Abstract
Activity of axillary meristems dictates the architecture of both vegetative and reproductive parts of a plant. In Arabidopsis thaliana, a model eudicot species, the transcription factor LFY confers a floral fate to new meristems arising from the periphery of the reproductive shoot apex. Diverse orthologous LFY genes regulate vegetative-to-reproductive phase transition when expressed in Arabidopsis, a property not shared by RFL, the homolog in the agronomically important grass, rice. We have characterized RFL by knockdown of its expression and by its ectopic overexpression in transgenic rice. We find that reduction in RFL expression causes a dramatic delay in transition to flowering, with the extreme phenotype being no flowering. Conversely, RFL overexpression triggers precocious flowering. In these transgenics, the expression levels of known flowering time genes reveal RFL as a regulator of OsSOC1 (OsMADS50), an activator of flowering. Aside from facilitating a transition of the main growth axis to an inflorescence meristem, RFL expression status affects vegetative axillary meristems and therefore regulates tillering. The unique spatially and temporally regulated RFL expression during the development of vegetative axillary bud (tiller) primordia and inflorescence branch primordia is therefore required to produce tillers and panicle branches, respectively. Our data provide mechanistic insights into a unique role for RFL in determining the typical rice plant architecture by regulating distinct downstream pathways. These results offer a means to alter rice flowering time and plant architecture by manipulating RFL-mediated pathways.
Keywords: axillary meristem, inflorescence branching, flowering transition, tillering
Arabidopsis thaliana LFY and its homologs encode an evolutionarily conserved land plant-specific transcription factor. Early studies on the expression pattern and phenotypes of loss-of-function mutations in LFY and FLO, homologs in two dicots A. thaliana and Antirrhinum majus, showed them to confer a floral fate to new meristems arising on the flanks of the shoot apex (1, 2). LFY homologs from species as diverse as gymnosperms, primitive land plants, and from many angiosperms retain the ability to at least partially complement Arabidopsis lfy mutants (3). These data show activation of floral meristem fate to be a conserved LFY function. Protein domains recognizable in all LFY homologs are an N-terminal proline-rich domain and a C-terminal domain; substitutions in these largely conserved DNA-binding domains are suggested to contribute to its potentially divergent functions (3). In fact, mutations in some LFY homologs show additional developmental roles (e.g., compound leaf development in pea and cell division in moss) (4, 5).
Unlike the simple inflorescence of Arabidopsis, grass inflorescences are striking in the multiple kinds of branch meristems made from the apical inflorescence meristem. In rice upon transition to reproductive phase, the vegetative apical meristem transforms to an inflorescence meristem. The latter terminates after making six to eight primary branch meristems. Primary branches produce two to four secondary branch meristems and terminate in a spikelet. Secondary branches also produce few spikelets. The branched inflorescence thus generated is called a panicle. Panicle-branching patterns in maize and wheat, two other crop plants of the grass family, differ from those in rice (6). Genetic loci that control panicle branching regulate spikelet (grain) number, an important yield trait. To unravel mechanisms regulating panicle architecture, approaches such as genetic analysis of inflorescence mutants, whole-genome microarray analysis, and understanding of gene interactions are required. These studies would enable the exploitation of inflorescence characteristics for improved yield (6).
Several lines of evidence implicate distinct functions for the rice LFY homolog, RFL. Examples are its inability to complement the phenotypes of Arabidopsis lfy mutants (7) and its deviant expression profile as compared with LFY or LFY orthologs from other grasses (8–11). LFY is expressed uniformly in floral meristem, but not in the apical inflorescence meristem (1). In contrast, RFL shows high-level and dynamic expression in apical inflorescence (panicle) meristem and is expressed in panicle branch primordia, but its expression is greatly diminished in the floral meristem (8, 10). This pattern also is distinct from maize ZFL1 and ZFL2, which are expressed in branching spikelet meristems and floret meristems, but not the inflorescence apex (11). To unravel regulatory actions of RFL and to correlate this with its expression profile, we studied the phenotypic consequences of RFL knockdown and overexpression in rice. We coupled these analyses with the effects on global gene expression. Our studies show that RFL controls two important traits in rice: flowering time and plant architecture as a whole. These functions are executed by regulating the expression of distinct transcription factors and hormone-dependent-signaling pathways that implicate functions for RFL not predicted from studies of its other homologs.
Results
RFL Promotes the Transition of the Vegetative Apical Meristem to an Inflorescence Meristem.
