Abstract
Human α-synuclein is a small soluble protein abundantly expressed in neurons. It represents the principal constituent of Lewy bodies, the main neuropathological characteristic of Parkinson's disease. The fragment corresponding to the region 61–95 of the protein, originally termed NAC (non-amyloid-β component), has been found in amyloid plaques associated with Alzheimer's disease, and several reports suggest that this region represents the critical determinant of the fibrillation process of α-synuclein. To better understand the aggregation process of α-synuclein and the role exerted by the biological membranes, we studied the structure and the topology of the NAC region in the presence of SDS micelles, as membrane-mimetic environment. To overcome the low solubility of this fragment, we analyzed a recombinant polypeptide corresponding to the sequence 57–102 of α-synuclein, which includes some charged amino acids flanking the NAC region. Three distinct helices are present, separated by two flexible stretches. The first two helices are located closer to the micelle surface, whereas the last one seems to penetrate more deeply into the micelle. On the basis of the structural and topological results presented, a possible pathway for the aggregation process is suggested. The structural information described in this work may help to identify the appropriate target to reduce the formation of pathological α-synuclein aggregation.
Keywords: non-amyloid-βcomponent, α-synuclein, Alzheimer's disease, Parkinson's disease, NMR, micelles
Alzheimer's disease (AD) and Parkinson's disease (PD) are the most common neurodegenerative disorders. An effective cure for these pathologies has not been found, and the existing drugs and therapies offer only marginal and transient symptomatic benefits. However, progress in characterizing insoluble protein aggregates associated with these neurodegenerative diseases has led to new insights that may be relevant to understand the molecular mechanisms of the disorders. A paradigmatic example is the discovery of the protein α-synuclein as the principal constituent of the Lewy bodies that are the cytopathological hallmark of dopaminergic neuronal degeneration in PD (Spillantini et al. 1997; Baba et al. 1998).
Human α-synuclein is a small, 140-residue soluble protein that is abundantly expressed in neurons, where it is localized at presynaptic terminals, particularly in the neocortex, hippocampus, striatum, thalamus, and cerebellum (Nakajo et al. 1994; Iwai et al. 1995a). The α-synuclein sequence is characterized by the presence of an imperfect 11-residue periodicity in the first 89 residues, with a highly conserved hexamer motif (KTKEGV). These repeats are typical of the lipid-binding domain of apolipoproteins (Clayton and George 1998). The peptide corresponding to the sequence 61–95, originally termed NAC (non-amyloid-β component), was observed in amyloid plaques associated with Alzheimer's disease (Ueda et al. 1993). Finally, the C-terminal region (residues 96–140) is rich in acidic residues and prolines.
Structurally, α-synuclein has a random conformation in water under physiological conditions (Weinreb et al. 1996). On the other hand, the first ∼100 residues interact with sodium dodecyl sulfate (SDS) micelles or acidic small unilamellar vesicles (SUV), undergoing a conformational transition to a helical state (Eliezer et al. 2001). The three-dimensional structure of α-synuclein, determined in SDS micelles by NMR, has been described as two curved α-helices (residues 3–37 and 45–92) connected by a well-ordered extended linker (Ulmer et al. 2005). Other conformational models also exist: The N-terminal portion of the molecule associated with SUV or SDS micelles has been described as an extended helix (Ramakrishnan et al. 2003; Jao et al. 2004) or as a noncanonical conformation, the α11/3 helix (Bussell and Eliezer 2003; Bussell et al. 2005). Recently, we suggested that the NAC region of the protein might be partially inserted into the membrane (Bisaglia et al. 2005a).
The mechanisms responsible for Lewy body formation are still poorly understood. Fibril assembly seems to be accompanied by the transition from a random-coil conformation to a β-pleated sheet (Serpell et al. 2000). In the fibrils found in the Lewy bodies, α-synuclein is indeed in a β-sheet structure. Some reports indicate that membranes accelerate the fibrillation of α-synuclein (Lee et al. 2002) and that the aggregation process may occur on the membrane surface (Cole et al. 2002). However, the effect of membranes on α-synuclein aggregation seems to be very sensitive to both relative concentration and membrane composition, making it difficult to predict what the behavior in vivo might be (Zhu et al. 2003; Madine et al. 2004).
It has been proposed that the NAC region of the protein could promote the formation of β-amyloid in vivo (Ueda et al. 1993; Li et al. 2002). Several indications point to this region as the highly amyloidogenic part of the molecule. In aqueous solution, the NAC peptide self-aggregates via a nucleation-dependent kinetic mechanism (Han et al. 1995) and the aggregates possess a distinct fibrillar morphology, as observed by electron microscopy of negatively stained samples (Han et al. 1995; Iwai et al. 1995b; El-Agnaf et al. 1998). As in the case of wild-type α-synuclein, fibril formation is a time-, concentration-, and temperature-dependent process (Iwai et al. 1995b), and metals, such as Zn (II) and Cu (II), accelerate the formation of fibrils (Khan et al. 2005). Moreover, the NAC fragment induces apoptotic cell death in human dopaminergic neuroblastoma SH-SY5Y cells by formation of β-sheet and amyloid-like filaments (El-Agnaf et al. 1998).
To better understand the possible pathway for the aggregation process and the role of membranes in fibril formation, we decided to investigate the micelle-associated structure and the topology of the NAC region in SDS micelles. The approach of using negatively charged micelles to mimic the molecular environment of biological membranes has been successfully applied to many peptides and small proteins (Henry and Sykes 1994; Opella et al. 1994; Sanders and Oxenoid 2000). As structural studies of this fragment are strongly hampered by the low solubility of NAC in water (Weinreb et al. 1996) and by its tendency to aggregate, we decided to analyze a recombinant polypeptide fragment of α-synuclein (αsyn57–102) that includes charged amino acids flanking the NAC region.
Results
Effects of detergents on asyn57–102 secondary structure
α-Synuclein is characterized by the presence of an amphipathic N-terminal region with a net predominance of basic residues over acidic ones. This charge distribution could explain why the protein interacts with acidic synthetic membranes. On the contrary, the αsyn57–102 fragment encompasses the most hydrophobic portion of the protein, with a slight predominance of basic amino acids only at the two termini of the sequence (Fig. 1). Therefore, we decided to analyze here the effects of zwitterionic and acidic micelles on the αsyn57–102 fragment by far-UV circular dichroism spectroscopy.
Figure 1.
Amino acid sequence of αsyn57–102. The NAC fragment, which constitutes the more hydrophobic region of α-synuclein, is in bold. The extra G-S-H-M sequence at the N terminus (see Materials and Methods) is shown in gray. Positively and negatively charged residues are indicated. Nt and Ct indicate the charged N and C termini of αsyn57–102, respectively.
