Abstract
Mucin-type O-glycans are the most typical O-glycans found in mammalian cells and assume many different biological roles. Here, we report a genetic engineered yeast strain capable of producing mucin-type sugar chains. Genes encoding Bacillus subtilis UDP-Gal/GalNAc 4-epimerase, human UDP-Gal/GalNAc transporter, human ppGalNAc-T1, and Drosophila melanogaster core1 β1–3 GalT were introduced into Saccharomyces cerevisiae. The engineered yeast was able to produce a MUC1a peptide containing O-glycan and also a mucin-like glycoprotein, human podoplanin (hPod; also known as aggrus), which is a platelet-aggregating factor that requires a sialyl-core1 structure for activity. After in vitro sialylation, hPod from yeast could induce platelet aggregation. Interestingly, substitution of ppGalNAc-T1 for ppGalNAc-T3 caused a loss of platelet aggregation-inducing activity, despite the fact that the sialyl-core1 was detectable in both hPod proteins on a lectin microarray. Most of O-mannosylation, a common modification in yeast, to MUC1a was suppressed by the addition of a rhodanine-3-acetic acid derivative in the culture medium. The yeast system we describe here is able to produce glycoproteins modified at different glycosylation sites and has the potential for use in basic research and pharmaceutical applications.
Keywords: glycosylation engineering, mucin-type glycan, podoplanin
Mucin-type glycosylation is one of the most abundant posttranslational modifications. The modification is initiated by O-linked N-acetylgalactosamine (GalNAc) to Ser or Thr residues on a peptide backbone and is involved in a variety of important biological processes, such as processing of hormones (1), endocytosis (2), and sorting of apical proteins in the Drosophila embryo (3). Mucin-type glycans are sometimes clustered, forming the “mucin domain” found on both membrane-bound (4) and secreted mucins (5), which function as a selective molecular barrier at the epithelial surface (6) and are involved in morphogenetic signal transduction (7). It is also known that changes in expression of mucin and in their glycosylation state are closely associated with the development of cancer and cancer-related processes such as cell growth, differentiation, adhesion, invasion, and immune surveillance (8). Immunohistochemical studies have identified several tumor-associated antigens (TAAs) of adenocarcinoma (9), and most TAAs on mucins were originally found as sialylated mucin-type glycans (10). Podoplanin (also called aggrus) (11, 12), one of mucin-type glycoproteins, acts as a platelet-aggregating factor for cancer cells, a finding that has attracted recent attention because it is well known that the platelet-aggregating activity of cancer cells influences tumor metastasis. In fact, the anti-human podoplanin antibody NZ-1 has an inhibitory effect on lung colonization of human podoplanin-transfected cells (13). Additionally, podoplanin is a potential diagnostic marker for many tumors, including testicular tumors, several squamous cell carcinomas, and brain tumors, and may be associated with malignancy (14, 15).
Mucins and mucin-like glycoproteins have the potential to be novel cancer markers and thus a new technology for production of a large amount of mucin-like glycoprotein for induction of specific antibodies could prove very useful. Production of a large amount of recombinant mucin-like glycoprotein may also help researchers to analyze the relationship between mucin-type glycans and their functions. Thus, we have attempted to create a yeast strain capable of producing mucin-type O-glycan. We first created a system for in vivo production of MUC1a peptides containing O-linked GalNAc and core1 structure (Galβ1–3GalNAcα1-O-Ser/Thr). We then engineered yeast capable of producing functional podoplanin and showed that yeast-produced podoplanins possess platelet aggregation activity after in vitro sialylation. A combination of glycosyltransferases introduced into yeast cells allowed us to analyze the structure–function relationship of the O-glycans on podoplanin.
Results
Construction of Yeast Strains Capable of Producing Mucin-Type Glycopeptides.
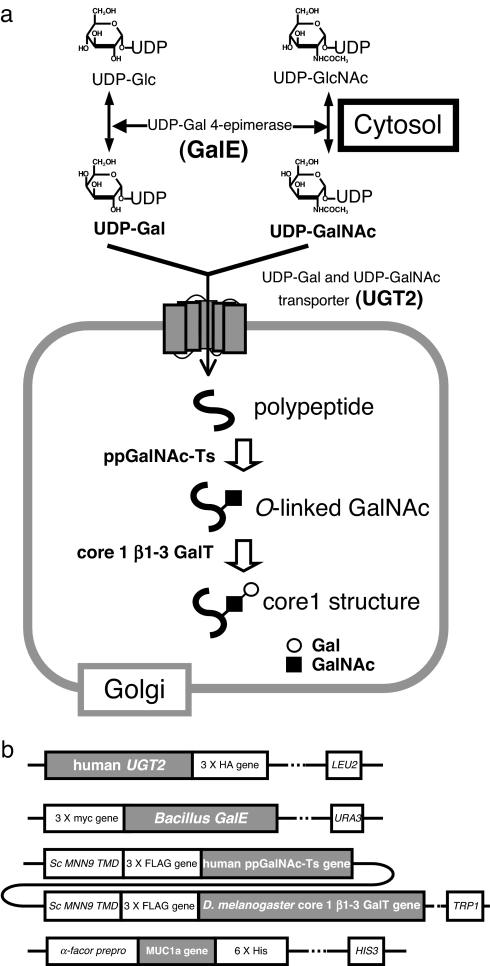
