Abstract
We report a method for single-molecule detection and biomolecular structural mapping based on dual-color imaging and automated colocalization of bioconjugated nanoparticle probes at nanometer precision. In comparison with organic dyes and fluorescent proteins, nanoparticle probes such as fluorescence energy-transfer nanobeads and quantum dots provide significant advantages in signal brightness, photostability, and multicolor-light emission. As a result, we have achieved routine two-color superresolution imaging and single-molecule detection with standard fluorescence microscopes and inexpensive digital color cameras. By using green and red nanoparticles to simultaneously recognize two binding sites on a single target, individual biomolecules such as nucleic acids are detected and identified without target amplification or probe/target separation. We also demonstrate that a powerful astrophysical method (originally developed to analyze crowded stellar fields) can be used for automated and rapid statistical analysis of nanoparticle colocalization signals. The ability to rapidly localize bright nanoparticle probes at nanometer precision has implications not only for ultrasensitive medical detection but also for structural mapping of molecular complexes in which individual components are tagged with color-coded nanoparticles.
Keywords: colocalization, multicolor, superresolution
Recent advances in superresolution optical imaging have made it possible to detect and locate single fluorescent molecules and single-nanoparticle emitters at spatial resolutions far beyond the optical diffraction limit and have led to a burst of activity in subdiffraction optical microscopy (1–18). For single-molecule imaging in living cells, Hell and coworkers (10–13) have developed stimulated emission depletion microscopy and achieved focal-plane resolutions as high as 10–20 nm. Single molecules and single particles have also been detected at nanometer accuracy by fitting their fluorescence-intensity profiles to a two-dimensional Gaussian function or point-spread function (1–9). Given that a sufficient number of photons can be detected from a single molecule, Thompson et al. (14) were the first to show that the precision of localization increases as the inverse square root of the number of photons in the spot for the shot-noise-limited case or as the inverse of the number of photons for the background-noise-limited case. Also on the basis of photon statistical analysis, Betzig et al. (1) and Hess et al. (9) developed photoactivation localization microscopy to localize single protein molecules to >2 nm, beating the diffraction limit by nearly two orders of magnitude. Most recently, Zhuang and coworkers (18) achieved multicolor superresolution imaging at 20–50 nm by using photoswitchable fluorescence probes. This burst of activity could lead to a new era in light microscopy in which single-molecule sensitivity is achieved at nanometer resolution.
Here, we report the development of color-coded nanoparticles and automated image-processing algorithms for single-molecule detection and nanometer-scale structural mapping of biomolecular complexes. The key concept is that two color-coded nanoparticles recognize two binding sites on a single target molecule and are colocalized within nanometer distances after target binding. A major advantage is that bioconjugated nanoparticles such as energy-transfer nanobeads and quantum dots are bright and photostable fluorescent labels for biological applications (19–24). In comparison with organic dyes and fluorescent proteins, these nanoparticle probes are nearly 100 times brighter (as judged by the rate of photon emission at the same flux of excitation light) and two to three orders of magnitude more stable against photobleaching (25–27). In addition, nanoparticles with different emission colors can be simultaneously excited with a single light source, avoiding the problems of par-focality and image registration that are often encountered in multicolor fluorescence imaging (5, 28, 29). For nanoparticle colocalization analysis, we recognize that nanoparticles spread on a glass surface are similar to distant (dim) stars in the night sky because both of them are point-light emitters with Gaussian distribution profiles. Indeed, past work has shown that the two-dimensional profiles of stars can be fitted with a Gaussian distribution, which allows their locations to be determined accurately in the image plane (30). Furthermore, an astrophysical photometry system called DAOPHOT has been developed for locating and analyzing closely spaced stars in dense clusters (30). In a serendipitous fashion, we show that this image-processing method can also be used to locate color-coded nanoparticle probes at nanometer precision. The optimized algorithm is rapid and accurate and allows >1 million to 10 million nanoparticle pairs to be processed per minute.