In the presence of DPC, no significant structuring effect can be observed even when the detergent is above its critical micellar concentration (cmc) of ∼1.5 mM (Fig. 2A). Only at significantly higher concentrations of DPC can the formation of a partial helical structure be observed. This behavior suggests the presence of an equilibrium between free and micelle-bound peptides. The free state seems to be largely favored so that only a large amount of detergent partially shifts the equilibrium toward the micelle-bound, ordered form. In these experimental conditions, no β structure is present, as indicated by the well-defined isodichroic point, which reveals an equilibrium between only two conformational states, i.e., a random coil and a helical structure. The results obtained in the presence of SDS are quite different (Fig. 2B). Low concentrations of detergent promote the formation of a β structure in the peptide. At concentrations below its cmc, SDS often promotes β-sheet structure (Zhong and Johnson 1992; Waterhous and Johnson 1994), and a similar behavior was previously observed in our and other laboratories for other nonfibrillogenic peptides (Bairaktari et al. 1990; Kanaori et al. 1997). Above the cmc (∼8 mM), αsyn57–102 specifically interacts with SDS micelles and adopts a helical conformation. From the ellipticity at 222 nm, the helical content can be estimated to be ∼50% (Greenfield and Fasman 1969).
Figure 2.
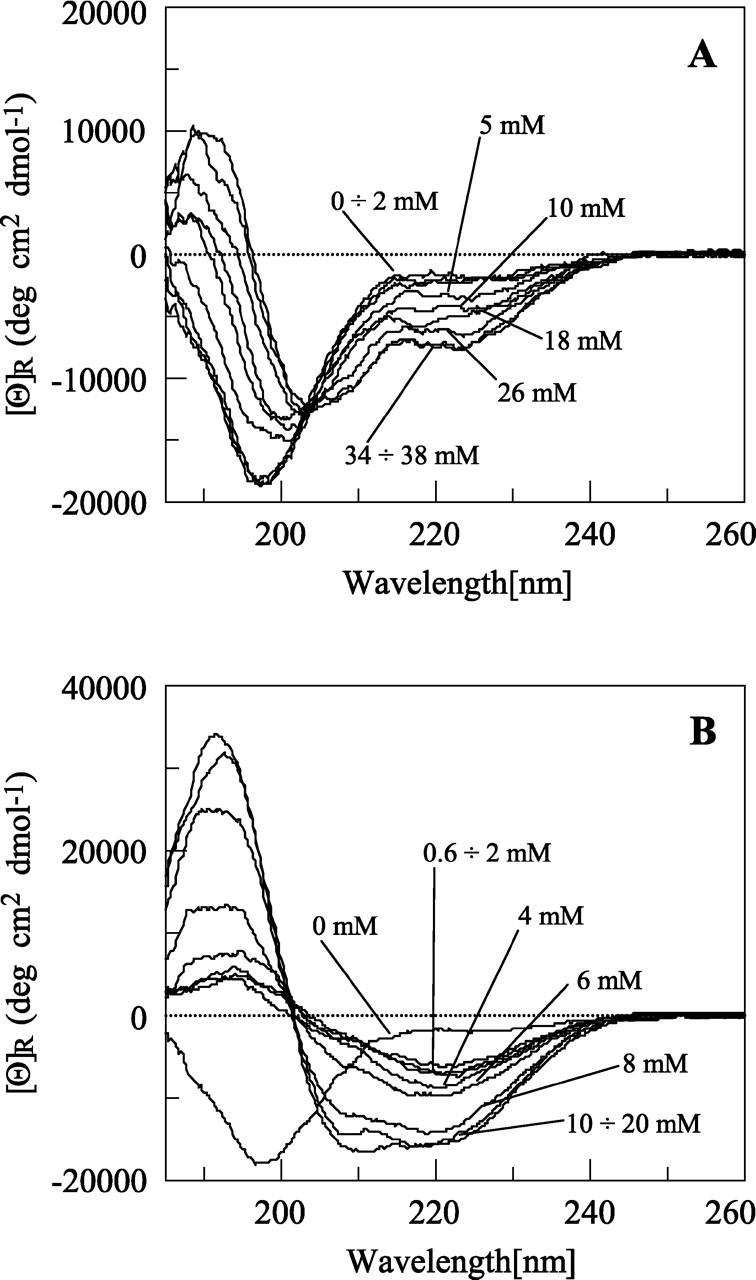
Effects of detergents on αsyn57–102 folding. Far-UV CD spectra of 10 μM αsyn57–102 after the addition of an increasing amount of DPC (A) and an increasing amount of SDS (indicated) (B). The fragment is random in water under physiological conditions and adopts a helical conformation after binding to micelles. The interaction is more pronounced with negatively charged micelles.
Resonance assignment of αsyn57–102
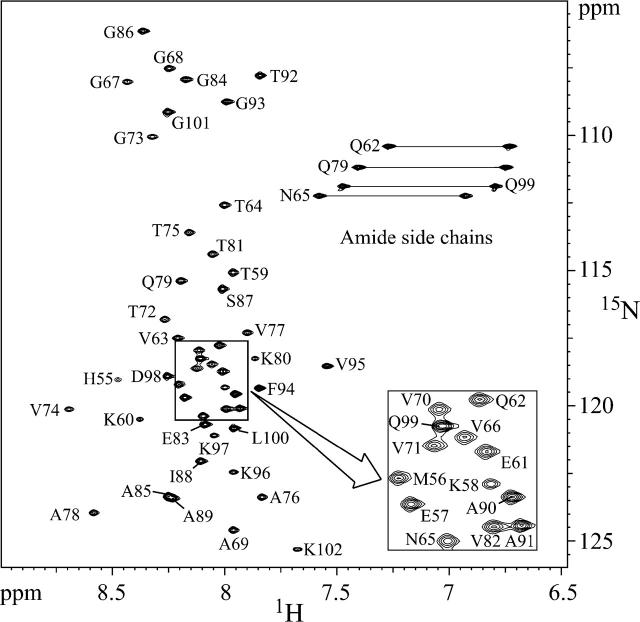
Based on the CD studies, we decided to use SDS micelles as the membrane-mimetic environment. αSyn57–102 was soluble at millimolar concentration in a water/SDS micelle solution and yielded good quality NMR spectra. There was no evidence of gelling or precipitation of the sample over several weeks. One-dimensional (1D) spectra acquired at different times confirmed the sample stability, indicating that SDS micelles provide an environment amenable to detailed structural analysis by high-resolution NMR methods. The SDS concentration in the sample was 125 times higher than that of αsyn57–102. Given that each SDS micelle consists of ∼60–70 molecules (Henry and Sykes 1994), the solution contained an ∼2:1 ratio of micelles to protein molecules, minimizing protein–protein interactions. The severe overlap of several peaks in the amide region of the homonuclear 2D spectra greatly impaired the analysis. In order to improve the peak dispersion, 3D heteronuclear experiments on a 15N-labeled sample were recorded. As shown in Figure 3, all the 15N-HSQC peaks of αsyn57–102 are accounted for in the spectrum. The 1H and NH resonances were assigned using 3D TOCSY-HSQC and NOESY-HSQC experiments.
