Abstract
Although the majority of colorectal cancers exhibit chromosome instability (CIN), only a few genes that might cause this phenotype have been identified and no general mechanism underlying their function has emerged. To systematically identify somatic mutations in potential CIN genes in colorectal cancers, we determined the sequence of 102 human homologues of 96 yeast CIN genes known to function in various aspects of chromosome transmission fidelity. We identified 11 somatic mutations distributed among five genes in a panel that included 132 colorectal cancers. Remarkably, all but one of these 11 mutations were in the homologs of yeast genes that regulate sister chromatid cohesion. We then demonstrated that down-regulation of such homologs resulted in chromosomal instability and chromatid cohesion defects in human cells. Finally, we showed that down-regulation or genetic disruption of the two major candidate CIN genes identified in previous studies (MRE11A and CDC4) also resulted in abnormal sister chromatid cohesion in human cells. These results suggest that defective sister chromatid cohesion as a result of somatic mutations may represent a major cause of chromosome instability in human cancers.
Keywords: CDC4, MRE11A, somatic mutation
Genetic instability is believed to play a critical role in the development of cancer (1–4). In colorectal cancers, this instability is most often due to chromosomal instability (CIN), resulting in losses and gains of entire chromosomes or large regions thereof. Another form of instability, in which microsatellite sequences are often affected, occurs in ≈15% of sporadic colorectal cancers and in all hereditary non-polyposis colorectal cancer patients (5, 6). Soon after the initial descriptions of microsatellite instability in human cancers, it was recognized that yeast cells with mismatch repair (MMR) defects exhibited a similar instability in simple sequence repeats (7). This realization, coupled with a cross-species candidate gene approach, rapidly led to the identification of human mismatch repair gene defects as the causes of microsatellite instability in colorectal cancers (8–12).
The importance of the CIN phenotype was demonstrated when it was shown that many colorectal cancers exhibit 10- to 100-fold higher rates of chromosome missegregation than normal cells or cells with mismatch repair defects (13). Accordingly, CIN cancers are generally aneuploid whereas MMR-deficient cancers are generally nearly diploid. Although the molecular basis for CIN in human cancers is still largely obscure, several studies have suggested potential mechanisms for it (4, 14, 15). For example, mice heterozygous for null alleles of the mitotic spindle checkpoint genes Mad2, BubR1, or Bub3 have an enhanced susceptibility to spontaneous or carcinogen-induced tumors (16, 17). Heterozygous CENPE+/− mice have been shown to develop aneuploidy both in vitro and in vivo, and develop spontaneous lymphomas and lung tumors in older animals (18).
To date, only a handful of genes thought to be important for maintaining chromosomal stability have been identified as having somatic mutations in human cancers (19–21). APC gene mutations have also been suggested to be responsible for CIN (22, 23). However, because many colorectal tumors with MMR-deficiency have APC gene mutations, yet remain diploid and do not manifest CIN, APC is unlikely to be the primary determinant of CIN (24).
The pathways involved in faithful chromosome transmission are conserved throughout eukaryotic evolution and much of what is known about the molecular basis of other types of genetic instability has been determined by studying model organisms, particularly yeast. By definition, CIN genes identified in model organisms can establish cross-species candidate genes that might be altered in human cancer and cause CIN during tumorigenesis. In this regard, one of our major goals has been to identify all genes (both essential and nonessential) that give rise to a CIN phenotype when disrupted in yeast. Such a comprehensive list of yeast CIN genes would allow the cognate set of human genes to be identified by “homology probing” (25).
In the current study, 102 human genes highly related to 96 yeast CIN genes were identified through bioinformatic approaches. We then determined the sequence of each of these 102 genes in a panel of colorectal cancers. Remarkably, all but one of the genes that were mutated encoded proteins directly involved in sister chromatid cohesion. In addition to the ability of these genes to cause CIN in yeast when disrupted, we found that down-regulation of these genes in human cells caused CIN in conjunction with defective sister chromatid cohesion. Analyses of the major genes previously hypothesized to play a role in CIN revealed similar effects on chromatid cohesion. These results suggest that abnormalities in sister chromatid cohesion play a major role in the CIN phenotype in human colorectal tumors.
Results
Somatic Mutations of Yeast CIN Gene Homologs in Human Colorectal Cancers.