The detection and identification of single molecules is considered the ultimate sensitivity limit in chemical analysis and mechanistic biophysical studies (31–36). By removing heterogeneous ensemble averaging, single-molecule measurements also provide insights into the dynamics of ligand-receptor binding, signal transduction, and intracellular transport. These advantages have been realized in direct imaging and dynamic studies of single molecules such as DNA, RNA, and proteins (37–39), as well as molecular motors and multiprotein assemblies (40, 41). For chemical analysis and medical diagnostics, however, the concept of single-molecule detection still has not reached its full potential and is currently used in only a small number of specialized laboratories. A problem is that target molecules often need to be chemically derivatized with a fluorophore, which is a difficult task for low-abundance genes and proteins. Another challenge is the need to discriminate bound targets from excess unbound probes in complex mixtures. Furthermore, current methods based on confocal imaging cannot handle real-world samples, because their sampling volumes (i.e., volume throughput) are often limited to nanoliters (10−9 liters) or less.
We recently reported that these long-standing problems can be overcome by using color-coded nanoparticle probes in a two-site sandwich binding format in which two nanoparticle probes of different colors simultaneously recognize two binding sites on a single target molecule (42). However, the previous work was based on the use of a confocal laser beam coupled with a capillary flow channel, and its volume throughput was limited to a few nanoliters. In addition, the overall detection efficiency was ≈40% (i.e., only 40% of the molecules flowing in the capillary channel were detected). Here, we demonstrate a detection format based on dual-color imaging and nanometer-scale colocalization. Target binding brings two color-coded nanoparticles together, and this binding event is detected by distance measurement at nanometer precision. Unbound particles are distributed randomly and do not show spatial correlation when spread and imaged on a solid surface. Nanoparticle colocalization (i.e., spatial correlation) allows one to differentiate the bound targets from excess unbound probes without a washing or separation step. This wide-field two-color imaging approach solves the practical problems associated with the confocal-detection and capillary-flow methods. As discussed in detail below, this method will be important to ultrasensitive detection of disease biomarkers and intact infectious agents such as viruses and bacteria in large-volume (ml) samples.
Results and Discussion
Single-Molecule Detection by Two-Site Sandwich Binding.
The basic principles of single-molecule detection using dual-color fluorescence imaging and automated colocalization analysis are illustrated in Fig. 1. A key feature is that two bioconjugated nanoparticles are designed to recognize each other (direct-binding mode) or recognize the same target molecule at two antigenic sites or nucleic acid sequences (indirect-binding mode). This sandwich binding event brings two color-coded nanoparticles to colocalize within a measurable distance. This distance and its statistical distribution carry important information about the size and binding geometry of a target molecule. A “colocalized” nanoparticle pair emits green and red fluorescent light simultaneously when it is excited by a laser beam and is imaged on a far-field microscope. In contrast, unbound green and red particles are not correlated in space and will be distributed randomly when they are spread and imaged on a surface. Thus, this type of spatial correlation allows one to differentiate the bound targets from excess unbound probes.
Fig. 1.
Schematic diagrams showing the design of color-coded nanoparticle probes for detecting single biomolecules in two different binding modes. (a) Direct binding between two bioconjugated nanoparticles leading to a separation distance of d1. In this work, this mode of binding was used to construct rigid molecular structures (molecular rulers) for verification/validation studies of the precision in distance measurements. (b) Indirect sandwich-type binding in which two nanoparticles recognize the same target molecule at two different sites. This indirect mode of binding allows native biomolecules such as genes to be recognized and detected at the single-molecule level.
By fitting their fluorescence-intensity profiles to a two-dimensional Gaussian function, the center positions (centroids) of individual nanoparticle probes can be determined with high precision. In fact, the errors in distance determination are determined mainly by the number of photons from each nanoparticle and can be made <1 nm as more photons are accumulated (1–6). By using image-processing software, the distances between all red and green nanoparticles are computed, and their probability of occurrence is plotted as a function of separation distance (histogram plots). A peak in the histogram indicates the most probable distance and is used to calculate the target size and the most favorable structure of the target–nanoparticle complex. The observed peak width arises from several factors including different molecular conformations, differences in nanoparticle-pair orientations on the surface, variations in the nanoparticle size, and errors in distance measurement. The total peak area indicates the number of colocalized nanoparticle pairs in a specific field of view. After subtracting the number of colocalized signals caused by random events (which can be calculated statistically and measured experimentally by using control samples under identical experimental conditions), the number of colocalized signals should correspond to the number of detected target molecules.
Image Processing and Colocalization Analysis.