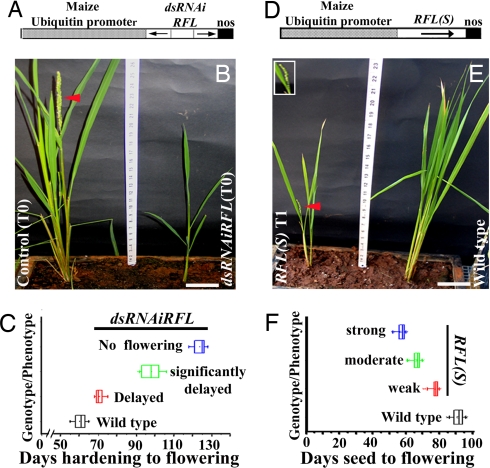
The functional relevance of RFL expression in the inflorescence meristem, from its inception and during branching, was investigated by knockdown and overexpression of RFL. Twenty-four independent transgenic lines expressing hairpin loop RNAs for RFL (Fig. 1A) showed a significant delay in flowering and had drastically reduced height (Fig. 1 B and C). The average time taken for flowering in these tissue culture-regenerated plants was 100 days at height ≈38 cm (Fig. 1C). Control wild-type-regenerated plants initiate panicles in ≈60 days at height ≈65 cm (Fig. 1 B and C). The weakest, yet statistically significant, RFL knockdown phenotype occurred in five lines where flowering took place ≈70 days after hardening (Fig. 1C). Strikingly, six other lines did not produce an inflorescence meristem even after 120 days (Fig. 1C) and eventually died without forming a panicle. Together these data show a critical function for RFL in promoting transition of the vegetative growth apex to an inflorescence meristem. A 20-fold decrease in endogenous RFL transcript levels was achieved in young dsRNAiRFL panicles [supporting information (SI) Fig. 6A], implicating the severe reduction in RFL expression as causal in the extremely delayed flowering. Similarly, we find that knockdown of RFL through antisense RNAs, despite not being fully effective for knockdown, still delays flowering by ≈15 days in T1 plants (data not shown). These results show that RFL expression in the panicle meristem is a critical determinant of its fate.
Fig. 1.
Phenotypes of RFL(S) and dsRNAiRFL plants. (A) Schematic diagram of dsRNAiRFL transgene. The ubiquitin promoter transcribes hairpin loop RNAs for RFL exon 1 and exon 2 segments. (B) Morphology of a flowering wild-type plant (Left, red arrowhead) regenerated through tissue culture and a dwarf nonflowering dsRNAiRFL (Right) plant of same age. (C) Distribution of days to flowering in dsRNAiRFL T0 plants. Flowering time (x axis) is plotted against phenotype (y axis). The statistical significance is P < 0.0001for all phenotypic groups. (D) Schematic diagram of Ubi::RFL transgene. (E) A RFL(S) plant (Left) with early panicle heading (Inset, red arrowhead with closeup), compared with a wild type of same age (Right). (F) Distribution of flowering time in RFL(S) T1 plants showing strong, moderate, and weak phenotypes.
Importantly, we observe a complementary early flowering phenotype on RFL overexpression from the Ubi::RFL transgene (Fig. 1D). Flowering time in 35 independent RFL(S) T1 lines was measured as days taken for the formation of young panicles. These plants flowered precociously with compromised vegetative growth (Fig. 1E). Ten lines with severe phenotypes made panicles (0.1–0.3 cm) in ≈54 days when plants were only ≈41 cm tall (Fig. 1 E and F). This contrasts with ≈90 days taken for wild-type plants to attain a similar developmental stage when the plants are ≈70 cm tall (Fig. 1 E and F). Eleven lines displayed moderate phenotypes; they flowered in 65 days at a height of ≈56 cm (Fig. 1F). Even the weakest phenotype (40% of the lines) was early flowering in 78 days (Fig. 1F). Ectopic expression of RFL in leaves and overexpression in young panicles of RFL(S) transgenics was quantitated (SI Fig. 6 A and B). Thus, we find that RFL overexpression triggers precocious flowering.
Relationship Between RFL and Activators and Repressors of Flowering.
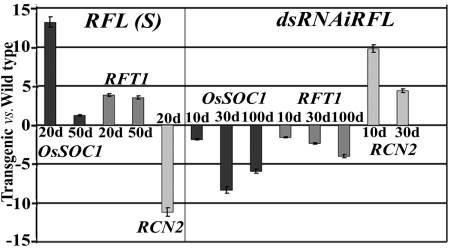
We have interrogated whether ubiquitous expression of RFL in Ubi::RFL transgenics promotes a change of the indeterminate apical vegetative meristem to determinate branched reproductive (panicle) meristem by affecting flowering time genes. Expression levels for some rice flowering time regulators were measured in transgenics with deregulated RFL expression. Transcript levels for OsSOC1/OsMADS50, a positive regulator of flowering, were measured in leaves of wild-type plants 20, 50, and 80 days after germination. Very low-level expression seen in 20-day-old leaves increases by day 50, as is known from previous work on OsSOC1 (SI Fig. 7A) (12). In 20-day-old transgenics that ectopically overexpress RFL, we find OsSOC1 transcripts levels are much higher than in wild-type plants of the same age (Fig. 2). This temporally early high-level OsSOC1 expression achieved in RFL overexpression lines with extremely precocious flowering is not transient. Expression is maintained in 50-day-old RFL(S) transgenics that are near flowering, wherein transcript levels are marginally higher than in 50-day-old wild-type plants that are still to attain flowering (Fig. 2). Concordant with these results are the complementary effects seen on RFL knockdown through RNA interference (RNAi). Leaves of young tissue culture-regenerated dsRNAiRFL T0 plantlets, 10 and 30 days, after hardening show markedly reduced OsSOC1 expression (Fig. 2), compared with control wild-type-regenerated plants of similar ages. We also analyzed the expression levels of RFT1 encoding a predicted signaling factor closely related to Hd3a (13). This was taken up because Hd3a is not expressed in the variety used for transformation (SI Fig. 7B) as also is the case with other varieties with a reduced photoperiod response (14). RFT1 is expressed in wild-type leaves, and RFL overexpression up-regulates RFT1 expression, but to a lesser extent than OsSOC1. Knockdown of RFL reduces RFT1 transcript levels with the effect persisting in 100-day-old plants, which is well beyond the time taken for flowering in control plants (Fig. 2).