Recent comprehensive genome-wide screens of the yeast nonessential gene deletion set for CIN mutants (26) and previously identified essential yeast CIN genes identified by traditional random mutagenesis (27) were used to generate a list of yeast CIN genes. This list was in turn used to identify a prioritized list of human homologues based on the extent of yeast/human protein sequence similarity and phenotype strength of the corresponding yeast CIN gene. In all, 102 human genes highly related to 96 yeast CIN genes were selected for somatic mutation analysis [see supporting information (SI) Table 3]. Before this work, a set of 108 candidate CIN genes that did not overlap with the 102 genes analyzed in this study had been sequenced in colorectal tumors (21). The prior set was chosen on the basis of different criteria than those used here and only a minority represented the most highly ranked human homologs of a yeast CIN gene (SI Table 5).
Primer pairs were designed to amplify the coding exons for each of the 102 human candidate genes encoding ≈220 Kb of ORF. PCR products were generated for each exon and amplicons were sequenced by using nested primers (see SI Materials and Methods). For each exon, 36 tumor DNA samples were analyzed in the first phase of sequencing. At this depth of sequencing, we would expect to detect somatic mutations in over three quarters of genes that were altered in 4% of cancers. After excluding known polymorphisms present in the human genomic database, all variants were reamplified and resequenced by using DNA from the tumor sample to exclude sequence artifacts. Sequencing of matched normal DNA from the same patient was used to distinguish true somatic mutations from germ line polymorphisms. In the first round of sequencing, six somatic mutations were identified in five genes: SMC1L1 (two independent mutations), CSPG6, NIPBL, STAG3, and RNF20 (Table 1). Notably, four of these five genes have been implicated in sister chromatid cohesion.
Table 1.
Mutated CIN genes
| Human gene name | Yeast gene name | e-value | Human mRNA accession no. | Human protein accession no. | Gene description | No. of mutations | No. of tumors analyzed | Exon no. | Nucleotide change | Amino acid change | Tumor |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SMC1L1 | SMC1 | 1.00E−35 | NM_006306.2 | NP_006297.2 | Structural maintenance of chromosomes 1A | 4 | 132 | 7 | 1186T > CT | 396F > L/F | CO94 |
| 8 | 1300C > T | 434R > W | HX8 | ||||||||
| 10 | 1680C > CG | 560I > I/M | CX3 | ||||||||
| 24 | 3556G > AG | 1186V > I/V | HX129 | ||||||||
| NIPBL | SCC2 | 3.00E−19 | NM_015384.3 | NP 597677.2 | Nipped-B homolog (Drosophila) | 4 | 132 | 8 | 1435C > CT | 479R > R/X | HX7 |
| 9 | 2967_2968 het_insT | Frameshift after 992 | CO71 | ||||||||
| 9 | 1660C > CT | 554Q > Q/X | HX168 | ||||||||
| 28 | 5378T > TA | 1793M > M/K | MX24 | ||||||||
| CSPG6 | SMC3 | 1.00E−45 | NM_005445.3 | NP 005436.1 | Structural maintenance of chromosomes 3 | 1 | 130 | 23 | 2635C > CT | 879R > R/X | MX13 |
| STAG3 | SCC3 | 3.00E−13 | NM_012447.2 | NP_036579.2 | Cohesin subunit SA-3 | 1 | 130 | 22 | 24117T > CT | 795I > T/I | CO71 |
| RNF20 | BRE1 | 5.00E−26 | NM_019592 | NP_062538.5 | Ring finger protein 20 | 1 | 36 | 3 | 370C > CT | 124R > R/X | HX88 |
To more accurately determine the mutation frequency of these five genes, we examined them in an additional 95 tumors. We thereby identified two more mutations in SMC1L1 and three more in NIPBL. In SI Table 4, we provide three different background (“passenger”) rates based on the analysis of mutations in noncoding regions or nonsynonymous mutations in a similar group of tumors. The lowest of these three estimates is likely an underestimate, and the highest of the estimates is an overestimate of the true passenger mutation rate, as explained in Wood et al. (28). SMC1L1 was mutated at statistically significant frequency at any of these three rates, whereas NIPBL and STAG3 were mutated at statistically significant levels when the lower and mid-rates rates were used.