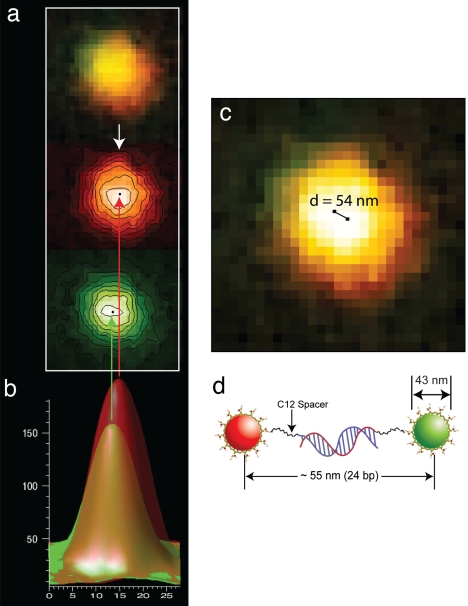
With the experimental data from a double-stranded DNA construct (see below), Fig. 2 depicts the major steps in determining the distance between colocalized nanoparticles from a dual-color fluorescence image. The first step is to separate the color image into its individual components (red and green), which then are convolved with a two-dimensional Gaussian matrix (called a kernel). In the convolution process, the amount of overlap between two functions is calculated as one function and is shifted over the other, and the two functions are then “blended” together. Next, a Gaussian point-spread function is derived from the experimental data and used to determine the green and red nanoparticle locations through a fitting procedure. Additional details on image processing and colocalization analysis are provided in supporting information (SI) Text. The localization precision achieved with this approach is expected to be >1 nm because of the low background and large number of photoelectrons (>1 million) detected from each particle (with the overall signal-to-noise ratios exceeding 60).
Fig. 2.
Automated processing of dual-color fluorescence images and nanoparticle colocalization at nanometer precision. (a) Dual-color fluorescence image of a red/green nanoparticle pair and separation into its individual color components (red and green). (b) Single-color fluorescence-intensity profiles after convolution and curve fitting with a two-dimensional Gaussian kernel function. (c) Determination of the distance between the red and green nanoparticles when mapped to the original color image. (d) Schematic diagram of the structure of a rigid DNA/nanoparticle construct and its expected contour length. Note that the experimentally determined distance (54 nm) is in close agreement with the expected length (55 nm).
As noted by Thompson et al. (14), the localization error depends on several factors including the nanoparticle brightness, the background noise, the pixel size of the camera, the magnification of the imaging microscope, and the model used to fit the intensity profiles. This error can be estimated by using the following equation:
where, Δx is the error in centroid localization, a is the pixel size (≈40 nm for our microscope, determined experimentally), s is the standard deviation of the Gaussian kernel (≈4.3 times the pixel size or ≈170 nm for our system), b is the background noise (< 5 CCD intensity units, corresponding to ≈500 photoelectrons), and N is the total number of photoelectron counts (≈1.0 million to 1.5 million) accumulated from each particle. Using these numbers, we calculated that the localization errors are 0.4 nm for the red particles (which emit more photons than the green particles) and 0.7 nm for the green particles. As discussed below, these calculated values are in agreement with the experimental data obtained from rigid double-stranded DNA constructs.
Validation Studies with Rigid Molecular Rulers.
To experimentally evaluate the localization precision that is achievable with nanoparticles, we constructed two rigid molecular rulers: a 24-bp duplex DNA and a 40-bp duplex DNA. Through the process of direct hybridization of two complementary oligonucleotide probes, these structures are covalently linked to a pair of red and green nanoparticles, one at each end (i.e., in a head-to-tail configuration; see Fig. 2d). To minimize steric and electrostatic interferences, the oligonucleotides are separated from the nanoparticles by a short 12-carbon spacer. These hybridized DNA constructs are believed to behave as a rigid rod, because the double-stranded DNA segment is shorter than the persistence length (≈50 bp) and the carbon linkers are able to rotate but are resistant to bending or shortening (43, 44). Thus, the overall lengths (measured from the red and green particle centers) are calculated to be 55.0 and 60.5 nm for the 24- and 40-bp constructs, respectively.
