Abstract
Over the last decades, bacterial chemotaxis in Escherichia coli has emerged as a canonical system for the study of signal transduction. A remarkable feature of this system is the coexistence of a robust adaptive behavior observed at the population level with a large fluctuating behavior in single cells [Korobkova E, Emonet T, Vilar JMG, Shimizu TS, Cluzel P (2004) Nature 428:574–578]. Using a unified stochastic model, we demonstrate that this coexistence is not fortuitous but a direct consequence of the architecture of this adaptive system. The methylation and demethylation cycles that regulate the activity of receptor-kinase complexes are ultrasensitive because they operate outside the region of first-order kinetics. As a result, the receptor-kinase that governs cellular behavior exhibits a sigmoidal activation curve. We propose that the steepness of this kinase activation curve simultaneously controls the behavioral variability in nonstimulated individual bacteria and the duration of the adaptive response to small stimuli. We predict that the fluctuating behavior and the chemotactic response of individual cells both peak within the transition region of this sigmoidal curve. Large-scale simulations of digital bacteria suggest that the chemotaxis network is tuned to simultaneously maximize both the random spread of cells in the absence of nutrients and the cellular response to gradients of attractant. This study highlights a fundamental relation from which the behavioral variability of nonstimulated cells is used to infer the timing of the cellular response to small stimuli.
Keywords: agent-based, fluctuation–dissipation, noise, ultrasensitivity
Molecular noise (i.e., stochastic fluctuations) has been largely reported as one important source of phenotypic variability in a number of biological systems as diverse as gene expression and signal transduction in prokaryotes and eukaryotes (1–3). A standard method to characterize noise in biological systems is to analyze the distribution of behaviors across a population of cells at steady state. This approach has been powerful in identifying specific molecular mechanisms responsible for controlling phenotypic variability. Alternatively, the temporal evolution of noise within a single cell also contains key information about underlying cellular dynamics, but this approach is seldom used to characterize cellular behavior outside the steady-state regime (4). It is conceivable, however, that biological systems that are sensitive to intracellular spontaneous noise are also sensitive to small extracellular perturbations such as a sudden change of environmental conditions (5). We wish to investigate whether there exists in bacterial chemotaxis a general relationship between the fluctuation of cellular behavior in single cells and the timing of the cellular response to a small external stimulus.
Bacterial chemotaxis, a cellular locomotion system, has become a classic model for signal transduction. Although the chemotaxis network consists of just a few molecular species, it can perform complex cellular functions such as adaptation in response to environmental changes. Escherichia coli bacteria swim using flagella activated by rotary motors. When motors spin counterclockwise (CCW), the flagella form a corkscrew bundle whose rotation propels the cell in a smooth trajectory called a run. Clockwise (CW) spinning of motors favors the disruption of the bundle and causes the cell to tumble. The trajectory of a swimming cell resembles that of a random walk, which consists of a succession of runs and tumbles. When exposed to a sudden increase of chemical attractant, the cell lengthens the time interval between consecutive tumbles. This modulation of tumbling rate allows bacteria to bias their random walk toward a source of attractants and to perform chemotaxis.
A combination of experiments and models has demonstrated that, at the population level, E. coli bacteria display robust exact adaptation: After an initial response to a chemical stimulus, the average tumbling rate returns precisely to its steady prestimulus level (6–8). Remarkably, these studies showed that the property of exact adaptation is robust to variations of biochemical parameters such as the concentration and reaction rates of the chemotaxis proteins (7, 8). In contrast with population measurements, recent single-cell experiments have revealed that the switching behavior of a single motor, from a cell adapted to a homogeneous environment, exhibits large temporal fluctuations (9, 10). It was found that the characteristic time scale of these fluctuations was so large that a steady tumbling rate could be defined only when taking time averages longer than several hundred seconds. The amplitude of these fluctuations, hereafter called noise, exceeded that expected from Poisson statistics. Very long time series of switching events from individual flagellar motors exhibited distributions of run (CCW) intervals with long tails. These distributions are in general not exponentially distributed as it was previously believed (9, 10).
Here, we develop a unified stochastic model of adaptation in bacterial chemotaxis and use the fluctuation–dissipation theorem (11, 12) to interpret how the noise in nonstimulated cells is related to the cellular response to a small chemical stimulus. We show that the design of the activity-dependent (de)methylation cycles that provide robust adaptation is equivalent to the architecture of a covalent modification cycle (13). We demonstrate how the large behavioral variability observed experimentally in single nonstimulated bacteria arises from the amplification of molecular noise in the ultrasensitive covalent modification cycle. As a result, the steady-state activity of the kinase as a function of the ratio of the concentrations of the modifying enzymes CheR and CheB is sigmoidal (Hill coefficient >1). We therefore analyze how the slope of the kinase activation curve simultaneously controls both the amplitude of spontaneous fluctuations and the relaxation time of the system in response to a small stimulus. The prediction emerging from this analysis is that the noisiest cells should also exhibit the largest chemotactic response.
Results
Kinetics of the Adaptation Module.