Fig. 2.
Expression status of flowering activators and a repressor in RFL(S) and dsRNAiRFL plants. Quantitative RT-PCR showing fold change, with respect to wild type, in expression for OsSOC1 and RFT1 in leaves and RCN2 in the culm of RFL(S) and dsRNAiRFL plants of various ages.
In Arabidopsis, the mutually antagonistic relationship between repressors of flowering such as TFL1 and activators of floral meristem fate (LFY and AP1) controls phase transition (15). The constitutive overexpression of the rice TFL1 homologs, RCN1 or RCN2, delays vegetative-to-reproductive phase transition (16), suggesting that RCN overexpression may extend the vegetative phase of the apical meristem. But whether this occurs through changes in expression of flowering activators is not known. We measured RCN2 transcript levels in the vegetative shoot apex of regenerated wild-type plantlets (10 and 30 days after hardening) and compared the levels to those in RFL knockdown plantlets of similar age. We find that RCN2 expression is up-regulated (Fig. 2) in young vegetative apices of RFL knockdown plants showing a reciprocal relationship between RCN and RFL, a promoter of panicle fate. Consistent with these data, RCN2 expression is much reduced in shoot apices of young T1 plants overexpressing RFL (Fig. 2). These data indicate that the antagonistic interaction between RFL and this flowering repressor is conserved with regard to transition from the vegetative to the reproductive phase.
Functions for RFL in Panicle Development.
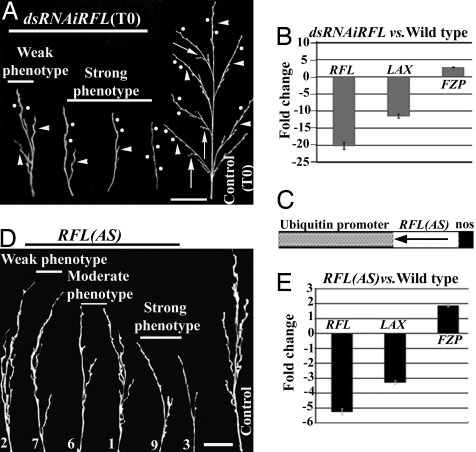
Transgenic plants with near-complete knockdown of RFL expression in the panicle were extremely delayed for flowering or did not flower at all. Further, panicles when produced were severely compromised for growth and branching (Fig. 3A). Less than two primary branches were made, if at all, instead of the six to eight branches in wild-type panicles. In these severely affected dsRNAiRFL panicles, the stunted main rachis or the stunted primary branches bear a few spikelets (Fig. 3A and SI Table 1). Further secondary branches are not produced in any panicles severely knocked down for RFL.
Fig. 3.
Panicle growth and branching in RFL knockdown plants. (A) Mature deseeded wild-type and dsRNAiRFL panicles displayed for rachis length and branching. The primary branches (arrowheads), secondary branches (arrows), and spikelet pedicels (solid dots) are marked at representative positions. (B) Fold change in expression of branching regulators in dsRNAiRFL panicles compared with wild-type panicles determined by quantitative RT-PCR. (C) Schematic diagram of Ubi::RFL(AS) transgene. (D) Progressive reduction in the panicle branching and no secondary branches in these plants (line numbers at the bottom of each panicle). (E) Normalized fold change in the expression of branching regulators in RFL(AS) panicles. (Scale bars: 1.0 cm.)
The effects on panicle architecture also were analyzed in transgenic lines expressing RFL antisense RNAs (Fig. 3 C and D) because we could examine the effects of varying degrees of RFL knockdown. Twenty-two independent transgenics have been characterized over three generations (T0, T1, and T2). Based on the degree of branching defects, these lines can be classified as strong, moderate, and weak (Fig. 3D). The graded effects on branching correlate with the level of antisense RNAs expressed in these plants (data not shown); progressively stronger phenotypes occur with increased antisense RNAs. Panicles in plants with strong and moderate phenotypes have no secondary branches (Fig. 3D and SI Table 2), but primary branches are made with a few fertile spikelets. The extent of endogenous RFL down-regulation was measured in T2-generation plants by quantitative RT-PCR where a marked 5-fold down-regulation was seen (Fig. 3E) in lines with the strongest phenotypes. The down-regulation is lesser than that attained through RNAi (Fig. 3 B vs. E). Importantly, the most severe panicle-branching phenotypes observed in the antisense RFL transgenic lines were nearly identical to the moderate branching defects of some RFL knockdown lines. These data confirm that the expression of endogenous RFL transcripts in the incipient branch primordia is a prerequisite for their formation. Surprisingly, we also observed poor panicle branching in transgenic lines overexpressing RFL, the indications of which are in Discussion section 2.