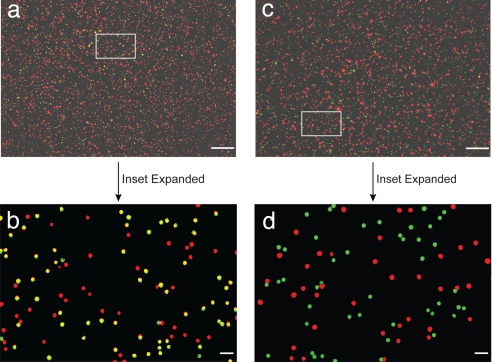
Fig. 3 a and d show wide-field color fluorescence images obtained from the 24-bp DNA construct and from a control sample under the same experimental conditions. Note that in the control sample the oligonucleotides are not complementary to each other and do not hybridize. Selected areas in the wide-field images are expanded (Fig. 3 b and e), which allows direct visualization of colocalized signals (yellow) and isolated red or green signals. Furthermore, a graphical rendering of the colocalized and isolated nanoparticles is shown in Fig. 3 c and f. These results indicate that colocalized signals arise from specific DNA hybridization.
Fig. 3.
Dual-color imaging and colocalization of red/green (R-G) nanoparticles attached to rigid DNA molecular rulers. (a and d) Wide-field fluorescence images obtained from a 24-bp DNA construct and a control (noncomplementary) sample under the same experimental conditions. (b and e) Expanded views of two selected areas in a and d (indicated by the boxes) showing colocalized signals (yellow) and isolated red and green signals. (c and f) Graphical rendering of colocalized red/green particle pairs and isolated particles corresponding to the signals observed in b and e. (Scale bars: a and d, 10 μm; b and e, 1 μm).
For quantitative detection, it is important to consider the random nature of nanoparticle distribution on the slide surface and nonspecific nanoparticle binding or aggregation, both of which could cause false colocalization signals. For this purpose, we have developed a statistical method to sort out the true and false colocalization signals. The occurrence (events) of nanoparticle colocalization was plotted as a function of the measured distance between the nanoparticle pairs for a nonhybridizing control sample and for the 24- and 40-bp DNA constructs and is shown in SI Fig. 6. The results reveal that a small number (<50) of colocalization events are detected in the control sample because of random particle distribution and nonspecific binding or aggregation. This value provides a baseline for background subtraction. For the 24- and 40-bp DNA constructs, the raw numbers of colocalization events are 307 and 637 (after background subtraction), respectively. These numbers correspond to overall detection efficiencies of 80–85% (defined as the ratio of the number of pairs detected and the number of events expected to appear in the area being imaged; i.e., >80% of the target molecules in the original sample are actually counted after many steps of sample preparation and binding).
As indicated by the histogram peaks, the most probable distances for the 24- and 40-bp DNA constructs occur at 54 and 61 nm, respectively. These values are remarkably similar to the rigid contour lengths for these structures (within ±1 nm). These distance values are statistically significant, because they are derived from ≈ 1 million nanoparticle pairs. This calculation covers all possible combinatorial pairs, a number that is much larger than the actual colocalized nanoparticle pairs. The widths of distance distribution (full width at half maximum) are in the range of 25 to 30 nm, which most likely are caused by molecular conformations, differences in nanoparticle-pair orientations on the surface, variations in the nanoparticle size, and errors in distance measurement. In addition, the histogram plots have a slightly asymmetric shape, with a faster decay on the longer-distance side than on the short-distance side. Because the most probable distances are already equal to the maximum contour lengths of rigid and hard-to-stretch DNA constructs, any variations in molecular conformations and nanoparticle orientations cannot exceed these distances. Thus, the measured distances that are longer than the average value can only arise from localization errors and variations in the particle size (e.g., the 43-nm beads have a size distribution of 11%), which causes a sharper decline at longer distances. At shorter distances, on the other hand, variations in molecular conformations and nanoparticle orientations are also contributing factors, which leads to a broader distribution. According to statistical theory, the variation in bead size should not limit the accuracy of distance determination if a large enough number of particle pairs are sampled. In practice, however, the variation in bead size is likely a limiting factor, because molecular linkers that are smaller than the bead-size standard deviation could remain hidden if only a small number of bead pairs are analyzed.
We have conducted additional experimental and data-processing work to address the question of whether nanoparticle pairs of the same color (e.g., green–green and red–red) could be localized beyond the optical diffraction limit. In particular, we varied the software parameters to test whether noncircular or nonsharp points containing two particles of the same color were thrown out or otherwise ignored by the software. None of the combinations of the sharpness and roundness parameters resulted in detection of same-color pairs. There was no difference observed between the samples with green–green or red–red pairs and pure green or pure red beads. The results showed that the same-color pairs cannot be resolved beyond the optical diffraction limit. In fact, recent work by our own group (A.A. and S.N., unpublished data) has indicated that nanometer-scale colocalization is only possible when a pair of two fluorophores or nanoparticle emitters can be differentiated by different colors, by photobleaching, or by on/off blinking intensities.