An analysis of functional groups showed that the group of 26 homologues predicted to be directly involved in sister chromatid cohesion was more likely to harbor mutations than predicted by the background mutation rate (P = 0.0002, 0.0053, or 0.2049 depending on the assumed passenger rate). Most importantly, the difference between mutation prevalence in genes known or not known to affect sister chromatid cohesion was highly significant (P = 0.02, two-sample binomial test; P = 0.005, Wilcoxon). These data are consistent with conclusions made from large scale sequencing of protein-encoding genes in human cancers (28, 29). Such studies suggest that there are many genes that, when mutated, result in similar phenotypes and it can be more informative to evaluate gene groups and pathways than individual genes.
Reduced Protein Expression of Mutated Genes Results in CIN.
We postulated that CIN likely arises through reduced activity of the gene products that were found to be mutated. We therefore used RNA-interference to decrease wild-type protein levels encoded by two of the five genes identified in the current study (SMC1L1 and CSPG6). We similarly down-regulated STAG2, a human gene closely related to one of the mutated genes (STAG3) and for which an antibody to monitor the extent of knock-down was available. Finally, we attempted to down-regulate the protein encoded by MRE11A, which represented the most frequently mutated candidate CIN gene identified in a previous study (21). In each case, we assayed chromosome stability using flow cytometry and the evaluation of mitotic chromosome spreads. The HCT116 cell line was chosen as the experimental system because it is a chromosomally stable, near diploid colorectal cell line that does not inherently exhibit CIN.
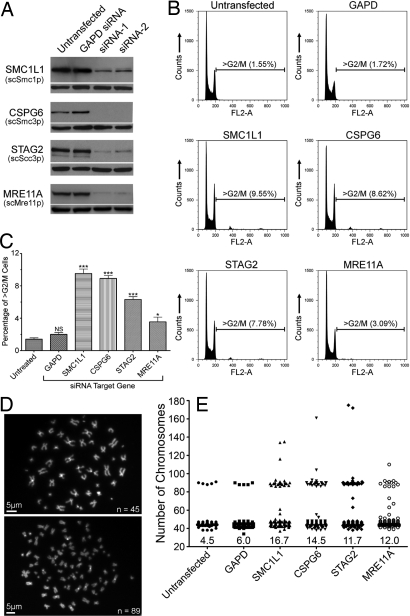
All four potential human CIN gene targets (SMC1L1, CSPG6, STAG2 and MRE11A) exhibited significantly reduced expression after transfection with siRNAs that targeted them compared with untransfected or cells transfected with siRNAs targeting a control gene (GAPD). For each target, we chose the two siRNA duplexes that demonstrated the greatest degree of protein knockdown for further experiments. A decrease in protein expression was observed as quickly as 24 h after transfection and minimum levels of proteins were observed two days thereafter (Fig. 1A). Protein levels remained decreased for a total of 7–10 days after transfection (data not shown).
Fig. 1.
Down-regulation of mutated gene products results in CIN in human cells. (A) Protein expression levels were examined by Western blot after siRNA-mediated knockdown of the genes indicated at the left (yeast protein names are shown in parentheses for reference purposes). Reprobing the same plots with anti-α-tubulin antibodies confirmed equal loading (shown below each targeted protein blot). (B) Asynchronous HCT116 cells were labeled with propidium iodide and subjected to flow cytometry. The bars delineate the G2/M population of cells with DNA contents greater than expected for cells in the G2/M phase of the cell cycle. The FL2-A channel has been left shifted (G0/G1 FL2-A ≈ 100; G2/M FL2-A ≈ 200) to include both the relatively small tetraploid (FL2-A ≈ 375) and octaploid (FL2-A ≈ 750) cell populations. (C) Graphs of the mean percentage of >G2/M cells (± SEM) after siRNA-induced knockdown of GAPD, SMC1L1, CSPG6, STAG2 and MRE11A. ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, P > 0.05. (D) Images of DAPI-stained mitotic spreads from untransfected cells (Upper, n = 45 chromosomes) and a markedly aneuploid cell after treatment with CSPG6 siRNA (Lower, n = 89 chromosomes). (E) Scatter plot depicting the total chromosome number distribution for cells treated with the indicated siRNA. At least 300 mitoses were evaluated for each scatter plot. Percentage of mitotic spreads with >46 chromosomes is indicated at the base of each column.
