Abstract
We have previously demonstrated that the C-terminal part of Rpn11, a deubiquitinating enzyme in the lid of the proteasome, is essential for maintaining a correct cell cycle and normal mitochondrial morphology and function. The two roles are apparently unlinked as the mitochondrial role is mapped to the Carboxy-terminus, whereas the catalytic deubiquitinating activity is found within the N-terminal region. The mitochondrial defects are observed in rpn11-m1 (originally termed mpr1-1), a mutation that generates Rpn11 lacking the last 31 amino acids. No mitochondrial phenotypes are recorded for mutations in the MPN+/JAMM motif. In the present study, we investigated the participation of the last 31 amino acids of the Rpn11 protein by analysis of intragenic revertants and site-specific mutants. We identified a putative α-helix necessary for the maintenance of a correct cell cycle and determined that a very short region at the C-terminus of Rpn11 is essential for the maintenance of tubular mitochondrial morphology. Furthermore, we show that expression of the C-terminal part of Rpn11 is able to complement in trans all of the rpn11-m1 mitochondrial phenotypes. Finally, we investigate the mechanisms by which Rpn11 controls the mitochondrial shape and show that Rpn11 may regulate the mitochondrial fission and tubulation processes.
INTRODUCTION
Recently, important research has focused on the relevant differences in the shape and organization of the mitochondrial tubular system at different moments of the cell cycle and in connection with mitochondrial heredity (Boldogh et al., 2005; Okamoto and Shaw, 2005). The mitochondrion, an essential organelle of eukaryotic cells, plays key roles in energy metabolism, generation of anabolic intermediates, thermogenesis, calcium signaling, and apoptosis (Butow and Avadhani, 2004; Danial and Korsmeyer, 2004). In Saccharomyces cerevisiae, during the cell cycle, the mitochondrion is the first organelle to enter the bud (Warren and Wickner, 1996), and its transport is ensured by cytoskeleton related proteins (Simon et al., 1997; Boldogh et al., 2001). An unexpectedly high number of proteins is required for the inheritance and the maintenance of mitochondrial morphology and many mitochondrial proteins, which are components of the fusion, fission, and tubulation apparatus, have been identified (Shaw and Nunnari, 2002; Mozdy and Shaw, 2003; Detmer and Chan, 2007; Hoppins et al., 2007). Although mitochondria respond to environmental changes and metabolic demands, the regulation of the mitochondrial morphology during metabolic changes, cell cycle or other conditions is still elusive. The ubiquitin-proteasome pathway is one of the most potent and pervasive cellular regulators, directly influencing the levels of key proteins in numerous biological pathways or cellular compartments. Yet, so far, only anecdotic evidence points to the ubiquitin-proteasome pathway controlling some aspects of mitochondrial morphology. Fisk and Yaffe (1999) demonstrated that a mutated form of a ubiquitin-ligase produces mitochondrial aggregation. The F-box protein Mdm30 regulates the level of the mitofusin Fzo1, though involvement of ubiquitin or the proteasome was not documented (Fritz et al., 2003; Escobar-Henriques et al., 2006). In cells arrested with α-factor pheromone, Fzo1 was found to be down-regulated by ubiquitin-mediated proteasome-dependent degradation, though in this case, the mechanism of recognition has not been elucidated (Neutzner and Youle, 2005). A systematic screen of a yeast strain has identified 119 essential genes as required for the maintenance of mitochondrial morphology: among them, genes of the ubiquitin-proteasome pathway have been identified (Altmann and Westermann, 2005). An integral mitochondrial E3 was shown to promote ubiquitylation and proteasomal degradation of components of the mitochondrial fission apparatus in mammalian cells (Nakamura et al., 2006; Yonashiro et al., 2006; Karbowski et al., 2007). Together, these results point to a connection between the ubiquitin-proteasome system and mitochondria.
Participation of the proteasomal lid subunit Rpn11 in mitochondrial biogenesis and structure had been demonstrated by the study of the rpn11-m1, which has a frameshift in position 276, resulting in a truncated protein lacking the last 31 amino acids. This region was found to be essential for maintaining a correct cell cycle and normal mitochondrial morphology and physiology (Rinaldi et al., 1998, 2004). Here we investigate in detail the function of these last 31 amino acids of the Rpn11 protein by the analysis of intragenic revertants, mutagenesis, site-specific mutants, and complementation of the rpn11-m1 defects. Finally, we investigate the mechanisms by which Rpn11 controls the mitochondrial shape and show that Rpn11 may regulate the mitochondrial fission and tubulation processes.
MATERIALS AND METHODS
Strains and Plasmids
The strains used in this study were derived from S288C. The experiments were also done using the W303 genetic context, and the same results were obtained. The S288C derivative BY4743 strain (MATa/MATa; his3D1/his3D1; leu2D0/leu2D0; met15D0/MET15; LYS2/lys2D0; ura3D0/ura3D0; RPN11::kanMX4/RPN11; Brachmann et al., 1998) was sporulated to obtain an haploid strain in which the centromeric plasmid pYC-RPN11 or the plasmids with the rpn11 mutations complement the lethal phenotype of the chromosomal knockout of RPN11. Haploid deletion strains Δdnm1, Δmdv1, Δfis1, Δmmm2, and Δfzo1 are isogenic to BY4741 and were obtained from Open Biosystems (Huntsville, AL). The double mutants used in this study were generated by mating of haploid strains, sporulation, and tetrad dissection. Genotypes of haploid progeny were determined by phenotype and PCR.
Construction of RPN11-HA and rpn11-m1-HA Strains
The triple HA-KanMX cassette was generated by PCR using the plasmid pFA6a-3HA-KanMX6 (Longtine et al., 1998) as template, and oligonucleotides RPN11: 5-GTTTCTGTGCTGACGGCGGGTGTTAATTCAGTGGCAATTAAAcggatccccgggttaattaa-3); rpn11-m1: 5-GGTAGGCAAGATGCAAAAGAAGCACCTTTCCGAAACAGCAGAcggatccccgggttaat taa-3 (upper case letters mark the nucleotides that were homologous to the C-terminus of RPN11/rpn11-m1, 42 base pairs upstream of the stop codon; lower case letters indicate the nucleotides homologous to the plasmid pFA6a-3HA-KanMX6) and L11: 5-GAATTTTTAGAAAAATACAATATAATATATATGTAGTGGGCTgaattcgagctcgtttaaac-3 (upper case letters mark the nucleotides that were homologous to a region 40 base pairs downstream of the RPN11 open reading frame [ORF]; the lower case letters indicate the nucleotides homologous to the plasmid pFA6a-3HA-KanMX6). The resulting integration cassette was transformed into WT and rpn11-m1 strains, and correct integration was verified by PCR using external oligonucleotides.
