Abstract
The Cdc25 phosphatase promotes entry into mitosis by removing cyclin-dependent kinase 1 (Cdk1) inhibitory phosphorylation. Previous work suggested that Cdc25 is activated by Cdk1 in a positive feedback loop promoting entry into mitosis; however, it has remained unclear how the feedback loop is initiated. To learn more about the mechanisms that regulate entry into mitosis, we have characterized the function and regulation of Mih1, the budding yeast homologue of Cdc25. We found that Mih1 is hyperphosphorylated early in the cell cycle and is dephosphorylated as cells enter mitosis. Casein kinase 1 is responsible for most of the hyperphosphorylation of Mih1, whereas protein phosphatase 2A associated with Cdc55 dephosphorylates Mih1. Cdk1 appears to directly phosphorylate Mih1 and is required for initiation of Mih1 dephosphorylation as cells enter mitosis. Collectively, these observations suggest that Mih1 regulation is achieved by a balance of opposing kinase and phosphatase activities. Because casein kinase 1 is associated with sites of polar growth, it may regulate Mih1 as part of a signaling mechanism that links successful completion of growth-related events to cell cycle progression.
Introduction
The dramatic events of chromosome segregation and cell division are initiated by the activity of Cdk1 (Morgan, 2006). Activation of Cdk1 requires binding of mitotic cyclins, which are synthesized anew each cell cycle and accumulate gradually during G2/M. Cdk1 activation is delayed by the Wee1 kinase, which phosphorylates and inhibits Cdk1 (Russell and Nurse, 1987; Gould and Nurse, 1989). The Cdc25 phosphatase promotes entry into mitosis by removing the inhibitory phosphorylation placed on Cdk1 by Wee1 (Russell and Nurse, 1986; Dunphy and Kumagai, 1991; Gautier et al., 1991; Kumagai and Dunphy, 1991; Strausfeld et al., 1991).
In yeast cells, inhibitory phosphorylation of Cdk1 has been proposed to mediate a checkpoint that delays entry into mitosis until sufficient growth has occurred (Nurse, 1975; Fantes and Nurse, 1977; Rupes, 2002;Kellogg, 2003). Wee1 mutant cells enter mitosis prematurely and become abnormally small, whereas Cdc25 mutant cells undergo delayed entry into mitosis and become abnormally large (Nurse, 1975; Russell and Nurse, 1986; Jorgensen et al., 2002; Harvey and Kellogg, 2003; Harvey et al., 2005). However, it has been difficult to clearly demonstrate that Wee1 and Cdc25 play a direct role in the control of cell growth because mutants may indirectly cause cell size defects by allowing more or less time to grow. Moreover, there are no clearly defined molecular links between Cdk1 inhibitory phosphorylation and proteins known to be involved in the control of cell size or cell growth. Inhibitory phosphorylation of Cdk1 in yeast has also been proposed to mediate a checkpoint that monitors cell morphogenesis (Lew, 2000, 2003; Keaton and Lew, 2006). In vertebrates, inhibitory phosphorylation of Cdk1 is thought to function in a checkpoint that monitors the status of the DNA, or in regulatory feedback loops that allow the gradual synthesis of mitotic cyclins to be converted into an abrupt all-or-nothing activation of Cdk1 (Dunphy, 1994; Ferrell, 2002; Lew, 2003; Pomerening et al., 2003, 2005; Karlsson-Rosenthal and Millar, 2006; Keaton and Lew, 2006). These proposed roles for Cdk1 inhibitory phosphorylation are not mutually exclusive and the relative importance of each role may vary in different cell types.
A full understanding of Cdk1 inhibitory phosphorylation will require elucidation of the mechanisms that regulate Wee1 and Cdc25. Regulation of Cdc25 family members has been studied most extensively in vertebrates, which have three Cdc25 paralogues that are referred to as Cdc25A, B, and C. In Xenopus laevis oocyte extracts, Cdc25A becomes hyperphosphorylated in mitosis, and the hyperphosphorylated form of Cdc25A isolated from mitotic extracts shows an approximately fivefold increase in phosphatase activity (Izumi et al., 1992; Kumagai and Dunphy, 1992). Fission yeast Cdc25 also undergoes hyperphosphorylation during mitosis (Kovelman and Russell, 1996; Esteban et al., 2004; Wolfe and Gould, 2004a). Cdk1 associated with mitotic cyclins can phosphorylate Cdc25C in vitro, which causes a three- to fourfold increase in phosphatase activity (Hoffmann et al., 1993; Izumi and Maller, 1993). These observations have led to the idea of a positive feedback loop in which mitotic Cdk1 directly activates Cdc25. However, it has remained unclear how a small amount of Cdk1 escapes inhibitory phosphorylation to initiate the feedback loop. The simplest possibility is that the feedback loop is initiated when mitotic cyclins accumulate above a threshold set by Wee1 inhibition. A small amount of active Cdk1 would then be generated, thereby activating the feedback loop. A second possibility is that the feedback loop is initiated by a triggering kinase that activates Cdc25 to generate a small amount of active Cdk1. In X. laevis, polo kinase has been proposed to act as a trigger kinase that directly phosphorylates and activates Cdc25C (Kumagai and Dunphy, 1996; Qian et al., 1998, 1999, 2001). However, in fission yeast and budding yeast, polo kinase appears to be activated downstream of mitotic Cdk1 (Tanaka et al., 2001; Mortensen et al., 2005). It is therefore unclear whether regulation of Cdc25 by polo kinase is part of a highly conserved mechanism. Another possibility is that Cdc25 is activated by signaling networks that relay information regarding cell growth or other physiological parameters.
Cdc25 is also thought to be regulated by 14-3-3 proteins and protein phosphatases (PPs). Phosphorylation of X. laevis Cdc25C at serine 287 triggers binding of 14-3-3 proteins and inhibition of phosphatase activity (Margolis and Kornbluth, 2004; Wolfe and Gould, 2004b; Boutros et al., 2006). Mutation of serine 287 to an alanine prevents binding of 14-3-3 proteins and causes premature entry into mitosis. Dephosphorylation of serine 287 is thought to be performed by PP1 and requires phosphorylation of T138 by Cdk2 or other kinases (Margolis et al., 2006a,b). The role of serine 287 phosphorylation has been studied most extensively in the context of a checkpoint that delays entry into mitosis in response to DNA damage. A similar DNA damage checkpoint that operates through inhibitory phosphorylation of Cdk1 does not appear to operate in budding yeast (Amon et al., 1992; Sorger and Murray, 1992). The PP2A phosphatase is thought to oppose activating phosphorylation of Cdc25C (Izumi et al., 1992; Kumagai and Dunphy, 1992; Clarke et al., 1993; Dunphy, 1994; Margolis et al., 2006b). Inhibition of PP2A in interphase extracts leads to rapid hyperphosphorylation and activation of Cdc25C in the absence of significant Cdk1 activity, which suggests that a kinase other than Cdk1 plays an important role in regulating Cdc25C (Kumagai and Dunphy, 1992; Izumi and Maller, 1995). The identity of the kinase that phosphorylates Cdc25C in interphase is unknown. In fission yeast, the Cdc14/Clp1 phosphatase is required for dephosphorylation of Cdc25 during mitotic exit (Esteban et al., 2004; Wolfe and Gould, 2004a).
