK Hog1 to switch Sko1-Tup1-Ssn6 from a repressor to an activator complex. In this situation, CRH1 would be induced by zymolyase through Rlm1 in a double sko1Δ hog1Δ mutant strain. However, this was not the case (data not shown).
Moreover, induction of CRH1 by zymolyase was not decreased in mutants deleted in transcription factors regulated by the Hog1 MAPK like smp1Δ, hot1Δ, msn2Δ msn4Δ, and sko1 (Figure 7A). Accordingly, the effect on CRH1 transcription in this case does not seem to be mediated by any of the transcriptional factors regulated by Hog1. Consistent with these results, a Hog1-GFP fusion protein did not translocated from the cytoplasm to the nucleus in response to zymolyase (Figure 4), in contrast to what happens under osmotic stress conditions (Ferrigno et al., 1998).
Figure 7.
Transcriptional activation of CRH1 requires Rlm1 but not the Sko1, Hot1, Msn2/4, or Smp1 transcription factors. (A) The expression of CRH1-lacZ was determined in WT, hog1Δ, hot1Δ, msn2Δ msn4Δ, sko1Δ, and smp1Δ exponentially growing strains in the absence (□) or presence of zymolyase 100T (0.4 U/ml for 3 h; ■). (B) β-Galactosidase activity was measured in WT BY4741 and its mutant derivatives hog1Δ and slt2Δ transformed with plasmid pCYC-R2, which includes the Rlm1-binding box 2 from the CRH1 promoter fused to the lacZ gene.
The CRH1 promoter has three putative Rlm1-binding domains. The one located at −359 (Rlm1 box 2 from CRH1) seems to be the most important one for transcriptional induction of CRH1 (our unpublished results). To test whether the effect of the HOG pathway on the transcriptional induction of CRH1 was exerted through this Rlm1 binding box, a minimal promoter plasmid containing this box fused to LacZ (pCYC-R2) was transformed in WT, hog1Δ and slt2Δ strains, and lacZ transcription was measured in the presence of zymolyase. As shown in Figure 7B, the Rlm1-binding domain was sufficient to drive induction by zymolyase in WT cells. As expected, because Rlm1 is activated by Slt2, there was no induction of the lacZ reporter in the slt2Δ strain. Additionally, transcriptional activation was almost lost in hog1Δ cells (Figure 7B). All this results are in agreement with a model in which induction of CRH1 by zymolyase is regulated by the transcription factor Rlm1, and the effect of the HOG1 pathway on this induction is exerted upstream of Rlm1.
Phosphorylation of the MAPK Slt2 by Zymolyase Depends on the Presence of a Functional HOG1 Pathway
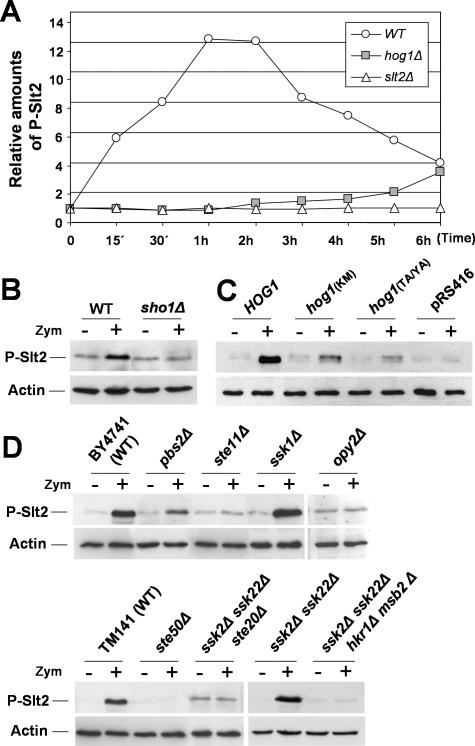
Because the absence of Hog1 seemed to impair CRH1 induction through a mechanism mediated by elements upstream from Rlm1, we decided to investigate whether Hog1 might be required for proper Slt2 overphosphorylation under activation conditions. To accomplish this, the kinetics of activation of Slt2 after addition of the zymolyase was analyzed by Western blotting in strains deleted in HOG1. As shown in Figure 8A, most of the phosphorylation of Slt2 due to zymolyase in the WT was lost in the hog1 strain. Moreover, Slt2 activation was completely abrogated in a sho1 mutant (Figure 8B).
Figure 8.