Effects of RFL Knockdown on Regulators of Panicle Branching.
Mutations in the bHLH transcription factor LAX abrogate secondary branch formation without affecting the establishment and growth of the main panicle axis (17). Conversely, mutations in the transcription factor FZP promote the formation of supernumerary axillary meristems, causing excessive panicle branches without any spikelet meristems (18). During panicle development, the expression of RFL precedes that of either LAX or FZP, which are spatially restricted, to distinct but small sets of cells. Therefore, we examined the relationship between RFL expression levels and these genetic regulators of panicle branching. Quantitative RT-PCR was used to measure LAX and FZP transcripts in RNA from both dsRNAiRFL and RFL(AS) transgenics (Fig. 3 B and E). A clear and reproducible down-regulation of LAX and an up-regulation of FZP occur in transgenics with strong panicle-branching phenotypes.
Down-Regulation of RFL Affects Vegetative Axillary Meristems.
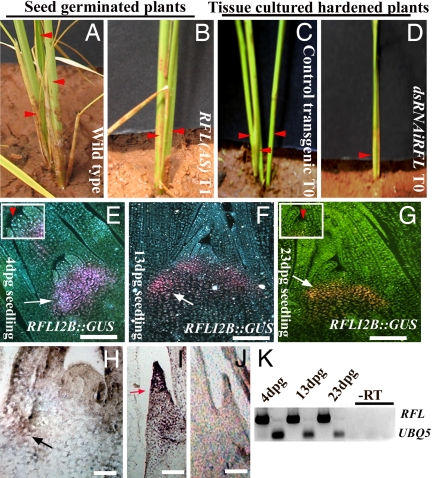
In rice, axillary meristems normally develop from the basal nodes of the plant to form tillers that generate the typical bushy plant architecture (Fig. 4A). The complete knockdown of RFL abolishes tiller development (Fig. 4 C vs. D), and tiller numbers are reduced on partial knockdown (Fig. 4 A vs. B). Closer inspection of the dissected culm in dsRNAiRFL plants show that, despite the normal number of nodes, tiller outgrowth is severely compromised. In contrast to the five to six buds seen in wild-type plants, the dsRNAiRFL transgenics of similar age either do not initiate tiller buds or, in some instances, generate one to two buds that fail to grow further (SI Fig. 8 D–F). Expression of RFL in tiller bud was not investigated in previous studies (8, 10). To ascertain any role for RFL in vegetative axillary meristem development, we have reexamined RFL expression at sites of tiller bud formation and in developing tiller buds using RFL promoter::GUS transcriptional fusions (10) and RNA in situ hybridization. The reporter constructs chosen (RFLI2B::GUS and its derivatives) were those that drive the normal spatially and temporally regulated RFL expression in the developing panicle. We note GUS expression at sites of new tiller primordia formation in young plants of various ages (Fig. 4 E–G and SI Fig. 8 A and B). The shoot apical meristem (SAM) in these plants does not express RFL (Fig. 4 E–G and Insets) as previously shown for SAM of yet older plants (8, 10). In addition to expression in incipient tiller primordia, we observe robust reporter expression at basal unelongated nodes and internodes of very young plants (Fig. 4E and SI Fig. 8A). RNA in situ hybridization confirms the presence of RFL transcripts in the axils of leaves that are sites for future tiller primordia (Fig. 4H) and in very young tiller primordia (Fig. 4I). Although the promoter::GUS fusions recapitulate some aspects of the RFL RNA expression patterns, it is perhaps insufficient to confer the entire profile in vegetative tissues. Further, semiquantitative RT-PCR also confirms RFL expression in the main culm enclosing the SAM of 4-, 13-, and 20-day-old plants (Fig. 4J). Together these expression analyses account for the phenotypes of poor or no tiller development upon RFL knockdown.
Fig. 4.
Tiller development in RFL knockdown plants. (A) Basal portion of a wild-type plant with tillers (red arrowheads). (B) RFL(AS) plant with few tillers. (C) Basal part of the tissue culture regenerated wild-type plant. (D) dsRNAiRFL plant with no side tillers. (E–G) Histochemical distribution of GUS activity in vegetative axillary meristems. Pink-orange fluorescence at sites of axillary/tiller bud initials shows reporter activity. Basal nodes (F and G, arrow) and internodes (E, arrow) of transgenics culms with RFL promoter::GUS fusions. (E and G Insets) Shoot apical meristem. (H–I) RFL mRNA localization in wild-type 23-day-old culms. (H and I) RNA expression at leaf axils (H, arrow) and in a young tiller bud (I, arrow). (J) Culm with a tiller bud probed with sense RNA. (Scale bars: E–H, 50 μm; I and J, 20 μm.) (K) Semiquantitative RT-PCR of RFL transcripts in 4-, 13-, and 23-day-old culms (Upper) and control UBQ5 transcripts (Lower).