To expand our knowledge of the mechanisms that regulate Cdc25, we have used the powerful approaches available in budding yeast to characterize the function and regulation of budding yeast Cdc25. The budding yeast homologues of Cdc25 and Wee1 are referred to as Mih1 and Swe1, and the primary mitotic cyclin that promotes entry into mitosis is called Clb2. We show that Mih1 undergoes dramatic changes in phosphorylation during the cell cycle and that Cdk1, PP2A, and casein kinase 1 each play a role in regulation of Mih1 phosphorylation. Because casein kinase 1 functions in the secretory pathway and is associated with sites of cell growth, it may provide a molecular link between control of cell growth and entry into mitosis.
Results
Loss of MIH1 causes delayed entry into mitosis and increased cell size
Previous studies reported that mih1Δ cells are larger than wild-type cells (Russell et al., 1989; Jorgensen et al., 2002; Harvey and Kellogg, 2003). We confirmed and extended these studies by using a Coulter counter to directly compare the sizes of wild-type, mih1Δ, swe1Δ, and mih1Δ swe1Δ cells (Fig. 1 A). These experiments demonstrated that mih1Δ cells are larger than wild-type cells and that mih1Δ swe1Δ cells are smaller than wild-type cells. To confirm that the large size of mih1Δ cells is a result of delayed entry into mitosis, we compared the timing of entry into mitosis in wild-type and mih1Δ cells. We assayed entry into mitosis by determining the percentage of cells with short mitotic spindles at 10-min intervals as cells progressed through the cell cycle after release from a G1 arrest. Previous work has shown that formation of short spindles is dependent on mitotic Cdk1 activity and is therefore a good marker for entry into mitosis (Fitch et al., 1992). The mih1Δ cells showed a 10–20-min delay in formation of short spindles (Fig. 1 B). Loss of MIH1 did not cause a delay in budding or in the accumulation of the S-phase cyclin Clb5, which demonstrates that the delay occurred during G2/M (Fig. 1, C and D). Collectively, these results demonstrate that Mih1 opposes the activity of Swe1 to promote entry into mitosis, and that the basic functions of fission yeast Cdc25 have been conserved in budding yeast Mih1.
Figure 1.
mih1Δ cells have an increased cell size and undergo delayed entry into mitosis. (A) Cells of the indicated genotypes were grown to log phase in YPD media at room temperature. Cell size was measured using a Z2 Coulter counter. (B) Wild-type and mih1Δ cells were synchronized with α-factor and released into fresh YPD media and samples were taken at 10-min intervals. Cells were fixed and stained with an anti-tubulin antibody and the percentage of cells with short spindles was determined. Over 200 cells were analyzed for each time point. The experiment was repeated at least three times with matched controls performed in parallel on the same day, and the same overall trend was observed. Error bars are not displayed because the timing of release from α-factor arrest varies from day to day. (C) Wild-type and mih1Δ cells were synchronized with α-factor and released into fresh YPD media and samples were taken at 10-min intervals. Cells were fixed and the percentage of cells with small buds was determined. Over 200 cells were analyzed for each time point. The experiment was repeated at least three times with matched controls performed in parallel on the same day, and the same overall trend was observed. Error bars are not displayed because the timing of release from α-factor arrest varies from day to day. (D) Wild-type and mih1Δ cells carrying CLB5-3XHA cells were synchronized with α-factor and released into fresh YPD media and samples were taken at 10-min intervals. Western blotting was used to monitor the behavior of Clb5-3XHA. (E) Wild-type cells were synchronized with α-factor and released into fresh YPD media and samples were taken at 10-min intervals. Western blotting was used to monitor the behavior of Swe1, Clb2, and Cdk1 tyrosine 19 phosphorylation. The exposures of the Western blots were normalized using background bands to allow direct comparison of the signals from each strain. The Swe1 protein migrates as a disperse group of phosphorylated forms above a 97 kD size standard, whereas Clb2 and Cdk1 migrate approximately at 50 and 35 kD, respectively.
To further characterize the phenotype of mih1Δ cells, we used Western blotting in synchronized cells to assay accumulation of the mitotic cyclin Clb2, inhibitory phosphorylation of Cdk1, and hyperphosphorylation of Swe1. Loss of Mih1 caused a significant delay in each of these events as well as increased levels of Cdk1 inhibitory phosphorylation (Fig. 1 E). The delayed accumulation of Clb2 is consistent with previous experiments, which found that activated Cdk1-Clb2 initiates a positive feedback loop that promotes further accumulation of Clb2 (Amon et al., 1993). Similarly, the delayed accumulation of Cdk1 inhibitory phosphorylation and delayed Swe1 hyperphosphorylation are consistent with previous experiments, which found that Cdk1-Clb2 hyperphosphorylates Swe1, which activates Swe1 to phosphorylate Cdk1 (Harvey et al., 2005). These results suggest that inhibitory phosphorylation of Cdk1 by Swe1 initially blocks the positive feedback loop that leads to increased accumulation of Clb2 and that Mih1 is required for activation of a small amount Cdk1 that initiates the positive feedback loop. Thus, in the absence of Mih1, accumulation of Clb2 and inhibitory phosphorylation of Cdk1 should both be delayed.
The fact that mih1Δ cells are able to exit mitosis suggested that an additional phosphatase may act redundantly with Mih1 to dephosphorylate Cdk1. PP2A appeared to be a good candidate because previous work in vertebrates and yeast implicated PP2A in regulation of mitosis (Goris et al., 1989; Izumi et al., 1992; Kumagai and Dunphy, 1992; Clarke et al., 1993; Lin and Arndt, 1995; Kinoshita et al., 1996; Minshull et al., 1996; Wang and Burke, 1997). PP2A is a heterotrimeric complex composed of a catalytic subunit and two regulatory subunits referred to as A and B subunits (Stark, 1996; Janssens and Goris, 2001). In budding yeast, there is a single A subunit called Tpd3 and two B subunits called Cdc55 and Rts1. Binding of Cdc55 and Rts1 to PP2A is mutually exclusive and deletion of their genes gives distinct phenotypes, which demonstrates that they mediate different functions of PP2A (Healy et al., 1991; Shu et al., 1997; Wang and Burke, 1997). The two different forms of PP2A are referred to as PP2ACdc55 or PP2ARts1. Previous work found that cdc55Δ causes a prolonged Swe1-dependent G2/M delay, which indicates that Cdc55 plays a role in regulating inhibitory phosphorylation of Cdk1 (Yang et al., 2000). We were unable to recover cdc55Δ mih1Δ cells from genetic crosses, which is consistent with the possibility that PP2ACdc55 is required for dephosphorylation of Cdk1 in mih1Δ cells. We were able to recover cdc55Δ mih1Δ swe1Δ cells, which further suggests that Mih1 and PP2ACdc55 act redundantly to dephosphorylate Cdk1. However, these experiments do not rule out the possibility that PP2ACdc55 is required for regulation of another phosphatase that directly dephosphorylates Cdk1 or that cdc55Δ is synthetically lethal with mih1Δ for other reasons. A GAL1-MIH1 cdc55Δ strain did not give a uniform G2/M arrest when shifted to dextrose, which suggests that PP2ACdc55 has diverse functions and does not act solely to regulate Cdk1 inhibitory phosphorylation at the G2/M transition (unpublished data).