Flexible Linkers and Nanoscale Structural Mapping.
Having validated our distance measurement with rigid molecular rulers, we demonstrate that flexible biomolecular structures can also be detected and mapped by using color-coded nanoparticles. In particular, a unique sequence of the breast cancer antiestrogen resistance gene (BCAR3) is rapidly detected at the level of single molecules. The significance for this selection is that antiestrogen agents such as tamoxifen are administered as a therapy for breast cancer, but some patients who carry the BCAR3 gene develop resistance to the therapy (45). Detection of this gene can help physicians administer alternative therapy or control the antiestrogen dosage and administration time. The two-site sandwich assay consists of two 15-base oligonucleotides linked to color-coded nanoparticles through a flexible 10-base poly(A) spacer and a 12-carbon linker. Each of the oligonucleotides recognizes half of a 30-base target (5′-CCCGAGAAACTGAAGAAGGAGCTGGAGGAC), part of the BCAR3 gene sequence. Because the poly(A) spacer remains single-stranded after target binding, its flexible structure allows the nanoparticle to adopt various configurations. Fig. 4 a and c shows wide-field color fluorescence images obtained from these flexible nanoparticle conjugates with and without the target gene sequence. Selected areas in the images (expanded in Fig. 4 b and d) reveal that colocalized signals (yellow) are observed only in the presence of the gene target. A number of red particles are not hybridized to the target and are observed as isolated signals because of an excess of red particles used in the hybridization solution. Note that the excess red spots arise from a stepwise hybridization procedure in which green nanoparticles were first allowed to hybridize with the BCAR3 gene fragment at an ≈1:1 ratio and red nanoparticle probes were allowed to hybridize at much higher concentrations.
Fig. 4.
Dual-color imaging and colocalization of red/green nanoparticles attached to flexible DNA structures. (a and c) Wide-field fluorescence images obtained from sandwich DNA hybridization assays in the presence of a cancer gene (BCAR3) sequence (a) and in the absence of this target gene sequence under the same experimental conditions (c). (b and d) Magnified images of selected areas (boxes in a and c) for direct visualization of colocalized signals (yellow) in the complementary sample and the absence of colocalized signals in the noncomplementary sample. The expanded images were processed by separating and leveling the red and green signals. (Scale bars: a and c, 10 μm; b and d, 1 μm.)
Statistical analysis shows a peak with the target gene sequence and essentially nothing without the target. In comparison with that of direct binding, the achieved detection specificity (lower background) is much higher with this two-site sandwich format. This result is not surprising, because two-site sandwich binding relies on a “double-selection” process to improve detection specificity. In contrast to rigid molecular structures, however, the histogram peak occurs at 45 nm, which indicates that the most probable distance between the nanoparticle pairs is far shorter than the expected contour length (62 nm) for this two-site hybridized structure. As illustrated in Fig. 5a, the flexible poly(A) spacers allow the nanoparticles to fold and approach each other at distances much shorter than the linear contour length.
Fig. 5.
Schematic drawings of two flexible structures in the head-to-tail configuration (a) and two constrained structures in the head-to-head configuration (b).
With both rigid and flexible biomolecular structures, we have shown that the use of bioconjugated nanoparticles allows single-molecule detection at nanometer precision. One step further, we ask the question of what the structure would be if biomolecular binding gives rise to a head-to-head configuration for the color-coded particles. For example, by tagging the 3′ end of one oligonucleotide with a red particle and the 5′ end of its complementary oligonucleotide with a green particle, the two particles should take on a head-to-head configuration after hybridization. In Fig. 5b are shown two possible head-to-head configurations. The head-to-head (or tail-to-tail) structure is considerably more constrained than the head-to-tail configuration because of repulsive interactions (electrostatic) between the nanoparticles (negatively charged) and the particles' relatively large sizes. Our colocalization data (not shown) reveal that the most favored distance between the nanoparticles is 54 nm in the head-to-head configuration. This value is larger than the distance (47 nm) expected from two nanoparticles in contact or in close proximity. On the basis of the measured distance, we estimate that 10–12 bases remain single-stranded in the most favored structure. This structure is less stable energetically than the fully hybridized structure, but it is favored because of the need to minimize both steric and electrostatic interferences.