Construction of DNM1-Green Fluorescent Protein Strains
The green fluorescent protein cassette¤ GFP(S65T)-TRP1, was generated by PCR on plasmid pFA6a-GFP(S65T)-TRP1 (Longtine et al., 1998) as template, using oligonucleotides DNM1-1: 5-AGTTTATAAAAAGGCTGCAACCCTTATTAGTAATATTCTGcggatccccgggttaat taa-3, (upper case letters mark the nucleotides that were homologous to the C-terminus of DNM1, 40 base pairs upstream of the stop codon; lower case letters indicate the nucleotides homologous to the corresponding plasmid) and DNM1-2 (5-GGAATAAACACTGACCTATAATCACGCCCGCAATGTTGAAgaattcgagctcgttta aac-3(upper case letters mark the nucleotides that were homologous to a region 30 base pairs downstream of the DNM1 ORF; the lower case letters indicate the nucleotides homologous to the corresponding plasmid). The resulting integration cassette was transformed into WT and rpn11-m1 strains, and correct integration was verified by PCR using external oligonucleotides.
Yeast Culture Media
YPD (1% bactopeptone, 1% yeast extract, and 2% glucose), YPG (1% bactopeptone, 1% yeast extract, and 2% glycerol), YPGal (1% bactopeptone, 1% yeast extract, and 2% galactose) were used as rich media. WO (0.17% yeast nitrogen base, 0.5% ammonium sulfate, and 2% glucose) was used as minimal medium. All media were supplemented with 2.3% bacto agar (Difco, Detroit, MI) for solid media, and WO was supplemented with the appropriate nutritional requirements according to the strains. Yeast cultures were grown at 26°C, if not indicated otherwise. For phenotypic test, 10-fold serial dilutions of each culture were plated onto YPD (Glucose) and YPG (Glycerol) and then incubated at different temperatures (26, 34, and 36°C). Pictures were taken after 2 d (glucose) and 3 d (glycerol) of incubation.
Plasmid Constructions
Rpn11 C-Terminal part was amplified by PCR from genomic DNA of the wild-type W303 or rpn11-m1 strains using the following oligonucleotides: 5′- CGGGATCCATTGATTATCATAAAACCGCG-3′ and 5′- CGGAATTCTGCATAATGACTTTATAAAATTTG-3′. The resulting DNA fragments were digested with BamHI and EcoRI restriction enzymes and ligated into the corresponding sites in BFG1 plasmid to generate BFG1 Cter-Rpn11 and BFG1 Cter-rpn11-m1, respectively, and sequenced.
All the constructions used for the pulldown assays were made as follows: plasmid pGEX-2T was digested by BamHI and EcoRI and ligated to PCR DNA fragments encoding C-ter-Rpn11, C-ter-rpn11-m1, and Rpn8, digested by the same enzymes (sequences of the oligonucleotides are available on request). Plasmids encoding GFP targeted to the mitochondrial matrix were kindly given by B. Westermann of the University of Bayreuth, Germany (Westermann and Neupert, 2000).
Isolation of Genetic Suppressors of the rpn11-m1 Strain
Spontaneous revertants were selected from strains carrying the rpn11-m1 allele: yeast cells were grown to late logarithmic phase, plated on glucose or glycerol media, and incubated at 36°C. The UV-induced revertants were UV-irradiated with 90 Joule/m2. The irradiated plates were incubated in the dark for 5 d at 28°C and then replica-plated both on glucose and on glycerol medium. Among 2 × 106 UV-mutagenized cells, a total of 21 revertants able to grow at 36°C on glucose or glycerol medium were selected. The revertants were crossed with the isogenic wild-type strain, and spore analysis was performed. In all the revertants that did not show the segregation of the suppression phenotype, the RPN11 locus was sequenced. In this article only the intragenic revertants are described.
Site-specific mutagenesis was performed with the Quik Change Site-directed Mutagenesis Kit (Stratagene, Agilent Technologies, Santa Clara, CA). Sequences of the oligonucleotides are available upon request.
Petites
Cells were grown at the density of 2 × 107 cells/ml on YPD medium and counted, and 200 cells were plated on YPD plates (three plates for each strain). After 3 d at 24°C, plates were replica plated on YPD and YPG. Colonies were counted to calculate vitality and petites production, respectively. Colonies grown on glucose but not on glycerol are petites.
Microscopy and Image Treatment
Cells were grown at 28°C to midlog phase in synthetic or complete media. DAPI was added to the final concentration of 2.5 μg/ml. Cells were further incubated at 28°C for 30 min and washed with 1× PBS. Finally, cells were resuspended in 1× PBS and analyzed by fluorescence microscopy. For in vivo localization studies, cells were grown to midlog phase in complete medium (YPG), transferred to slides, and immediately examined with a DMIRE2 microscope (Leica, Deerfield, IL). Filters for GFP (450/490-nm excitation and 500/550-nm emission), and DAPI (340/380-nm excitation and 450/490-nm emission) were used. Images were captured by a CCD camera (Roper Scientific, Tucson, AZ). Metamorph software (Universal Imaging, West Chester, PA) was used to deconvolute Z-series and treat the images.
Respiration Measurements
Exogenous respiration of the cells was determined with stationary-growth-phase cells (48 h at 28°C) from YPGal agar plates. Cells were suspended in 10 mM potassium phosphate buffer (pH 7.4) containing 20 mM glucose to give a cell density of 109 cells/ml. Oxygen consumption was measured at room temperature with an Oxytherm oxygraph (Hansatech Instruments, Norfolk, England). Respiration rates are expressed as nanomoles of oxygen consumed per milliliter per minute.
Pulldown Assays
Glutathione S-transferase (GST)-fusion proteins were expressed in Escherichia coli. A single colony of each bacterial clone expressing those constructions was selected, inoculated into 15 ml LB + ampicillin, and grown up overnight at 37°C. From each culture, 0.2 ml was diluted into 200 ml LBA media and incubated with vigorous shaking at 37°C for at least 6 h. Once the correct OD600 (0.6–0.8) had been reached, the culture was removed from the 37°C incubator and transferred to 30°C. The induction of the recombinant protein expression was performed by addition of IPTG to a final concentration of 1 mM. After vigorous shaking overnight (∼12 h), cells were pelleted by centrifugation at 4000 rpm for 20 min at RT. The pellets were resuspended into 15 ml TPG1X (20 mM Tris, pH 8, 100 mM NaCl, 1 mM EDTA, 0.5% NP40, 1 mM 2 β-mercapthoethanol, and protease inhibitor cocktail) and sonicated two times during 50 s. Lysates were spun at 13,000 rpm for10 min at 4°C. Glutathione beads (300 μl; glutathione Sepharose 4B, Boehringer, Indianapolis, IN) were incubated with each surpernatant on a roller drum at 4°C for 30 min. After extensive washes with TPG1X, yeast crude extract from different strains (Δrpn11::URA3/pFL39-RPN11, Δrpn11::URA3/pFL39-rpn11-m1, and rpn11-m1 transformed with BFG1 HA-Cter-Rpn11) were incubated with the different beads for 30 min on a roller drum at 4°C. Beads were washed again five times with TPG1X. The bound proteins were solubilized in Laemmli buffer, separated by SDS-PAGE, and analyzed by immunoblotting using anti-hemagglutinin (HA)–specific antibodies (12CA5, BabCO, Richmond, CA).