Figure 3.
Backbone assignment of αsyn57–102. Proton-nitrogen correlation spectrum (HSQC) of 2 mM αsyn57–102 sample in the presence of 250 mM SDS.
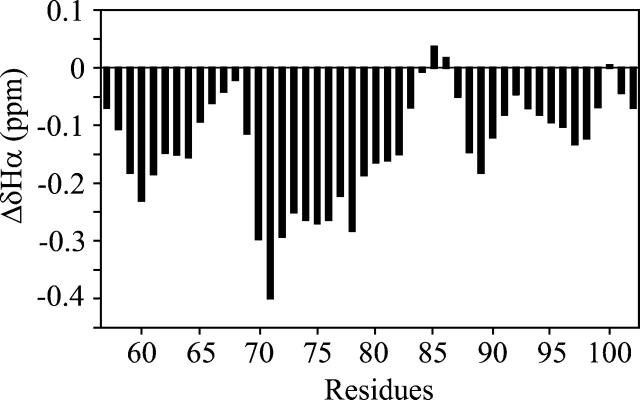
The measured Hα chemical shifts were compared with random coil shifts (Wüthrich 1986), and the deviations are shown in Figure 4. From the analysis of the plot, three regions show a propensity to adopt a helical conformation (residues 58–64, 69–82, and 88–98). The most dramatic deviations are in the central region, while the smaller effect occurs in the C-terminal one, suggesting that the latter adopts a less stable helix.
Figure 4.
Conformational shifts of αsyn57–102 bound to SDS. Plot of the Hα chemical shift deviations from a random-coil conformation observed for αsyn57–102 in SDS as a function of residue number. Each data point was averaged with its nearest neighbors to eliminate local effects (Pastore and Saudek 1990). Negative deviations are typical of helical conformations.
Three-dimensional structures
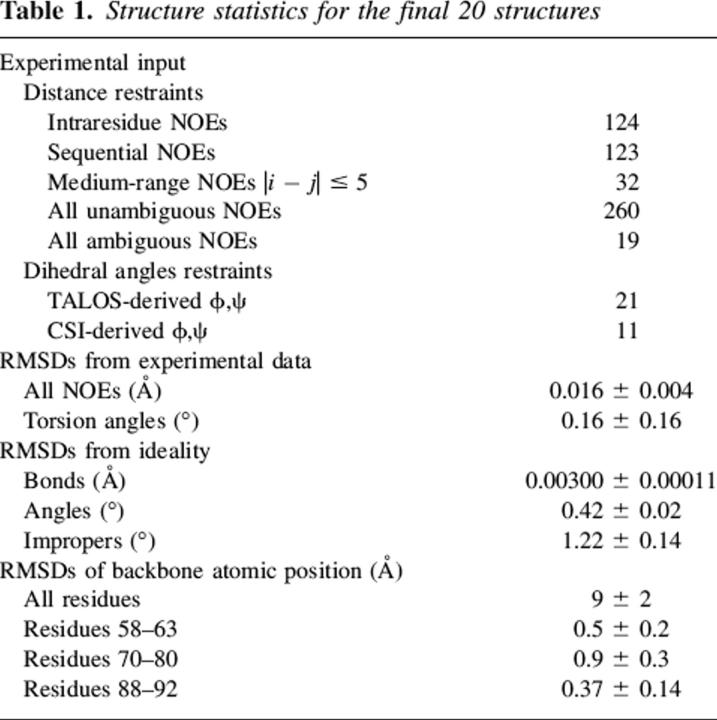
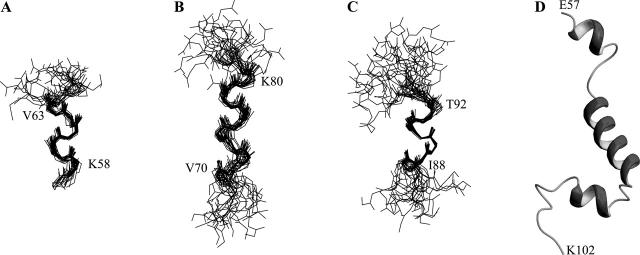
A total of 279 NOE-based distance restraints and 32 pairs of (ϕ,ψ) backbone dihedral angles derived from chemical shift analysis were used in structure calculation (Table 1). The 20 lowest energy structures selected for the final analysis show good agreement with the experimental data even if the convergence of the structures is globally very poor. Over the whole molecule, the RMSD for backbone atoms is very high (9.1 ± 2.2 Å) due to the unstructured portions of the αsyn57–102 fragment. On the final structure set, angular order parameters were calculated for the backbone torsion angles ϕ and ψ of all residues. High values of the order parameter, indicating well-defined torsion angles, were observed in three regions of the molecule, with dihedral angles characteristic of helical conformation: 58–63 (helix-1), 70–80 (helix-2), and 88–92 (helix-3). Backbone superposition of each of these three regions yielded good RMSD values: 0.54 ± 0.23 Å (helix-1), 0.95 ± 0.29 Å (helix-2), and 0.37 ± 0.14 Å (helix-3), indicating well-defined structural elements (Fig. 5). These helical segments are present in the large majority of the structures, although the length of each segment is slightly different in the different structures of the ensemble. From the calculated structures, the fraction of residues in helical conformation was calculated to be 48% ± 8%, in good agreement with CD results. This agreement indicates a high stability of the micelle-bound form and excludes the possibility of an equilibrium with the micelle-free state (Schievano et al. 2003). Sequential and medium-range connectivities expected for helical regions are present (or hidden by spectral overlap) along the mentioned stretches.
Table 1.
Structure statistics for the final 20 structures
Figure 5.
Structure of micelle-bound αsyn57–102. Backbone superposition of 20 minimized structures for α-syn57–102. (A) E57–V66 by superposing residues K58–V63; (B) G67–E83 by superposing residues V70–K80; (C) G84–D98 by superposing residues I88–T92. (D) α-Syn57–102 structure at minimal energy.
The NOE data alone cannot explain whether the poor definition of the two disordered regions between the helices is due to actual flexibility or to lack of sufficient experimental restraints. Nevertheless, these two regions correspond very well with portions of the entire α-synuclein (Ulmer et al. 2005) and of the 1–99 truncated mutant (Bisaglia et al. 2005a) that show increased backbone dynamics by 15N relaxation measurements performed in SDS.