To engineer yeast capable of producing mucin-type sugar chains, three enzymatic steps should be established. Namely, these are synthesis of UDP-GalNAc and UDP-Gal in the cytosol, transport of UDP-sugars from the cytosol to the Golgi lumen, and transfer of the sugars from the UDP-sugars to an acceptor (Fig. 1a). S. cerevisiae produces intrinsic UDP-GlcNAc, UDP-Glc, and GDP-Man, but UDP-Gal is produced only when galactose is used as a carbon source. Thus, we first introduced a UDP-galactose 4-epimerase encoded by the Bacillus subtilis GalE gene (16), which can bring about 4-epimerization of both UDP-GlcNAc and UDP-Glc, forming UDP-GalNAc and UDP-Gal, respectively. After transformation expressing B. subtilis GalE, yeast cells grown in yeast extract peptone adenine dextrose (YPAD) medium were analyzed by reverse-phase HPLC to detect the presence of UDP-sugars. Two new peaks not present in the parental strain appeared in the yeast strain with GalE and corresponded to authentic UDP-GalNAc and UDP-Gal [see supporting information (SI) Fig. 5]. Next, we introduced human UDP-Gal transporter, which is encoded by UGT2 to transport both UDP-GalNAc and UDP-Gal from cytosol to the Golgi lumen (17). Finally, we introduced either the human polypeptide:N-acetylgalactosaminyltransferase (ppGalNAc-T) 1 (18), 2 (18), or 3 (19) gene. ppGalNAc-Ts transfer GalNAc from UDP-GalNAc to serine or threonine residues in polypeptide, forming an alpha anomeric linkage. Human ppGalNAc-T family proteins comprise 20 members (20), and each member appears to have a different substrate specificity. For core1 structure (Galβ1–3GalNAcα1-O-Ser/Thr) synthesis, we chose Drosophila melanogaster core1 β1–3 galactosyltransferase (core1 β1–3GalT) (21), because unlike human core1 β1–3GalT (22), the Drosophila core1 β1–3GalT does not require chaperones (21). The transmembrane domain of S. cerevisiae MMN9 (23) (mannosyltransferase), which encodes a component of a Golgi resident mannosyltransferase complex, was fused to each of the glycosyltransferases (Fig. 1b) to ensure that they are localized to the correct cellular compartment. The yeast strains used in this study are listed in SI Table 2.
Fig. 1.
Engineering of a mucin-type glycosylation pathway in yeast. (a) Introduction of a mucin-type glycosylation pathway in yeast. UDP-Gal and UDP-GalNAc are synthesized from UDP-Glc and UDP-GlcNAc, respectively, by GalE protein in the cytosol. UGT2 protein transports UDP-Gal and UDP-GalNAc from the cytosol to the Golgi lumen. Next, ppGalNAc-Ts and core1 β1–3 GalT transfer GalNAc and Gal to polypeptides. (b) Construction of each gene integrated into the yeast genome. UGT2 protein was C-terminally 3× HA-tagged, and GalE protein had an N-terminal 3× myc tag. The glycosyltransferases were fused to the transmembrane domain of yeast MNN9, and a 3× FLAG-tag was inserted between the transmembrane domain and the catalytic domains of the transferases. MUC1a was fused to the α-factor prepro sequence for secretion into the medium, and a hexa-histidine tag was added to the C terminus.
Expression of Mucin-Type Glycopeptides in Yeast.
Our first attempt was to produce a mucin-type glycopeptide in yeast. The MUC1a peptide (AHGVT5SAPDTR) was selected as a model, and our construct included a hexa-histidine tag at the C terminus (Fig. 1b). Because at least in vitro a GalNAc residue can be transferred to Thr-5 in MUC1a by ppGalNAc-T1 (24), the yeast strain expressing the ppGalNAc-T1 gene seemed appropriate for production of the mucin-type glycopeptide form of MUC1a.
To test for expression and modification of the peptide, the MUC1a coding region was introduced into N-1, TN-1, and T-1 cells (see SI Table 2), and the resultant yeast strains were cultivated in YPAD medium for 2 d at 30°C. MUC1a peptides produced in these strains were then analyzed by matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) mass spectrometry (MS) (Fig. 2a). The protonated molecular ion of MUC1a produced by N-1 cells (MUC1a/N-1) was observed at m/z 1,933, which is identical to the estimated molecular mass of MUC1a peptide. Moreover, the protonated molecular ions of MUC1a produced by TN-1 cells and T-1 cells (MUC1a/TN-1 and MUC1a/T-1, respectively) were observed at m/z 2,136 and 2,298, respectively. The m/z value of MUC1a/TN-1 is identical to the estimated mass of MUC1a modified by addition of a single N-acetylhexosamine molecule (HexNAc), and the m/z value of MUC1a/T-1 is identical to the estimated mass of MUC1a modified by addition of a HexNAc and a hexose (Hex) molecule (Fig. 2a). More than 85% of MUC1a/TN-1 contained a HexNAc residue, and a total of ≈100 μg of HexNAc-MUC1a was obtained from the supernatant of a 1-liter culture after purification by HPLC. The transfer efficiency of Hex to HexNAc-MUC1a is 70%, and a total of ≈70 μg of Hex-HexNAc-MUC1a was obtained from the supernatant of a 1-liter culture after HPLC. The results of protein sequencing revealed that MUC1a/TN-1 contains a HexNAc at Thr-5 (see SI Fig. 6), indicating that ppGalNAc-T1 has the same substrate specificity in yeast as it does in in vitro analysis (24).
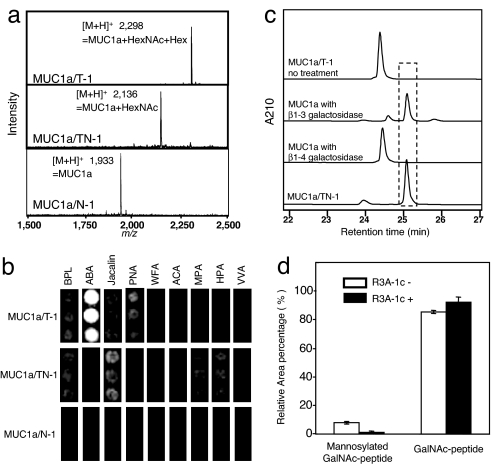
Fig. 2.