Mitochondrial Fusion Assay
Mitochondrial fusion during mating was observed as described (Nunnari et al., 1997). Cultures of (a/α) W303 and rpn11-m1 strains transformed with pYEF1mtGFP or pYEF1mtRFP were grown overnight in 2% galactose medium to log phase at 28°C with shaking. Cells were pelleted by centrifugation, washed in YPD, and resuspended in 4 ml of YPD medium (OD600 = 0.1–0.2/ml). Cells were grown 2–3 h on YPD medium, which inhibits further fluorescent protein expression, before mating. Same OD600 units of MATa and MATα cells were mixed and collected by centrifugation. Cells were resuspended in YPD medium and collected on a 0.45-μm pore, 25-mm nitrocellulose filter disk. The nitrocellulose membrane was then incubated on YPD medium at 28°C for 2.5–3 h. Zygotes were examined by fluorescence microscopy.
Miscellaneous
Cells from the RPN11-HA and rpn11-m1-HA strains were grown in YPGal medium (250 ml) to an 600OD = 1. Cellular fractionation and purification of mitochondria on a sucrose gradient were performed as described previously (Rowley et al., 1994). Half of the mitochondria fraction after sucrose cushion was directly TCA precipitated (M1), whereas the other half was further washed with 0.5M NH4Cl and then TCA-precipitated (M2).
RESULTS
Site-directed Mutagenesis of the Carboxyl Terminal Region of Rpn11
Mutagenesis followed by proteasome characterization studies on extragenic revertants (Rinaldi et al., 2002) has demonstrated that the two main phenotypes of the rpn11-m1 mutant, cell cycle arrest and mitochondrial defects, can be separated (Rinaldi et al., 2004). The mitochondrial defects of the rpn11-m1 strain consist of an aberrant mitochondrial morphology even at the permissive temperature, whereas respiration is defective (see below) and the growth on glycerol is absent at temperatures higher than 32°C. For a better understanding of the situation we decided to pinpoint the sequence motif responsible for the rpn11-m1 phenotypes. We first searched for spontaneous and UV-induced intragenic revertants that were able to grow on fermentable or nonfermentable carbon source (glucose or glycerol), at the nonpermissive temperature (36°C). Numerous intragenic revertants were isolated, and 12 of them were sequenced. In all cases, the mutation was found to restore the correct frame downstream Proline 276 and to rescue normal cell cycle and mitochondrial function (some of these revertant sequences are presented in Figure 1A). Only revertant rpn11-RevA5, in which the frameshift occurs seven amino acids downstream to Proline 276, is slightly thermosensitive on glycerol at 36°C (Figure 1A).
Figure 1.
Structure and phenotypes of the mutated Rpn11 proteins studied. (A) Alignment of the C-terminal amino acid sequences of Rpn11 and the rpn11-m1 mutant compared with intragenic revertants obtained from rpn11-m1 strain by spontaneous reversion or UV-induced mutagenesis. The amino acid sequence specific for rpn11-m1 is in blue. In green the amino acid sequence of intragenic revertants. (B) Alignment of C-terminal amino acid sequences of the rpn11 mutants obtained from the RPN11 wild-type gene by site-specific mutagenesis. The rpn11Δcoil has a wild-type Rpn11 C-terminal protein, but the predicted coiled coils domain at position 256–270 was deleted. I292f.sh has a frame shift at position 292 and the resulting new amino acid sequence is indicated in magenta. rpn11-L280stop and rpn11-V293stop have a stop codon before and after the α-helical region, respectively. (C) single site mutants in the Rpn11 protein. Eight amino acids were changed in the α-helical sequence in order to investigate the corresponding mitochondrial phenotype. For each mutant the phenotypes (the growth phenotype, the mitochondrial morphology, and the percentage of vitality and of petites production) are shown. WT, wild-type growth; 36 gly or glu + or −, positive or negative growth on glucose or glycerol at 36°C; MM−, mitochondrial morphology defects; %Vit and %Pet, the percentage of cell vitality and petites production, respectively.
We had previously used site-directed mutagenesis to introduce a deletion at position 256–270, a region corresponding to a coiled coil motif that might be considered important for Rpn11 interactions. A plasmid expressing the mutated version of Rpn11 lacking only this domain (rpn11-Δcoil) was introduced into rpn11-m1 and was found to be able to suppress the pleiotropic phenotypes associated with this mutant (temperature sensitivity and growth on nonfermentable carbon at elevated temperatures, Figure 1B) and even to support growth of a strain lacking the essential RPN11 gene (Δrpn11, data not shown, see Materials and Methods). This indicates that this coiled coil motif is not essential for growth even at high temperature. These results return our focus to the missing sequence in rpn11-m1 (31 amino acids), which alone may account for the mitochondrial and cell cycle phenotypes (Figure 1A).
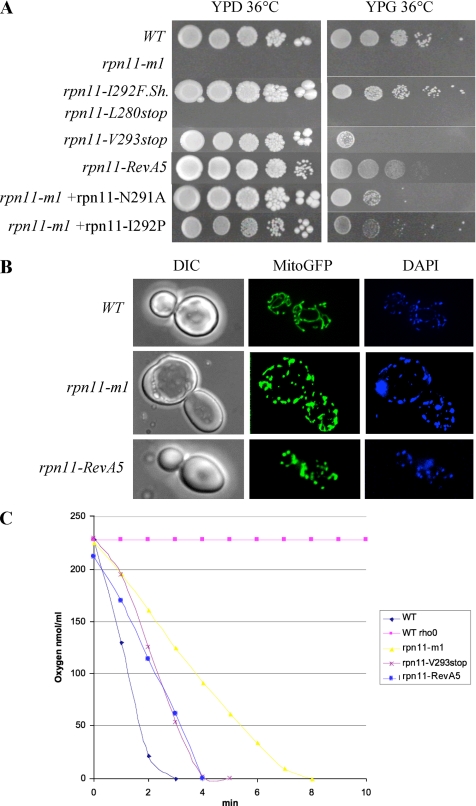
We then concentrated our analysis on the amino acid sequence downstream Proline 276. We identified a hypothetical amphipathic α-helical structure in this region from amino acid 280–293 (Figure 1). To test whether this helix has a functional role, the lack of which could account for the rpn11-m1 phenotypes, we produced a frameshift at the end of this motif at position 292 (Figure 1B; rpn11-I292f.sh). The Δrpn11 strain or the rpn11-m1 strain containing the plasmid encoding this mutant showed a wild-type phenotype (Figure 2A), indicating that this α-helical region is essential for proper function of the Rpn11 protein.