Mih1 is hyperphosphorylated early in the cell cycle and undergoes dephosphorylation as cells enter mitosis
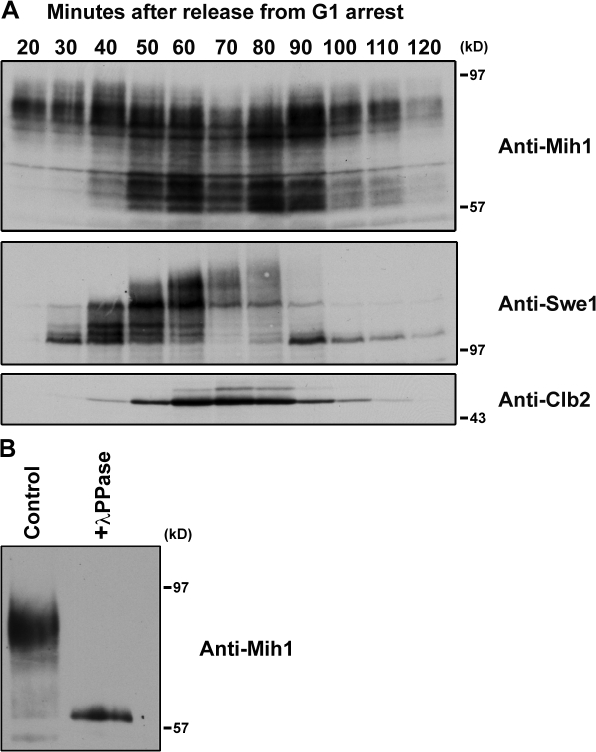
To begin to understand how Mih1 is regulated, we used Western blotting to follow the behavior of the Mih1 protein in synchronized cells. For comparison, we also followed the behavior of Swe1 and Clb2 in the same samples. Mih1 showed dramatic changes in posttranslational modification during the cell cycle (Fig. 2 A). Treatment of immunoprecipitated Mih1 with phosphatase eliminated all Mih1 modification, which indicated that the modification was caused by phosphorylation (Fig. 2 B). Mih1 was hyperphosphorylated early in the cell cycle and became dephosphorylated as cells entered mitosis. Mih1 dephosphorylation occurred concomitantly with Clb2 accumulation and Swe1 hyperphosphorylation. The extent of Mih1 hyperphosphorylation is unusual. Mih1 is a 63-kD protein and shifts as much as 30 kD in apparent electrophoretic mobility. There are 83 serines and 20 threonines in Mih1, and only nine of these match the minimal consensus phosphorylation site for Cdk1 (S/TP). This relatively small number of Cdk1 consensus sites suggests that additional sites must be phosphorylated to account for the large shift in the electrophoretic mobility of Mih1.
Figure 2.
Mih1 undergoes dramatic changes in phosphorylation state during the cell cycle. (A) Wild-type cells were synchronized with α-factor and released into fresh YPD media and samples were taken at 10-min intervals. Western blotting was used to monitor the behavior of Mih1, Swe1, and Clb2. The differently phosphorylated forms of Mih1 migrate between 57 and 97 kD size standards. (B) Phosphorylated 3XHA-Mih1 was immunoaffinity purified and treated with λ phosphatase. The phosphorylation state of Mih1 was monitored by Western blotting.
PP2A is required for dephosphorylation of Mih1 during mitosis
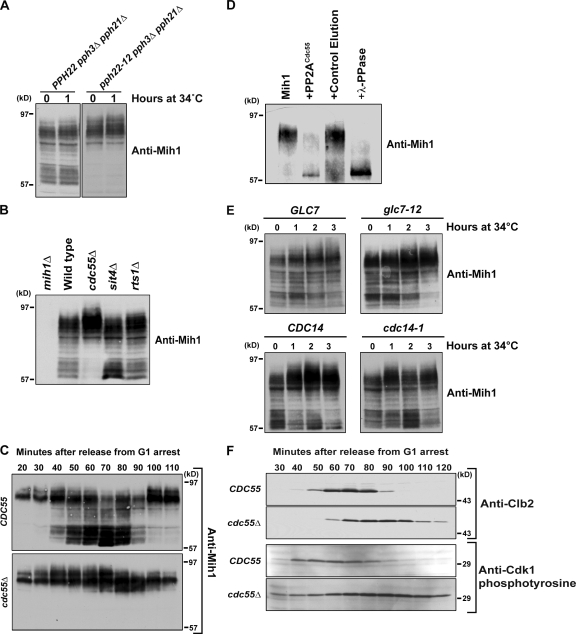
Because regulation of the phosphorylation state of Mih1 is likely to play a critical role in the mechanisms that control entry into mitosis, we sought to identify the phosphatases and kinases that regulate Mih1. We first used a candidate approach to identify the phosphatase that dephosphorylates Mih1. Because PP2A is known to be involved in the regulation of entry into mitosis, we first tested PP2A. In budding yeast, the catalytic subunit of PP2A is encoded by three partially redundant genes called PPH21, PPH22, and PPH3 (Evans and Stark, 1997). A temperature-sensitive allele of PPH22 in a pph21Δ pph3Δ background caused loss of the dephosphorylated forms of Mih1 and the appearance of hyperphosphorylated forms, even at the permissive temperature (Fig. 3 A). Similarly, loss of the Cdc55 regulatory subunit of PP2A caused a complete loss of dephosphorylated forms of Mih1 and the appearance of hyperphosphorylated forms, whereas loss of Rts1 had no effect (Fig. 3 B). Loss of Cdc55 also caused a failure to dephosphorylate Mih1 in synchronized cells (Fig. 3 C), and purified PP2ACdc55 was able to dephosphorylate Mih1 in vitro (Fig. 3 D). Finally, several other phosphatases implicated in regulation of the cell cycle were not required for the major downward shift in Mih1 electrophoretic mobility, including Glc7, Sit4, and Cdc14 (Fig. 3, B and E). A relatively slight dephosphorylation of Mih1 occurs at later time points in cdc55Δ cells, which suggests that another phosphatase may work on Mih1 (Fig. 3 C). Collectively, these experiments indicate that PP2ACdc55 is likely to directly dephosphorylate Mih1 as cells enter mitosis.