The adaptation system in bacterial chemotaxis is governed by a well defined module of specific interacting proteins (Fig. 1). This adaptation module consists of two antagonistic enzymes, CheR and phosphorylated CheB (CheB-P), which respectively add and remove methyl groups at multiple receptor residues of the receptor-kinase complex. This series of methylation and demethylation cycles controls the activity of the histidine kinase CheA in the complex. Methylation and demethylation are complex biochemical processes that involve the enhanced recruitment of CheR and CheB to the receptors by a tethering site that is distinct from methylation sites (14, 15). As in previous models of chemotaxis (7, 16–23), we coarse-grain the underlying biochemical details to capture the behavior of the full chemotaxis system obtained from experiments on living cells. In particular, we assume that receptor-kinase complexes can be either active or inactive (23), and we use the Barkai and Leibler activity-dependent feedback in the receptor modification system that yields robust exact adaptation (7, 8) (Fig. 1B). We use Michaelis–Menten kinetics for the methylation–demethylation steps of the complexes. Here, CheR (CheB-P) binds the methylation sites only of inactive (active) receptors (16–23). But in Sect. 4.2 of supporting information (SI) Appendix, we also consider two alternative hypotheses where CheR methylates the receptors in both active and inactive conformations, or where only the catalytic step of the methylation reaction is activity dependent. Only the latter assumption is incompatible with single-cell experiments. We report separately the concentration of free receptor complexes and the concentrations of receptor complexes bound to CheR and/or CheB-P. Under these conditions, the temporal evolution of the average methylation level, M, of free receptors throughout the methylation–demethylation cycles (19) obeys Eq. 1 (see Materials and Methods):
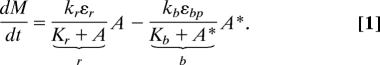 |
Here, A* and A are the concentrations of free active and inactive receptor complexes, and r and b are the rates of methylation and demethylation of inactive and active receptors complexes. The parameters εr and εbp are the concentrations of CheR and CheB-P. Kr and Kb are the effective Michaelis–Menten constants for the methylation–demethylation of a receptor complex, and kr and kb are the corresponding catalytic rates. The total concentration of receptor complexes is constant:
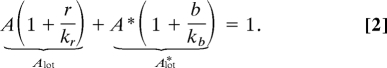 |
The first (second) term in Eq. 2 represents the total concentration of inactive (active) receptor complexes including those bound to CheR and CheB. Eq. 2 is equal to 1 because we normalize all of the concentrations (A, εr, Kr, etc.) with the concentration, N, of receptor complexes present in the system. The concentration of active and inactive receptors at steady state, Atot* and Atot, is independent from the ligand concentration (see Materials and Methods), which is consistent with the property of perfect adaptation (7, 18). We model the phosphorylation cascade using mechanisms and parameter values similar to those proposed by Sourjik and others (19, 20, 24): The rate of autophosphorylation of the kinase CheA is proportional to the total kinase activity Atot* and the phosphate transfer from the kinase to CheY and CheB is proportional to the concentration of phosphorylated kinases (Eqs. S6–S8 of SI Appendix).
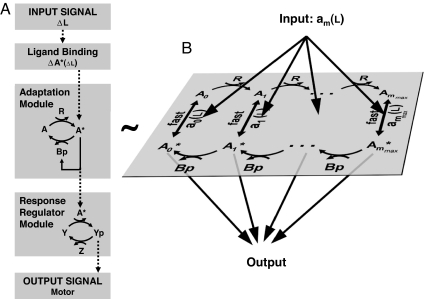
Fig. 1.
Modular representation of the chemotaxis system. (A) Transmembrane receptors bind the ligand (L) and control the activity of histidine kinases CheA (A). The kinase CheA phosphorylates the response regulator CheY (Y) into the active form CheY-P (Yp). CheY-P diffuses throughout the cell and interacts with the flagellar motors to induce clockwise rotation (tumble). The phosphatase CheZ (Z) dephosphorylates CheY-P (42). A sudden increase of ligands ΔL causes the kinase activity to decrease by ΔA*. The chemotaxis system is equipped with an adaptation module in which two antagonistic enzymes regulate the activity of the kinase-receptor complexes. The methyltransferase CheR (R) catalyzes the autophosphorylation of CheA by methylating the receptors. The active kinase A* phosphorylates the methylesterase CheB in CheB-P (Bp). CheB-P removes methyl groups from active receptor complexes, which catalyzes kinase deactivation. (B) The adaptation module consists of a series of slow (de)methylation reactions that modulate the activity of the receptor complexes. We use a two-state model where the probability am of a receptor complex to be in active conformation depends on the occupancy of its ligand binding sites and on the level of methylation of the receptors that ranges within m = 0, …, mmax (7, 17–19). mmax is the total number of methylation sites. We assume that CheR only methylates inactive complexes (43), whereas CheB-P only demethylates active complexes (44) (details on the model and alternative hypotheses in Sect. 4 of SI Appendix).