Figure 2.
The α-helical domain in the C-terminal part of Rpn11 is essential in maintaining a correct cell cycle, mitochondrial morphology, and respiration. (A) The growth of the Δrpn11 strain transformed with plasmids containing the rpn11-I292f.sh, rpn11-L280stop, and rpn11-V293stop, together with rpn11-m1 mutant transformed with N291A and I292P point mutations, is compared with the wild-type, rpn11-m1-1, and rpn11-RevA5 growth. Cultures containing identical number of cells were spotted at 10-fold dilutions on YPD or YPG medium. Plates were incubated at 36°C for 3 d. (B) Wild-type (W303), rpn11-m,1 and mpr1-RevA5 strains expressing mtGFP were grown to log phase in glucose-containing medium (YPD) and examined by fluorescence (middle), phase contrast (left), and DAPI staining (right) microscopy. (C) The wt and wt [rho°] strains, the rpn11-m1 mutant, and the mpr1-RevA5 and the rpn11-V293stop mutants were grown on galactose medium (YPGal) 2 d at 28°C, and oxygen consumption was followed in the oxygraph. The WT strain shows a normal respiration rate expressed as nanomoles of oxygen consumed per milliliter per minute. The rpn11-m1 mutant shows a lower respiration rate, whereas mpr1-RevA5 and the rpn11-V293stop mutants are intermediate. The wt [rho°] strain is used as a control.
We then produced two new truncated forms of the Rpn11 protein at either end of this α-helix (rpn11-L280stop and rpn11-V293stop). The Δrpn11 strain transformed with plasmids containing the rpn11-V293stop mutation, which retains the α-helical domain, or the rpn11-L280stop mutation, completely lacking the α-helical sequence, resulted in different phenotypes. Transformation with rpn11-L280stop produced a thermosensitive phenotype both on glucose- and glycerol-containing media at 36°C (Figure 2A). On the other hand, the plasmid rpn11-V293stop showed in the same context wild-type growth but was still heat sensitive on glycerol at 36°C (Figure 2A), pointing to some mitochondrial defects in this strain. The same phenotypes as the ones described for the Δrpn11 strain containing these plasmids were observed for the rpn11-m1 strain transformed with these constructions (data not shown). These results indicate that the α-helical domain is essential in maintaining the correct cell cycle and growth at elevated temperatures. Yet, the integrity of this α-helical domain alone is insufficient to maintain growth on the respiratory-restrictive medium glycerol at elevated temperatures pointing to some mitochondrial defects.
The phenotypes observed in the two truncated forms of Rpn11 (rpn11-L280stop and rpn11-V293stop) confirm that the cell cycle and mitochondrial defects can be separated. We suggest that the rpn11-m1 phenotypes reflect two independent functions of Rpn11. To pinpoint the source of the mitochondrial phenotype, we produced single amino acid substitutions in this α-helical domain at the following positions: L280A, E282A, A284P, D285A, L288P, E289K, N291A, I292P. Plasmids bearing the above rpn11 mutations were transformed into Δrpn11 and growth on glucose or glycerol was examined at 24 and 36°C. Growth was observed at all temperatures, but slightly slower growth on glycerol at 36°C could be observed for all the mutants tested (with the exception of rpn11-L288P; data not shown). These results indicate that all these mutants have a correct cell cycle and are able to utilize glycerol as sole carbon source. When we checked growth of rpn11-m1 mutant transformed with plasmids expressing the rpn11 point mutations we uncovered an interesting observation. Whereas L280A, E282A, A284P, D285A, L288P, and E289K mutated constructs of Rpn11 all completely suppressed the rpn11-m1 phenotypes, transformations with the plasmids encoding for N291A or I292P substitutions did not. The latter strains were unable to efficiently utilize glycerol (Figure 2A).
The incomplete suppression of rpn11-m1 as compared with restoration of normal growth of the Δrpn11 null strain, can be explained by the simultaneous presence of two populations of proteasomes, one bearing the rpn11-N291A or I292P substitution coexisting with proteasomes bearing the rpn11-m1 proteins. The abnormal tail in the rpn11-m1 protein may cause a dominant-negative effect on mitochondria and thus override the corrective effect of the single site substitution population. This is consistent with the heat sensitivity of glycerol growth observed for strains expressing Rpn11 proteins truncated immediately after the position 292 (rpn11-V293Stop). These results point to a sequence around position 290–292 as important for respiration.
Mitochondrial Alterations of the rpn11 Mutants
Mitochondrial Morphology
We next undertook to examine the mitochondrial morphology of all the mutant strains unable to grow on glycerol at 36°C (rpn11-RevA5, rpn11-L280stop, rpn11-V293stop). When grown on glucose at 28°C, all of these strains contained small round mitochondria, an example of such mitochondrial morphology defect is presented in Figure 2B. We then examined the mitochondrial morphology of all the mutants of the putative α-helical structure. In only two of them: rpn11-N291A and rpn11-I292P, fragmented mitochondrial morphology was observed, indicating that these two amino acids are important in maintaining a correct mitochondrial structure. These results point to sequences around positions 290–292 and 276–282 as essential for the mitochondrial structure.
Oxygen Consumption of the rpn11 Mutants
In several physiological conditions and when fission events predominate on fusion (see later) the tubular mitochondrial network is disrupted, resulting in fragmented, punctuate or round mitochondria. While, by now, hundreds of genes have been demonstrated to influence the maintenance of the tubular mitochondrial network (Altmann and Westermann, 2005), the physiological meaning of the fragmented versus tubular mitochondrial network in terms of respiration is still not clear. To link altered mitochondrial morphology in the studied mutants with their physiological state, we measured O2 consumption in rpn11-m1, rpn11-RevA5 and rpn11-V293stop mutants using an oxygraph on live cells at room temperature (Figure 2C). The rpn11-m1 mutant shows a slow rate of oxygen consumption when compared with the rpn11-V293stop and rpn11-RevA5 strains, which themselves are between wild-type and rpn11-m1. This result associates a physiological alteration in respiration to an altered mitochondrial morphology in these mutant strains.