Global Expression Profiling Shows RFL as a Master Regulatory Transcription Factor.
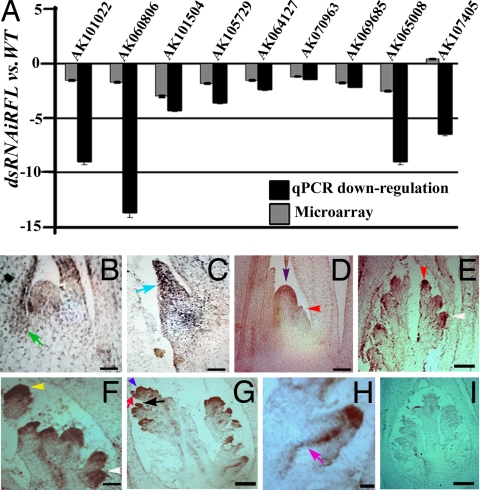
Functions for RFL as a regulator of meristem fate particularly during formation of the branched inflorescence and spikelet was explored through global gene expression profile analysis. RNA pools from young panicles that were wild type or knocked down for RFL were compared in rice Agilent 22,000 arrays. These competitive hybridizations were performed with two independent RNA pools from dsRNAiRFL and RFL(AS) transgenics and matched wild-type panicles. Briefly, 522 genes deregulated in both experimental hybridizations and in their reverse-labeling hybridizations were studied further. These genes were manually inspected and categorized based on the occurrence of predicted protein domains to assign them to functional categories (SI Table 3). We note a preponderance of transcription factors (9.4%) and signaling molecules (10.36%) among the transcripts affected on RFL knockdown (SI Fig. 9). In addition, genes involved in various aspects of metabolism (37%) are deregulated, significant among them are genes that may contribute to the synthesis or catabolism of plant hormones or metabolites (e.g., cytokinin oxidase, GA oxidase, cytochrome P450s, etc.). Nine representative candidate downstream genes were validated for their down-regulation in panicle RNAs of RFL knockdown transgenics (Fig. 5A) by quantitative RT-PCR. These data place RFL as a regulator of several unique genes encoding transcription factors; ethylene signaling factors (EIL3), auxin efflux facilitator (PIN3-like), and perhaps even hormone biogenesis/catabolism (Fig. 5A and SI Table 3). The latter is suggested from the down-regulation of many cytochrome P450s that may be involved in gibberellin, carotenoid, or brassinosteroid biogenesis and from the down-regulation of an ethylene biosynthesis enzyme (ACC synthase) and an enzyme in cytokinin metabolism (cytokinin oxidase) (Fig. 5A and SI Table 3). The data also hint at roles for factors affecting meristem function and emergence of lateral organs (19). One of the 18 rice AGO-like genes is down-regulated on RFL knockdown. The expression of this gene in the early stages of panicle and spikelet development (20) is consistent with a plausible role for RFL in regulating this AGO family member (Fig. 5A and SI Table 3). Notably, most of the predicted rice homologs of genes regulated by LFY in Arabidopsis inflorescence apices or genes regulated by the ectopic expression of LFY::GR in young Arabidopsis plants are not found in our dataset of genes affected on RFL knockdown (SI Table 4) (3, 21). This finding suggests that the global architecture of RFL regulatory action is different from its Arabidopsis counterpart and that RFL executes its functions through distinct pathways.
Fig. 5.
Genome-wide expression analysis of genes regulated by RFL. (A) Comparison of fold change in expression levels in wild-type versus dsRNAiRFL panicles for nine representative transcripts chosen from microarray data (SI Table 3). Data from microarrays are compared with that from quantitative RT-PCR analysis. (B–I) In situ RNA hybridization of an RFL-regulated transcript, AK101504 (PIN3-like). Transcripts at leaf axils (B, green arrow) and in a young tiller bud (C, cyan arrow) are indicated. (D) Transcripts in the panicle apex (purple arrowhead) and initiating primary branch (red arrowhead). (E) Expression at the apical end of a primary branch (red arrowhead) and in emerging secondary branches (white arrowhead). (F) Uniform expression in a young spikelet meristem (yellow arrowhead). (G) Transcripts in the emerging lemma (black arrow), palea (red arrow), and carpel anlagen (blue arrowhead) of spikelets with differentiating organs. (H) Expression in the vascular strands of an emerging primary branch (pink arrow). (I) Panicle probed with sense AK101504 RNA. (Scale bars: B–D, F, and I, 20 μm; E and G, 50 μm; H, 10 μm.)
Discussion
An Effect of RFL on Flowering Time Genes Places RFL as a Regulator of Vegetative to Inflorescence Meristem Transition.