Topological analysis
The positioning of αsyn57–102 with respect to the surface or the interior of the micelles was investigated by NMR, using the effects of two different spin-labeled stearates on the amide resonances in the assigned HSQC spectrum as structural probe. From 13C NMR experiments (Papavoine et al. 1994; Bussell et al. 2005), the nitroxide group of 5-doxyl stearic acid is localized close to the sulfate group of the micelle, while that of 16-doxyl stearic acid is found roughly in the center of the micelle. These paramagnetic probes are able to induce broadening of the NMR signals and a decrease of resonance intensity for residues close to the surface (5-DSA) or deeply buried in the micelle (16-DSA) (Hilty et al. 2004). To interpret the results in a quantitative manner, the percent reduction of the intensity of each amide peak in the presence of spin probes was measured. This method has been widely used (Piserchio et al. 2000; Lindberg et al. 2001; Bisaglia et al. 2005a) and provides reliable answers. The principal advantage of using the reduction of signal intensities instead of the line broadening is the much higher sensitivity.
The results obtained with 1 mM paramagnetic agents (micelle:spin probe ratio of 1) are reported in Figure 6A,B. The peak reductions observed confirm that αsyn57–102 interacts with SDS micelles. The effects are generally more pronounced in the presence of 5-DSA, suggesting that αsyn57–102 is positioned on the surface of the micelle. In both cases, the reduction in peak intensity is more pronounced at the C-terminal extremity of the NAC region (residues 88–96). These results suggest that the first two helices are positioned closer to the micelle surface, whereas the third helix penetrates more deeply into the micelle.
Figure 6.
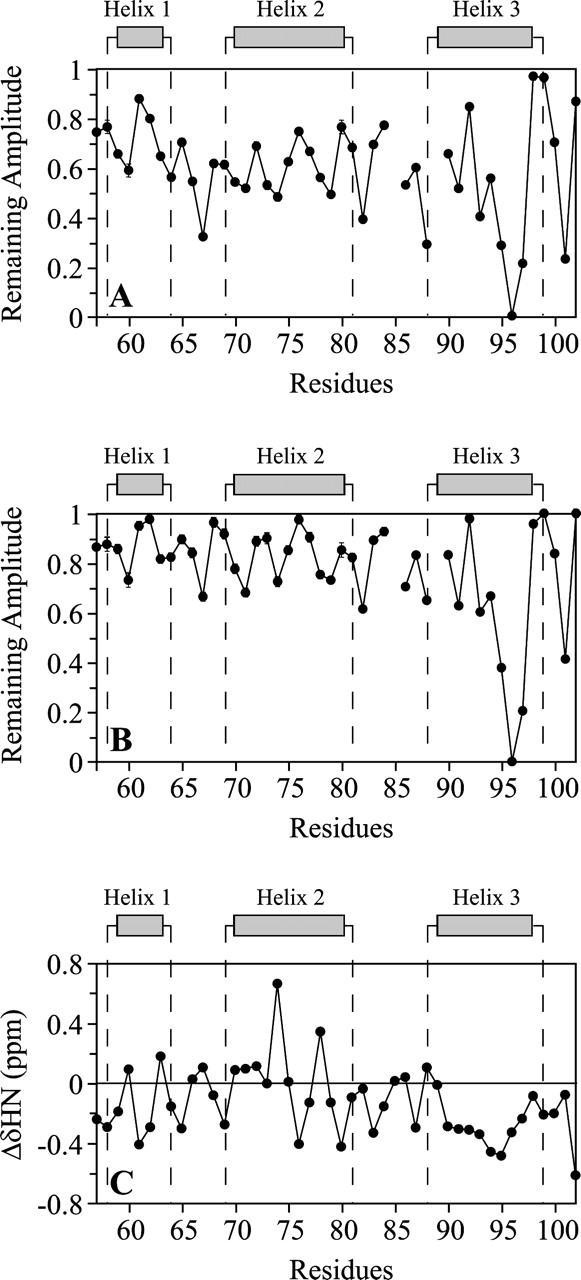
Topological analysis of αsyn57–102 in SDS micelles. (A,B) Plots of the remaining amplitude of the HSQC peaks of 0.3 mM αsyn57–102 in the presence of 50 mM SDS after the addition of 1 mM 5-DSA (A) or 1 mM 16-DSA (B). (C) Plot of the amide proton chemical shift deviations from a random-coil conformation. For bent helices, NH typically displays a 3–4 residue periodicity in chemical shifts.
An analogous picture emerges from the analysis of NH chemical shifts. Statistical analysis of protein secondary structures and amide proton chemical shifts indicated that, on average, the amide protons resonate upfield in α-helices and downfield in β-strands (Kuntz et al. 1991; Wishart et al. 1991). Moreover, the NH chemical shifts can be directly correlated with hydrogen bond lengths and therefore with α-helix bending. For bent α-helices, the NH chemical shifts typically display a three- to four-residue periodicity because the convex and concave sides of the α-helix promote upfield and downfield NH shifts, respectively (Kuntz et al. 1991; Zhou et al. 1992). Such a periodicity is clear in the amino acid regions encompassing the first two helices of αsyn57–102 (Fig. 6C). The micelle curvature can cause bending of these helices if they are positioned on the micelle surface and strongly interact with it. The spin-label analysis supports this interpretation. The residues in the helices that show more pronounced peak reductions in the presence of 5-DSA are located on the concave side, i.e., on the helical region facing the micelle. The behavior of the third purported helix is very different. The amide protons present an up-field deviation typical of a helical structure but without any periodicity. The absence of bending suggests that the third helix is not anchored to the micelle surface, and this result is in line with the spin-label studies that show a more pronounced immersion of this helix in the micelle.
Discussion
The NAC fragment of α-synuclein is the region responsible for the ability of the protein to fibrillate. Even if it constitutes the second major component of the amyloid plaques in AD, no structural study on this fragment has been published to date. This can probably be rationalized with its low solubility in water and with its tendency to aggregate. To overcome these limitations, we produced a recombinant polypeptide containing residues 57–102 of α-synuclein, which includes charged residues flanking the NAC region (Bisaglia et al. 2005b). This construct is more soluble than NAC itself, and it was possible to produce it enriched with 15N. This helped to resolve the peak overlap present in the homonuclear NMR spectra that greatly impaired the structural analysis. There are no significant differences in the HSQC spectra of the fragment and that of the entire α-synuclein (Bisaglia et al. 2005a), except at the two termini of the sequence. This indicates that truncation does not have important structural effects.
The structure of micelle-bound αsyn57–102 consists of three helices separated by two flexible stretches. The first two helices, residues 58–63 and 70–80, are well defined. The last one is slightly disordered and may exist as an ensemble of unfolded and helical structures, with a well-defined core formed by residues 88–92. The analysis of the NH chemical shift deviations and the spin-label experiments indicate that the first two helices are curved and remain close to the micelle surface, whereas the last one penetrates more deeply into the micelle and is not bent.