Petites Production in the rpn11 Mutants
Defective mitochondrial morphology is often correlated with a “petite” production phenotype (Hermann and Shaw, 1998). However, an altered mitochondrial network is not an obligatory prerequisite for a higher petites production. Petites are actually completely unable to respire and hence are unable to grow on respiratory mandatory media (glycerol), either due to complete absence of mitochondrial DNA (rho°) or due to partial mitochondrial DNA deletions (rho−). Therefore, we quantified the level of petites production in the following rpn11 mutations: L280stop, V293stop, I292f.sh, N291A, and I292P, as well as wild-type and rpn11-m1 for comparison (Figure 1). Despite the altered mitochondrial morphology, the rpn11-m1 mutant (like rpn11-RevA5) has a stable mitochondrial genome (3% petites). In contrast, rpn11-V293stop, rpn11-N291A or rpn11-I292P show a rate of petites production between 20 and 30%. But the most surprising phenotype is the very high rate of petites production in the rpn11-L280stop mutant: more than 80% of petites. Cells from this strain, which didn't grow on glycerol, were found to lack mitochondrial DNA by DAPI staining, indicating that the petites were in fact rho°. The same mutant in the rpn11-m1 background only produced 3% of petites, just like the wild-type strain. This result suggests that the presence of the mutated rpn11-m1 protein can suppress the petite phenotype of the rpn11-L280stop mutation. This surprising result is presently under investigation.
Effect of the Addition of an Isolated C-Terminal Domain of Rpn11
Suppression of the rpn11-m1 Phenotypes
Because the C-terminal domain of Rpn11 seems to play a major role in the mitochondrial morphogenesis and function, we investigated whether this region is sufficient to carry out a role independent of the rest of the protein by expressing it in trans. The wild-type and the rpn11-m1 strains were each transformed with a plasmid encoding the last 100 amino acids of Rpn11 or the last 79 amino acids of the mutated rpn11-m1 protein, tagged at their N-terminus with two HA epitopes. Growth on glucose or glycerol was examined at three different temperatures. No difference in growth rate on either medium was observed upon overexpression of these truncated C-terminal domains in the wild-type background (Figure 3A). In contrast, the isolated wild-type C-terminal domain of Rpn11 was completely able to restore growth of the rpn11-m1 strain at elevated temperatures on glucose and glycerol. Thus, results identify the C-terminal domain of Rpn11 as a functional domain involved in both cell cycle and mitochondrial function. This result corroborates our earlier observation that the Rpn11 protein plays a crucial nonredundant role in the maintenance of mitochondrial structure and function that is independent of its function as a deubiquitinase within the proteasome.
Figure 3.
Addition of the C-terminal domain of Rpn11 in trans rescues the rpn11-m1 defects. (A) Wild-type and rpn11-m1 cells were transformed with the plasmid BFG1-Cter-Rpn11 or BFG1-Cter-mpr1 encoding respectively the last 100 amino acids of Rpn11 or mpr1. Cells were grown overnight at 28°C in glucose medium, and then 10-fold serial dilutions were spotted onto glucose (YPD) or glycerol (YPG) plates and incubated 2 d for YPD and 3 d for YPG plates at the indicated temperatures. (B) Wild-type and rpn11-m1 strains expressing mtGFP were transformed or not with a plasmid encoding the last 100 amino acids of Rpn11 or 79 amino acids of mpr1 and grown to log phase in glucose-containing medium (YPD) and examined by fluorescence (middle), phase contrast (left), and DAPI staining (right) microscopy. (C) Interaction assays of the C-terminal domain of Rpn11. Interactions between GST fusion proteins and proteins from a yeast total extract were analyzed by immunoblotting by using specific antiserum directed against the HA epitope. (a) Total cell extract from Δrpn11 strain expressing HA-Rpn11 (S) incubated with different purified GST-fusion proteins: (1) GST, (2) GST-Rpn8, (3) GST-CterRpn11, and (4) GST-Ctermpr1. (b) Total cell extract from Δrpn11 strain expressing HA-Rpn11 (S1) or HA-mpr1 (S2) incubated with (1) GST and (2) GST-Rpn8. (c) Total cell extract from W303 strain expressing HA-Cter-Rpn11 (S) incubated with different purified GST-fusion proteins: (1) GST, (2) GST-Rpn8, (3) GST-Cter-Rpn11, and (4) GST-Cter-mpr1. (d) Total cell extract from Δrpn11 strain expressing HA-mpr1 (1) or HA-Cter-Rpn11 (2), mixed and incubated with GST-Rpn8 (#).
Restoration of the Mitochondrial Tubular Network
We then looked at mitochondrial morphology of strains coexpressing Rpn11 or rpn11-m1 and the C-terminal domain of Rpn11. For that purpose, we transformed these strains with a plasmid encoding GFP targeted to the mitochondria to allow visualization of the mitochondrial network. As seen in Figure 3B, the highly fragmented mitochondrial network of the rpn11-m1 strain resumed a tubular structure only when the last 100 amino acids of Rpn11 and not the carboxy terminal of rpn11-m1 were introduced in trans. Overexpression of this wild-type domain did not affect the tubular network of the wild-type cells. Thus, an isolated C-terminal domain of Rpn11 plays an independent role on mitochondrial morphology.
The C-Terminal Domain of Rpn11 Does Not Interact with Rpn11, rpn11-m1, or Rpn8
We have undertaken pulldown experiments in order to test whether the C-terminal domain of the Rpn11 protein can dimerize via its C-terminal domain. Specifically, we tested full-length Rpn11, Rpn8, and rpn11-m1 proteins versus the isolated C-terminal regions of wt Rpn11 or the rpn11-m1 mutation. Previously, Rpn8, another lid subunit, was shown to interact with Rpn11 in a two-hybrid experiment (Fu et al., 2001). Amino acids 1-179 of Rpn11 are sufficient to interact with Rpn8, whereas region 1-187 of Rpn8 is competent to recognize Rpn11. GST-fusion constructs fused to the wild-type or rpn11-m1 C-terminal domains were produced in E. coli and the corresponding proteins were immobilized on glutathione beads. As a control, the full-length Rpn8 protein fused to GST was also purified. Yeast crude extracts expressing HA-tagged Rpn11 (HA-Rpn11) or HA-rpn11-m1 (Figure 3C) were loaded onto immobilized GST-fusions of various Rpn11 or Rpn8 constructs. After extensive washing, the presence of the Rpn11 protein retained on the column was checked by SDS-PAGE gel and immunoblotting. We observed that HA-Rpn11 or HA-rpn11-m1 proteins were retained on immobilized GST-Rpn8, but were not retained on a control of GST alone, GST-C-ter-Rpn11, or GST-C-ter-rpn11-m1 (Figure 3C). These results biochemically confirm that Rpn11 dimerizes with Rpn8 to form a stable complex. This stable heterodimer may serve as the nucleating particle for the eight-subunit lid assembly or may fulfill an independent function (Sharon et al., 2006). To map the region of dimerization, yeast crude extract expressing HA-Cter-Rpn11 was incubated with purified GST, GST-Rpn8, GST-Cter-Rpn11, or GST-Cter-rpn11-m1. The Rpn11 C-terminal domain expressed in yeast was not retained on any of these constructs (Figure 3C). Thus the C-terminal domain of Rpn11 interacts with neither Rpn8 nor with itself.