Flow cytometric analysis of DNA content was performed on PI-labeled asynchronous cells generated from cell populations harvested 7-days after transfection. Because most colorectal cancers exhibit increases in chromosome numbers (3, 14), we focused our initial analyses on the population of cells with DNA contents beyond the diploid G2/M peak (i.e., >G2/M) rather than on subdiploid cells. Fig. 1B depicts DNA content plots for each of the control and experimental conditions and reveals that SMC1L1, CSPG6, STAG2 or MRE11A knockdown resulted in increases in the frequency of cells with >G2/M DNA content (SI Table 6). Furthermore, small discrete peaks corresponding to tetraploid (FL2-A ≈ 375) or octaploid (FL2-A ≈ 750) populations became evident upon SMC1L1, CSPG6 and STAG2 knockdown. Although MRE11A knockdown did not generate an octaploid population, it did generate increased DNA content just beyond the G2/M boundary, consistent with a heterogeneous collection of cells with extra chromosomes. One-way analysis of variance (ANOVA) coupled with a Tukey multiple comparison test identified statistically significant differences in the mean >G2/M populations observed after SMC1L1, CSPG6, STAG2 and MRE11A or control transfections (Fig. 1C and SI Tables 7 and 8).
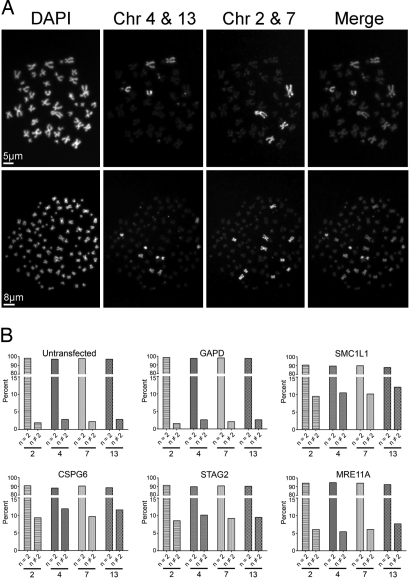
To establish that the increases in DNA content observed by flow cytometry actually reflected increases in chromosome numbers, we examined mitotic spreads after targeted knockdown. Increases in total chromosome numbers were indeed observed after SMC1L1, CSPG6, STAG2 or MRE11A silencing (SI Table 9). Furthermore, the increases in the near tetraploid and near octaploid populations that were observed in the flow cytometry data (Fig. 1B), were also observed in the chromosome spreads (Fig. 1 D and E). Chromosome paintings revealed increases in the numbers of all four of the assayed chromosomes (chromosomes 2, 4, 7 and 13) after silencing of SMC1L1, CSPG6, STAG2 or MRE11A but not of the GAPD control (Fig. 2).
Fig. 2.
Instability of specific chromosomes in si-RNA treated cells. (A) Representative images of a near diploid (Upper) and near tetraploid (Lower) cells. Chromosomes 4 and 13 were pseudocolored green whereas chromosomes 2 and 7 were pseudocolored red in the merged images. (B) Graphical representations of the fraction of mitotic spreads with normal and aberrant numbers of chromosomes in cells treated with the indicated siRNAs. Each painted chromosome (indicated at the bottom of each panel) was determined to occur at either the normal number (two per spread) or at an aneuploid number (different from 2).
Evidence for Defective Chromatid Cohesion.
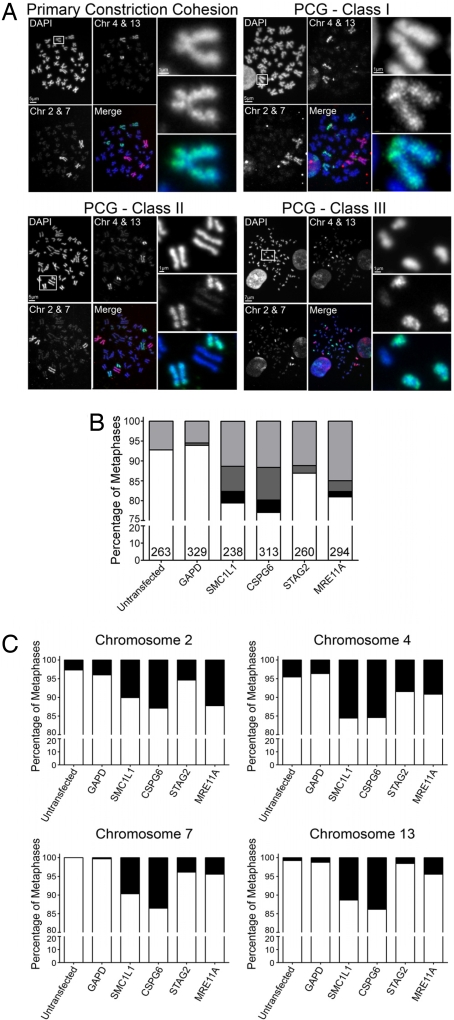
In yeast, mutations causing defective sister chromatid cohesion frequently cause chromosome instability in dividing cells (26, 30, 31). We wished to examine whether sister chromatid cohesion, as measured by primary constriction cohesion, was adversely affected after siRNA-mediated reduction of the four genes studied above. We defined primary constriction gaps (PCG) as a clear and distinct separation in DAPI signal between the sister chromatids at the centromere, where the primary constriction is normally located (centromere). PCG was evaluated in a minimum of 200 DAPI-stained, mitotic spreads from control cells or cells transfected with siRNAs. Reduction of SMC1L1, CSPG6, STAG2, MRE11A gene products, but not that of GAPD, resulted in striking increases in PCG frequencies compared with untransfected control cells (Fig. 3, Table 2). These increases were also reflected in the analysis of each painted chromosome (SI Table 10).