Analysis of MUC1a peptide expressed in engineered yeast cells. (a) MALDI-TOF MS spectra of each exogenously expressed MUC1a peptide. MUC1a produced by N-1, TN-1, and T-1 cells show MUC1a/N-1, MUC1a/TN-1, and MUC1a/T-1, respectively. The molecular mass of MUC1a is 1,932. (b) Lectin microarray. BPL, Bauhinia purpurea lectin; ABA, Agaricus bisporus agglutinin; Jacalin, Artocarpus integrifolia lectin; PNA, peanut agglutinin; WFA, Wisteria floribunda agglutinin; ACA, Amaranthus caudatus agglutinin; MPA, Maclura pomifera agglutinin; HPA, Helix pomatia agglutinin; VVA, Vicia villosa agglutinin. GalNAc binders: Jacalin, WFA, MPA, HPA, and VVA; core1 binders: ABA, PNA, and ACA; core1 and GalNAc binder: BPL. (c) Enzymatic digestion of MUC1a/T-1. The peak at 24.5 min corresponds to a core1-MUC1a peptide, whereas the peak at 25.1 min corresponds to the GalNAc-MUC1a peptide. (d) Inhibition of yeast O-mannosylation with a chemical reagent. MUC1a was expressed in TN-1 strains in the presence or absence of R3A-1c reagent. The total area of the recombinant MUCla peak on the chromatogram is set to 100%, and error bars show standard deviation of the mean (n = 3).
We next analyzed the glycan of the MUC1a peptides from each strain on a lectin microarray (Fig. 2b) (25). MUC1a/N-1 gave no signal, whereas MUC1a/TN-1 resulted in positive signals on Gal/GalNAc binders (Jacalin, BPL, and MPA) and GalNAc binders (WFA, HPA, and VVA). In contrast, MUC1a/T-1 resulted in positive signals on core1 binders (ABA, PNA, and ACA) but not on GalNAc binders. These results indicate that MUC1a/TN-1 and MUC1a/T-1 were successfully modified by addition of O-GalNAc and a core1 structure, respectively.
MUC1a/T-1 was further analyzed by exo-glycosidase digestion to confirm the sugar chain linkage. To do this, MUC1a/T-1 was treated with β1–4 galactosidase from Jack bean (26) or β1–3 galactosidase from Xanthomonas sp. (27). Untreated MUC1a/T-1 eluted at 24.5 min on reverse-phase HPLC (Fig. 2c); however, after β1–3 galactosidase digestion, the peak was shifted to 25.1 min, which is identical with what is observed for MUC1a/TN-1. The shift was not observed, however, after β1–4 galactosidase digestion. These results clearly indicate that the MUC1a/T-1 produced in the engineered yeast contains the core1 structure (Galβ1–3GalNAcα1-O-Ser/Thr).
Inhibition of Yeast O-Mannosylation by a Chemical Reagent.
Although most of the MUC1a/TN-1 in yeast cells contained a mucin-type glycan, the peptide was also partially modified by O-mannosylation (<8%). In eukaryotic cells, protein O-mannosylation occurs by transfer of a mannose from Dol-P-Man to a serine or threonine residue of polypeptide. The reaction can be catalyzed by at least six members of the protein O-mannosyltransferase (Pmt) family in yeast (28). Both Pmt and ppGalNAc-T recognize Ser and Thr residues as an acceptor substrate; therefore, Pmt may compete with ppGalNAc-T in yeast cells. Moreover, terminal modification of O-linked sugar chains by α-1,3-mannosyltransferase encoded by MNN1 sometimes evokes an immune response in human (29). Thus, suppression of yeast O-mannosylation is important to produce mucin-type glycoproteins as a therapeutic agent. We first constructed six pmt single disruptants (pmt1–pmt6) to inhibit O-mannosylation of MUC1a. However, MUC1a was still partially O-mannosylated in cells carrying these disruptions (data not shown). Next, we tried to suppress O-mannosylation via a rhodanine-3-acetic acid (R3A) derivative that was developed as an antifungal compound. The compound is known to act against C. albicans, and more specifically, it inhibits C. albicans Pmt1p activity (30). We also chose to use 5-(3,4-bis-phenylmethoxyphenylmethylene)-4-oxo-2-thioxo-3-thiazolidinacetic acid (R3A-1c) in the study because the compound is much easier to synthesize than other inhibitors of Pmt (30).
TN-1 cells with MUC1a were cultivated in YPAD medium for 3 d at 30°C with 10 μM R3A-1c. Cell growth and expression of the MUC1a peptide in TN-1 cells did not change in the presence or absence of 10 μM R3A-1c (data not shown). However, the peak of mannosylated GalNAc-MUC1a visible on the HPLC chromatogram all but disappeared after addition of R3A-1c (data not shown). Additionally, the ratio of GalNAc-MUC1a increased from 85% to 93%, suggesting that R3A-1c is effective in suppression of O-mannosylation in MUC1a (Fig. 2d).
ppGalNAc-T1, but Not ppGalNAc-T2 or -T3, Recognizes and Modifies the PLAG Domain of Podoplanin.