Importantly, we find that the equation (Eq. S15 of SI Appendix) defining the steady state of the kinase activity is identical to the equation obtained by Goldbeter and Koshland (13) for covalent modification of a common substrate by two antagonistic enzymes (compare figure 3 in ref. 13 with Eq. S15 of SI Appendix). Thus, the adaptation module (Fig. 1) should share some of the properties of a covalent modification cycle. As in ref. 13, the normalized constants, Kr and Kb, control the steepness of the activation curve of the kinase. When Kr and/or Kb are <1, the activation curve is steeper than a hyperbolic function (Hill coefficient >1). The ratio α = krεr/kbεbp of the maximal enzymatic velocities determines the steady-state activity of the kinase (13). Because the ratio α is proportional to the ratio of [CheR] to [CheB-P], it can be easily adjusted by changing the relative concentration of these two proteins.
Fig. 3.
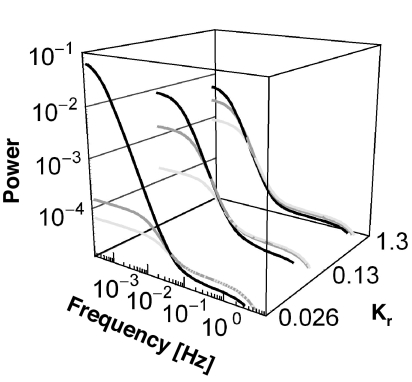
Power spectra of the fluctuations of the output signal (CheY-P) from nonstimulated cells. Shown are 1-fold (black), 2-fold (gray), and 4-fold (light gray) wild-type levels of CheR for a fixed wild-type level of [CheB]. For Kr and Kb, the spectra are in agreement with experiments in ref. 10. For Kr and Kb 5 times smaller or 10 times larger, the differences between the power spectra of wild-type and mutants are too large or too small in comparison with those in ref. 10.
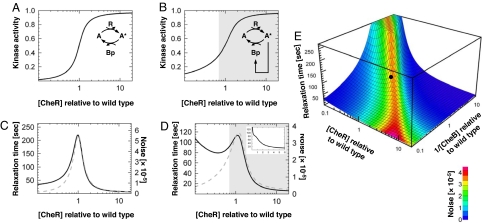
Using recent biochemical parameters (Table S1 of SI Appendix), we plotted the steady-state activity of the kinase, Atot*, as a function of [CheR] and for [CheB] fixed at wild-type level (Fig. 2A). In the absence of the CheB-P feedback loop, the kinase activation curve exhibits an effective Hill coefficient of 3.5. In the presence of the CheB-P feedback loop, the curve is less steep (Hill coefficient ≈2.5) (Figs. 2B). This feedback loop also helps maintaining the kinase activity within the narrow functioning range of the rotary motor (20). The kinase activity as a function of 1/[CheB], and [CheR] fixed at wild-type level, exhibits a Hill coefficient >1 as well (data not shown). These results suggest that the adaptation module in bacterial chemotaxis is more sensitive, namely ultrasensitive (13), to small variations of [CheR] and [CheB-P] than a standard hyperbolic activation curve. Outside of the transition region of the sigmoid (Fig. 2B), the kinase would be either fully active or fully inactive. Thus, we expect the ratio α of the methylation and demethylation velocities from wild-type cells to lie within the transition region of the sigmoid where the activity of the kinase can vary (see alternative models in Sect. 4 of SI Appendix) (16).
Fig. 2.
Sensitivity of the adaptation module without (A and C) and with (B, D, and E) the CheB-P feedback loop on CheA. (A) Total kinase activity Atot* as a function of [CheR] for a fixed wild-type level of [CheB] (model parameters in Table S1 of SI Appendix). Hill coefficient, H ∼ 3.5. (C) Relaxation time τa (black) and variance σa2 (gray) of the noise associated with the total kinase activity. (B and D) Same as A and C with the CheB-P feedback loop on CheA. H ∼ 2.5. Gray-shaded area, functioning range of the motor [1.5 < [CheY-P] < 4.5 μM (45)]. (Inset) Relaxation time averaged from a population of cells (104) with cell-to-cell variability in the levels of CheR and CheB (see Sect. 5 of SI Appendix). (E) Relaxation time (surface) and variance of the noise (color) associated with the total kinase activity as a function of [CheR] and 1/[CheB]. The black dot indicates the wild-type cell (Table S1 of SI Appendix).
Stochastic Model of the Adaptation Module.
Recent experiments showed that individual E. coli bacteria adapted to a homogeneous environment exhibit large temporal variations in their behavior (10). It was hypothesized that the observed behavioral variability was due to the fluctuations in kinase activity. More specifically, the slow methylation–demethylation processes in the adaptation module govern the fluctuations in kinase activity. Three complementary experiments in which cells did not display large fluctuations at long time scales support this hypothesis: (i) when [CheY-P] was not regulated by the chemotaxis network but substituted by the active mutant CheYD13K stably expressed from an inducible plasmid; (ii) in mutant cells whose receptors have a fixed methylation level; (iii) behavioral variability was found tunable in ΔcheR mutant cells complemented with various levels of [CheR]. Behavioral variability decreased from its maximal to minimal value when [CheR] increased from 1- to 4-fold wild-type level (10).