Fig. 3.
Down-regulation of mutated gene products induces cohesion defects. (A) Representative images of mitotic spreads demonstrating normal primary constriction cohesion (Upper Left) and various classes of defective cohesion; PCGI (mild), PCGII (moderate), and PCGIII (severe). Each cohesion category (quadrant) is composed of a low resolution image of the entire mitotic spread in which the DAPI (blue), FITC (green), Texas Red (red), and merged images are presented. Each DAPI image contains a white box that defines the region magnified and presented in the three right-hand panels of each quadrant. For illustrative and comparative purposes, the high-resolution images all contain an FITC-labeled chromosome 4. The high resolution merged image (Bottom) has the DAPI (Top) and FITC (Middle) channels pseudocolored blue and green, respectively. (B) Graphs of the fractions of cells with normal or defective cohesion of the three different classes (PCGI, light gray; PCGII, dark gray; and PCGIII, black). The total number of mitoses scored in each experiment is denoted at the bottom of each bar. (C) The fraction of metaphases with cohesion that was normal (white column) or abnormal (any of the three classes, black column) is presented for each of the four chromosomes studied in cells treated with the indicated siRNAs.
Table 2.
Increased primary constriction gaps after siRNA-induced knockdown
| Treatment | n* | Normal† | Primary constriction gaps (PCG) |
Total†‡ | FITotal§ | ||
|---|---|---|---|---|---|---|---|
| PCGI† | PCGII† | PCGIII† | |||||
| Untransfected | 263 | 92.78 | 7.22 | 0.00 | 0.00 | 7.22 | 1.0 |
| GAPD | 329 | 93.92 | 5.47 | 0.61 | 0.00 | 6.08 | 0.84× |
| SMC1L1 | 238 | 79.41 | 11.34 | 6.30 | 2.94 | 20.59 | 2.85× |
| CSPG6 | 313 | 77.04 | 11.64 | 8.18 | 3.14 | 22.96 | 3.18× |
| STAG2 | 260 | 86.92 | 11.15 | 1.92 | 0.00 | 13.08 | 1.81× |
| MRE11A | 294 | 80.95 | 14.97 | 2.72 | 1.36 | 19.05 | 2.64× |
*Number of mitotic spreads included in the analyses.
† Values represent percentages.
‡Total represents the sum of PCGI + PCGII + PCGIII.
§FITotal represents the relative fold increase of the total PCG for the indicated condition over that of the untransfected control population.
To further characterize the severity of the PCG phenotype, PCG was further subdivided into three distinct classes. PCGIII was defined as those spreads in which no semblance of cohesion could be identified, PCGII was defined as those in which three or more of the eight painted chromosomes exhibited gaps, and PCGI were defined as spreads in which one or two painted chromosomes exhibited gaps (examples of each class are illustrated in Fig. 3A). Reduction of all four yeast CIN gene homologs resulted in increases in PCGI and PCGII relative to the GAPD knockdown or untransfected control cells, and three of the four resulted in spreads with PCGIII (Fig. 3 B and C and Table 2).
Cells Lacking CDC4 Exhibit CIN and Have an Underlying Cohesion Defect.