The mucin-type glycoprotein podoplanin (11) acts as a platelet-aggregating factor on cancer cells. Podoplanin has a platelet aggregation-stimulating (PLAG) domain, and we previously reported that a sialylated core1 structure at Thr-52 in the PLAG domain is essential for induction of platelet aggregation (12). However, it is unclear which ppGalNAc-T is responsible for the transfer of GalNAc to Thr-52 in mammalian cells. To help address this, we next examined the specificity of ppGalNAc-Ts by producing the PLAG domain fused to GST in TN-1, TN-2, and TN-3 yeast cells (Fig. 3a).
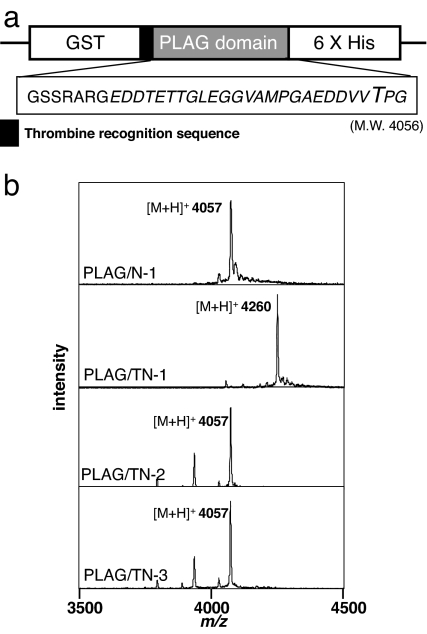
Fig. 3.
ppGalNAc-T1 transfers GalNAc to the PLAG peptide. (a) PLAG peptide was expressed with an N-terminal GST tag and a C-terminal hexa-histidine tag. The construct contains a thrombin recognition sequence (LVPRGS) between GST and the PLAG-peptide coding region. MALDI-TOF MS analysis of the PLAG peptide was done after thrombin digestion. The amino acid sequence of PLAG peptide after thrombin digestion is GSSRARGEDDTETTGLEGGVAMPGAEDDVVTPG. (b) The molecular mass of the PLAG peptide is 4,056. PLAG/N-1, PLAG/TN-1, PLAG/TN-2, and PLAG/TN-3 refer to PLAG peptide produced by the N-1, TN-1, TN-2, and TN-3 yeast strains, respectively.
In MALDI-TOF MS analysis, the protonated molecular ion of the PLAG peptide produced by TN-1 cells was m/z 4,260, whereas those from N-1, TN-2, and TN-3 cells were m/z 4,057 (Fig. 3b). The former corresponds to the mass of a PLAG peptide modified by addition of one HexNAc and the latter to the mass of unmodified PLAG peptide. Thus, the PLAG peptide produced by TN-1 cells contained GalNAc; consequently, we can conclude that ppGalNAc-T1 is able to transfer GalNAc to the PLAG domain in yeast cells. By peptide sequence analysis, we detected a peak of N-acetylgalactosaminyl-Thr at Thr-31 of the PLAG peptide, which corresponds to Thr-52 of the full-length human podoplanin (SI Fig. 7). The peptide preference of ppGalNAc-T1, which was previously reported by Gerken et al. (31), is in good agreement with our result in yeast cells.
Production of Mucin-Type Glycosylated Human Podoplanin in Yeast.
Next, we attempted to produce a mucin-type glycoprotein, human podoplanin (hPod), which serves as a model glycoprotein (Fig. 4a). As described above, ppGalNAc-T1 transfers GalNAc to Thr-52 in PLAG domain in yeast cells. Thus, we used TN-1 and T-1 cells for production of hPod and TN-3 and T-3 cells for controls. The yeast strains were cultivated in YPAD medium for 3 d at 20°C, and production of hPod was confirmed by immunoblotting with an anti-hPod antibody (NZ-1) (14). The hPods produced in each strain had different electrophoretic mobility, presumably due to the addition of different mucin-type glycans (Fig. 4b). We also performed glycan profiling of the hPods produced by yeast strains on a lectin microarray. The results suggest that the hPods were modified by addition of mucin-type glycans, such as the O-linked GalNAc and core1 structure, regardless of the type of ppGalNAc-T that was introduced (SI Fig. 8). Little O-mannosylation of the hPods was observed on lectin microarray (SI Fig. 8).
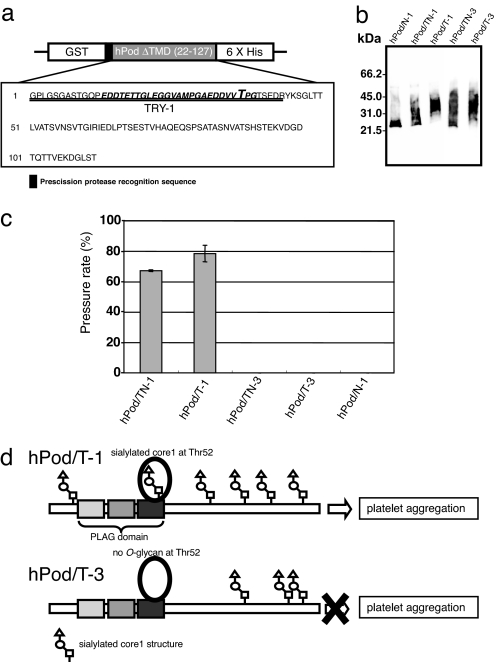
Fig. 4.