Elements of the SHO1 branch of the HOG pathway are required for Slt2 activation by zymolyase. (A) Time course of Slt2 activation in hog1Δ cells exposed to zymolyase (0.4 U/ml) for the indicated times compared with the WT and slt2Δ strains. Relative amounts of phosphorylated Slt2 after densitometric analysis, of the Slt2-P are represented. Each data point was first normalized to the total amount of Act1 in the sample and then to the value at time 0 h. (B) Activation of Slt2 after 2 h of treatment with zymolyase is completely lost in a sho1Δ strain. (C) Slt2 activation by zymolyase is lost in the presence of inactivated forms of the Hog1 MAPK. The amount of phosphorylated Slt2 was examined in hog1Δ cells transformed with pRS416 (empty vector), the WT HOG1 allele (pRS416-HOG1), or inactive mutant hog1 alleles (hog1 TA/YA and hog1 KM) after 2 h of zymolyase treatment. (D) Slt2 activation due to zymolyase depends on the presence of SHO1-branch elements of the HOG pathway. The Slt2 phosphorylation status was investigated in WT and strains deleted in different elements of the SHO1 and SLN1 branches: the WT BY4741 and its mutant derivatives pbs2Δ, ste11Δ, ssk1Δ, and opy2Δ, and the WT TM141 and its derivatives ste50Δ, ssk2Δ ssk22Δ, ssk2Δ ssk22Δ ste20Δ, and ssk2Δ ssk22Δ hkr1Δ msb2Δ.
To test whether only the presence of Hog1 was necessary for the hyperphosphorylation of Slt2 by zymolyase or whether the catalytic activity of the protein was also required, Slt2 phosphorylation was measured in hog1Δ cells transformed with either WT, TA/YA, or KM hog1 alleles. Although Slt2 was clearly overphosphorylated by zymolyase in a hog1Δ strain expressing the WT HOG1, phosphorylation was not significantly increased in the other two strains (Figure 8C). All these results clearly indicate that adequate Slt2 phosphorylation by zymolyase is dependent not only on the presence of Hog1 but also on the catalytic activity of this protein, and therefore they suggest a new connection between the HOG and the cell integrity pathway.
To identify the elements of the HOG1 pathway necessary for the overphosphorylation of Slt2 in response to zymolyase, this phosphorylation was monitored after zymolyase treatment in mutant strains deleted in different elements of the HOG pathway. As shown in Figure 8D, deletion of PBS2 or STE11 resulted in a strong decrease or the absence of Slt2 activation, respectively. Additionally, in accordance with the CRH1 transcriptional data, deletant strains in elements of the Sho1 branch of the HOG pathway such as STE50 or STE20 clearly abrogated Slt2 overphosphorylation, whereas mutations in elements of the Sln1 branch, such as ssk1Δ or ssk2/22Δ, did not impair Slt2 activation (Figure 8D). New elements of the SHO1 branch of the HOG pathway have been recently described: Opy2, a transmembrane protein that targets Ste50 to the membrane (Wu et al., 2006; Tatebayashi et al., 2007) and a couple of mucin-like proteins, Hkr1 and Msb2, identified as potential osmosensors in the SHO1 branch (Tatebayashi et al., 2007). To investigate the possible involvement of these proteins in sensing zymolyase stress, Slt2 phosphorylation by zymolyase was characterized in strains lacking these elements. Activation of Slt2 was completely abrogated in the opy2Δ mutant (Figure 8D). Additionally, overphosphorylation of Slt2 was also blocked by simultaneous deletion of HKR1 and MSB2 in a ssk2/22Δ strain (Figure 8D).
DISCUSSION
Yeast MAPK pathways respond to different activating signals developing specific physiological responses, but evidence of interactions between such pathways is emerging. We provide evidence here that the HOG pathway is not only necessary for survival under hyperosmotic conditions but also is involved in adaptation to cell wall stress. Although cell wall interfering compounds like Congo Red, a compound that binds to chitin, and zymolyase (β-1,3 glucanase activity) elicit common genome-wide transcriptional responses related to the set of genes involved in cell remodeling and signaling (García et al., 2004), adaptation responses to these two cell wall stresses are differentially regulated. The response to Congo Red only depends on the CWI pathway, whereas the Sho1 branch of the HOG pathway, in addition to the CWI pathway, is also responsible for remodeling the cell wall in response to specific cell wall damage to the β-1,3 glucan network caused by zymolyase.