The structural information described here for αsyn57–102 can be compared to the structural models proposed for the same fragment in the entire α-synuclein. Unfortunately, most of them are at low resolution (Bussell and Eliezer 2003; Chandra et al. 2003; Ramakrishnan et al. 2003; Jao et al. 2004; Bussell et al. 2005). Only one NMR study reported the 3D structure of the protein, determined in SDS micelles, using mainly structural constraints derived from residual dipolar couplings (Ulmer et al. 2005). The investigators described the α-synuclein structure as composed of two curved α-helices (residues 3–37 and 45–92) connected by a short linker in an antiparallel arrangement. Unlike this representation, our results indicate the presence of two well-defined helical breaks at residues 64–69 and 81–87. The identification of these breaks could be ascribed to the reduced number of restraints in our structures. However, several investigators suggested that around residues 65–68 and 83–86, the helix is less stable or more flexible (Bussell and Eliezer 2003; Chandra et al. 2003; Bisaglia et al. 2005a; Ulmer et al. 2005).
The topological analysis presented here is consistent with a model recently proposed for the 1–99 fragment of α-synuclein (Bisaglia et al. 2005a), although the idea of a transmembrane helix, also suggested by the high hydrophobicity of the NAC region, is not supported by the present data. Possibly, insertion into the membrane of a small region of α-synuclein is necessary to anchor the protein more strongly to the membrane in order to exert its biological function.
Because of their similar size, their ability to form fibrils, and their coexistence in the Alzheimer's plaques, we compared the solution structure of αsyn57–102 with that of Aβ peptide, determined in similar experimental conditions (Coles et al. 1998; Shao et al. 1999; Crescenzi et al. 2002). Despite some differences in the topology of the helices, it is interesting to note that both of these natively unstructured peptides adopt a predominantly helical conformation upon binding to the micelles without a well-defined tertiary structure.
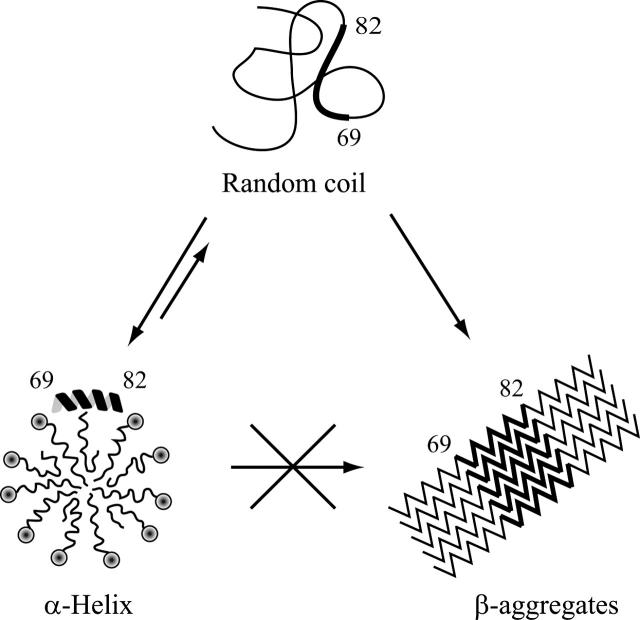
Recent studies have defined the NAC region as responsible for α-synuclein aggregation and β-sheet formation. Region 68–78 of α-synuclein was shown to aggregate to form fibrils and to be neurotoxic, and region 68–76, although not forming fibrils, is the shortest peptide that exhibits neurotoxicity (Bodles et al. 2001). Region 72–84 forms the core of α-synuclein filaments, and a synthetic peptide corresponding to this region self-aggregates to form filaments and promotes fibrillation of full-length human α-synuclein in vitro (Giasson et al. 2001). Very recently, a method to identify the most amyloidogeic regions of a protein, applied to α-synuclein, confirmed a high intrinsic aggregation propensity for the NAC region, with the highest propensity for region 69–79 (Pawar et al. 2005). Interestingly, our results position these residues within the second helical stretch. As already suggested for the Aβ peptide (Coles et al. 1998), the α-syn57–102 stability in micelle/water solution at relatively high concentrations (up to 2 mM) and 35°C seems to indicate that the SDS micelle-bound, helical structure is unable to evolve to the β-structures typical of both protofibrils and fibrils (Fig. 7). Membranes could exert a protective role simply by decreasing the cytosolic concentration of the protein and preventing its aggregation, which is a concentration-dependent process. A more intriguing possibility is that specific residues involved in promoting the aggregation process are prevented from intermolecular interactions because they are inserted into the micelle. The analysis of the NH chemical shift deviations, together with the results obtained using paramagnetic probes, indicates residues V71, V74, A78, and V82 as the ones located on the concave side of the helix, directed toward the interior of the micelle. These residues are the most likely ones to be involved in the fibrillation process.
Figure 7.
Proposed model for the protective role exerted by the membrane on the aggregation process of α-synuclein. In the cytosolic state, the unfolded α-synuclein can undergo a conformational transition to form β-aggregates. The presence of SDS micelles or negatively charged biological lipids organized in micelles or SUVs decreases the cytosolic concentration of the protein and prevents its aggregation. The α-helix to β-structures transition may be hampered because the residues responsible for the aggregation process are protected by the membrane.
Recently, Zweckstetter and coworkers (Bertoncini et al. 2005) have suggested that part of the NAC region and residues at the C terminus can form transient hydrophobic clusters that could protect the natively unfolded α-synuclein from oligomerization. A more specific identification of the amyloidogenic residues could be important in defining targets against aggregation. In the case of AD, the use of peptidomimetics or small molecules to inhibit amyloid aggregation was suggested (Schenk et al. 1995; Ghanta et al. 1996). An N-methylated NAC analog (residues 68–78) was shown to be nonamyloidogenic and to reduce fibril formation (Bodles et al. 2004). The results described here suggest that NAC analogs including up to residue 82 could be considered to improve efficacy.
Materials and methods
Production of αsyn57–102
Histidine-tagged αsyn57–102 was produced as described elsewhere (Bisaglia et al. 2005b). Briefly, overexpression of the protein was achieved by transferring 40 mL of pre-culture in Luria Bertani broth to 1 L of M9 minimal medium (supplemented with 1 g/L 15N ammonium chloride for 15N-labeled protein) and growing cells at 37°C to an OD600 of ∼0.7, followed by induction with 1 mM isopropyl-β-thiogalactopyranoside for 4 h. The protein was purified on a Cobalt-agarose resin (Clontech) using the manufacturer's recommended protocol. To eliminate the His-tag, the purified protein was digested with thrombin (Amersham Pharmacia), using the manufacturer's protocol. After cleavage, the His-tag was separated from the protein through a passage in the cobalt resin, and the protein was lyophilized. The final protein contains an extra G-S-H-M sequence at the N terminus. From the solid powder, the protein was extracted with three 500-μL aliquots of 1,1,1,3,3,3-hexafluoro-2-propanol (Sigma) to desalt the sample; it was again lyophilized and stored frozen.