To investigate whether this approach might be applicable to “real-world” samples in complex environments, we have conducted single-molecule studies in cell lysate samples (which contain a mixture of proteins, nucleic acids, and lipids). The results showed single-molecule detection in complex cellular extracts and a detection dynamic range over three orders of magnitude (10−14 to 10−11 M DNA) (42). The theoretical detection limit is calculated to be ≈13 aM (24 targets in 3 μl), but in practice we found that the target signals could not be differentiated from background noises (e.g., random colocalization events and nonspecific binding) at target concentrations <10–20 fM. Thus, we believe that there is still considerable room for improvement by optimizing the experimental conditions and the data-analysis parameters. For nanometer-scale mapping studies, it will be important to use color-coded nanoparticles to study the structures of protein–protein and DNA–protein complexes, as well as the distribution and clustering of receptor proteins on fixed cell membranes. These studies, however, need to first be validated by using well defined model structures as controls, similar to the use of rigid DNA–nanoparticle complexes as reported here.
In conclusion, we have reported a simple method for rapid detection of single native biomolecules based on dual-color imaging and automated colocalization of nanoparticle probes at nanometer precision. By taking advantage of the attractive properties of energy-transfer nanobeads (e.g., signal brightness, photostability, and multicolor-light emission), we have achieved routine two-color imaging and single-molecule detection with standard fluorescence microscopes and inexpensive digital color cameras. Using a “star-gazing” software system, we are able to analyze the locations of a large number of nanoparticle pairs. In comparison with previous confocal and flow-based methods that detect one molecule at a time (serial), the dual-color imaging technology allows high-speed detection in a parallel fashion involving much larger sample volumes (microliter to milliliters). In addition to ultrasensitive detection of disease biomarkers and intact infectious agents such as viruses and bacteria, use for nanoparticle colocalization microscopy should be found in structural mapping studies of biomolecular complexes and assemblies.
Materials and Methods
Materials.
Polyl-lysine hydrobromide (molecular weight = 350,000), trizma base [Tris(hydroxymethyl)aminomethane], trizma hydrochloride [Tris(hydroxymethyl)amino-methane hydrochloride], 2-mercaptoethanol, 1-ethyl-3-(3-dimethyl-aminopropyl)-carbodiimide, and BSA were purchased from Sigma. Carboxylate-modified energy-transfer nanoparticles (43-nm diameter) (Fluosphere 505/515, product no. F-8795 and TransFluosphere 488/685, product no. T-8868) were purchased from Molecular Probes. Microscope coverslips (0.17-mm thick) were purchased from Corning. Ultrapure water was prepared by using a Milli-Q purification system (Millipore).
Nanoparticle Bioconjugation.
Oligonucleotides were conjugated covalently to nanoparticles by using standard conjugation chemistry (46). For energy-transfer nanoparticles (43 nm), the number of oligonucleotide or antibody ligands per particle was approximately one (determined experimentally by using fluorescently labeled oligonucleotides or antibodies; see below). These nearly “monovalent” nanoparticles produced excellent binding results while avoiding the problem of target/probe aggregation. Thus, a solution of green nanoparticles (40 nm, λex = 505 nm, λem = 515 nm, 53 pmol) (Molecular Probes) was mixed with oligo-1 (5′-CTTCAGTTTCTCGGG-A10-NH2, 52.8 pmol) or oligo-2 (5′-NH2-A10-CTCCTCCAGCTCCTT-3′, 37.4 pmol) (Sigma Genosys) together with an amine activation reagent (50 μg, sulfo-NHS; Pierce) in a pH 5.75 buffer (0.1 M morpholino ethanesulfonic acid). To this mixture was added a carbodiimide cross-linking agent [50 μg, 1-ethyl-3-(3-dimethyl-aminopropyl)-carbodiimide; Pierce], which was thoroughly mixed and sonicated during a 1-h period at room temperature. Bioconjugated nanoparticles were purified by centrifugation with S-400 microspin columns (Amersham Bioscience) at 740 × g. The bioconjugation efficiencies were determined by using fluorescently labeled amine-oligonucleotides (amine-A10-CTCCTCCAGCTCCTT-Oregon green; Sigma Genosys) and plain (nonfluorescent) 40-nm beads. After the cross-linking reaction, free unreacted oligonucleotides were separated from conjugated oligonucleotides by centrifugation. Fluorescence-intensity measurements of the two fractions showed that ≈11% of oligonucleotides were conjugated to nanoparticles under our experimental conditions. The fraction of fluorescent beads obtained after passing through an identical reaction scheme was also determined by fluorescence measurements before and after the reaction. Next, stock concentration for the probes and beads and knowledge of the fraction of both probes and the beads retained during reactions were used to determine the number of conjugated ligands per nanoparticle (1.2 ligands per particle).