Unlike other grass LFY genes, such as Lolium LtLFY and maize ZFL1 and ZFL2, rice RFL shows robust expression in the early reproductive shoot (panicle) apex, but not in the vegetative apical meristem (8–11). The drastic effects on flowering time that we see on the deregulation of RFL are concordant with a role for the unique expression profile of this gene. These flowering time effects are far more pronounced than the mild flowering delay of the maize zfl1 and zfl2 mutants (11). The flowering time phenotypes, seen on perturbations in RFL expression, are similar to the precocious flowering triggered by LFY overexpression in Arabidopsis or other species, such as aspen (22).
Arabidopsis flowering time genes that promote transition of the vegetative apical meristem to an inflorescence meristem act through multiple pathways that are integrated by transcriptional up-regulation of FT and SOC1. The latter activate floral meristem genes on new lateral primordia (23). The rice SOC1 ortholog, OsSOC1/OsMADS50, can accelerate flowering in rice upon overexpression, and its knockdown delays flowering (12). Our data of precocious transcriptional up-regulation of OsSOC1 upon RFL overexpression and of its delayed activation on RFL knockdown strongly support a new role for RFL as a regulator of OsSOC1. OsSOC1 is thought to act downstream of or function parallel to other rice flowering time genes, such as Hd1 (CO ortholog) and OsGI, to eventually activate expression of the FT-like gene Hd3a (12). FT is a potent photoperiod-dependent mobile activator of flowering in both Arabidopsis and rice (23). In addition to Hd3a, nine other FT-like rice genes are known (13). Hd3a is expressed at very low levels even in inductive conditions in varieties like Taichung65 that are mutant for Hd1 and EHD1 and show poor photoperiod response (14). The variety Taipei TP309, used in our studies, does not express Hd3a (SI Fig. 7B), whereas RFT1 (an FT-like gene) is expressed in these growth conditions. We find that RFT1 expression is regulated by RFL, but to a lesser extent than OsSOC1. These data indicate that changing RFL levels alters OsSOC1 expression whose effects on flowering time may be mediated by other members of the rice FT family. Unlike LFY, which functions downstream of SOC1 (23), our data show that RFL acts upstream of OsSOC1 and RFT1 to promote flowering in a photoperiod-insensitive variety. Establishing a regulatory and possibly even a feedback relationship between RFL and OsSOC1 awaits the analysis of overexpression of OsSOC1 in RFL knockdown lines and vice versa.
Dynamic RFL Expression Profile, Unlike That in Other Species, Regulates Plant Architecture.
The diversity of inflorescence branching patterns and vegetative axillary shoot development seen in grasses (6) presents an interesting hypothesis that changes in inflorescence architecture and plant form may arise, at least in part, from changes in expression pattern of conserved regulators. We now demonstrate that RFL is expressed at sites of vegetative axillary meristems and in very young tiller buds, which is required for their outgrowth. Expression in axillary buds, or even the initials of axillary meristems, is not known for LFY or its other homologs, except pea UNI and tobacco NFL, which are expressed in developing lateral shoot primordia (4, 24). Interestingly, the expression of RFL in leaf axils is similar to that of STM (25) and may relate to a role in maintaining a zone of meristematic cells.
RFL expression in the branching panicle is dynamic, with the expression in incipient lateral branch primordia being high but transient (8, 10). The dynamic pattern of RFL expression, in the panicle, bears similarity to rice KN1-type and Arabidopsis STM homeodomain transcription factor genes (25, 26). By comparison of RFL overexpression and knockdown phenotypes, we infer that this profile, in the panicle, may first support a meristematic state and act later for the formation of inflorescence branches. These inferences agree with a recent study on the evolution of inflorescence forms (27). They predict that in panicles all lateral meristems are first in a transient vegetative state, where brief LFY expression is followed by its repression, thereby resulting in branch meristems. Subsequently, these meristems are fully committed to form flowers by entering a different state. The expression levels of critical regulators such as LFY and TFL and their mutual interactions determine these two meristem states (27). The phenotypic effects of poor inflorescence branching that occur on changes in RFL expression are consistent with their model as argued below. RFL knockdown fails to provide the initial high-level expression required to maintain meristems, and the overexpression fails to repress RFL needed for the transition to another meristem state. Our study clearly demonstrates how a diverged RFL expression pattern in incipient vegetative and reproductive lateral branch primordia, which is unlike other LFY-like genes, regulates their initiation and growth, thereby controlling the architecture of entire rice plant.
RFL Regulates Panicle-Branching Regulators.
The graded phenotypic effects on panicle branching seen on the gradual reduction in RFL expression levels, together with the failure of inflorescence meristem specification and growth on complete RFL knockdown, indicate critical functions for RFL in determining panicle morphology. Our data are consistent with RFL promoting panicle branch primordia by activating positively acting branching regulators such as LAX. LAX expression is restricted in the inflorescence meristem to boundary cells adjacent to sites of new lateral meristems (17). This profile overlaps with the broader expression of RFL in the branching inflorescence. Excessive panicle branching occurs in fzp mutants, where supernumerary axillary meristems are formed in axils of bracts in young spikelet meristems (18). Our data of FZP overexpression and lack of axillary meristems in RFL knockdown panicles agree with the hypothesis that FZP represses axillary meristem formation (18). These data attribute an upstream position for RFL in the genetic network controlling panicle architecture. Regulators of rice panicle architecture are conserved in maize, where BRANCHED SILKLESS1 (BD1) is the homolog for FZP and BA1 is the homolog of LAX. However, their relationship to maize LFY genes ZFL1 and ZFL2 is unknown (28).