Next, we wondered whether the Rpn11 C-terminal domain was able to bind a pre-existing complex between rpn11-m1 and Rpn8. For that purpose, GST-Rpn8 fusion was purified on glutathione beads and incubated with a mixed-yeast extracts from a strain expressing HA-rpn11-m1 and a strain producing the HA-Rpn11 C-terminal domain. As seen in Figure 3C, the Rpn11 C-terminal domain was not able to bind the Rpn8/rpn11-m1 complex.
Taken together, these results indicate that Rpn8 is able to interact in vitro with Rpn11 supporting previous observations (Fu et al., 2001). Rpn8 is also able to interact with rpn11-m1, but the Rpn11 C-terminal domain itself is unable to interact with Rpn11, rpn11-m1 or Rpn8, or even with a pre-existing complex rpn11-m1/Rpn8. This important observation suggests that the positive role of the isolated C-terminal region in suppressing mitochondrial defects in trans is probably not due to proteasome incorporation, but stems from an independent function of Rpn11.
Mitochondrial Fusion Still Occurs in the rpn11-m1 Mutant
Fragmentation of mitochondria in the rpn11-m1 mutant suggests a decrease in mitochondrial fusion activity or an increase in mitochondrial fission activity. To distinguish between these two possibilities we used yeast-mating assays for mitochondrial fusion. Mitochondrial fusion and content mixing has been shown to occur in zygotes soon after cell fusion (Azpiroz and Butow, 1993). In mating between wild-type cells, one expressing GFP addressed to mitochondria, and the other produced the RFP targeted to mitochondria, the two fluorescent markers rapidly and completely colocalized in zygotes, indicating that the parental mitochondrial membranes had fused and mitochondrial contents had mixed. Mitochondrial fusion also occurred in rpn11-m1 zygotes, indicating that Rpn11 does not play an essential role in mitochondrial fusion (Figure 4).
Figure 4.
Mitochondrial fusion still occurs in rpn11-m1 mutant. Wild-type cells (A–D) and rpn11-m1 (E–H) cells of opposite mating type were labeled with the mtGFP or mtRFP and mated at 28°C. Deconvolution microscopy was used to score the distribution of mtGFP and RFP. Fusion and mixing of mitochondrial content was evaluated in merged mtGFP and mtRFP (D and H). The zygote bud is marked with an asterisk.
Genetic Interaction of RPN11 with DNM1 and Mitochondrial Localization of the Dnm1 Protein in the rpn11-m1 Mutant
Because the mitochondrial fusion process does not seem to be affected in the mutant, we have investigated the possibility of an exacerbated fission activity in the rpn11-m1 mutant strain. For this purpose, we first investigated the rpn11-m1/Δdnm1 double mutant and found that the double mutation (i.e., the absence of dynamin) altered the mitochondrial fragmentation pattern observed in rpn11-m1, as shown in Figure 5. Eighty-five percent of the double mutant cells presented several elongated mitochondria and collapsed mitochondria-retaining mtDNA nucleoids, nothing like the “net” of interconnected tubules of the Δdnm1 parent. The observed pattern was different from the fragmented mitochondria observed in rpn11-m1, but also different from a Δfzo1/Δdnm1 double mutant, shown as a control in the same figure. This mitochondrial morphology was found for all the fission mutations associated with rpn11-m1 allele. Fragmented mitochondria could only occasionally be seen, and no wild-type-like mitochondrial tubular network was observed. It is important to note that the temperature-sensitive growth on glucose and the respiratory deficiency of the rpn11-m1 mutant was not rescued by deletion of the DNM1 gene.
Figure 5.
Deletion of DNM1 prevents mitochondrial fragmentation of the rpn11-m1 strain. Δdnm1, rpn11-m1, and rpn11-m1/Δdnm1 mutant strains expressing mtGFP were grown to log phase in glucose-containing medium (YPD) and examined by fluorescence (right) and phase-contrast (left) microscopy. Δfzo1 and Δfzo1/Δdnm1 strains are shown as controls.
We then started to look at the Dnm1-GFP mitochondrial localization. DNM1-GFP was integrated at the DNM1 locus in the wild-type and rpn11-m1 mutant strains. As previously shown (Legesse-Miller et al., 2003), we observed Dnm1-GFP in punctuate structures associated with mitochondria. In the rpn11-m1 strain the Dnm1-GFP formed smaller puncta throughout the cytoplasm and colocalized with mitochondria to a lesser extent compared with the wild-type strain (Figure 6). The overall GFP signal did not change between the two genetic contexts, indicating that DNM1-GFP expression was not affected. Taken together, the above result point to an interaction of rpn11-m1 with Dnm1 localization and function, even if we still do not know whether an altered equilibrium between the cytoplasmic and mitochondrial pools of Dnm1 in rpn11-m1 is involved in the aberrant morphology, as a cause or as a consequence.
Figure 6.
The distribution of the Dnm1-GFP protein is different in the rpn11-m1 strain compared with the wild-type strain. The wild-type and rpn11-m1 strains containing a chromosome-tagged DNM1-GFP were grown to log phase in glucose-containing medium (YPD) and examined by fluorescence and phase-contrast microscopy. Mitochondria were visualized using RFP targeted to mitochondria.
The rpn11-m1 Mutation Mimics a Tubulation Defect When Associated with Mitochondrial Fission Mutations
To investigate deeper the rpn11-m1 mitochondrial morphology defect, we turned our attention on the tubulation apparatus. We first constructed double mutants between rpn11-m1 and outer membrane tubulation mutants (Δmdm10, Δmdm12, Δmmm1, and Δmmm2). We reasoned that only the outer membrane proteins would be accessible to the Rpn11 protein. We observed a severe synthetic growth defect on glucose medium at the permissive temperature only when rpn11-m1 was associated with deletion of the MMM2 gene (data not shown). This result prompted us to analyze the effect of the absence of MMM2 in combination with fission mutants. Interestingly, we observed that the double mutants Δfis1/Δmmm2, Δmdv1/Δmmm2, and Δdnm1//Δmmm2 (Figure 7) exhibit a mitochondrial phenotype very similar to that of the rpn11-m1/Δdnm1 (Figure 4) and Δrpn11-m1/Δmdv1 (Figure 7). This result strongly suggests that the Rpn11 protein is involved in the mitochondrial tubulation process in addition to its role in the Dnm1 localization.
Figure 7.
The Rpn11 protein is involved in the mitochondrial tubulation process. The double mutant strains Δfis1/Δmmm2, Δmdv1/Δmmm2, Δdnm1/Δmmm2, and rpn11-m1/Δmdv1 and single mutants expressing mtGFP were grown to log phase in glucose-containing medium (YPD) and examined by fluorescence (right) and phase-contrast (left) microscopy.