CD experiments
CD measurements were carried out on a JASCO J-715 spectropolarimeter interfaced with a personal computer. The CD spectra were acquired and processed using the J-700 program for Windows. The experiments were carried out at room temperature using HELLMA quartz cells with Suprasil windows and an optical pathlength of 0.1 cm. All spectra were recorded in the 185- to 260-nm wavelength range, using a bandwidth of 2 nm and a time constant of 4 sec at a scan speed of 50 nm/min. The signal-to-noise ratio was improved by accumulating four scans. Spectra were acquired on 10 μM solutions of αsyn57–102 in the presence of 10 mM phosphate buffer (pH 6.0) and with the addition of increasing amounts of dodecylphosphocholine (DPC) or SDS. The spectra are reported in terms of mean residue molar ellipticity [Θ]R (deg·cm2/dmol). Peptide concentration was determined by measuring the absorbance at 205 nm (Scopes 1974).
NMR assignment
All NMR experiments were carried out at 35°C on a Bruker Avance DMX600 spectrometer equipped with a gradient triple resonance probe. The spectra were processed using the software GIFA (Pons et al. 1996) and analyzed using the program XEASY (Bartels et al. 1995) on a Silicon Graphics workstation. NMR samples used for the assignment of α-syn57–102 contained ∼2 mM protein in H2O:D2O [90:10 (v/v)], 20 mM phosphate buffer (pH 6.0), 250 mM SDS-d25, and 0.05% NaN3. Assignment of protons and amide groups was obtained by means of 3D-TOCSY-HSQC using a DIPSI2 isotropic mixing pulse sequence (Shaka et al. 1988), and 3D-NOESY-HSQC experiments with 70-msec and 100-msec mixing times, respectively. The 3D experiments were acquired with 64 complex data points in the 15N dimension; 128 or 160 points in the 1H indirect dimension; and 512 points in the 1H direct dimension. Spectral widths were 1796 Hz (1H-direct), 1338 Hz (15N), and 6000 Hz (1H-indirect). The frequency offsets were 4800 Hz (1H-direct), 7053.5 Hz (15N), and 2325.8 Hz (1H-indirect). The time domain data were multiplied by a 90° shifted sine function in all dimensions before Fourier transformation.
Structure calculation
Structural NOE data for the αsyn57–102 fragment were extracted from the 3D-NOESY-HSQC. The program ARIA1.2 (Linge et al. 2001), as an extension of CNS1.1, was used to compute the solution structure. ARIA enables the incorporation of ambiguous NOE distance restraints into structure calculation as well as calibration of the NOE distance restraints using a structure-based NOE back-calculation. This approach was chosen in order to deal with the low dispersion of the resonances and consequent severe spectral overlap present in our data. A total of 279 NOE-based distances were introduced in the calculation, 209 of which had been manually assigned, while the remaining ones were automatically assigned during the structure calculation. Dihedral angle restraints were derived using both TALOS (Cornilescu et al. 1999) and CSI (Wishart et al. 1992) analysis of the chemical shifts. The dihedral angle restraints were taken to be ±2 standard deviations or at least ±20° for the average values predicted by TALOS and ±30° for the values predicted by CSI. ARIA runs were performed using the default parameters supplied in the program. The initial conformation for the simulated annealing protocol was extended. In the final iteration, 50 structures were calculated, and the 20 with the lowest energy were selected for the final analysis. The energy of the final structure ensemble was minimized through a short molecular dynamics simulation (Linge et al. 2003). Structures were analyzed with MOLMOL (Koradi et al. 1996).
Paramagnetic relaxation experiments
The HSQC experiments were recorded at 35°C on 0.3 mM αsyn57–102 samples, in 20 mM phosphate buffer (pH 6.0), 50 mM SDS-d25. Two series of paramagnetic relaxation experiments were acquired with a final spin-probe concentration of 0, 1, and 5 mM, respectively. Each titration point was obtained by transferring the sample from the NMR tube into a polypropylene 1.5-mL vial containing the desired amount of 5-doxyl-stearic acid (5-DSA) or 16-doxyl-stearic acid (16-DSA) (Aldrich) and transferring it back into the NMR tube. All recording parameters were kept rigorously constant, the only modification concerning probe tuning and field shimming. The experiments (256 increments of 512 time points each) were acquired with 48 transients each. The spectral widths were 1796 Hz (1H) and 1338 Hz (15N), and the frequency offset 4800 Hz (1H) and 7053.5 Hz (15N). Prior to Fourier transformation, the data were multiplied by a 90° shifted sine function in both dimensions. Baseplane correction was applied before peak integration.
The NH amide peak intensities were measured before and after the addition of the paramagnetic probes and compared. The results are reported in terms of remaining amplitude (RA) defined as RA = A(probe)/A(0), where A(probe) is the amplitude of the peak measured after the addition of the paramagnetic agent and A(0) is the amplitude in the absence of paramagnetic agent. Uncertainties on peak intensity were determined by evaluating the noise level in a peak-free region of the spectra.
Electronic supplemental material
The Supplemental Material contains a table with the 1H and NH resonance assignment of αsyn57–102 and two figures. The first figure shows a survey of the NOE distance restraints versus the αsyn57–102 sequence and the angular order parameters calculated over the final structure set for the backbone angles ϕ and ψ of all residues. The second figure shows a summary of NOESY connectivities of αsyn57–102.
Acknowledgments
This work was supported by a grant from the University of Padova (Progetto di Ateneo 2002) and from the Italian Ministry of Education, University and Research (MIUR-PRIN 2004).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Stefano Mammi, Department of Chemical Sciences, University of Padova, Via Marzolo 1, 35131 Padova, Italy; e-mail: stefano.mammi@unipd.it; fax: +390498275239.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.052048706.
Abbreviations: AD, Alzheimer's disease; CD, circular dichroism; cmc, critical micellar concentration; DPC, dodecyl phosphocholine; DSA, doxyl-stearic acid; EPR, electron paramagnetic spectroscopy; HSQC, heteronuclear single quantum coherence; NAC, non-amyloid-β component; NMR, nuclear magnetic resonance; NOESY, nuclear Overhauser effect spectroscopy; PD, Parkinson's disease; SDS, sodium dodecyl sulfate; SUV, small unilamellar vesicles; TOCSY, total correlation spectroscopy.