Figure 3.
PP2ACdc55 is required for dephosphorylation of Mih1. (A) PPH22 pph3Δ pph21Δ and pph22-12 pph3Δ pph21Δ cells were grown to log phase in YPD media at room temperature. After taking a sample (0-min time point), the cells were shifted to the restrictive temperature and samples were collected after 1 h. The phosphorylation state of Mih1 was followed by Western blotting. (B) Western blot analysis of Mih1 phosphorylation in log-phase populations of wild-type, cdc55Δ, sit4Δ, and rts1Δ cells. As a control, mih1Δ cells were included in the analysis. The phosphorylation state of Mih1 was monitored by Western blotting. (C) Wild-type and cdc55Δ cells were synchronized with α-factor and released into fresh YPD media and samples were taken at 10-min intervals. Western blotting was used to monitor the behavior of Mih1. (D) Immunoaffinity-purified phosphorylated 3XHA-Mih1 was treated with immunoaffinity purified Cdc55-3XHA. For controls, purified 3XHA-Mih1 was also incubated with λ phosphatase or a control elution from a nonspecific immunoaffinity column. The phosphorylation state of Mih1 was monitored by Western blotting. (E) Cells of the indicated genotypes were grown to log phase at room temperature. After taking a 0-min time point, the cells were shifted to the restrictive temperature. Samples were collected at 1, 2, and 3 h after shifting to 34°C. Western blotting was used to monitor the behavior of Mih1. The apparent upward shift in mobility of Mih1 in glc7-12 cells at 3 h was not reproducible and may have been caused by nutrient limitation after 3 h in culture. (F) Wild-type and cdc55Δ cells were synchronized with α-factor and released into fresh YPD media and samples were taken at 10-min intervals. Western blotting was used to monitor the behavior of Clb2 and Cdk1 tyrosine 19 phosphorylation. The exposures of the Western blots were normalized using background bands to allow direct comparison of the signals from each strain.
Previous studies reported that cdc55Δ causes a Swe1-dependent G2/M delay and an elongated cell phenotype (Healy et al., 1991; Yang et al., 2000). We found that cdc55Δ caused delayed accumulation of Clb2, a prolonged G2/M delay, and a large increase in inhibitory tyrosine phosphorylation of Cdk1 (Fig. 3 F). These observations demonstrate that PP2ACdc55 is required for regulation of Cdk1 inhibitory phosphorylation; however, they cannot distinguish whether PP2ACdc55 works via activation of Mih1, inhibition of Swe1, or both. The phenotype of cdc55Δ cells could also be caused by a direct role for PP2ACdc55 in removing Cdk1 inhibitory phosphorylation.
A systematic screen for kinases that regulate the phosphorylation state of Mih1
We next used a systematic approach to identify kinases that act directly or indirectly to regulate hyperphosphorylation of Mih1. Western blotting was used to screen 118 strains carrying deletions of genes encoding nonessential kinases or known kinase activators for effects on Mih1 hyperphosphorylation (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200711014/DC1). A single kinase deletion (mck1Δ) caused a decrease in Mih1 hyperphosphorylation, and three kinase deletions (sky1Δ, cla4Δ, and ste20Δ) caused an increase in Mih1 hyperphosphorylation (Fig. 4 A). No single kinase deletion caused a complete loss of Mih1 hyperphosphorylation.
Figure 4.
Mih1 phosphorylation is dependent on the Yck1 and 2 kinases. (A) Western blot analysis of Mih1 phosphorylation in log-phase populations of wild-type, mck1Δ, sky1Δ, ste20Δ, and cla4Δ cells. (B) Purified dephosphorylated 3XHA-Mih1 was incubated with purified TAP-tagged kinases at 30°C for 45 min in the presence of ATP. For each kinase, a control reaction was performed in which no phosphatase was added to ensure that the purified kinase did not introduce background bands to the anti-Mih1 Western blot. The phosphorylation state of Mih1 was monitored by Western blotting. (C) YCK1 YCK2, YCK1 yck2Δ, and yck2-2 yck1Δ cells were grown to log phase in YPD media. After taking a sample, the cells were shifted to 34°C and samples were collected at 1 and 2 h. The phosphorylation state of Mih1 was followed by Western blotting. (D) Purified 3XHA-Mih1 was incubated with the indicated combinations of purified Cdk1-Clb2-3XHA, Yck1-TAP, and dephosphorylated Mih1 at 30°C for 1 h in the presence of ATP. The phosphorylation state of Mih1 was monitored by Western blotting.
Essential kinases were purified as TAP-tagged fusion proteins and tested for their ability to phosphorylate Mih1 in vitro (Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200711014/DC1). To prepare Mih1 substrate for these assays, 3XHA-Mih1 was purified by immunoaffinity chromatography and treated with λ phosphatase to remove endogenous phosphorylation. In addition to the 22 essential kinases, we tested Yck1/2 and Cka1/2, two pairs of redundant kinases that are the budding yeast homologues of casein kinases 1 and 2. We also used TAP-tagged versions of Cln2 (G1 cyclin), Clb5 (S cyclin), and Clb2 to test the ability of Cdk1 associated with various cyclins to phosphorylate Mih1. Kinases that gave positive results in the screen are shown in Fig. 4 B. Yck1, Yck2, Cka1, Cka2, Sgv1, and Cdk1-Clb2 were able to induce partial hyperphosphorylation of Mih1. Another member of the casein kinase family (Hrr25) was not able to induce hyperphosphorylation of Mih1, whereas Yck3, a nonessential member of the casein kinase family, was able to induce partial hyperphosphorylation. Mck1 was unable to hyperphosphorylate Mih1 in vitro, which suggests that it regulates Mih1 hyperphosphorylation indirectly or that it is inactive when purified as a TAP-tagged fusion protein. Cdc5/polo kinase, which is thought to phosphorylate Mih1 homologues in other organisms, was unable to hyperphosphorylate Mih1. However, this assay would not identify kinases that phosphorylate Mih1 but fail to induce an electrophoretic mobility shift, or kinases that are inactive when purified by the TAP-tag purification protocol.