Expression and analysis of PLAG domain and human podoplanin (hPod). (a) A construct that expresses hPod with an N-terminal GST tag and a C-terminal hexa-histidine tag. The resultant recombinant protein contains a PreScission Protease recognition sequence (LEVLFQGP) between GST and hPod. The amino acid sequence of the TRY-1 trypsin fragment is double-underlined, and the PLAG domain is in italics. Big T, the essential Thr residue for platelet-aggregation inducing activity. (b) Immunoblot analysis of recombinant hPod. hPod/TN-1, hPod/T-1, hPod/TN-3, hPod/T-3, and hPod/N-1 shows hPod produced by TN-1 cells, T-1 cells, TN-3 cells, T-3 cells, and N-1 cells. (c) Platelet aggregation-inducing activity of recombinant hPod after in vitro sialylation. Before sialylation, no sample induced platelet aggregation (data not shown). Platelet aggregation was measured using WBA carna with the screen-filtration method. Pressure means a value of a pressure required for sample to pass through a filter. The pressure rate that a sample cannot pass through the filter shows 100%. The normalized mean ± SD of three independent experiments is shown. (d) Both hPod/T-1 and hPod/T-3 are modified with mucin-type O-glycan, although they are glycosylated at different sites. Thus, hPod/T-1, but not hPod/T-3, contains sialylated mucin-type O-glycan at Thr-52, and hPod/T-1 can induce platelet aggregation whereas hPod/T-3 cannot. All O-glycosylation sites of podoplanin except at Thr-52 are estimated O-glycosylation sites.
Peptide mass fingerprinting (PMF) was done to confirm transfer of mucin-type glycans to the PLAG domain present in hPod. To do this, the hPods from each yeast strain were digested with trypsin and the resultant peptide fragments were analyzed by MALDI-TOF MS. TRY-1 fragments contain PLAG domain, and those produced by N-1, TN-3, or T-3 cells (hPod/N-1, hPod/TN-3, and hPod/T-3, respectively) were observed at m/z 4,177 (Table 1), suggesting that these hPods did not contain a mucin-type glycan in the PLAG domain. The presence of a molecular ion at m/z 4,177 and m/z 4,380 in the TRY-1 fragment of hPod from TN-1 cells (hPod/TN-1) suggests that in this strain some of the hPod is modified by the addition of O-linked GalNAc. MS analysis of TRY-1 from hPod produced by T-1 cells (hPod/T-1) revealed four peaks, at m/z 4,380, 4,542, 4,745, and 4,907. This suggests the presence of four different types of glycans on TRY-1: (i) one HexNAc, (ii) one HexNAc and one Hex, (iii) two HexNAc and one Hex, and (iv) two HexNAc and two Hex, respectively. Thus, we can conclude that the TRY-1 fragments from hPod/T-1 contain both O-linked GalNAc and a core1 structure. With the exception of hPod/N-1, all hPods contained mucin-type glycans; however, it appears that the O-GalNAc modification of the PLAG domain occurs only in strains that express ppGalNAc-T1.
Table 1.
Peptide mass fingerprinting of recombinant hPod from yeast cells
| Peptide name | Theoretical m/z [M + H]+ | Observed MALDI-TOF | Expected glycopeptide |
|---|---|---|---|
| TRY-1 from hPod/N-1 | 4,161.22 | 4,177* | |
| TRY-1 from hPod/TN-2 | 4,161.22 | 4,177* | |
| TRY-1 from hPod/TN-3 | 4,161.22 | 4,177* | |
| TRY-1 from hPod/TN-1 | 4,161.22 | 4,177* | |
| 4,380* | TRY-1 with HexNAc | ||
| TRY-1 from hPod/T-1 | 4,161.22 | 4,380* | TRY-1 with HexNAc |
| 4,542* | TRY-1 with one HexNAc and one Hex | ||
| 4,745* | TRY-1 with two HexNAc and one Hex | ||
| 4,907* | TRY-1 with two HexNAc and two Hex |
*TRY-1 possesses one methionine residue, and this residue was oxidized. hPod/TN-1, hPod/T-1, hPod/TN-3, hPod/T-3, and hPod/N-1 show hPod produced by TN-1 cells, T-1 cells, TN-3 cells, T-3 cells, and N-1 cells.
Next, we attempted to transfer N-acetylneuraminic acid (Neu5Ac) to hPods in vitro using human α2–6 sialyltransferase (ST6GalNAc-I) (32) and human α2–3 sialyltransferase (ST3Gal-I) (33), followed by an assay of platelet aggregation-inducing activity. Purified hPod/TN-1 and hPod/TN-3 were successfully sialylated by ST6GalNAc-I, and hPod/T-1 and hPod/T-3 were also modified by ST3Gal-I. The transfer of Neu5Ac to hPod was confirmed on a lectin microarray, and the results revealed that after sialylation hPod/TN-1 has a sialyl-Tn structure (Neu5Acα2–6GalNAc-O-Ser/Thr) and hPod/T-1 has a α2–3 sialyl core1 structure (Neu5Acα2–3Galβ1–3GalNAc-O-Ser/Thr) (SI Fig. 8). Sialylation of hPod produced by TN-3 and T-3 cells was also confirmed on a lectin microarray (SI Fig. 8).
We investigated whether sialylated hPods were capable of inducing platelet aggregation (Fig. 4c). None of the hPods was able to induce aggregation before sialylation; however, after sialylation, hPods produced by T-1 cells but not T-3 cells did induce aggregation of mouse platelets (Fig. 4c). Platelet aggregation was also measured using human platelets as a substrate. In this assay, hPod produced by T-1 cells induced platelet aggregation, whereas hPod produced by T-3 cells did not (data not shown). The results in a previous report showed that the presence of a sialylated core1 structure at Thr-52 is important for the platelet aggregation-inducing activity of hPod (12). The results in this study also confirm the significance of having a sialic acid in the nonreducing end of the O-glycan that is initiated by ppGalNAc-T1, in the PLAG domain of hPod. Moreover, we found that hPod with a sialyl-Tn structure in the PLAG domain (hPod/TN-1) was able to induce platelet aggregation, although it is not clear whether this type of modification occurs at Thr-52 in humans cells. We also performed sialylation of PLAG peptide produced by T-1 cells, but a PLAG peptide with a sialylated core1 modification alone was not sufficient to induce platelet aggregation (data not shown). This result suggested that not only was PLAG domain with sialyl-core1 structure required for induction of platelet-aggregation by podoplanin, but so was the C-terminal domain or a structural conformation of podoplanin.