In this section, we wish to discuss how the steep kinase activation curve (Hill coefficient >1) (Fig. 2B) could amplify the spontaneous stochastic fluctuations associated with the methylation and demethylation reactions. We show that the amplification of fluctuations in kinase activity is large enough to produce the large behavioral variability observed experimentally in nonstimulated cells (10). Chemotaxis assays are usually performed in media that do not support growth but that provide in excess the essential elements to produce energy. Consequently, we consider bacteria that are adapted to a homogenous environment to have reached the equilibrium (25). Under this condition, thermal fluctuations cause spontaneous fluctuations of the rate constants in the chemotaxis network (11, 26).
We use a linear noise approximation (see Sect. 2.5 of SI Appendix) (11, 12, 27, 28) to derive the spontaneous noise in the kinase activity (and in [CheY-P]) around the steady state (Fig. 2 A and B). This approximation allows us to compute the relaxation time, τa, and the power spectrum of the spontaneous fluctuations in the output signal of the chemotaxis system. The relaxation time is the typical time scale that defines the steady state of a nonstimulated cell. At time scales shorter than τa, the spontaneous fluctuations of the system are strongly correlated and the behavior is unsteady.
For the sake of simplicity, we first ignore the CheB phosphorylation step. The spontaneous fluctuations in kinase activity, δA*, include “fast” fluctuations in receptor activity due to the binding and unbinding of ligand and also slow fluctuations βδM associated with the methylation–demethylation reactions (see Materials and Methods). At long time scales, only the slow fluctuations are relevant and δA* ≅ βδM. Insertion of the latter relation into Eq. 1 and the use of the linear noise approximation yield the noise, δAtot*, associated with the total kinase activity in nonstimulated cells:
Here, δηa is a source of stochastic white noise and Da is the strength of the spontaneous fluctuations associated with (de)methylation reactions of the receptors. Within the linear noise approximation, the individual methylation and demethylation steps follow simple Poisson statistics. Summing up the contributions from all of the methylation and demethylation steps, we find that Da is proportional to the sum of the methylation and demethylation velocities (see Sect. 2.5 of SI Appendix):
Interestingly, Da is independent of the slope (i.e., the Hill coefficient) of the kinase activation curve [bA* is approximately kbεbp/(1 + 2Kb) ≅ kbεbp in the middle of the transition]. By contrast, we find that the relaxation time, τa, is proportional to the slope of the kinase activation curve (see Materials and Methods):
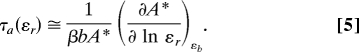 |
Eq. 3 is analogous to the Langevin equation γẋ = −κx + f(t) that describes the fluctuations of a mass-spring system in a viscous fluid, with spring constant κ = 2kBT/(τaDa), damping constant γ = 2kBT/Da, and random force f(t). Fluctuations and dissipation are related through the fluctuation–dissipation theorem 〈f(t)f(t′)〉 = 2kBTγδ(t − t′), where T is the temperature and kB is the Boltzmann constant (11). As a consequence, the variance of the spontaneous fluctuations in kinase activity δAtot* is σa2 = kBT/κ = τaDa/2. Thus, increasing the Hill coefficient of the kinase activation curve is equivalent to decreasing the spring constant κ in the mass-spring system without changing the damping constant γ. The higher the Hill coefficient, the larger the fluctuations of the kinase activity, σa2, and the longer the relaxation time, τa. Importantly, we find that τa and 1/σa2 calculated for the full adaptation module are proportional to the relaxation time and the inverse of the noise for one covalent modification cycle [e.g., the Goldbeter and Koshland system (13)] with the proportionality factor 1/β.
A practical way to study the behavioral variability of a nonstimulated bacterium is to plot the power spectrum associated with the spontaneous fluctuations in the output of the full chemotaxis system. The Fourier transform of Eq. 3 gives the power spectrum of the spontaneous fluctuations of the kinase activity. Similarly, we also calculate the power spectrum of the fluctuations of [CheY-P] from the full pathway that includes the CheB-P feedback loop (see Sect. 2.6 of SI Appendix). In Fig. 3, we plot the power spectra of the fluctuations of [CheY-P] for various values of [CheR] and the Michaelis–Menten constants Kr and Kb. Using recently published biochemical parameters (29), we find that this model best reproduces single-cell experiments (power spectra) (10) when the following two conditions are fulfilled: (i) Kr and Kb are ≈10−1 and (ii) the methylation and demethylation catalytic rates kb and kr are adjusted such that wild-type cells, like in experiments, exhibit the noisiest behavior (see Table S1 of SI Appendix). The values of these parameters are also in agreement with biochemical data and parameters used in earlier models (see Sects. 3.2 and 4 of SI Appendix). By contrast, when Kr, Kb ≈ 1, we find that the behavioral variability is not as sensitive to variations in [CheR] as in experiments (10). Similarly, when Kr, Kb ≈ 10−2, the difference in behavioral variability between wild-type and mutants cells that express 4-fold wild-type level of [CheR] is too large in comparison with experiments (10).