Casein kinase 1 activity is required for Mih1 hyperphosphorylation in vivo
As a secondary screen of the essential kinases, we determined whether kinases that can hyperphosphorylate Mih1 in vitro are required for Mih1 hyperphosphorylation in vivo. Inactivation of Cka1/2, Sgv1, or Hrr25 with temperature-sensitive alleles had no significant effect on Mih1 hyperphosphorylation (unpublished data). Similarly, yck3Δ had no effect on Mih1 phosphorylation in the screen for nonessential kinases that phosphorylate Mih1. In contrast, there was a quantitative loss of Mih1 hyperphosphorylation when yck2-2 yck1Δ cells were shifted to the restrictive temperature (Fig. 4 C). Mih1 hyperphosphorylation was also significantly decreased at the permissive temperature. A subset of Mih1 phosphorylation was unaffected in yck2-2 yck1Δ cells, which suggests that an additional kinase phosphorylates Mih1 or that the yck2-2 allele does not fully eliminate kinase activity. Deletion of the YCK3 gene in yck2-2 yck1Δ cells did not further reduce phosphorylation of Mih1, which demonstrates that Yck3 does not act redundantly with Yck1 and 2 (unpublished data). To determine if the remaining phosphorylation of Mih1 might be caused by residual yck2-2 kinase activity at the restrictive temperature, we attempted to fully eliminate Yck1 and 2 by creating a yck2-degron yck1Δ strain. Levels of the Yck2-degron fusion protein were reduced at the restrictive temperature; however, the protein was still present after 4 h and Mih1 remained partially phosphorylated (unpublished data).
Purified Yck1 and 2 were not able to induce the large shift in Mih1 electrophoretic mobility that occurred in vivo (Fig. 4 B). A possible explanation for this result is that we did not purify sufficient amounts of TAP-tagged Yck1 or 2 to induce quantitative hyperphosphorylation of Mih1. Yck1 and 2 are palmitoylated and would therefore be found primarily in the membrane fraction during the TAP-tag purification, which could significantly decrease yields. We therefore scaled up the purification and used higher levels of detergent to solubilize Yck1. Using this preparation, we found that Yck1 was able to induce a shift in the electrophoretic mobility of Mih1 that was similar in extent to the shift observed in vivo, although we were still unable to induce quantitative hyperphosphorylation of Mih1 (Fig. 4 D). Previous work has suggested that casein kinase 1 may preferentially phosphorylate proteins that have been primed by previous phosphorylation by other kinases (Knippschild et al., 2005). We found that phosphorylation of Mih1 with Cdk1-Clb2 in vitro did not significantly enhance phosphorylation of Mih1 by Yck1 (Fig. 4 D).
Casein kinase 1 is required for regulation of Cdk1 inhibitory phosphorylation
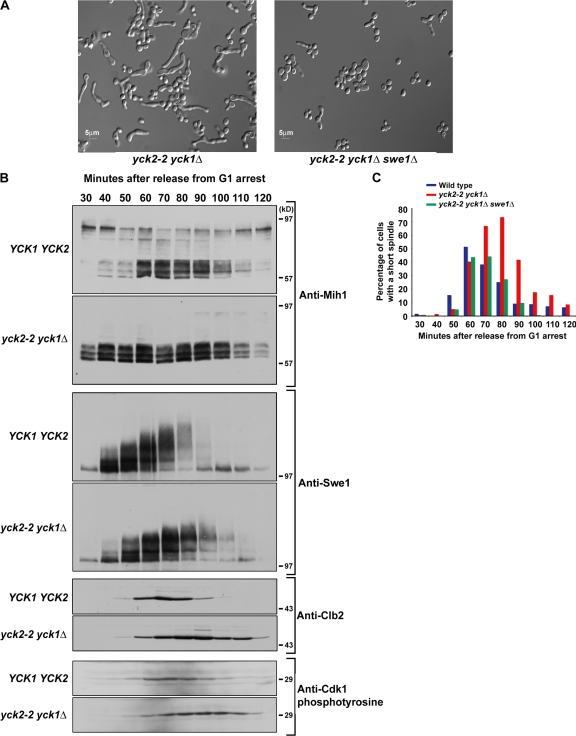
Several observations support the idea that Yck1 and 2 are required for the regulation of Cdk1 inhibitory phosphorylation in vivo. First, a fraction of yck2-2 yck1Δ cells become abnormally large and elongated at the restrictive temperature (Robinson et al., 1993). Previous work has shown that misregulation of Cdk1 inhibitory phosphorylation can lead to a similar elongated cell phenotype (Ma et al., 1996; Kellogg, 2003). We found that deletion of SWE1 in yck2-2 yck1Δ cells largely eliminated the elongated cell phenotype, which confirmed that the phenotype is caused by misregulation of Cdk1 inhibitory phosphorylation (Fig. 5 A). We also tested the effects of yck2-2 yck1Δ on the cell cycle at 30°C, a temperature at which cells are viable but Mih1 hyperphosphorylation is significantly reduced. This experiment revealed that a partial loss of Yck1 and 2 function caused increased accumulation of Cdk1 inhibitory tyrosine phosphorylation and delayed accumulation of Clb2, which indicates delayed entry into mitosis. In addition, Swe1 failed to undergo full hyperphosphorylation. Because dephosphorylation of Cdk1 is thought to help initiate full hyperphosphorylation of Swe1, this observation is consistent with reduced Mih1 activity (Harvey et al., 2005). The yck2-2 yck1Δ cells delayed at the short spindle stage of mitosis and swe1Δ eliminated the delay (Fig. 5 C). Finally, previous work found that cdc55Δ is synthetically lethal with yck2-2 yck1Δ (Robinson et al., 1993). Because cdc55Δ is also synthetically lethal with mih1Δ, this observation is consistent with the idea that Yck1 and 2 regulate Mih1. Collectively, these results demonstrate that Yck1 and 2 play important roles in regulation of Cdk1 inhibitory phosphorylation, most likely via Mih1. They do not rule out the possibility that Yck1 and 2 also regulate Cdk1 inhibitory phosphorylation via Swe1.
Figure 5.
Yck1 and 2 are required for Mih1 hyperphosphorylation. (A) yck2-2 yck1Δ and yck2-2 yck1Δ swe1Δ cells were grown to log phase at room temperature in YPD media and shifted to 34°C for 6 h. (B) YCK1 YCK2 and yck2-2 yck1Δ cells were synchronized with α-factor and released into fresh YPD media at 30°C and samples were taken at 10-min intervals. Western blotting was used to monitor the behavior of Mih1, Swe1, Cdk1 tyrosine 19 phosphorylation, and Clb2. The exposures of the Western blots were normalized using background bands to allow direct comparison of the signals from each strain. (C) Isogenic YCK1 YCK2, yck2-2 yck1Δ, and yck2-2 yck1Δ swe1Δ cells were synchronized with α-factor and released into fresh YPD media at 30°C and samples were taken at 10-min intervals. Cells were fixed and stained with an anti-tubulin antibody and the percentage of cells with short spindles was determined. Over 200 cells were analyzed for each time point. The experiment was repeated at least three times with matched controls performed in parallel on the same day, and the same overall trend was observed. Error bars are not displayed because the timing of release from α-factor arrest varies from day to day.