The effect of Hog1 on the induction by zymolyase of CRH1, one of the genes induced in the adaptation response to cell wall stress, does not seem to be exerted through the transcriptional factors Sko1, Msn2/Msn4, Hot1, and Smp1, all of them regulated by this MAPK. In contrast, the induction of CRH1 by zymolyase is fully mediated by Rlm1, the transcription factor of the CWI pathway. The phosphorylation of Hog1 under osmotic stress leads to a rapid concentration of Hog1 in the nucleus, whereas in the absence of stress Hog1 appears evenly distributed between the cytosol and the nucleus (Ferrigno et al., 1998). Although Hog1 is phosphorylated under zymolyase stress, the activation of Hog1 under these circumstances does not lead to the accumulation of this MAPK in the nucleus. This would be in agreement with a mechanism different from the transcriptional regulation mediated by the MAPK Hog1. In fact, we demonstrated that a functional Sho1 branch of the HOG pathway is necessary for proper Slt2 activation upon zymolyase stress. Induction of the cell wall integrity pathway by other stresses like the hydrogen peroxide is also reduced in a hog1Δ mutant (Staleva et al., 2004). However, in that case disruption of HOG1 did not abolish the phosphorylation of Slt2, suggesting that the Hog1-mediated regulation of Rlm1 and Slt2 activation by hydrogen peroxide have different upstream regulators.
The cell wall stress conditions studied to date, including Calcofluor White, heat shock, or the presence of β-1,3 glucan inhibitors, have been shown to activate the CWI pathway through the main sensors of this pathway, Mid2 and Wsc1 (Levin, 2005). However, our results suggest that these proteins are apparently not involved in sensing the cell wall damage elicited by zymolyase. Interestingly, the sensors of the CWI pathway are not only dispensable for Slt2 activation in response to zymolyase but also to other stresses such as actin depolymerization (Harrison et al., 2001) or rapamycin treatment (Krause and Gray, 2002). Moreover, activation of Slt2 by zymolyase is not deficient in rom2Δ, rom1Δ, and tus1Δ. The most logic hypothesis concerning the view of a complete lack Slt2 overphosphorylation and CRH1 induction in a sho1Δ strain is that the signal transmitted by zymolyase would be sensed directly through the Sho1 branch, activating the MAPKKK Ste11, the MAPKK Pbs2, and the Hog1 MAPK. Activation of this complex would be necessary for proper Slt2 phosphorylation, on the basis of the existence of a new connection between the CWI and HOG pathways. Elements of the CWI pathway such as the MAPKKs, Mkk1 and Mkk2, the MAPKKK Bck1 and Pkc1 are also required for proper Slt2 activation. It is interesting to note that although the overphosphorylation of Slt2 by zymolyase is completely lost in sho1Δ, ste50Δ, ste20Δ, and ste11Δ mutants, pbs2Δ and hog1Δ mutants still show a slight phosphorylation of this MAPK, suggesting the existence of an additional direct activation branch from Ste11 to the cell wall integrity pathway. In agreement with the importance of this level of regulation for cell survival under these circumstances, mutants deleted in elements of both pathways are hypersensitive to zymolyase (Alonso-Monge et al., 2001 and this work). The phenotype of hypersensitivity to zymolyase is, however, more severe in the pbs2Δ and hog1Δ mutants than in the sho1Δ, ste50Δ, and ste11Δ deleted strains. This could be explained by differences in the cell wall architecture of pbs2Δ and hog1Δ mutants (García-Rodríguez et al., 2000). Indeed, the expression of several genes involved in cell wall biogenesis under basal growth conditions is dependent on Hog1 (O'Rourke and Herskowitz, 2004).
It is worth noting that although both zymolyase and osmostress activate the Hog1 MAPK, only zymolyase is able to activate Slt2. The intracellular signals that couple external stimuli to the MAPK may determine the effects of each stimulus. It has been recently suggested that Cdc37, an Hsp90 chaperone, is involved in the functionality of both MAPKs, Slt2 and Hog1 (Hawle et al., 2007), and therefore it could be a putative candidate. The different kinetics of induction and the low level of induction of Hog1 in response to zymolyase when compared with osmostress (Figure 4 and data not shown) could explain the specificity of the outcome in response to the two stimuli.