RFL Targets Are Putative Hormone Signaling, Metabolic, and Transcription Factors.
The effects of RFL on panicle regulators LAX and FZP implicate a likely mechanism of RFL action during inflorescence branching. Our global microarray analysis of gene expression profiles, in the branching panicle, provides further mechanistic insights into RFL regulatory action. A large proportion of the genes deregulated in the absence of RFL are predicted signaling molecules and transcription factors. In addition, we anticipate that many deregulated genes currently hypothesized to perform metabolic roles may influence the levels of signaling molecules. Our attempt to understand downstream signaling molecules also is motivated by recent studies showing the STM and KNOX homeodomain factors to orchestrate meristem function by simultaneously activating cytokinin and repressing gibberellin biosynthetic pathways in Arabidopsis (29). Besides the down-regulation of molecules involved in hormone biogenesis/catabolism, a notable finding is the down-regulation of AK101504, which is a predicted homolog of Arabidopsis PIN3, an auxin efflux facilitator. The spatial distribution of AK101504 transcripts overlaps with RFL in the very young panicle apex (Fig. 5D) (8, 10). In branch meristems, its expression persists in domains that overlap with RFL and in adjacent cells that do not (Fig. 5 E–H) (8, 10), hinting at signaling-mediated interactions. Strikingly, AK101504 transcripts also are expressed in leaf axils and in young tiller buds (Fig. 5 B and C), as is RFL. This raises the possibility that the AK101504 gene could contribute to some extent to the panicle-branching and axillary meristem defects of RFL knockdown plants. Our hypothesis agrees with the critical role played by PIN-dependent auxin transport during axillary meristem initiation in Arabidopsis (30). Recently, one of the rice PIN1-like genes has been implicated in tiller bud outgrowth and adventitious root initiation (31), but its contributions to inflorescence structure are not known. Furthermore, the maize BIF2 (co-ortholog of PINOID-like serine/threonine kinase) regulates the initiation of axillary meristem and lateral primordia (32), underscoring the importance of auxin signaling for primordia emergence in grasses. Our data provide starting points for further investigations on RFL mechanism of action. Altogether, we demonstrate functions for RFL as a regulator of plant architecture through its effects on apical and axillary meristems throughout the growth of the rice plant.
Materials and Methods
Transgenic Plants.
Transformation of rice calli was carried out as in ref. 10. The construction of pUbi::RFL, pUbi::RFL(AS), and pUbi::dsRNAiRFL plasmids is given in SI Materials and Methods.
Flowering Time Measurements.
RFL(S) and wild-type 8-day-old aseptically grown T1 seedlings were moved to clay, and the date when panicles (0.1–0.3 cm) were formed was recorded. For dsRNAiRFL and control transgenics, tissue-cultured plantlets that regenerated at about the same time were hardened together in soilrite and moved to clay. The time taken from hardening to make panicles (0.1–0.3 cm) was noted.
RT-PCRs.
Quantitative RT-PCR analysis of specified transcripts was done as in ref. 33. Panicle RNAs isolated with a plant RNeasy minikit (Qiagen) or RNA from main culm or leaf lamina extracted with TriReagent was used for reverse transcription with SuperScript III or cloned AMV enzymes (Invitrogen). Then 25 to 150 ng of the cDNA was taken for each quantitative PCR with SYBR green kit (Finnzymes) and detected in an ABI prism 7000 system. Transcript levels normalized to UBQ5, in three to six PCRs from two biological samples, were used to determine the difference in the cT values between transgenic and wild-type RNAs. This was used to compute mean and standard deviation for fold change in gene expression. Primer sequences are detailed in SI Table 5.
Expression Profiling Using DNA Microarrays.
The RFL(AS) (0.1–0.3 cm) and dsRNAiRFL (0.1–0.5 cm) panicle RNA pools isolated with the RNeasy plant minikit (Qiagen) were compared with two matched pools of wild-type RNAs. For microarray analysis, Agilent Technologies custom rice (22,000) arrays were hybridized with Cy3- and Cy5-labeled cRNAs in dye-swap experiments according to the manufacturer's instructions. The data (GEO database accession no. GSE-10098) were analyzed by using Genespring GX. An average ratio of the mutant to wild type of <0.5 for a given gene was taken as the criterion for its differential expression.
GUS Assays and in Situ Hybridizations.
Briefly, 4-, 13-, and 23-day-old culms with shoot apices were processed for GUS assays as in ref. 10. Then 10-μm paraffin longitudinal sections were observed in dark field illumination (Axioscop2 microscope; Zeiss). RNA in situ hybridization was performed according to ref. 33. Riboprobes nucleotides +1 to +764 for RFL or +1,777 to +1,982 for AK101504 were prepared from cDNA clones.