References
- Baba M., Nakajo S., Tu P.H., Tomita T., Nakaya K., Lee V.M., Trojanowski J.Q., Iwatsubo T. 1998. Aggregation of α-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am. J. Pathol. 152 879–884. [PMC free article] [PubMed] [Google Scholar]
- Bairaktari E., Mierke D.F., Mammi S., Peggion E. 1990. Conformational studies by circular dichroism, 1H NMR, and computer simulations of bombolitins I and III in aqueous solution containing surfactant micelles. Biochemistry 29 10090–10096. [DOI] [PubMed] [Google Scholar]
- Bartels C., Xia T.H., Billeter M., Guntert P., Wüthrich K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6 1–10. [DOI] [PubMed] [Google Scholar]
- Bertoncini C.W., Jung Y.S., Fernandez C.O., Hoyer W., Griesinger C., Jovin T.M., Zweckstetter M. 2005. Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc. Natl. Acad. Sci. 102 1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaglia M., Tessari I., Pinato L., Bellanda M., Giraudo S., Fasano M., Bergantino E., Bubacco L., Mammi S. 2005a. A topological model of the interaction between α-synuclein and SDS micelles. Biochemistry 44 329–339. [DOI] [PubMed] [Google Scholar]
- Bisaglia M., Trolio A., Tessari I., Bubacco L., Mammi S., Bergantino E. 2005b. Cloning, expression, purification, and spectroscopic analysis of the fragment 57–102 of human α-synuclein. Protein Expr. Purif. 39 90–96. [DOI] [PubMed] [Google Scholar]
- Bodles A.M., Guthrie D.J., Greer B., Irvine G.B. 2001. Identification of the region of non-Aβ component (NAC) of Alzheimer's disease amyloid responsible for its aggregation and toxicity. J. Neurochem. 78 384–395. [DOI] [PubMed] [Google Scholar]
- Bodles A.M., El-Agnaf O.M., Greer B., Guthrie D.J., Irvine G.B. 2004. Inhibition of fibril formation and toxicity of a fragment of α-synuclein by an N-methylated peptide analogue. Neurosci. Lett. 359 89–93. [DOI] [PubMed] [Google Scholar]
- Bussell R. Jr. and Eliezer D. 2003. A structural and functional role for 11-mer repeats in α-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329 763–778. [DOI] [PubMed] [Google Scholar]
- Bussell R. Jr., Ramlall T.F., Eliezer D. 2005. Helix periodicity, topology, and dynamics of membrane-associated α-synuclein. Protein Sci. 14 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S., Chen X., Rizo J., Jahn R., Sudhof T.C. 2003. A broken α-helix in folded α-synuclein. J. Biol. Chem. 278 15313–15318. [DOI] [PubMed] [Google Scholar]
- Clayton D.F. and George J.M. 1998. The synucleins: A family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci. 21 249–254. [DOI] [PubMed] [Google Scholar]
- Cole N.B., Murphy D.D., Grider T., Rueter S., Brasaemle D., Nussbaum R.L. 2002. Lipid droplet binding and oligomerization properties of the Parkinson's disease protein α-synuclein. J. Biol. Chem. 277 6344–6352. [DOI] [PubMed] [Google Scholar]
- Coles M., Bicknell W., Watson A.A., Fairlie D.P., Craik D.J. 1998. Solution structure of amyloid β-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry 37 11064–11077. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Crescenzi O., Tomaselli S., Guerrini R., Salvadori S., D'Ursi A.M., Temussi P.A., Picone D. 2002. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 269 5642–5648. [DOI] [PubMed] [Google Scholar]
- El-Agnaf O.M., Jakes R., Curran M.D., Middleton D., Ingenito R., Bianchi E., Pessi A., Neill D., Wallace A. 1998. Aggregates from mutant and wild-type α-synuclein proteins and NAC peptide induce apoptotic cell death in human neuroblastoma cells by formation of β-sheet and amyloid-like filaments. FEBS Lett. 440 71–75. [DOI] [PubMed] [Google Scholar]
- Eliezer D., Kutluay E., Bussell R. Jr., Browne G. 2001. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 307 1061–1073. [DOI] [PubMed] [Google Scholar]
- Ghanta J., Shen C.L., Kiessling L.L., Murphy R.M. 1996. A strategy for designing inhibitors of β-amyloid toxicity. J. Biol. Chem. 271 29525–29528. [DOI] [PubMed] [Google Scholar]
- Giasson B.I., Murray I.V., Trojanowski J.Q., Lee V.M. 2001. A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J. Biol. Chem. 276 2380–2386. [DOI] [PubMed] [Google Scholar]
- Greenfield N. and Fasman G.D. 1969. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8 4108–4116. [DOI] [PubMed] [Google Scholar]
- Han H., Weinreb P.H., Lansbury P.T. Jr. 1995. The core Alzheimer's peptide NAC forms amyloid fibrils which seed and are seeded by β-amyloid: Is NAC a common trigger or target in neurodegenerative disease? Chem. Biol. 2 163–169. [DOI] [PubMed] [Google Scholar]
- Henry G.D. and Sykes B.D. 1994. Methods to study membrane protein structure in solution. Methods Enzymol. 239 515–535. [DOI] [PubMed] [Google Scholar]
- Hilty C., Wider G., Fernandez C., Wuthrich K. 2004. Membrane protein–lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. Chembiochem 5 467–473. [DOI] [PubMed] [Google Scholar]
- Iwai A., Masliah E., Yoshimoto M., Ge N., Flanagan L., de Silva H.A., Kittel A., Saitoh T. 1995a. The precursor protein of non-A β component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron 14 467–475. [DOI] [PubMed] [Google Scholar]
- Iwai A., Yoshimoto M., Masliah E., Saitoh T. 1995b. Non-A β component of Alzheimer's disease amyloid (NAC) is amyloidogenic. Biochemistry 34 10139–10145. [DOI] [PubMed] [Google Scholar]
- Jao C.C., Der-Sarkissian A., Chen J., Langen R. 2004. Structure of membrane-bound α-synuclein studied by site-directed spin labeling. Proc. Natl. Acad. Sci. 101 8331–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaori K., Takai M., Nosaka A.Y. 1997. Comparative study of chicken and human parathyroid hormone-(1–34)-peptides in solution with SDS. Eur. J. Biochem. 249 878–885. [DOI] [PubMed] [Google Scholar]
- Khan A., Ashcroft A.E., Higenell V., Korchazhkina O.V., Exley C. 2005. Metals accelerate the formation and direct the structure of amyloid fibrils of NAC. J. Inorg. Biochem. 99 1920–1927. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wuthrich K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1429–32 51–55. [DOI] [PubMed] [Google Scholar]
- Kuntz I.D., Kosen P.A., Craig E.C. 1991. Amide chemical shifts in many helices in peptides and proteins are periodic. J. Am. Chem. Soc. 113 1406–1408. [Google Scholar]