Although Hog1 was already known to be activated exclusively by osmotic stress, recent works have demonstrated that this MAPK is also involved in the response to many other stress conditions (Singh, 2000; Winkler et al., 2002; Lawrence et al., 2004; Thorsen et al., 2006). The role of the two branches (Sho1 and Sln1) in the activation of the HOG pathway seems to be different, depending on the input. Both the Sln1 and the Sho1 branches are involved in the phosphorylation of Hog1 under osmostress conditions, although the existence of a certain specificity has been suggested on the basis that moderate osmolyte concentrations seem to activate the Sln1 branch (Maeda et al., 1994), whereas under higher solute concentrations, the Sho1 branch would be mainly involved in the activation of Hog1 (Maeda et al., 1995). Regarding other inputs that lead to the activation of the HOG1 pathway, Hog1 phosphorylation requires Sho1, but not Sln1, under heat stress (Winkler et al., 2002) and zymolyase treatments (this work), whereas phosphorylation by citric acid stress (Lawrence et al., 2004), As (III) (Thorsen et al., 2006), or low temperature (Panadero et al., 2006) depends on the Sln1 branch but not Sho1. The different dependence on Sln1 and Sho1 for HOG1 activation associated to different stimuli suggests some membrane protein specificity to perceive and sense different input signals. Sln1 has been proposed to function as a sensor responding to changes in turgor pressure (Reiser et al., 2003). More recently, the work by Saito and colleagues (Tatebayashi et al., 2007) elucidated the existence of a couple of transmembrane mucin-like proteins, Hkr1 and Msb2, that might directly monitor osmotic changes functioning as putative osmosensors of the SHO1 branch of the HOG pathway. Interestingly, a double mutant strain deleted in HKR1 and MSB2 does not respond to zymolyase, suggesting that these mucins are not only sensors for osmotic changes but also for zymolyase-mediated cell wall stress conditions.
It has been described that complete removal of the cell wall by severe zymolyase treatment gives rise to Hog1 activation in spheroplasts (Reiser et al., 2003). However, in contrast to the mild treatments of zymolyase used in our work, Hog1 activation under those other conditions, in which most of the cell wall is removed, depends on the SLN1 but not the SHO1 branch (Reiser et al., 2003). The different sensitivities and responsiveness of the two branches under these conditions can be envisioned as a mechanism used by the cell to sense different physical stimuli, such as membrane stretching or cell wall stress. An interesting question is that of the nature of the damage caused by zymolyase. What does the SHO1 branch detect that triggers signaling through the HOG pathway? The main activity in the zymolyase mixture corresponds to that of a β-1,3 glucanase, but some protease activity is also present (Zlotnik et al., 1984). Therefore, although it is more likely that transmembrane mucins together with Sho1 would sense damage to the β-1,3 glucan network, the possibility that alterations of cell wall mannoproteins due to protease activity might activate the SHO1 branch of the HOG pathway cannot be ruled out. The characterization of the precise mechanism by which these mucins are able to detect zymolyase-mediated cell wall damage will require further studies.
Our data clearly suggest a model in which different mechanisms are involved in the activation of the MAPK Slt2 under different cell wall stress conditions. In agreement with all these findings, Harrison et al. (2004) have suggested that activation of the yeast cell integrity pathway by different stresses such as heat shock, hypo-osmotic shock, or actin perturbation, rather than operating in a linear “top-down” manner, would provide lateral inputs that impact this regulatory pathway at different levels. Although further work will be necessary to identify new elements of the connection described here between the HOG and SLT2 pathways and to fully understand how cells are able to discriminate between osmotic signals and cell wall damage caused by zymolyase in order to respond in different ways, our results describe novel aspects of the interaction between these MAPK pathways to regulate stress in yeast.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. A. de la Torre (Universitat de Lleida, Spain) for the pkc1 strain and to J. Heinisch (Universität Osnabrük, Germany) and D. Levin (Johns Hopkins School of Public Health, MD) for double wsc1Δ mid2Δ mutants and to H. Saito (University of Tokyo, Japan) for hkr1Δ msb2Δ mutant. We thank all members from the Genomic Unit UCM-PCM for DNA sequencing and Q-PCR experiments. We also thank to the other members of the Research Unit 4 as well as to M. Molina, H. Martín, and V. Cid at the Department of Microbiology and Y. Ohya (University of Tokyo, Japan) for helpful discussions. M. Molina is acknowledged for critical reading of the manuscript. This work was supported by projects BIO2004-06376 and BIO2007-67821 from the MEC (Spanish government) and EU STREP FUNGWALL LSBH-CT-2004-511952 to J.A. and grants from the MEC (Spanish government) and EURYI program to F.P. C.N. is head of the Merck Sharp & Dohme special chair in genomics and proteomics.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0742) on January 9, 2008.