Discussion
We have generated S. cerevisiae strains capable of producing mucin-type O-glycans. We confirmed that the sialyl-core1 structure at Thr-52 of podoplanin is essential for the platelet aggregation-inducing activity by comparing cases of hPod/T-1 and hPod/T-3 (Fig. 4d). Generally, site-directed substitution or deletion of amino acid sequences has been used for functional analysis of the glycan on a glycoprotein (34, 35); however, in this work we were able to show the importance of a glycan by using a newer method. The biological roles of N-glycosylation are well elucidated at the molecular level; for example, they play roles in both quality control of glycoproteins (36) and stability of glycoproteins in the blood (37). By contrast, the biological roles of O-glycosylation have been less well understood. It is expected that we are able to control the site of glycosylation and the number of glycans added to a given glycoprotein through manipulation of exogenously introduced ppGalNAc-T in the near future. The availability of recombinant glycoproteins with different patterns of glycosylation may be useful for discovery or validation of novel biological roles for O-glycans.
It is anticipated that in vitro synthesis of mucin-type glycoprotein by sequential glycosyltransferase reactions is expensive and low-throughput. Hamilton et al. (38) have already reported in vivo addition of sialic acid to N-glycan in yeast; therefore, it may be possible to produce sialylated mucin-type glycan in vivo in the same way, and the addition of sialic acid to mucin-type glycan in vivo may lead to widespread adoption of our technique. In our study, however, sialic acid was transferred to podoplanin by in vitro reaction because the introduction of another six genes is required to produce a sialylated mucin-type glycan in vivo and this may reduce the productive efficiency of the sialylated glycan.
Our results show that in yeast the presence of appropriate ppGalNAc-T(s) for specific acceptor proteins is important for the production of functional mucin-type glycoproteins. In the previous work ppGalNAc-T2 had the strongest activity (39) toward the IgA hinge region, in which alteration of an O-glycan structure is involved in selective deposition of IgA1 in the renal glomerular mesangium (40). In mammalian cells, a competition among ppGalNAc-Ts will be influenced by many factors, not only apparent Km but also the different enzymes that may be spatially localized in discrete regions of the Golgi. Although it is difficult to produce the recombinant glycoprotein that has the same O-glycan structure in the same position as the mammalian one in yeast cells, for example, the ppGalNAc-T2-expressing yeast cell is the first choice to investigate the function of O-glycans for IgA nephropathy.
O-mannosylation should be eliminated in therapeutic use because yeast O-mannose is highly immunogenic for mammals, and abnormal O-mannosylation reduces the biological activity of an antibody produced in yeast cells (41). In this study, R3A-1c was used to suppress O-mannosylation; however, it is hard to inhibit O-mannosylation activity completely even if R3A-1c is used because O-mannosylation is required for the survival of yeast cells. We assume that the use of other derivatives of R3A in combination with six pmt single-gene disruptions (pmt1 to pmt6) may be effective for the further elimination of residual O-mannosylation.
Qualitative and quantitative changes in O-glycosylation are a consistent feature of cancer cells. Therefore, alternation of glycans is expected to serve as a tumor-specific epitope, and indeed, tumor-associated carbohydrate antigens such as CA19–9 (42) and CA125 (43) are widely used in serum diagnostic assays. Similarly, a somatic mutation in Cosmc (core1 β3-Gal-T-specific molecular chaperone) abolished function of core1 β1–3GalT, disrupting core1 synthesis and creating a tumor-specific glycopeptidic neo-epitope consisting of O-linked GalNAc and a specific amino acid sequence of podoplanin (44). A novel anti-mouse podoplanin antibody, 237 mAb, recognizes the aberrant glycoform of podoplanin and is a specific marker for Ag104A cells, which are a highly aggressive fibrosarcoma cell type (44). Tumor-specific antigen creation should be expanded to include antigens modified by tumor-specific, mucin-type glycan. The yeast system described here can be used to produce glycoproteins containing many types of glycoforms. Some of these may be specific to cancer cells, and once identified these distinct glycoforms could serve as tumor-specific antigens, which can be important not only for diagnosis but also in the development of therapeutic agents, such as anti-epitope antibodies. We have already succeeded in creating a strain of the methylotrophic yeast Ogataea minuta that can produce the mucin-type glycopeptide GalNAc-MUC1a in this yeast (data not shown). Use of a methylotrophic yeast-based approach makes it easy to produce larger amounts of recombinant protein. Production of mucin-type glycoproteins in yeast will further the course of basic research and pharmaceutical applications in the near future.
Materials and Methods
Glycan Profiling by Lectin Microarray.
Interaction of lectins with glycopeptides was analyzed by using a lectin microarray as described in ref. 25. Glycopeptides were purified by HPLC and labeled with Cy3-succimidyl ester in PBST (PBS, 1% Triton X-100) for 1 h. Cy3-labeled peptides (25 nM) were applied to each well of a glass slide, and the slides were then incubated at 20°C for 2 h. A fluorescence image of the array was acquired by using an evanescent-field fluorescence scanner (GTMASScan III, Nippon Laser and Electronics Laboratory) as described in ref. 25.
Inhibition of PMT Family Proteins with a Chemical Reagent.