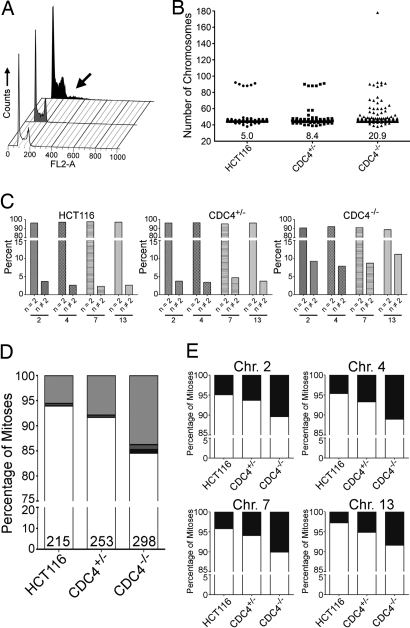
The results described above demonstrated that the homologs of yeast CIN genes that were mutated in human tumors were connected through the control of chromosome cohesion. We therefore were interested in determining whether reduced expression of CDC4 (FBXW7), which has been reported to be the most frequently mutated gene associated with CIN (32), might also cause aberrant cohesion. Previous studies have shown that disruption of CDC4 in human cells caused CIN in association with decreases in its cyclin E substrate, but no evidence associating CDC4 with chromatid cohesion has heretofore been available (20). As a result of the availability of HCT116 cells in which the CDC4 gene was disrupted through targeted homologous recombination, we were able to assay cohesion in paired cell lines with or without CDC4 alleles (20). Flow cytometry conducted on logarithmically growing cell populations revealed an increase in the proportion cells in the >G2/M population in the CDC4−/− cells compared with the control CDC4+/− cells or the parental HCT116+/+ cells (Fig. 4A and SI Table 11). The flow cytometric data indicated that the increased DNA content of the CDC4−/− cells was more likely to reflect an increase in a portion of the chromosomes (i.e., near diploid aneuploidy) than in all chromosomes (which would have resulted in near tetraploid cells). This result was confirmed by the analysis of mitotic chromosome spreads and whole chromosome paintings. Fig. 4B shows that the proportion of cells with chromosome numbers between 46 and 80 were increased in CDC4−/− cells vs. CDC4−/+ or CDC4+/+ cells, in agreement with previous results (20). Chromosome specific paints showed that individually labeled chromosomes also were present in greater than or less than two copies per cell at increased frequencies (Fig. 4C). Finally, a 2.55-fold increase in the occurrence of primary constriction gaps was observed in the CDC4−/− cells (Fig. 4D and SI Table 12). Unlike the case in cells with SMC1L1 or MRE11A knockdown, only a very modest number of PCGII or PCGIII mitotic spreads were identified (Fig. 4D). The increases in primary constriction gaps were confirmed in each tested chromosome by using paints (Fig. 4E).
Fig. 4.
Cells with disrupted CDC4 exhibit cohesion defects. (A) Flow cytometric analyses of HCT116 control cell and those with heterozygous (CDC4+/−, gray) or homozygous (CDC4−/−, black) null alleles. The arrow highlights the CDC4−/− cells that exhibited increases in DNA content (i.e., aneuploidy). (B) Scatter plots depicting the total chromosome number distribution for at least 300 mitotic spreads from cells with the indicated genotype. Percentage of mitotic spreads with >46 chromosomes is indicated at the base of each column. (C) Bar graphs indicating the fraction of mitotic spreads with normal (n = 2) or abnormal (n ≠ 2) chromosome numbers as identified by whole chromosome paints. (D) Graphs of the fractions of cells with normal (white) or defective cohesion of the three different classes (PCGI, light gray; PCGII, dark gray; and PCGIII, black). The total number of mitoses scored in each experiment is denoted at the bottom of each bar. (E) The fraction of metaphases with cohesion that was normal (white column) or abnormal (any of the three classes, black column) is presented for each of the four chromosomes studied in cells with the indicated genotypes.
Discussion
Ten of the 11 mutations (91%) we identified in this study occurred in homologs of genes that directly contribute to sister chromatid cohesion in yeast. Because only 25% of the 102 genes studied were cohesion-related, this result was highly significant. To provide evidence that these human genes actually control sister chromatid cohesion in human cells, we tested the effect of their down-regulation upon siRNA treatment. We found that three genes predicted to be homologs of the SMC1, SCC3, and SMC3 genes in yeast each resulted in chromatid cohesion defects and chromosome instability in human cells when down-regulated. We also tested the effects of down-regulation of MRE11 and CDC4, the two most commonly mutated human candidate CIN genes identified in previous sequencing studies. Knockdown of MRE11 as well as knockout of CDC4 resulted in unambiguous cohesion defects despite the fact that these proteins have been postulated to play other roles in cell biology. Taken together, our results suggest that genes involved in sister chromatid cohesion are common mutational targets whose disruption leads to chromosome instability in colorectal cancers.