REFERENCES
- Agarwal A. K., Rogers P. D., Baerson S. R., Jacob M. R., Barker K. S., Cleary J. D., Walker L. A., Nagle D. G., Clark A. M. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:34998–35015. doi: 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- Aguilera J., Rodríguez-Vargas S., Prieto J. A. The HOG MAP kinase pathway is required for the induction of methylglyoxal-responsive genes and determines methylglyoxal resistance in Saccharomyces cerevisiae. Mol. Microbiol. 2005;56:228–239. doi: 10.1111/j.1365-2958.2005.04533.x. [DOI] [PubMed] [Google Scholar]
- Alonso-Monge R., Real E., Wojda I., Bebelman J. P., Mager W. H., Siderius M. Hyperosmotic stress response and regulation of cell wall integrity in Saccharomyces cerevisiae share common functional aspects. Mol. Microbiol. 2001;41:717–730. doi: 10.1046/j.1365-2958.2001.02549.x. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Burke D. J., Strathern J. N. New York: John Inglis; 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. [Google Scholar]
- Baetz K., Moffat J., Haynes J., Chang M., Andrews B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 2001;21:6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorsma A., De Nobel H., ter Riet B., Bargmann B., Brul S., Hellingwerf K. J., Klis F. M. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast. 2004;21:413–427. doi: 10.1002/yea.1109. [DOI] [PubMed] [Google Scholar]
- Cabib E., Blanco N., Grau C., Rodríguez-Peña J. M., Arroyo J. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1–6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 2007;63:921–935. doi: 10.1111/j.1365-2958.2006.05565.x. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ruiz M. A., Torres J., Ariño J., Herrero E. Sit4 is required for proper modulation of the biological functions mediated by Pkc1 and the cell integrity pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:33468–33476. doi: 10.1074/jbc.M203515200. [DOI] [PubMed] [Google Scholar]
- de Nadal E., Alepuz P. M., Posas F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002;3:735–740. doi: 10.1093/embo-reports/kvf158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Casadome L., Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell. Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nobel H., Ruiz C., Martín H., Morris W., Brul S., Molina M., Klis F. M. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology. 2000;146(Pt 9):2121–2132. doi: 10.1099/00221287-146-9-2121. [DOI] [PubMed] [Google Scholar]
- Ferrigno P., Posas F., Koepp D., Saito H., Silver P. A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García R., Bermejo C., Grau C., Perez R., Rodríguez-Peña J. M., Francois J., Nombela C., Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- García-Rodríguez L. J., Duran A., Roncero C. Calcofluor antifungal action depends on chitin and a functional high-osmolarity glycerol response (HOG) pathway: evidence for a physiological role of the Saccharomyces cerevisiae HOG pathway under noninducing conditions. J. Bacteriol. 2000;182:2428–2437. doi: 10.1128/jb.182.9.2428-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J. C., Bardes E. S., Ohya Y., Lew D. J. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 2001;3:417–420. doi: 10.1038/35070104. [DOI] [PubMed] [Google Scholar]
- Harrison J. C., Zyla T. R., Bardes E. S., Lew D. J. Stress-specific activation mechanisms for the “cell integrity” MAPK pathway. J. Biol. Chem. 2004;279:2616–2622. doi: 10.1074/jbc.M306110200. [DOI] [PubMed] [Google Scholar]
- Hawle P., Horst D., Bebelman J. P., Yang X. X., Siderius M., van der Vies S. M. Cdc37p is required for stress-induced high-osmolarity glycerol and protein kinase C mitogen-activated protein kinase pathway functionality by interaction with Hog1p and Slt2p (Mpk1p) Eukaryot. Cell. 2007;6:521–532. doi: 10.1128/EC.00343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Sánchez M., Cid V. J., Molina M. Retrophosphorylation of Mkk1 and Mkk2 MAPKKs by the Slt2 MAPK in the yeast cell integrity pathway. J. Biol. Chem. 2007;282:31174–31185. doi: 10.1074/jbc.M706270200. [DOI] [PubMed] [Google Scholar]
- Jung U. S., Levin D. E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- Ketela T., Green R., Bussey H. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 1999;181:3330–3340. doi: 10.1128/jb.181.11.3330-3340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S. A., Gray J. V. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 2002;12:588–593. doi: 10.1016/s0960-9822(02)00760-1. [DOI] [PubMed] [Google Scholar]
- Lagorce A., Hauser N. C., Labourdette D., Rodríguez C., Martín-Yken H., Arroyo J., Hoheisel J. D., Francois J. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:20345–20357. doi: 10.1074/jbc.M211604200. [DOI] [PubMed] [Google Scholar]