The Pmt inhibitor used in this study—5-(3,4-bis-phenylmethoxyphenylmethylene)-4-oxo-2-thioxo-3-thiazolidinacetic acid, designated as R3A-1c—was prepared according to the method described in ref. 30. The R3A-1c compound was dissolved in 10 mM dimethyl sulfoxide and added into YPAD to a final concentration of 10 μM. Yeasts were then grown at 30°C for 2 d. Glycopeptides were purified as described above and then analyzed by HPLC.
Sialylation of hPod.
Each putative catalytic domain of ST3GalI and ST6GalNAcI was expressed as a secreted protein with a FLAG peptide in human embryonic kidney (HEK) 293T cells. To prepare hPod modified by a sialylGalNAc (Neu5Acα2–6GalNAcβ1-) residue or a sialylcore1 (Neu5Acα2–3Galβ1–3GalNAcβ1-) residue, 25 mM Hepes buffer (pH 7.0) containing the acceptor substrate (25 μg), 10 mM MnCl2 and an appropriate concentration of CMP-Neu5Ac (Sigma) was used. Purified ST6GalNAcI and ST3GalI were added to the reaction mixture, respectively, and incubated at 37°C for 20 h. Supernatant from the reaction mixture was subjected to a platelet aggregation assay after dialysis against PBS.
Platelet Aggregation Assay.
Platelet aggregation was measured by WBA Carna (IMI, Saitama, Japan), using the screen filtration pressure method as described in refs. 12–14. Briefly, 200-μl samples of mouse whole blood were preincubated for 2 min, followed by the addition of 12 μl (1.2 μg) of each partially purified protein. After 5 min, samples were aspirated to detect aggregation pressure.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Drs. N. Uchiyama and Y. Kubo (Research Center for Medical Glycoscience, National Institute of Advanced Industrial Science and Technology) for help in preparing the lectin microarray and Dr. T. Kubota for helpful discussion. This work was supported in part by the New Energy and Industrial Technology Development Organization.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710412105/DC1.
References
- 1.Birch NP, Estivariz FE, Bennett HP, Loh YP. Differential glycosylation of N-POMC1–77 regulates the production of gamma 3-MSH by purified pro-opiomelanocortin converting enzyme. FEBS Lett. 1991;290:191–194. doi: 10.1016/0014-5793(91)81257-9. [DOI] [PubMed] [Google Scholar]
- 2.Atiya-Nasagi Y, Cohen H, Medalia O, Fukudan M, Sagi-Eisenberg R. O-glycosylation is essential for intracellular targeting of synaptotagmins I, II in non-neuronal specialized secretory cells. J Cell Sci. 2005;118:1363–1372. doi: 10.1242/jcs.01710. [DOI] [PubMed] [Google Scholar]
- 3.Tian E, Ten Hagen KG. A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J Biol Chem. 2007;282:606–614. doi: 10.1074/jbc.M606268200. [DOI] [PubMed] [Google Scholar]
- 4.Gendler SJ, Spicer AP. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem. 1999;274:31751–31754. doi: 10.1074/jbc.274.45.31751. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenberger LM. The hydrophobic barrier properties of gastrointestinal mucus. Annu Rev Physiol. 1995;57:565–583. doi: 10.1146/annurev.ph.57.030195.003025. [DOI] [PubMed] [Google Scholar]
- 7.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Hollingsworth MA, Swanson BJ. Mucins in cancer: Protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 9.Goldenberg DM, Pegram CA, Vazquez JJ. Identification of a colon-specific antigen (CSA) in normal and neoplastic tissues. J Immunol. 1975;114:1008–1013. [PubMed] [Google Scholar]
- 10.Kawa S, et al. Clinical evaluation of pancreatic cancer-associated mucin expressing CA19–9, CA50, Span-1, sialyl SSEA-1, and Dupan-2. Scand J Gastroenterol. 1992;27:635–643. doi: 10.3109/00365529209000132. [DOI] [PubMed] [Google Scholar]
- 11.Kato Y, et al. Molecular identification of Aggrus/T1alpha as a platelet aggregation-inducing factor expressed in colorectal tumors. J Biol Chem. 2003;278:51599–51605. doi: 10.1074/jbc.M309935200. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko MK, et al. Functional glycosylation of human podoplanin: Glycan structure of platelet aggregation-inducing factor. FEBS Lett. 2007;581:331–336. doi: 10.1016/j.febslet.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 13.Kato Y, et al. Molecular analysis of the pathophysiological binding of the platelet aggregation-inducing factor podoplanin to the C-type lectin-like receptor CLEC-2. Cancer Sci. 2008;99:54–61. doi: 10.1111/j.1349-7006.2007.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato Y, et al. Inhibition of tumor cell-induced platelet aggregation using a novel anti-podoplanin antibody reacting with its platelet-aggregation-stimulating domain. Biochem Biophys Res Commun. 2006;349:1301–1307. doi: 10.1016/j.bbrc.2006.08.171. [DOI] [PubMed] [Google Scholar]
- 15.Mishima K, et al. Increased expression of podoplanin in malignant astrocytic tumors as a novel molecular marker of malignant progression. Acta Neuropathol. 2006;111:483–488. doi: 10.1007/s00401-006-0063-y. [DOI] [PubMed] [Google Scholar]
- 16.Soldo B, Scotti C, Karamata D, Lazarevic V. The Bacillus subtilis Gne (GneA, GalE) protein can catalyse UDP-glucose as well as UDP-N-acetylglucosamine 4-epimerisation. Gene. 2003;319:65–69. doi: 10.1016/s0378-1119(03)00793-5. [DOI] [PubMed] [Google Scholar]