- Lee H.J., Choi C., Lee S.J. 2002. Membrane-bound α-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J. Biol. Chem. 277 671–678. [DOI] [PubMed] [Google Scholar]
- Li H.T., Du H.N., Tang L., Hu J., Hu H.Y. 2002. Structural transformation and aggregation of human α-synuclein in trifluoroethanol: Non-amyloid component sequence is essential and β-sheet formation is prerequisite to aggregation. Biopolymers 64 221–226. [DOI] [PubMed] [Google Scholar]
- Lindberg M., Jarvet J., Langel U., Graslund A. 2001. Secondary structure and position of the cell-penetrating peptide transportan in SDS micelles as determined by NMR. Biochemistry 40 3141–3149. [DOI] [PubMed] [Google Scholar]
- Linge J.P., O'Donoghue S.I., Nilges M. 2001. Automated assignment of ambiguous nuclear Overhauser effects with ARIA. Methods Enzymol. 339 71–90. [DOI] [PubMed] [Google Scholar]
- Linge J.P., Williams M.A., Spronk C., Bonvin A., Nilges M. 2003. Refinement of proteins structures in explicit solvent. Proteins 50 496–506. [DOI] [PubMed] [Google Scholar]
- Madine J., Doig A.J., Middleton D.A. 2004. Studies of the aggregation of an amyloidogenic α-synuclein peptide fragment. Biochem. Soc. Trans. 32 1127–1129. [DOI] [PubMed] [Google Scholar]
- Nakajo S., Shioda S., Nakai Y., Nakaya K. 1994. Localization of phosphoneuroprotein 14 (PNP 14) and its mRNA expression in rat brain determined by immunocytochemistry and in situ hybridization. Brain Res. Mol. Brain Res. 27 81–86. [DOI] [PubMed] [Google Scholar]
- Opella S.J., Kim Y., McDonnell P. 1994. Experimental nuclear magnetic resonance studies of membrane proteins. Methods Enzymol. 239 536–560. [DOI] [PubMed] [Google Scholar]
- Papavoine C.H., Konings R.N., Hilbers C.W., van de Ven F.J. 1994. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry 33 12990–12997. [DOI] [PubMed] [Google Scholar]
- Pastore A. and Saudek V. 1990. The relationship between chemical shift and secondary structure in proteins. J. Magn. Reson. 90 165–176. [Google Scholar]
- Pawar A.P., Dubay K.F., Zurdo J., Chiti F., Vendruscolo M., Dobson C.M. 2005. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J. Mol. Biol. 350 379–392. [DOI] [PubMed] [Google Scholar]
- Piserchio A., Bisello A., Rosenblatt M., Chorev M., Mierke D.F. 2000. Characterization of parathyroid hormone/receptor interactions: Structure of the first extracellular loop. Biochemistry 39 8153–8160. [DOI] [PubMed] [Google Scholar]
- Pons J.L., Malliavin T.E., Delsuc M. 1996. Gifa V.4: A complete package for NMR data set processing. J. Biomol. NMR 8 445–452. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan M., Jensen P.H., Marsh D. 2003. α-Synuclein association with phosphatidylglycerol probed by lipid spin labels. Biochemistry 42 12919–12926. [DOI] [PubMed] [Google Scholar]
- Sanders C.R. and Oxenoid K. 2000. Customizing model membranes and samples for NMR spectroscopic studies of complex membrane proteins. Biochim. Biophys. Acta 1508 129–145. [DOI] [PubMed] [Google Scholar]
- Schenk D.B., Rydel R.E., May P., Little S., Panetta J., Lieberburg I., Sinha S. 1995. Therapeutic approaches related to amyloid-β peptide and Alzheimer's disease. J. Med. Chem. 38 4141–4154. [DOI] [PubMed] [Google Scholar]
- Schievano E., Mammi S., Monticelli L., Ciardella M., Peggion E. 2003. Conformational studies of a bombolitin III-derived peptide mimicking the four-helix bundle structural motif of proteins. J. Am. Chem. Soc. 125 15314–15323. [DOI] [PubMed] [Google Scholar]
- Scopes R.K. 1974. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 59 277–282. [DOI] [PubMed] [Google Scholar]
- Serpell L.C., Berriman J., Jakes R., Goedert M., Crowther R.A. 2000. Fiber diffraction of synthetic α-synuclein filaments shows amyloid-like cross-β conformation. Proc. Natl. Acad. Sci. 97 4897–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaka A.J., Lee C.J., Pines A. 1988. Iterative schemes for bilinear operators; application to spin decoupling. J. Magn. Reson. 77 274–293. [Google Scholar]
- Shao H., Jao S., Ma K., Zagorski M.G. 1999. Solution structures of micelle-bound amyloid β-(1–40) and β-(1–42) peptides of Alzheimer's disease. J. Mol. Biol. 285 755–773. [DOI] [PubMed] [Google Scholar]
- Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. 1997. α-Synuclein in Lewy bodies. Nature 388 839–840. [DOI] [PubMed] [Google Scholar]
- Ueda K., Fukushima H., Masliah E., Xia Y., Iwai A., Yoshimoto M., Otero D.A.C., Kondo J., Ihara Y., Saitoh T. 1993. Molecular cloning of cDNA encoding an unrecognised component of amyloid in Alzheimer's disease. Proc. Natl. Acad. Sci. 90 11282–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer T.S., Bax A., Cole N.B., Nussbaum R.L. 2005. Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 280 9595–9603. [DOI] [PubMed] [Google Scholar]
- Waterhous D.V. and Johnson W.C. Jr. 1994. Importance of environment in determining secondary structure in proteins. Biochemistry 33 2121–2128. [DOI] [PubMed] [Google Scholar]
- Weinreb P.H., Zhen W., Poon A.W., Conway K.A., Lansbury P.T. Jr. 1996. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 35 13709–13715. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Sykes B.D., Richards F.M. 1991. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222 311–333. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Sykes B.D., Richards F.M. 1992. The Chemical Shift Index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31 1647–1651. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. In NMR of protein and nucleic acids. . 1986. John Wiley, New York.
- Zhong L. and Johnson Jr. W.C. 1992. Environment affects amino acid preference for secondary structure. Proc. Natl. Acad. Sci. 89 4462–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N.E., Kay C.M., Hodges R.S. 1992. Synthetic model proteins: The relative contribution of leucine residues at the nonequivalent positions of the 3-4 hydrophobic repeat to the stability of the two-stranded α-helical coiled-coil. Biochemistry 31 5739–5746. [DOI] [PubMed] [Google Scholar]
- Zhu M., Li J., Fink A.L. 2003. The association of α-synuclein with membranes affects bilayer structure, stability, and fibril formation. J. Biol. Chem. 278 40186–40197. [DOI] [PubMed] [Google Scholar]