Cdk1 is required for phosphorylation and dephosphorylation of Mih1
Because Cdk1-Clb2 was identified in the screen for kinases that phosphorylate Mih1, we performed further tests for a role of Cdk1 in regulating Mih1. The in vitro phosphorylation reactions performed in the screen suggested that Cdk1-Clb2 preferentially phosphorylates Mih1 when compared with Cdk1-Cln2 (Fig. 4 B). We tested this more carefully using Cdk1-Clb2-3XHA and Cdk1-Cln2-3XHA purified by immunoaffinity chromatography. Mih1 was preferentially phosphorylated by Cdk1-Clb2-3XHA under conditions where equal amounts of Cdk1-Clb2-3XHA and Cdk1-Cln2-3XHA were added (unpublished data). A caveat to this experiment is that previous work found that Cdk1-Clb2 has an ∼25-fold greater specific activity than Cdk1-Cln2 (McCusker et al., 2007). For technical reasons, it was not possible to add enough Cdk1-Cln2-3XHA to the reaction to allow comparison of the ability of Cdk1-Cln2-3XHA and Cdk1-Clb2-3XHA to phosphorylate Mih1 under conditions of equal kinase activity.
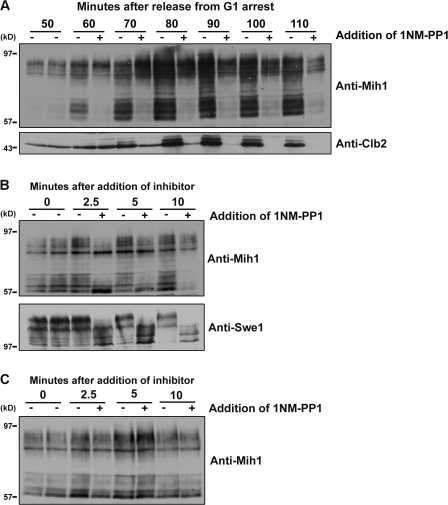
We next tested whether Cdk1 activity is required for phosphorylation of Mih1 in vivo. For these experiments, we took advantage of the cdk1-as allele, which can be rapidly and specifically inhibited in vivo with 1NM-PP1. Cells carrying the cdk1-as allele were released from a G1 arrest and divided into two aliquots. 1NM-PP1 was added to one aliquot and samples were taken periodically during the cell cycle. The phosphorylation state of Mih1 and accumulation of Clb2 were assayed by Western blotting. Inhibition of cdk1-as in G1 (addition of 1NM-PP1 30 min after release from α-factor arrest) had no effect on the phosphorylation state of Mih1 over a period of 20 min (unpublished data). However, inhibition of cdk1-as when Clb2 was beginning to accumulate at the G2/M transition (addition of 1NM-PP1 50 min after release from α-factor arrest) eliminated Mih1 dephosphorylation (Fig. 6 A). Mih1 dephosphorylation is therefore dependent on Cdk1 activity.
Figure 6.
Cdk1 regulates Mih1 phosphorylation and dephosphorylation. (A) Log-phase cdk1-as cells were synchronized in G1 with α-factor and released into fresh YPD lacking supplemental adenine. At the indicated times, the cells were split into two aliquots, 1NM-PP1 was added to one aliquot, and samples were collected at 10-min intervals. For the control, an equivalent amount of DMSO was added. The phosphorylation state of Mih1 was monitored by Western blotting. (B) cdk1-as cells were grown to log phase at room temperature and synchronized in G1 with α-factor. The cells were then released into fresh YPD lacking supplemental adenine. At 80 min, the culture was divided in half. 1NM-PP1 was added to one half, and samples were collected at 0, 2.5, 5, and 10 min after the addition of 1NM-PP1. For the control, an equivalent amount of DMSO was added. The phosphorylation state of Mih1 was monitored by Western blotting. (C) Wild-type cells were grown to log phase at room temperature and synchronized in G1 with α-factor. The cells were then released into fresh YPD lacking supplemental adenine. At 80 min, the culture was divided in half. 1NM-PP1 was added to one half, and samples were collected at 0, 2.5, 5, and 10 min after the addition of 1NM-PP1. For the control, an equivalent amount of DMSO was added. The phosphorylation state of Mih1 was monitored by Western blotting.
The finding that inhibition of Cdk1 leads to a failure to dephosphorylate Mih1 suggested that Cdk1 does not phosphorylate Mih1 in vivo. To examine this issue more carefully, we added 1NM-PP1 to synchronized cells at 80 min, after cells had entered mitosis and Mih1 dephosphorylation had been initiated. We then collected samples at 2.5, 5, and 10 min and assayed Mih1 phosphorylation. Mih1 showed a striking behavior in this experiment. At short times after inhibition of cdk1-as, Mih1 appeared to show a transient loss of a subset of phosphorylations; however, by 10 min there was a recovery of Mih1 phosphorylation (Fig. 6 B). The addition of 1NM-PP1 to wild-type control cells had no effect (Fig. 6 C). A possible explanation for this result is that Cdk1 phosphorylates Mih1 on a subset of sites and is also required for PP2A activity. Inhibition of cdk1-as would initially cause a loss of Mih1 phosphorylation but would also cause inactivation of PP2A, which would lead to hyperphosphorylation of Mih1 by Yck1 and 2.
Discussion
Mih1 promotes entry into mitosis in budding yeast
Pioneering work in fission yeast demonstrated that loss of Cdc25 causes delayed entry into mitosis and increased cell size, whereas loss of Wee1 causes premature entry into mitosis and decreased cell size (Nurse, 1975; Nurse et al. 1976; Russell and Nurse, 1986, 1987; Gould and Nurse, 1989). Early studies in budding yeast concluded that swe1Δ does not cause reduced cell size or premature entry into mitosis and that Swe1 and Mih1 mediate a morphogenesis checkpoint that is activated in response to cytoskeletal stress (Sia et al., 1996, 1998; Lew, 2003; McNulty and Lew, 2005; Keaton and Lew, 2006). Here, we have shown that mih1Δ causes delayed entry into mitosis, increased cell size, and increased levels of Cdk1 inhibitory phosphorylation. Previous studies also reached the conclusion that mih1Δ causes delayed entry into mitosis and increased levels of Cdk1 inhibitory phosphorylation (Booher et al., 1993; Rudner et al., 2000). Recent work has shown that swe1Δ causes premature entry into mitosis and a reduced cell size (Jorgensen et al., 2002; Harvey and Kellogg, 2003; Harvey et al., 2005). Previous results that swe1Δ does not cause reduced cell size (McNulty and Lew, 2005) have been revised after more recent measurements with a Coulter counter (J. Bean and F. Cross, personal communication). We have also shown that inhibitory phosphorylation of Cdk1 occurs in wild-type cells growing under unstressed conditions (Fig. 1 C; Harvey et al., 2005). Together, these observations demonstrate that Swe1 actively delays entry into mitosis and is required for cell size control in the absence of cytoskeletal stress and that the basic functions of fission yeast Cdc25 and Wee1 are likely to have been conserved in budding yeast Mih1 and Swe1. These experiments do not rule out a role for Swe1 and Mih1 in mediating a morphogenesis checkpoint.