- 17.Segawa H, Kawakita M, Ishida N. Human and Drosophila UDP-galactose transporters transport UDP-N-acetylgalactosamine in addition to UDP-galactose. Eur J Biochem. 2002;269:128–138. doi: 10.1046/j.0014-2956.2001.02632.x. [DOI] [PubMed] [Google Scholar]
- 18.White T, et al. Purification and cDNA cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1995;270:24156–24165. doi: 10.1074/jbc.270.41.24156. [DOI] [PubMed] [Google Scholar]
- 19.Bennett EP, Hassan H, Clausen H. cDNA cloning and expression of a novel human UDP-N-acetyl-alpha-D-galactosamine. Polypeptide N-acetylgalactosaminyltransferase, GalNAc-t3. J Biol Chem. 1996;271:17006–17012. doi: 10.1074/jbc.271.29.17006. [DOI] [PubMed] [Google Scholar]
- 20.Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 21.Muller R, et al. Characterization of mucin-type core-1 beta1–3 galactosyltransferase homologous enzymes in Drosophila melanogaster. FEBS J. 2005;272:4295–4305. doi: 10.1111/j.1742-4658.2005.04838.x. [DOI] [PubMed] [Google Scholar]
- 22.Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA. 2002;99:16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wandall HH, et al. Substrate specificities of three members of the human UDP-N-acetyl-alpha-D-galactosamine:Polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J Biol Chem. 1997;272:23503–23514. doi: 10.1074/jbc.272.38.23503. [DOI] [PubMed] [Google Scholar]
- 25.Kuno A, et al. Evanescent-field fluorescence-assisted lectin microarray: A new strategy for glycan profiling. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 26.Li Y-T. Studies on the glycosidases in jack bean meal. I. Isolation and properties of alpha-mannosidase. J Biol Chem. 1967;242:5474–5480. [PubMed] [Google Scholar]
- 27.Wong-Madden ST, Landry D. Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology. 1995;5:19–28. doi: 10.1093/glycob/5.1.19. [DOI] [PubMed] [Google Scholar]
- 28.Gentzsch M, Tanner W. The PMT gene family: Protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996;15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- 29.Yip CL, et al. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc Natl Acad Sci USA. 1994;91:2723–2727. doi: 10.1073/pnas.91.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orchard MG, et al. Rhodanine-3-acetic acid derivatives as inhibitors of fungal protein mannosyl transferase 1 (PMT1). Bioorg Med Chem Lett. 2004;14:3975–3978. doi: 10.1016/j.bmcl.2004.05.050. [DOI] [PubMed] [Google Scholar]
- 31.Gerken TA, Raman J, Fritz TA, Jamison O. Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem. 2006;281:32403–32416. doi: 10.1074/jbc.M605149200. [DOI] [PubMed] [Google Scholar]
- 32.Ikehara Y, et al. Cloning and expression of a human gene encoding an N-acetylgalactosamine-alpha2,6-sialyltransferase (ST6GalNAc I): A candidate for synthesis of cancer-associated sialyl-Tn antigens. Glycobiology. 1999;9:1213–1224. doi: 10.1093/glycob/9.11.1213. [DOI] [PubMed] [Google Scholar]
- 33.Gillespie W, Kelm S, Paulson JC. Cloning and expression of the Gal beta 1,3GalNAc alpha 2,3-sialyltransferase. J Biol Chem. 1992;267:21004–21010. [PubMed] [Google Scholar]
- 34.Shi S, et al. The threonine that carries fucose, but not fucose, is required for cripto to facilitate nodal signaling. J Biol Chem. 2007;282:20133–20141. doi: 10.1074/jbc.M702593200. [DOI] [PubMed] [Google Scholar]
- 35.Delacour D, et al. Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic. 2007;8:379–388. doi: 10.1111/j.1600-0854.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- 36.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 37.Ashwell G, Harford J. Carbohydrate-specific receptors of the liver. Annu Rev Biochem. 1982;51:531–554. doi: 10.1146/annurev.bi.51.070182.002531. [DOI] [PubMed] [Google Scholar]
- 38.Hamilton SR, et al. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- 39.Iwasaki H, et al. Initiation of O-glycan synthesis in IgA1 hinge region is determined by a single enzyme, UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2. J Biol Chem. 2003;278:5613–5621. doi: 10.1074/jbc.M211097200. [DOI] [PubMed] [Google Scholar]
- 40.Kokubo T, et al. Protective role of IgA1 glycans against IgA1 self-aggregation and adhesion to extracellular matrix proteins. J Am Soc Nephrol. 1998;9:2048–2054. doi: 10.1681/ASN.V9112048. [DOI] [PubMed] [Google Scholar]
- 41.Kuroda K, et al. Efficient antibody production with the suppression of O-mannosylation in yeast. Appl Environ Microbiol. 2008;74:446–453. doi: 10.1128/AEM.02106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnani JL, Steplewski Z, Koprowski H, Ginsburg V. Identification of the gastrointestinal and pancreatic cancer-associated antigen detected by monoclonal antibody 19-9 in the sera of patients as a mucin. Cancer Res. 1983;43:5489–5492. [PubMed] [Google Scholar]
- 43.Klug TL, Bast RC, Jr, Niloff JM, Knapp RC, Zurawski VR., Jr Monoclonal antibody immunoradiometric assay for an antigenic determinant (CA 125) associated with human epithelial ovarian carcinomas. Cancer Res. 1984;44:1048–1053. [PubMed] [Google Scholar]
- 44.Schietinger A, et al. A mutant chaperone converts a wild-type protein into a tumor-specific antigen. Science. 2006;314:304–308. doi: 10.1126/science.1129200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.