SMC1 and SMC3, together with SCC1 and SCC3, form the essential cohesin complex required for cohesion of sister chromatids and for accurate chromosome segregation in yeast (33). The human homologs of SMC1 (SMC1L1), SMC3 (CSPG6), and SCC3 (STAG3) were all found to be mutated in colorectal cancers. SCC2, together with SCC4, forms part of an essential complex that loads cohesin onto chromosomes (34), and the human homolog of SCC2 (NIPBL) was also found to be mutated in colorectal cancers. Thus, components of both of the key complexes required for assembling and loading cohesin on chromatids were found to undergo genetic alterations. Recently, NIPBL and SMC1L1 were found to be mutated in Cornelia de Lange (CdL) syndrome, characterized by facial dysmorphisms, upper limb abnormalities, growth delay and cognitive retardation (35–37). The phenomenon of precocious sister chromatid separation has been described in CdL syndrome, Roberts syndrome, mosaic variegated aneuploidy, and various cancers (38). SCC-112 is a PDS5-like protein that may play a role in stable maintenance of cohesin-mediated cohesion by modulating the interaction of cohesin with chromatin (39). In yeast, alterations of the MRX complex genes (including a homolog of the human MRE11 gene) have been shown to lead to defective chromatid cohesion, although this property of MRE11 has not been previously investigated in human cells. Similarly, CDC4 was known to play a role in chromosome stability and to have a variety of other functions, but there were no previous data linking CDC4 mutations to cohesion defects. Our study demonstrates that down-regulation of these genes leads to cohesion defects in human cells, and also identifies somatic mutations in human cancers in genes required for sister chromatid cohesion.
The results presented here are consistent with the idea that individual genes within the same pathway can each be mutated in relatively small fractions of cancers but in aggregate point to a pathway that is commonly altered (40). All genes putatively implicated in CIN, including those analyzed herein, have been found to be individually mutated in only a minority of cancers (19–21). In sum, however, at least one of the genes affecting proper chromatid cohesion (SMC1L1, CSPG6, NIPBL, STAG3, MRE11A, and CDC4) is mutated in >20% of colorectal cancers. BUB1, although mutated very infrequently, also controls cohesion, either indirectly, through maintenance of the spindle checkpoint (19, 41), or directly, through recruitment of SgoI to kinetochores (42). It is tempting to speculate that CIN is a direct consequence of mutations in these genes in a subset of colorectal cancers. However, it will require larger studies evaluating all cohesin homologs in a larger panel of tumors to know the true prevalence of mutations in this pathway in colorectal cancers. Moreover, although four of the 10 mutations observed in the genes noted above clearly disrupt function through the creation of stop codons or frameshift mutations (Table 2) the six missense mutations identified do not necessarily have functional effects. Further experiments, particularly knockins of mutant alleles, will be required to definitively address this question. Because CIN is a hallmark of many forms of malignancy in addition to those of the colon, it will also be informative to evaluate cohesion-controlling genes in other common solid tumors.
Recognition that genes involved in chromatid cohesion are often mutated in colorectal cancers has practical as well as basic scientific implications. In particular, it is conceivable that defects in cohesion can be exploited, exposing an “Achilles heel” not found in normal tissues (26). In yeast, it has been shown that combinations of mutations lead to synthetic lethality even when the individual mutations are not essential for growth (43, 44). For example, mutations in four nonessential genes (ctf4Δ, ctf8Δ, ctf18Δ, and dcc1Δ) are synthetically lethal when combined with mutations in five different CIN gene homologues (mre11, smc1, smc3, scc2, and pds1). The homologs of each of the latter five genes are mutated in colorectal cancers (Table 2). The yeast data indicate that it would be worthwhile to search for human genes that, when inactivated, result in the death of cells with a mutation in a chromatid cohesion gene. Such genes could be identified bioinformatically using cross-species comparisons with the yeast genes that form synthetic lethal partners. Alternatively, experimental screens for such synthetic lethality could be performed with siRNA libraries (45). The protein products of such genes, when inhibited by a drug, would specifically kill tumor cells and thereby be attractive drug targets for the treatment of CIN tumors.