The power spectra in Fig. 3 exhibit two characteristic (knee) frequencies. At low frequency, the fluctuations of kinase activity govern the fluctuating behavior of the full system. The position of the knee at long time scales reflects the magnitude of σa2 and τa, which depend on [CheR] and [CheB-P]. At short time scales, the second knee frequency corresponds to the spontaneous fluctuations occurring within the phosphorylation cascade, and its position depends only on the time scale associated with the phosphorylation of CheY (Fig. 1A). Next, we plotted the noise in kinase activity, σa2, and the relaxation time, τa, as a function of [CheR], in the absence or presence of the CheB-P feedback loop (Fig. 2 C and D). As expected for a covalent modification cycle with a Hill coefficient >1 (13, 30–32), σa2 and τa peak within the transition region of the kinase activation curve (Fig. 2C). This effect is similar to the one observed in refs. 30 and 32. In the presence of the CheB-P feedback loop, the overall profiles of the variance and relaxation time as a function of [CheR] are conserved, with the exception that τa decreases at very low levels of CheR (Fig. 2D). We also find that the CheB-P feedback loop slightly reduces the noise and the relaxation time (see detailed analysis in Sect. 2.4 of SI Appendix). We obtain similar results when we compute σa2 and τa as a function of 1/[CheB]. In Fig. 2E, we plot σa2 and τa as a function of both [CheR] and 1/[CheB] for the whole system including the CheB-P feedback loop. The presence of the crest over the hyperbolic surface is the signature of the deviation from first-order kinetics. As expected, large fluctuations in kinase activity coincide with long relaxation times.
Figs. 2 and 3 suggest that in wild-type cells, the ratio α of the methylation and demethylation velocities is tuned to the transition region of the kinase activation curve (Fig. 2B) to exhibit fluctuations (Fig. 3) similar to those observed experimentally (10). This model is also compatible with the experimental observation in single cells where behavioral variability is maximal when CheR is expressed at wild-type level and decreases to the variability expected from Poisson statistics when [CheR] is expressed at 4-fold wild-type level (10).
Relationship Between Behavioral Variability in Nonstimulated Cells and the Timing of the Chemotactic Response to Small Stimuli.
The critical hypothesis in this section is that the behavioral variability observed in nonstimulated single cells is fundamentally related to the timing of the response of stimulated cells. Because stimuli encountered by the bacterium in its natural habitat may be small, we calculate the response to stimulus using a linear perturbation analysis of the kinetic system. Although we chose to keep our study independent from the actual chemical stimulus present in the environment, the chemotactic response in the real system also depends on the initial amplification of the input stimulus mediated by complex allosteric mechanisms taking place in the receptor-kinase complex (17, 22, 33–37). In this model, we consider that a small external perturbation, such as a sudden exposure to attractant, causes an “instantaneous” change of the receptor activity, ΔAinput*. During the subsequent adaptation, the changes ΔM in the methylation level of the receptors govern the changes in kinase activity. We linearize Eq. 1 and eliminate ΔM (see Materials and Methods) to obtain the input–output relationship of the adaptation module solely in terms of changes in total kinase activity ΔA*input:
Within this linear approximation, the pathway relaxes with the same time scale τa in response to either an external stimulus or internal spontaneous noise (Eqs. 3 and 6). Thus, one can infer the sensitivity of the chemotactic response ΔAtot*(t) to a small external perturbation ΔAinput* by characterizing the time correlation of the noise, δAtot*, in nonstimulated cells. In this picture, a large τa will cause on average long runs following a small step stimulus ΔAinput*. Consequently, we interpret the relaxation time τa as a relative measure of the sensitivity of the chemotaxis system in response to a small external perturbation ΔAinput*. We expect the chemotactic response to peak with the relaxation time τa at about wild-type level of [CheR] and 1/[CheB] (Fig. 2 D and E). For small or large values of [CheR] and/or 1/[CheB], it is conceivable that [CheY-P] becomes too small or too large to fall within the narrow functioning range of the motor. Under this extreme condition, the motor does not switch and the system is not chemotactic. In population measurements (8), it is difficult to observe the peaking of the relaxation time because of the inherent cell-to-cell variability of cheR and cheB expression levels (Fig. 2D Inset and Sect. 5 of SI Appendix), but this prediction should be testable by the means of single-cell experiments.
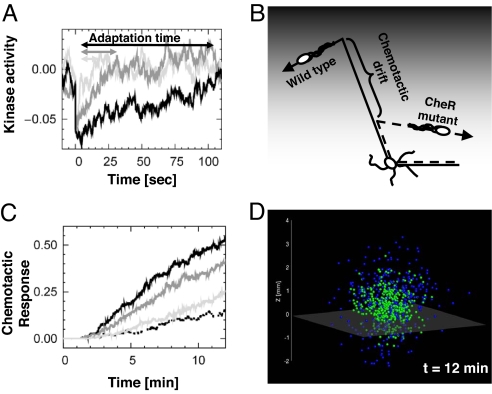
To highlight the significance of the relaxation time for chemotaxis, we hypothesize that the average duration of runs varies like the time τa when cells respond to small input stimuli (Fig. 4A). We anticipate that the chemotactic drift associated with cells swimming up-gradient of nutrients will be larger for wild-type cells with a larger τa than that of CheR mutants (Fig. 4B). We tested this hypothesis by performing large-scale computer simulations with the agent-based simulator AgentCell (38) (www.agentcell.org). We find that the chemotaxis response decreases with small variations in [CheR] relative to wild-type level (Fig. 4C). Small changes in the concentration of CheB from the wild-type level produced similar results (Fig. S8 of SI Appendix). In these simulations, we adjusted the narrow functioning range of the motor so that the CW bias would remain the same in all populations expressing various levels of [CheR] (CW bias = 0.23). In this way, we ensured that the changes in chemotactic response were solely caused by the changes of the relaxation time and not by the differences in tumbling rate.