Mih1 is regulated by casein kinase 1 and PP2ACdc55
Mih1 is hyperphosphorylated in interphase and undergoes dephosphorylation as cells enter mitosis. The dramatic changes in Mih1 phosphorylation suggest that phosphorylation plays an important role in regulation of Mih1. Yck1 and 2 are responsible for the bulk of Mih1 hyperphosphorylation, whereas PP2ACdc55 is responsible for dephosphorylation of Mih1 as cells enter mitosis. Although it is clear that PP2ACdc55 and casein kinase 1 regulate Mih1 phosphorylation, the exact roles that they play in regulating Mih1 remain unclear. Phosphorylation of Mih1 could regulate phosphatase activity, localization, or association with other proteins. Paradoxically, genetic tests seem to suggest that both the dephosphorylated and hyperphosphorylated forms of Mih1 are required for full Mih1 activity. For example, the cdc55Δ phenotype suggests that hyperphosphorylated forms of Mih1 have reduced activity. Conversely, the yck2-2 yck2Δ phenotype suggests that dephosphorylated forms of Mih1 have reduced activity. However, the complexity of mitotic regulation makes it difficult to make strong conclusions regarding the relative activities of differently phosphorylated forms of Mih1 from mutant phenotypes. For example, it is possible that PP2ACdc55 or Yck1 and 2 also function upstream of Swe1, which would complicate the interpretation of phenotypes. Moreover, it is possible that there are additional kinases or phosphatases that regulate Mih1 that were missed in our screens.
The finding that the phosphorylation state of Mih1 is regulated by casein kinase 1 and PP2Acdc55 suggests that Mih1 is not regulated by relatively simple positive feedback loops that only involve core regulators of the cell cycle. The discovery that casein kinase 1 is a regulator of Mih1 is of particular interest and provides an important step toward understanding the regulation and physiological functions of inhibitory phosphorylation of Cdk1. Yck2 is a plasma membrane–associated protein that is concentrated at sites of polar cell growth and cytokinesis (Robinson et al., 1999; Babu et al., 2002). Loss of Yck1 and 2 causes defects in the pattern of cell growth as well as in endocytosis and septin organization (Robinson et al., 1993, 1999; Panek et al., 1997). Key targets of Yck1 and 2 that mediate these functions have remained unknown. However, the fact that Yck1 and 2 regulate Mih1 and are associated with sites of cell growth suggests that they are well-positioned to relay signals regarding the status of growth-related events to the cell cycle control machinery. The discovery of a role for casein kinase 1 in regulation of Mih1 may therefore provide a key missing link between the mechanisms that regulate entry into mitosis and mechanisms that regulate aspects of cell growth. Such links must exist to allow coordination of cell growth and cell division but have remained elusive.
Cdk1 is required for initiation of Mih1 dephosphorylation
Previous work found that X. laevis Cdc25 is regulated by Cdk1 (Izumi et al., 1992; Kumagai and Dunphy, 1992; Izumi and Maller, 1993). Several experiments demonstrated that Mih1 is also regulated by Cdk1. For example, Cdk1-Clb2 preferentially phosphorylated Mih1 in vitro when compared with Cdk1-Cln2. In addition, inhibition of cdk1-as in cells that are entering mitosis caused a failure to dephosphorylate Mih1, and inhibition of cdk1-as after entry into mitosis caused a transient dephosphorylation of Mih1. A possible explanation for these results is that Cdk1 phosphorylates Mih1 on a subset of sites and also activates dephosphorylation of Mih1 by PP2ACdc55. In this model, inhibition of cdk1-as during entry into mitosis would prevent Mih1 dephosphorylation, whereas inhibition of cdk1-as during mitosis would initially lead to a rapid loss of Cdk1-dependent phosphorylation of Mih1 but would also lead to inactivation of PP2ACdc55, which would allow hyperphosphorylation of Mih1 by Yck1 and 2. Another possibility is that Cdk1-Clb2 initiates dephosphorylation of Mih1 by inhibiting phosphorylation of Mih1 by Yck1 and 2. In this case, inhibition of cdk1-as would again cause a loss of Cdk1-dependent phosphorylation of Mih1 but also an increase in phosphorylation of Mih1 by Yck1 and 2. It is also possible that the response of Mih1 to the inhibition of cdk1-as involves more complex signaling networks that include additional kinases and phosphatases.
Previous work in X. laevis suggested that Cdk1 phosphorylates and activates Cdc25 during entry into mitosis (Izumi et al., 1992; Kumagai and Dunphy, 1992; Izumi and Maller, 1993). Thus far we have not been able to purify sufficient quantities of tyrosine-phosphorylated Cdk1-Clb2 and differentially phosphorylated forms of Mih1 to carefully test whether phosphorylation regulates Mih1 activity.
Is entry into mitosis regulated by highly conserved mechanisms?
The mechanisms that regulate Mih1 seem to provide a striking contrast to the mechanisms that regulate Cdc25. Mih1 is hyperphosphorylated in interphase and undergoes dephosphorylation when cells enter mitosis, whereas vertebrate Cdc25 is dephosphorylated in interphase and undergoes hyperphosphorylation when cells enter mitosis (Izumi et al., 1992; Kumagai and Dunphy, 1992). Yet the mechanisms that regulate entry into mitosis in eukaryotic cells are of fundamental importance and it seems likely that core components of these mechanisms have been conserved. Several observations suggest that the mechanisms that regulate Cdc25 and Mih1 may be more similar than superficial comparisons might suggest. Both Mih1 and Cdc25 undergo Cdk1-dependent regulation, and Cdk1 appears to help initiate feedback loops that trigger changes in their phosphorylation states (Dunphy, 1994). There is also good evidence that both Mih1 and Cdc25 are regulated by a balance of opposing kinase and phosphatase activities in which PP2A plays a central role. For example, inhibition of PP2A in interphase X. laevis extracts that lack significant Cdk1 activity rapidly induces full hyperphosphorylation of Cdc25C. This suggests that a kinase other than Cdk1 can hyperphosphorylate Cdc25C and that PP2A opposes hyperphosphorylation of Cdc25C (Kumagai and Dunphy, 1992; Izumi and Maller, 1995). Although Cdk1 and polo kinase have been implicated in regulation of Cdc25, no experiments have found that these kinases are capable of directly inducing the full hyperphosphorylation of Cdc25 that is observed in vivo, which again suggests the existence of another kinase that phosphorylates Cdc25 (Izumi and Maller, 1995). The activity of the kinases that phosphorylate X. laevis Cdc25 is high in mitosis and low in interphase, whereas the activity of the phosphatases that dephosphorylate Cdc25 is high in interphase and low in mitosis (Kumagai and Dunphy, 1992). Collectively, these experiments suggest that Cdc25 is regulated by a balance of opposing kinase and phosphatase activities (Dunphy, 1994). The identity of the kinase that phosphorylates X. laevis Cdc25 in interphase extracts is unknown, but analysis of Mih1 regulation suggests that it may be casein kinase 1.