Supplementary Material
ACKNOWLEDGMENTS.
We thank laboratory members for their help and discussions and anonymous reviewers for their critical input. This work was supported by Department of Biotechnology (DBT) Government of India grants, University Grant Commission to Department of Microbiology and Cell Biology, a DBT and Council of Scientific and Industrial Research Associateship (to N.N.R.), and a DBT Research Associateship (to P.R.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE10098).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709059105/DC1.
References
- 1.Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- 2.Coen ES, et al. Floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- 3.Maizel A, et al. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science. 2005;308:260–263. doi: 10.1126/science.1108229. [DOI] [PubMed] [Google Scholar]
- 4.Hofer J, et al. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol. 1997;7:581–587. doi: 10.1016/s0960-9822(06)00257-0. [DOI] [PubMed] [Google Scholar]
- 5.Tanahashi T, Sumikawa N, Kato M, Hasebe M. Diversification of gene function: homologs of the floral regulator FLO/LFY control the first zygotic cell division in the moss Physcomitrella patens. Development. 2005;132:1727–1736. doi: 10.1242/dev.01709. [DOI] [PubMed] [Google Scholar]
- 6.Kellogg EA. Floral displays: Genetic control of grass inflorescences. Curr Opin Plant Biol. 2007;10:26–31. doi: 10.1016/j.pbi.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Chujo A, Zhang Z, Kishino H, Shimamoto K, Kyozuka J. Partial conservation of LFY function between rice and Arabidopsis. Plant Cell Physiol. 2003;44:1311–1319. doi: 10.1093/pcp/pcg155. [DOI] [PubMed] [Google Scholar]
- 8.Kyozuka J, Konishi S, Nemato K, Izawa T, Shimamoto K. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci USA. 1998;95:1979–1982. doi: 10.1073/pnas.95.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gocal GF, et al. Evolution of floral meristem identity genes. Analysis of Lolium temulentum genes related to APETALA1 and LEAFY of Arabidopsis. Plant Physiol. 2001;125:1788–1801. doi: 10.1104/pp.125.4.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasad K, Kushalappa K, Vijayraghavan U. Mechanism underlying regulated expression of RFL, a conserved transcription factor, in the developing rice inflorescence. Mech Dev. 2003;120:491–502. doi: 10.1016/s0925-4773(02)00457-4. [DOI] [PubMed] [Google Scholar]
- 11.Bomblies K, et al. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development. 2003;130:2385–2395. doi: 10.1242/dev.00457. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Kim J, Han JJ, Han MJ, An G. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004;38:754–764. doi: 10.1111/j.1365-313X.2004.02082.x. [DOI] [PubMed] [Google Scholar]
- 13.Izawa T, et al. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doi K, et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ratcliffe OJ, et al. A common mechanism controls the life cycle and architecture of plants. Development. 1998;125:1609–1615. doi: 10.1242/dev.125.9.1609. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313x.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu K, et al. LAX and SPA: Major regulators of shoot branching in rice. Proc Natl Acad Sci USA. 2003;100:11765–11770. doi: 10.1073/pnas.1932414100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130:3841–3850. doi: 10.1242/dev.00564. [DOI] [PubMed] [Google Scholar]
- 19.Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Furutani I, Sukigawa S, Kyozuka J. Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J. 2006;46:503–511. doi: 10.1111/j.1365-313X.2006.02703.x. [DOI] [PubMed] [Google Scholar]
- 21.William DA, et al. Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA. 2004;101:1775–1780. doi: 10.1073/pnas.0307842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi Y, Weigel D. Move on up, it's time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- 24.Kelly AJ, Bonnlander MB, Meeks-Wagner DR. NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell. 1995;7:225–234. doi: 10.1105/tpc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis. Dev Biol. 2000;218:341–353. doi: 10.1006/dbio.1999.9572. [DOI] [PubMed] [Google Scholar]
- 26.Sentoku N, et al. Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell. 1999;11:1651–1664. doi: 10.1105/tpc.11.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prusinkiewicz P, Erasmus Y, Lane B, Hardner LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- 28.McSteen P, Leyser O. Shoot branching. Annu Rev Plant Biol. 2005;56:353–374. doi: 10.1146/annurev.arplant.56.032604.144122. [DOI] [PubMed] [Google Scholar]
- 29.Jasinski S, et al. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol. 2005;15:1560–1565. doi: 10.1016/j.cub.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 30.Leyser O. Dynamic integration of auxin transport and signalling. Curr Biol. 2006;16:R424–R433. doi: 10.1016/j.cub.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Xu M, Zhu L, Shou H, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005;46:1674–1681. doi: 10.1093/pcp/pci183. [DOI] [PubMed] [Google Scholar]
- 32.McSteen P, et al. barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 2007;144:1000–1011. doi: 10.1104/pp.107.098558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yadav SR, Prasad K, Vijayraghavan U. Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics. 2007;176:283–294. doi: 10.1534/genetics.107.071746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.