Fig. 4.
Relationship between relaxation time and chemotactic drift. (A) Temporal evolution of the kinase activity relative to steady state upon sudden deactivation of active receptor complexes for 1-fold (black), 2-fold (gray), and 4-fold (light gray) wild-type level of [CheR] and fixed wild-type level of [CheB]. We normalized the kinase activity with that of wild-type cells. Increasing [CheR] causes a reduction of the relaxation time τa. In all cases, the initial perturbation is ≈4.4% of the wild-type steady-state kinase activity (≈100 receptor complexes). Simulations of a single covalent modification cycle (Fig. 1A) using Stochastirator (http://web.mit.edu/endy/www/sections/resources.html) (parameters are in Table S1 of SI Appendix). (B) Role of the relaxation time in chemotaxis. Cells with longer relaxation time swim farther along the gradient of attractant (gray shade). (C) Effect of variations of [CheR] on the chemotactic response of a bacterial population of 400 cells. Digital swimming bacteria are exposed to a constant gradient of aspartate [dL/dz = 10−8 M/μm, L(z = 0) = 1 μM]. Shown is the percentage of cells in the gradient that are above z = 1 mm as a function of time: 1-fold (black), 2-fold (gray), and 4-fold (light gray) wild-type [CheR] level. Dashed line, wild-type response without gradient. The CW bias for mutant and wild-type cells is 0.23. The initial position of the bacteria is z = 0 mm. (D) Position of the cells from C with 1-fold (blue) and 4-fold (green) wild-type level of CheR after 12 min. Below the gray transparent plane (z = −0.1 mm) there is no nutrient, and bacteria perform an unbiased random walk. Above the plane the random walk is biased upward the gradient of aspartate.
Because wild-type cells display the largest behavioral variability, they spread farther than mutant cells in the direction perpendicular to the gradient of nutrient (Fig. 4D). It is then conceivable that the nonlinearity (i.e., Hill coefficient >1) of the adaptation module (10) confers two advantages. First, it enhances the chemotactic drift by producing a long relaxation time. Second, it produces a large behavioral variability that allows a population of bacteria to explore a wider area. These two predictions should be testable experimentally. In real bacterial populations, cell-to-cell variations of [CheR] and [CheB] arise naturally. Given the fine-tuning of the chemotactic response on [CheR] and [CheB-P], we anticipate the existence of molecular mechanisms to limit the variation of the ratio of the methylation–demethylation velocities. For example, cheR and cheB genes are adjacent on the multicistronic meche operon, a strategy known to reduce independent variations between the expression of the two genes (Fig. S6 of SI Appendix). Another mechanism that helps maintaining the [CheR]/[CheB-P] ratio is the existence of the negative-feedback loop on CheB as illustrated in ref. 20.
This study reconciles the presence of large behavioral variability observed at the single-cell level with the chemotactic response of a cellular population. From the design analysis of the chemotaxis system, we showed that the nonlinearity of the adaptation mechanism simultaneously amplifies both the noise and the relaxation time in the chemotaxis system. This approach highlights a key property of the adaptive system in chemotaxis, which consists of a relationship between cellular response and behavioral variability. Large-scale simulations of digital bacteria suggest that the chemotaxis network is tuned to simultaneously maximize both the random spread of cells in the absence of attractants and the cellular response to gradients of attractant. The ability to infer the timing of cellular response to small stimuli from the noise in nonstimulated cells should be applicable to a wide range of biological systems.
Materials and Methods
Kinetic System.
Derivation of the full kinetic and stochastic models is available in Sect. 2 of SI Appendix. Ligand binding and conformational changes are much faster than the (de)methylation reactions. Therefore, we describe the activity of receptor complexes using equilibrium probabilities am(L) (Fig. 1B) that are functions of the concentration of ligand in the external medium. The subscript m = 0, …, mmax indicates the methylation level of the receptor complex. To ensure perfect adaptation, we assume that a0(L) = 0 and ammax(L) = 1 (18). The methylation–demethylation steps and the time evolution of the concentrations Xm of free (not bound to enzyme) receptor complexes with m methyl groups (Fig. 1B) are governed by (19)
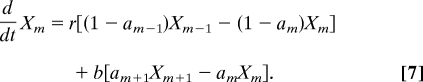 |
Multiplying Eq. 7 with m and summing over all m's gives Eq. 1, where A* = ΣmamXm, A = Σm(1 − am)Xm, and M = ΣmmXm.