A Subpopulation of Rpn11 Is Associated with Mitochondria
We previously reported that Rpn11-GFP encoded in a high copy number plasmid was present in the cytoplasm (Rinaldi et al., 1998). On the other side, we have shown that an overexpressed Rpn11 protein could be stable and found as a subpopulation outside the proteasome (Rinaldi et al., 2004). To test if a subpopulation of Rpn11 was associated with mitochondria, we performed cellular subfractionation of mitochondria of wild-type and rpn11-m1 strains carrying a chromosomal 3XHA epitope-tagged version of Rpn11 or rpn11-m1. Exponentially growing cells were split by differential centrifugation, and mitochondria were purified on sucrose cushions. We first found that the rpn11-m1 protein was largely less expressed compared with the wild-type Rpn11 protein in the same condition (Figure 8). Most of the Rpn11 protein is found in the cytosolic fraction, whereas a very small portion of the protein is associated with mitochondria. In contrast, a larger portion of the rpn11-m1 protein remained associated with mitochondria. These results suggest that a subpopulation of Rpn11 may interact dynamically with mitochondria and that rpn11-m1 is associated more stably than the full-length protein. This localization data support an important function of Rpn11 in the vicinity of the mitochondria involved in mitochondrial dynamics.
Figure 8.
rpn11-m1 and to a lesser extend Rpn11, are found associated with mitochondria. Wild-type RPN11 and rpn11-m1 strains containing a triple HA epitope integrated at the chromosomal locus (Rpn11-HA, rpn11-m1-HA) were fractionated into cytosol (C) and mitochondria. Mitochondria were further purified on a sucrose step gradient (M1) and further washed with NH4Cl 0.5 M (M2). Each fraction was analyzed by immunoblotting. Markers were porin for mitochondria and eRF3 for cytosol.
DISCUSSION
This article details the involvement of the Rpn11 proteasomal lid protein in mitochondrial morphology and function. As previously demonstrated, a proteolytic motif located in the N-terminal portion of this protein is responsible for the deubiquitination function and for the stability of the proteasomal structure (Maytal-Kivity et al., 2002; Verma et al., 2002; Yao and Cohen, 2002). The present data add to our knowledge of Rpn11 in showing that many of the pleiotropic phenotypes of the rpn11-m1 mutant are due to complex interactions of a domain in the carboxy-terminal part of the Rpn11 protein, which may not be due to its role as a deubiquitinase within the proteasome. The current study highlights four points that open the way to new perspectives in the complex mechanisms linking the UPS and mitochondrial biogenesis.
First, the α-helical region within the C-terminal region (amino acids 280-292) of Rpn11, which is lacking in the rpn11-m1 mutant, takes part in several of the pleiotropic functions of the Rpn11 carboxy-terminal domain, including cell growth and mitochondrial morphology.
The essential function of this region is demonstrated by the study of the intragenic revertants: all restore the correct frame after the Proline 276, mutated in the rpn11-m1 strain (Figure 1A), resulting in normal growth and tubular mitochondrial morphology. The only exception is the revertant rpn11-RevA5, for which the correct reading frame is restored after amino acid 282. This mutant has a normal cell cycle but presents a respiratory defect at 28°C (Figure 2C), does not grow on glycerol above 36°C, and has an altered mitochondrial morphology even at 26°C (Figure 2B). Moreover, by site-directed mutagenesis we have identified two amino acids (291 and 292) that upon mutation cause a mitochondrial morphology defect. We conclude that the two short sequences located at the borders of the α-helix (a.a. 276-282 and a.a. 291-292) are essential for maintenance of the mitochondrial morphology and respiration (Figures 1 and 2).
Second, the region downstream Proline 276 has a special importance for mitochondrial DNA retention. Although the rpn11-m1 mutant exhibits a physiological rate of petites production (3%), the rpn11-L280stop mutant has petites production higher than 80% (rho° cells) and mutants in position 291 and 292 have a petites production around 30%. As a whole, these rather surprising characteristics of the studied mutants point to a very delicate interaction of the C-terminal fragment of the Rpn11 protein with substrates involved in mitochondrial fusion, fission, and tubulation processes, with potential consequences on respiration and DNA retention.
Third, the activity of the C-terminal part of Rpn11 can be studied independently when expressed in trans by the use of a plasmid expressing the last 100 amino acids of the wild-type Rpn11 protein. Suppression of the rpn11-m1 phenotypes by the overexpression of the wild-type C-terminal Rpn11 region, lacking the proteasomal interaction domain (MPN domain), could support the hypothesis of a possible role of Rpn11 in maintaining a wild-type mitochondrial morphology in a proteasomal independent pathway. We could not detect any interaction between the C-terminal domain of Rpn11 with itself, with full-length Rpn11 or rpn11-m1, or with its natural proteasomal partner Rpn8 (Figure 3C). These results are in complete agreement with our previous experiments with the chimerical proteins (Rpn8–Rpn11; Rinaldi et al., 2004). We found that the N-terminal part of Rpn11 (responsible for lid protein interactions) was not necessary for the suppression of the mitochondrial morphology defect by the C-terminal part of Rpn11. It is also interesting to notice that Rpn11 has been shown to be stable outside the 26S proteasome (Rinaldi et al., 2004). We must also stress that the paralogue protein of Rpn11 (Csn5), a subunit of the Cop9 signalosome, is stable in a free monomeric form (Kwok et al., 1998). Moreover, an updated interaction map for lid subunits of the proteasome highlights a Rpn5:6:8:9 subcomplex forming the core scaffold of the lid, which only subsequently recruits Rpn11 and the other subunits (Sharon et al., 2006). A “more external” position of Rpn11 could be in agreement with a possible dissociation of Rpn11 from the proteasome and an independent role of Rpn11, further supported by the finding that Rpn11 could be associated with mitochondria (Figure 8).
Fourth, Rpn11, and in particular its C-terminal part, can be considered as one of the many factors required for maintaining the delicate balance between fusion, fission, and tubulation events determining the mitochondrial morphology. This is consistent with the previous observation (Rinaldi et al., 2004) that a subpopulation of stable Rpn11 subunits can exist independently from the proteasome and is supported by the finding that a high proportion of the poorly expressed rpn11-m1 protein copurifies with mitochondria. Now, mitochondrial fragmentation is typically observed in mutants defective in mitochondrial fusion, but here we show that mitochondrial fusion still takes place in the mutant (Figure 4). This is consistent with the fact that rpn11-m1 strain does not loose its mtDNA, a hallmark of mitochondrial fusion mutants. On the other side, we observed a delocalization of the Dnm1 protein in the mutant. Whether Rpn11 directly influences the localization of Dnm1 or other components of the fission process (Fis1, Mdv1, Caf4, and Num1) cannot be concluded as yet.