- Lawrence C. L., Botting C. H., Antrobus R., Coote P. J. Evidence of a new role for the high-osmolarity glycerol mitogen-activated protein kinase pathway in yeast: regulating adaptation to citric acid stress. Mol. Cell. Biol. 2004;24:3307–3323. doi: 10.1128/MCB.24.8.3307-3323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. N., Elion E. A. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc. Natl. Acad. Sci. USA. 1999;96:12679–12684. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorberg A., Jacoby J. J., Schmitz H. P., Heinisch J. J. The PH domain of the yeast GEF Rom2p serves an essential function in vivo. Mol. Genet. Genomics. 2001;266:505–513. doi: 10.1007/s004380100579. [DOI] [PubMed] [Google Scholar]
- Maeda T., Takekawa M., Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S. M., Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 19; 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Martín H., Rodríguez-Pachón J. M., Ruiz C., Nombela C., Molina M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:1511–1519. doi: 10.1074/jbc.275.2.1511. [DOI] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panadero J., Pallotti C., Rodríguez-Vargas S., Randez-Gil F., Prieto J. A. A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:4638–4645. doi: 10.1074/jbc.M512736200. [DOI] [PubMed] [Google Scholar]
- Popolo L., Gualtieri T., Ragni E. The yeast cell-wall salvage pathway. Med. Mycol. 2001;39(Suppl 1):111–121. [PubMed] [Google Scholar]
- Posas F., Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Witten E. A., Saito H. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1998;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Elion E. A. MAP kinase pathways. J. Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- Rajavel M., Philip B., Buehrer B. M., Errede B., Levin D. E. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:3969–3976. doi: 10.1128/mcb.19.6.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinoso-Martín C., Schuller C., Schuetzer-Muehlbauer M., Kuchler K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell. 2003;2:1200–1210. doi: 10.1128/EC.2.6.1200-1210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V., Raitt D. C., Saito H. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J. Cell Biol. 2003;161:1035–1040. doi: 10.1083/jcb.200301099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Reiser V., Gartner U., Thevelein J. M., Hohmann S., Ammerer G., Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña J. M., Cid V. J., Arroyo J., Nombela C. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 2000;20:3245–3255. doi: 10.1128/mcb.20.9.3245-3255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña J. M., Pérez-Díaz R. M., Alvarez S., Bermejo C., García R., Santiago C., Nombela C., Arroyo J. The ‘yeast cell wall chip,’ a tool to analyse the regulation of cell wall biogenesis in Saccharomyces cerevisiae. Microbiology. 2005;151:2241–2249. doi: 10.1099/mic.0.27989-0. [DOI] [PubMed] [Google Scholar]
- Roncero C., Duran A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K. K. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 2000;29:1043–1050. doi: 10.1016/s0891-5849(00)00432-9. [DOI] [PubMed] [Google Scholar]
- Staleva L., Hall A., Orlow S. J. Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent manner. Mol. Biol. Cell. 2004;15:5574–5582. doi: 10.1091/mbc.E04-02-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straede A., Heinisch J. J. Functional analyses of the extra- and intracellular domains of the yeast cell wall integrity sensors Mid2 and Wsc1. FEBS Lett. 2007;581:4495–4500. doi: 10.1016/j.febslet.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Tatebayashi K., Tanaka K., Yang H. Y., Katsuyoshi Y., Matsushita Y., Tomida T., Imai M., Saito H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHOi branch of yeast HOG pathway. EMBO J. 2007;26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen M., et al. The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Mol. Biol. Cell. 2006;17:4400–4410. doi: 10.1091/mbc.E06-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verna J., Lodder A., Lee K., Vagts A., Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Takaesu G., Hagiwara M., Irie K., Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall P. J., Ballon D. R., Thorner J. When the stress of your environment makes you go HOG wild. Science. 2004;306:1511–1512. doi: 10.1126/science.1104879. [DOI] [PubMed] [Google Scholar]
- Winkler A., Arkind C., Mattison C. P., Burkholder A., Knoche K., Ota I. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell. 2002;1:163–173. doi: 10.1128/EC.1.2.163-173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Jansen G., Zhang J., Thomas D. Y., Whiteway M. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 2006;20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik H., Fernandez M. P., Bowers B., Cabib E. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J. Bacteriol. 1984;159:1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.