Perturbation Analysis of the Adaptation Module.
The perturbation of the kinase activity ΔA* around steady-state Ā* is
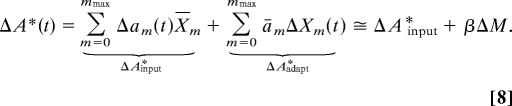 |
To keep our analysis independent from various models of receptors, we use ΔAinput* directly as the input of the adaptation module without detailing the relationship of Δam to changes in ligand concentration (Fig. 1B). The term ΔAadapt* represents the change in kinase activity due to small changes in the methylation levels of the receptors at constant external concentration of ligand, L. In previous models of bacterial chemotaxis (16, 19, 20), the probability of activation of receptors complexes at steady state increases approximately linearly with the number of methyl groups when the external concentration of ligand is small (Fig. S7 of SI Appendix). In our linear perturbation analysis, we take ām ≈ m/mmax. The second term in Eq. 8 becomes ΔAadapt* = βΔM with β = 1/mmax. When considering the slow stochastic fluctuations in nonstimulated cell we have δA* = δAinput* + βδM ≅ βδM. This linear approximation simplifies the analytical treatment while capturing the basic dependence of the kinase activity on methylation level as established by biochemical data (20, 34, 39). We validate our analytical results with stochastic simulations that include nonlinear activation probabilities am(L) (see Sects. 4 and 7 of SI Appendix). In the cases where the activation probability deviates significantly from ām ≈ m/mmax, we can obtain an estimate for the parameter β by expanding the activation probability around the mean steady-state methylation, ām = ā(M) + ā′(M)(m − M) + … Inserting the expansion in Eq. 8 and neglecting the contributions from small changes in the second and higher moments of the distribution of methyl groups, we obtain β = ā′(M) = (∂a/∂m)L=L̄, m=M̄ (see Sect. 2.3 of SI Appendix).
Relaxation Time.
For the case without the CheB-P feedback loop, the linear expansion of Eqs. 1 and 2 yields dδM/dt = −δA*/τGK, where τGK−1 = krθr(1 + θb)/(1 + θr) + kbθb with θr = Krεr/(Kr + A)2, θb = Kbεb/(Kb + A*)2 is the inverse relaxation time for only one modification cycle [e.g., the Goldbeter and Koshland system (13)]. Using δA* ≅ βδM calculated above, we then obtain the relaxation time τa−1 = βτGK−1, as well as Eqs. 3 and 6. The relationship between the relaxation time τa and the slope of the kinase activation curve (Eq. 5) follows from considering the equilibrium relation rA = bA* and its total derivative τGK−1dA* ≅ bA*(d ln εr − d ln ε̄bp) (Eq. S30 of SI Appendix). Without the CheB-P feedback loop and at fixed [CheB], we have d ln εbp = 0 and therefore τGK ≅ (∂A*/∂ ln εr)εbp/(bA*) from which Eq. 5 follows. In the system with the CheB-P feedback loop, Eq. 5 remains valid, but now τa−1 ≅ β(τGK−1 + bμa), where μa represents the strength of the CheB-P feedback loop (Eqs. S26 and S33 of SI Appendix). As in Eq. 5, we can calculate how the relaxation time depends on [CheB] for fixed [CheR]: τa(εb−1) ≅ (∂A*/∂ ln εb−1)εr/(βbA*μb) (Eq. S34 of SI Appendix). We neglected the receptor complexes bound to CheR and CheB-P. The full derivations are in Sect. 2.4 of SI Appendix.
Filtering Properties of the Pathway.
Eq. 6 represents a negative integral feedback system (40). The Fourier transform of Eq. 6 indicates that the frequency response of the adaptation module resembles that of a high-pass filter with a cutoff frequency τa−1. Similarly, the response regulator module (Fig. 1A) behaves like a low-pass filter with cutoff frequency τy−1, where τy is the relaxation time of the response regulator module (see Sect. 2.6 of SI Appendix). Thus, the chemotaxis system resembles a bandpass filter. The value of the low cutoff frequency τa−1 predicted by our model for wild-type level of [CheR] is in agreement with measurements from tethered cells (25). A recent analysis of the noise-filtering properties of the chemotaxis system suggested that a lower cutoff frequency τa−1 corresponds to longer time integration of the input signal and therefore better removal of high-frequency fluctuations (21). However, that study did not take into account the slow fluctuations in kinase activity due to stochastic fluctuations in the methylation–demethylation process.
Large-Scale Simulations.
Within each digital cell, the chemotaxis network was simulated by using stochastic methods (16, 38) with rates as in Table S1 of SI Appendix. Digital cells for each population were first adapted to a constant external concentration of aspartate of 1 μM for >1,000 s. Then, at time point t = 0, cells were placed at z = 0 in a linear gradient of attractant. This gradient (10−8 M/μm) increases along the z direction (L = 1 μM at z = 0) in a 3D infinite medium. The trajectories of individual cells were subject to rotational diffusion with diffusion coefficient Dr = 0.062 rad2/s (41).