DNA Hybridization.
The 24-bp duplex has the sequences of NH2-C12–5′-AGGACCCAGA ATTCGATAGA CCCG-3′ and 3′-TCCTGGGTCT TAAGCTATCT GGGC-5′-C12-NH2, whereas the 40-bp duplex has the sequences of NH2-C12–5′-GATCCAGTTT CCATTAGAGA CCCAGAGTCC GATATTACCG-3′ and 3′-CTAGGTCAAA GGTAATCTCT GGGTCTCAGG CTATAATGGC-5′-C12-NH2. For hybridization, complementary strands of DNA (at ≈0.3 to 3 nM) coupled with red or green beads were allowed to hybridize for 75 min at 45°C in pH 7.0, 8× standard saline citrate buffer (i.e., 0.75 M NaCl). As a control, beads conjugated with noncomplementary DNA sequences (24- and 40-base DNA sequences) were incubated in the hybridization buffer at similar concentrations. A similar protocol was followed for hybridization of probes with the target in the sandwich assay. The main difference in the sandwich-assay hybridization was that the green nanoparticle probes were incubated with the target at 1:1 concentration (≈100 pM) for 45 min followed by incubation with 10- to 20-fold excess of red nanoparticle probes for an additional 45 min. The hybridization temperature was kept low to prevent leaching out of the fluorescent dye molecules trapped in the polystyrene nanoparticles. Hybridized product was diluted 2-fold for imaging in the case of 24- and 40-bp experiments and 10-fold in case of the sandwich assay.
Dual-Color Fluorescence Imaging.
An aliquot (1–3 μl) of the nanoparticle probe solution (hybridized or control) was spread between two clean coverslips (no. 1 cover glass; Corning) and was allowed to spread and dry over 4 h in a desiccator. One coverslip was then removed and placed on an epifluorescence microscope (Nikon Eclipse TE 300 or Olympus IX-71) equipped with mercury-lamp excitation, a Nikon D1 color digital camera, and a ×100 objective (N.A. = 1.25) on a vibration-free optical table (Newport). True-color fluorescence images were obtained by using a 488-nm excitation filter and a long-pass filter (505 nm; Chroma Technology). The exposure times were 0.2–2.0 s.
Image Processing and Colocalization.
We used the DAOPHOT astrophysical method (30) for image processing and colocalization analysis (also see SI Fig. 6, SI Table 1, and SI Text). To evaluate whether other computing programs could be used for rapid colocalization analysis of nanoparticle pairs, we explored a number of computing algorithms ranging from simple Gaussian fitting to robust Kalman tracking. However, we found that it took 36 h for a single CPU machine to process a two-color nanoparticle image of 6 million pixels. The processing time was reduced to 4 h by using a 68-CPU Itanium2 Linux cluster (running on software codes optimized for parallel computing), which is still far slower than the DAOPHOT method running on a personal computer. Thus, the final choice was to user the astrophysical image-processing method on a desktop computer.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Michael Crenshaw (Georgia State University) for helpful discussions. This work was supported in part by National Institutes of Health Grants P20 GM072069, R01 CA108468, and R01 GM60562 (to S.N.), the Emory-Georgia Tech Nanotechnology Center, the Department of Energy Genomes to Life Program (S.N.), and the Georgia Cancer Coalition Distinguished Cancer Scholars Program (S.N. and M.D.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712351105/DC1.