Most interestingly, we found a further functional relationship between MMM2 and rpn11-m1. Mmm2 has been identified as a protein that participates in the tubulation process (Dimmer et al., 2002; Youngman et al., 2004). The deletion of the MMM2 gene is semisynthetic lethal in combination with rpn11-m1, indicating that these two proteins may interact or act on the same pathway. Moreover, when the fission is defective together with the absence of Mmm2 (Figure 7), the mitochondria resemble those of the rpn11-m1/Δdnm1 (Figure 5). This indicates that Rpn11 may control a step in the tubulation pathway. It has been proposed that the tubulation complex, controlling mitochondrial shape and inheritance, could act upstream of the fusion, and division apparatus as these important complexes are intimately coordinated (Dimmer et al., 2005). In the light of our results we suggest that the effect of Rpn11 is primarilary on tubulation process and that the effect on the delocalization of the Dnm1 might be a consequence of an altered cross-talk between tubulation and fission apparatus. Altogether, our results point to an important role of Rpn11 in the regulation of the mitochondrial dynamic equilibrium. It is interesting to notice that the Mmm2 protein seems to interact transiently with the tubulation machinery (Mmm1, Mdm10, and Mdm12) and that the protein Mmm2 is also located in a yet unknown separate complex in the mitochondrial outer membrane (Youngman et al., 2004).
What is the molecular role of Rpn11 in the vicinity of mitochondria? Up to date, we do not have evidence concerning the direct involvement of the proteasome activity in mitochondrial morphology in vegetative growth. Recently, it was shown that two distinct pathways were responsible for Fzo1 degradation. The canonical proteasomal pathway degrades the Fzo1 protein in response to α-factor pheromone (Neutzner and Youle, 2005), whereas the second pathway seems to be proteasome-independent and involves the F-box protein Mdm30 (Fritz et al., 2003; Altmann and Westermann, 2005, Dürr et al., 2006, Escobar-Henriques et al., 2006). Considering that the deubiquitinating activity of Rpn11 is not necessary for a correct mitochondrial biogenesis (Rinaldi et al., 2004) and considering the diversity of the mitochondrial phenotypes showed by different point mutations in the last 31 amino acids of Rpn11 (mitochondrial morphology, mtDNA stability and respiration), we propose the existence of a complex network of interactions between Rpn11 and different substrates and/or adaptators. The present results also suggest that Rpn11, in addition to its role in the 26S proteasome, plays an important role in the regulation of localization and function of key proteins of the outer mitochondrial membrane.
ACKNOWLEDGMENTS
We thank Dr. B. Westermann for the kind gift of the plasmid PYX-mtGFP. We thank Dr. Monique Bolotin-Fukuhara for continuous support, many discussions, and critical comments on the manuscript. We are grateful to the microscopy station of the “Institut de Génétique et Microbiologie” and to M. H. Cuif and M. Prigent for their useful advice. This work was supported by MIUR 2003, University of Rome “La Sapienza” and Fondo Investimenti Ricerca di Base (code RBNE01KMT9) to L.F. M.G. was supported by grants from the Israel Science foundation (ISF) and the USA-Israel Binational Science Foundation (BSF).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0717) on January 2, 2008.
REFERENCES
- Altmann K., Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R., Butow R. A. Patterns of mitochondrial sorting in yeast zygotes. Mol. Biol. Cell. 1993;4:21–36. doi: 10.1091/mbc.4.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I. R., Yang H. C., Pon L. A. Mitochondrial inheritance in budding yeast. Traffic. 2001;2:368–374. doi: 10.1034/j.1600-0854.2001.002006368.x. [DOI] [PubMed] [Google Scholar]
- Boldogh I. R., Fehrenbacher K. L., Yang H. C., Pon L. A. Mitochondrial movement and inheritance in budding yeast. Gene. 2005;354:28–36. doi: 10.1016/j.gene.2005.03.049. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Butow R. A., Avadhani N. G. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Danial N. N., Korsmeyer S. J. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Detmer S. A., Chan D. C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Dimmer K. S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer K. S., Jakobs S., Vogel F., Altmann K., Westermann B. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J. Cell Biol. 2005;168:103–115. doi: 10.1083/jcb.200410030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr M., Escobar-Henriquez M., Merz S., Geimer S., Langer T., Westermann B. Non-redundant roles of mitochondria-associated F-Box proteins, Mfb1 and Mdm30, in maintenance of mitochondria morphology in yeast. Mol. Biol. Cell. 2006;17:3745–3755. doi: 10.1091/mbc.E06-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M., Westermann B., Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk H. A., Yaffe M. P. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S., Weinbach N., Westermann B. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell. 2003;14:2303–2313. doi: 10.1091/mbc.E02-12-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Reis N., Lee Y., Glickman M. H., Vierstra R. D. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G. J., Shaw J. M. Mitochondrial dynamics in yeast. Annu. Rev. Cell. Dev. Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., Youle R. J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S. F., Solano R., Tsuge T., Chamovitz D. A., Ecker J. R., Matsui M., Deng X. W. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10:1779–1790. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A., Massol R. H., Kirchhausen T. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell. 2003;14:1953–1963. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K., Glickman M. H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;20:3–28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A. D., Shaw J. M. A fuzzy mitochondrial fusion apparatus comes into focus. Nat. Rev. Mol. Cell Biol. 2003;4:468–478. doi: 10.1038/nrm1125. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A., Youle R. J. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall W. F., Straight A., Murray A., Sedat J. W., Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Rinaldi T., Ricci C., Porro D., Bolotin-Fukuhara M., Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol. Biol. Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi T., Ricordy R., Bolotin-Fukuhara M., Frontali L. Mitochondrial effects of the pleiotropic proteasomal mutation mpr1/rpn11, uncoupling from cell cycle defects in extragenic revertants. Gene. 2002;286:43–51. doi: 10.1016/s0378-1119(01)00799-5. [DOI] [PubMed] [Google Scholar]
- Rinaldi T., Pick E., Gambadoro A., Zilli S., Maytal-Kivity V., Frontali L., Glickman M. H. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem. J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley N., Prip-Buus C., Westermann B., Brown C., Schwarz E., Barrell B., Neupert W. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77:249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- Sharon M., Taverner T., Ambroggio X. I., Deshaies R. J., Robinson C. V. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:1314–1323. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. M., Nunnari J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002;12:178–184. doi: 10.1016/s0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V. R., Karmon S. L., Pon L. A. Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil. Cytoskelet. 1997;37:199–210. doi: 10.1002/(SICI)1097-0169(1997)37:3<199::AID-CM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:549–552. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Westermann B., Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Warren G., Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Yao T., Cohen R. E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Youngman M. J., Hobbs A. E., Burgess S. M., Srinivasan M., Jensen R. E. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 2004;164(5):677–88. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R., et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]