Materials and Methods
By using the Refseq database and BLASTp, yeast CIN genes identified in comprehensive genome-wide screens (26) were used as queries to search for top-ranked human homologues. Genes that previously had been evaluated in human colorectal cancers were excluded (19, 21, 46), resulting in a collection of 102 genes. PCR amplification, sequencing, and sequence analysis of all coding exons of these genes were performed on tumor DNA from 36 cancers as described in ref. 29. When a gene was found to harbor a mutation in at least one of the 36 cancers, it was subsequently analyzed in 95 additional cancers. siRNA duplexes targeting SMC1L1, CSPG6, STAG2, and MRE11A were purchased from Dharmacon (Lafayette, CO). Western blots were conducted on proteins extracted from asynchronous and subconfluent cells 1–10 days posttransfection, essentially as described elsewhere (47). Triplicate populations of asynchronous and subconfluent cells were harvested seven days after transfection and prepared for flow cytometry as detailed (47, 48). Mitotic spreads were prepared and imaged by using methods based on those described in ref. 39, 49, and 50.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Abcam for generously providing the antibodies used in this study. This work was supported by the Virginia and D. K. Ludwig Fund for Cancer Research; The Clayton Fund; the National Colorectal Cancer Research Alliance; The Pew Charitable Trusts; National Institutes of Health Grants CA 43460 (to B.V.), CA121113 (to V.E.V.), GM62368 (to F.S.), and CA016519 (to P.H.); and Canadian Institutes of Health Research (CIHR) Grant MOP-38096 (to P.H.). K.M. is supported by fellowships from CIHR and the Michael Smith Foundation for Health Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712384105/DC1.
References
- 1.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 2.Duesberg P, et al. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887–4906. [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto H, Imai K, Perucho M. Gastrointestinal cancer of the microsatellite mutator phenotype pathway. J Gastroenterol. 2002;37:153–163. doi: 10.1007/s005350200015. [DOI] [PubMed] [Google Scholar]
- 6.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 7.Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 8.Leach FS, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 9.Fishel R, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulos N, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 11.Bronner CE, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 12.Nicolaides NC, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 13.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 14.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 15.Doxsey S. Duplicating dangerously: Linking centrosome duplication and aneuploidy. Mol Cell. 2002;10:439–440. doi: 10.1016/s1097-2765(02)00654-8. [DOI] [PubMed] [Google Scholar]
- 16.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Cahill DP, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 20.Rajagopalan H, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.can-04-0587. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan KB, et al. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 23.Fodde R, et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 24.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: Cutting through the mystery. Nat Rev Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 25.Hieter P, Bassett DE, Jr, Valle D. The yeast genome: A common currency. Nat Genet. 1996;13:253–255. doi: 10.1038/ng0796-253. [DOI] [PubMed] [Google Scholar]
- 26.Yuen KW, et al. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci USA. 2007;104:3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 29.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 30.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storchova Z, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 32.Rajagopalan H, Lengauer C. hCDC4 and genetic instability in cancer. Cell Cycle. 2004;3:693–694. doi: 10.4161/cc.3.6.940. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 34.Ciosk R, et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 35.Musio A, et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 36.Krantz ID, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 38.Kaur M, et al. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet A. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, et al. A multidimensional analysis of genes mutated in breast and colorectal cancers. Genome Res. 2007;17:1304–1318. doi: 10.1101/gr.6431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perera D, et al. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 43.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 44.Pan X, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 45.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 46.Wang TL, et al. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McManus KJ, Biron VL, Heit R, Underhill DA, Hendzel MJ. Dynamic changes in histone H3 lysine 9 methylations: Identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J Biol Chem. 2006;281:8888–8897. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- 48.Bickar D, Reid PD. A high-affinity protein stain for western blots, tissue prints, and electrophoretic gels. Anal Biochem. 1992;203:109–115. doi: 10.1016/0003-2697(92)90049-d. [DOI] [PubMed] [Google Scholar]
- 49.Gimenez-Abian JF, et al. Regulated separation of sister centromeres depends on the spindle assembly checkpoint but not on the anaphase promoting complex/cyclosome. Cell Cycle. 2005;4:1561–1575. doi: 10.4161/cc.4.11.2146. [DOI] [PubMed] [Google Scholar]
- 50.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.