References
- 1.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Ram S, Ward ES, Ober RJ. Proc Natl Acad Sci USA. 2006;103:4457–4462. doi: 10.1073/pnas.0508047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu X, Wu D, Mets L, Scherer NF. Proc Natl Acad Sci USA. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon MP, Ha T, Selvin PR. Proc Natl Acad Sci USA. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacoste TD, Michalet X, Pinaud F, Chemla DS, Alivisatos AP, Weiss S. Proc Natl Acad Sci USA. 2000;97:9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michalet X, Weiss S. Proc Natl Acad Sci USA. 2006;103:4797–4798. doi: 10.1073/pnas.0600808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherer NF. Nat Nanotechnol. 2006;1:19–20. doi: 10.1038/nnano.2006.79. [DOI] [PubMed] [Google Scholar]
- 8.Rust MJ, Bates M, Zhuang XW. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess ST, Girirajan TP, Mason MD. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnert G, Keller J, Medda R, Andrei MA, Rizzoli SO, Luhrmann R, Jahn R, Eggeling C, Hell SW. Proc Natl Acad Sci USA. 2006;103:11440–11445. doi: 10.1073/pnas.0604965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willig KI, Kellner RR, Medda R, Hein B, Jakobs S, Hell SW. Nat Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 12.Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 13.Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al. Science. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- 14.Thompson RE, Larson DR, Webb WW. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 16.Yildiz A, Selvin PR. Acc Chem Res. 2005;38:574–582. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- 17.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Proc Natl Acad Sci USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates M, Huang B, Dempsey GT, Zhuang XW. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 20.Chan WCW, Nie SM. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 21.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 22.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 23.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 24.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 25.Chan WCW, Maxwell DJ, Gao XH, Bailey RE, Han MY, Nie SM. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 26.Alivisatos AP. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 27.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li HT, Zhou D, Browne H, Balasubramanian S, Klenerman D. Anal Chem. 2004;76:4446–4451. doi: 10.1021/ac049512c. [DOI] [PubMed] [Google Scholar]
- 29.Li HT, Ying LM, Green JJ, Balasubramanian S, Klenerman D. Anal Chem. 2003;75:1664–1670. doi: 10.1021/ac026367z. [DOI] [PubMed] [Google Scholar]
- 30.Stetson PB. Publ Astron Soc Pac. 1987;99:191–222. [Google Scholar]
- 31.Nie SM, Zare RN. Annu Rev Biophys Biomol Struct. 1997;26:567–596. doi: 10.1146/annurev.biophys.26.1.567. [DOI] [PubMed] [Google Scholar]
- 32.Yeung ES. Annu Rev Phys Chem. 2004;55:97–126. doi: 10.1146/annurev.physchem.54.011002.103820. [DOI] [PubMed] [Google Scholar]
- 33.Xie XS, Trautman JK. Annu Rev Phys Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 34.Moerner WE, Orrit M. Science. 1999;283:1670–1676. doi: 10.1126/science.283.5408.1670. [DOI] [PubMed] [Google Scholar]
- 35.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 36.Wabuyele MB, Farquar H, Stryjewski W, Hammer RP, Soper SA, Cheng YW, Barany F. J Am Chem Soc. 2003;125:6937–6945. doi: 10.1021/ja034716g. [DOI] [PubMed] [Google Scholar]
- 37.Ha TJ, Ting AY, Liang J, Caldwell WB, Deniz AA, Chemla DS, Schultz PG, Weiss S. Proc Natl Acad Sci USA. 1999;96:893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ha TJ. Biochemistry. 2004;43:4055–4063. doi: 10.1021/bi049973s. [DOI] [PubMed] [Google Scholar]
- 39.Zhuang XW, Bartley LE, Babcock HP, Russell R, Ha TJ, Herschlag D, Chu S. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 40.Asbury CL, Fehr AN, Block SM. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 42.Agrawal A, Zhang C, Byassee T, Tripp RA, Nie SM. Anal Chem. 2006;78:1061–1070. doi: 10.1021/ac051801t. [DOI] [PubMed] [Google Scholar]
- 43.Nelson P. Biological Physics. New York: Freeman; 2003. [Google Scholar]
- 44.Hagerman PJ. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 45.van Agthoven T, van Agthoven TLA, Dekker A, van der Spek PJ, Vreede L, Dorssers LCJ. EMBO J. 1998;17:2799–2808. doi: 10.1093/emboj/17.10.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermanson GT. Bioconjugate Techniques. San Diego: Academic; 1996. pp. 173–176. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.