Supplementary Material
ACKNOWLEDGMENTS.
We thank H. Park, S. Needham, C. Guet, P. Oikonomou, T. Shimizu, and R. Alexander for insightful discussions, C. Macal and M. North for computing time on the ANL Jazz Linux cluster; and Eric Lyons for providing Stochastirator. This work was supported by the National Institutes of Health and joint research funding under H.28 of U.S. Department of Energy Contract W-31-109-ENG-38.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705463105/DC1.
References
- 1.Pedraza JM, van Oudenaarden A. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 2.Raser JM, O'Shea EK. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raser JM, O'Shea EK. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin DW, Allen MS, McCollum JM, Dar RD, Wilgus JR, Sayler GS, Samatova NF, Cox CD, Simpson ML. Nature. 2006;439:608–611. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- 5.Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 6.Block SM, Segall JE, Berg HC. Cell. 1982;31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 7.Barkai N, Leibler S. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 8.Alon U, Surette MG, Barkai N, Leibler S. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 9.Fahrner KA. Cambridge, MA: Harvard Univ; 1995. PhD Thesis. [Google Scholar]
- 10.Korobkova E, Emonet T, Vilar JMG, Shimizu TS, Cluzel P. Nature. 2004;428:574–578. doi: 10.1038/nature02404. [DOI] [PubMed] [Google Scholar]
- 11.Bialek W, Setayeshgar S. Proc Natl Acad Sci USA. 2005;102:10040–10045. doi: 10.1073/pnas.0504321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kampen NG v. Stochastic Processes in Physics and Chemistry. Amsterdam: North-Holland; 1992. [Google Scholar]
- 13.Goldbeter A, Koshland DE. Proc Natl Acad Sci USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Li J, Li G, Long DG, Weis RM. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Hazelbauer GL. Molecular microbiology. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 16.Morton-Firth CJ, Shimizu TS, Bray D. J Mol Biol. 1999;286:1059–1074. doi: 10.1006/jmbi.1999.2535. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu TS, Aksenov SV, Bray D. J Mol Biol. 2003;329:291–309. doi: 10.1016/s0022-2836(03)00437-6. [DOI] [PubMed] [Google Scholar]
- 18.Mello BA, Tu Y. Biophys J. 2003;84:2943–2956. doi: 10.1016/S0006-3495(03)70021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao CV, Kirby JR, Arkin AP. PLoS Biol. 2004;2:E49. doi: 10.1371/journal.pbio.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kollmann M, Lovdok L, Bartholome K, Timmer J, Sourjik V. Nature. 2005;438:504–507. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- 21.Andrews BW, Yi TM, Iglesias PA. PLoS Comput Biol. 2006;2:e154. doi: 10.1371/journal.pcbi.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endres RG, Wingreen NS. Proc Natl Acad Sci USA. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asakura S, Honda H. J Mol Biol. 1984;176:349–367. doi: 10.1016/0022-2836(84)90494-7. [DOI] [PubMed] [Google Scholar]
- 24.Sourjik V, Berg HC. Proc Natl Acad Sci USA. 2002;99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Block SM, Segall JE, Berg HC. J Bacteriol. 1983;154:312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulsson J. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y-D. Adv Chem Phys. 1978;37:67–97. [Google Scholar]
- 28.Elf J, Ehrenberg M. Genome Res. 2003;13:2475–2484. doi: 10.1101/gr.1196503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li MS, Hazelbauer GL. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berg OG, Paulsson J, Ehrenberg M. Biophys J. 2000;79:1228–1236. doi: 10.1016/S0006-3495(00)76377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Detwiler PB, Ramanathan S, Sengupta A, Shraiman BI. Biophys J. 2000;79:2801–2817. doi: 10.1016/S0006-3495(00)76519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elf J, Paulsson J, Berg OG, Ehrenberg M. Biophys J. 2003;84:154–170. doi: 10.1016/S0006-3495(03)74839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duke TAJ, Bray D. Proc Natl Acad Sci USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sourjik V, Berg HC. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mello BA, Tu Y. Proc Natl Acad Sci USA. 2005;102:17354–17359. doi: 10.1073/pnas.0506961102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Proc Natl Acad Sci USA. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sourjik V, Berg HC. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 38.Emonet T, Macal CM, North MJ, Wickersham CE, Cluzel P. Bioinformatics. 2005;21:2714–2721. doi: 10.1093/bioinformatics/bti391. [DOI] [PubMed] [Google Scholar]
- 39.Borkovich KA, Alex LA, Simon MI. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi TM, Huang Y, Simon MI, Doyle J. Proc Natl Acad Sci USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berg HC, Brown DA. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 42.Bourret RB, Stock AM. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 43.Shapiro MJ, Panomitros D, Koshland DE., Jr J Biol Chem. 1995;270:751–755. doi: 10.1074/jbc.270.2.751. [DOI] [PubMed] [Google Scholar]
- 44.Sanders DA, Koshland DE., Jr Proc Natl Acad Sci USA. 1988;85:8425–8429. doi: 10.1073/pnas.85.22.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cluzel P, Surette M, Leibler S. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.