Mih1 also appears to be regulated by a balance of opposing kinase and phosphatase activities. Loss of PP2ACdc55 causes Mih1 to shift to hyperphosphorylated forms, whereas loss of Yck1 and 2 causes Mih1 to shift to dephosphorylated forms. In budding yeast, the balance may be shifted more toward the kinase during interphase so that Mih1 is found primarily in the hyperphosphorylated form. In vertebrates, the balance may be shifted more toward the phosphatase in interphase so that Cdc25 is primarily in the dephosphorylated form. Thus, it is possible that Mih1 and Cdc25 are both regulated by a conserved mechanism that is based on the opposing activities of kinases and phosphatases. Regulation of a protein by opposing kinase and phosphatase activities can potentially generate switch-like changes in the activity of the protein (Goldbeter and Koshland, 1981, 1984; LaPorte and Koshland, 1983; Ferrell and Xiong, 2001; Ferrell, 2002). Regulation of Mih1 and Cdc25 by opposing kinase and phosphatase activities may therefore play an important role in the mechanisms that convert gradually increasing mitotic cyclin levels to an abrupt switch-like activation of Cdk1 at the G2/M transition.
Materials and methods
Yeast strains and culture conditions
The gene deletions and TAP-tagged kinases used to screen for kinases that regulate Mih1 were obtained from Open Biosystems. The strains used for this study are listed in Table I. In most cases, cells were grown in yeast extract-peptone-dextrose (YPD) media supplemented with 40 mg/liter adenine. The cdk1-as1 strain was grown in YPD lacking supplemental adenine. GP16 was generated by integrating the GAL1 promoter and a 3XHA tag upstream of MIH1 in strain DK186 using standard protocol (oligos: CATTGGCACATTCATCTTCAGTTCCATGAAA-TATATTGTTGCACTGAGCAGCGTAATCT and CAGGACTAAGGATGAA-GAGGGGCTTCAATGTTTTATATGGGAATTCGAGCTCGTTTAAAC; Longtine et al., 1998). GP28 was generated by sporulating the diploid sit4Δ∷kanMX4/SIT4 strain (diploid deletion collection; Open Biosystems). DK1053 was generated by amplifying swe1Δ∷HIS5 (Kluyveromyces lactis) from strain SH181 with oligos (GCGACGCGACGCGAAAAAAATGC and AATGCTTGAAGCGGCTGTACTT) and transforming it into LRB756. Strains DK1121 and 1138 were generated by integrating 3XHA tag at the N-terminal ends of Clb5 in HT179 and DK186 using standard protocol (oligos: CGAAATGCATAGCAACTTTCAAAATCT-ATTTAATCTTAAGCGGATCCCCGCGTTAATTAA and GTAAAGAGTATGCGAATTCATGAGCATTACTAGTACTAATGAATTCG-AGCTCGTTTAAAC; Longtine et al., 1998).
Table I. Strains used in this study.
| Strain | Genotype | Strain background | Reference or source |
|---|---|---|---|
| DK186 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + | W303 | Altman et al. (1997) |
| DK303 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + CLB2-3XHA∷TRP1 | W303 | Harvey et al. (2005) |
| DK354 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + zds1Δ∷LEU2 zds1Δ∷TRP1 CDC55-3XHA∷HIS5 (K. lactis) | W303 | H. Tjandraa |
| DK1053 | MATa, his3 leu2 ura3-52 yck1-1∷URA3 yck2-2 swe1Δ∷HIS5 (K. lactis) | Unknown | This study |
| DK1121 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + mih1Δ∷URA3 Clb5-3XHA∷HIS | W303 | This study |
| DK1138 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + Clb5-3XHA∷HIS | W303 | This study |
| DEY213 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + PPH22∷URA3 pph3Δ1∷LYS2 pph21Δ∷HIS3 | W303 | Evans and Stark (1997) |
| DEY214 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + pph22-12∷URA3 pph3Δ1∷LYS2 pph21Δ∷HIS3 | W303 | Evans and Stark (1997) |
| GP16 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + GAL1-3XHA-MIH1∷HIS5 (K. lactis) | W303 | This study |
| GP28 | MATa, his3Δ1 leu2Δ0 URA3Δ0 lys2 sit4Δ∷kanMX4 | BY4743 | This study |
| HT179 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + mih1∷URA3 | W303 | H. Tjandra |
| JAU05 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + cdc28∷cdc28-as1 | W303 | Bishop et al. (2000) |
| JC33a | MATa,cdc5-1 bar1∷URA3, leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 GAL + | W303 | D. Morganb |
| JM82 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 GAL + cdc55Δ∷HIS3 | W303 | A. Rudnerc |
| KKY387 | MATa,ura3-52 lys2-801amber ade2-101ochre trp1Δ63 his1-Δ200 leu2-Δ hrr25Δ∷loxP-kanMX-loxP (pKK204 2μ pGAL-3XHA-HRR25degron) | YPH499 | Kafadar et al. (2003) |
| LRB756 | MATa, his3 leu2 ura3-52 yck1-1∷URA3 yck2-2 | Unknown | Panek et al. (1997) |
| LRB758 | MATa, his3 leu2 ura3-52 | Unknown | Panek et al. (1997) |
| LRB1039 | MATa,his3 leu2 ura3-52 yck2∷kanMX | Unknown | L. Robinsond |
| PAY704-1 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + glc7Δ∷LEU2 trp1∷GLC7∷TRP1 | W303 | Andrews and Stark (2000) |
| PAY701-2 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + trp1∷glc7-12∷TRP1 | W303 | Andrews and Stark (2000) |
| RJD1229 | MATα,cdc14-1 pep4∷TRP1 bar1∷LEU2 ade2-1 can1-100 his3-11,15 leu 2-3,112 trp1-1 ura3-1 | W303 | R. Deshaiese |
| SH24 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + swe1Δ∷URA3 | W303 | Harvey et al. (2005) |
| SH113 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + mih1Δ∷URA3 swe1Δ∷HIS3MX6 (K. lactis) | W303 | S. Harveya |
| SH181 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11,15 trp1-1 bar1Δ GAL + ura3-1 swe1Δ∷HIS3MX6 (K. lactis) | W303 | S. Harvey |
| SH650 | MATa,leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1 bar1Δ GAL + + cdc55Δ∷KanMX6 | W303 | S. Harvey |
| YDH6 | MATa,ade2-101his3-Δ200 leu2-Δ1 lys2-801amber trp1-Δ1 ura3-52 cka1- Δ1∷HIS3 cka2-Δ1TRP1 (pCEN6/ARSH4 LEU2 CKA2) | YPH250 | Hanna et al. (1995) |
| YDH8 | MATa,ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 cka1- Δ1∷HIS3 cka2-Δ1∷TRP1 (pCEN6/ARSH4 LEU2 cka2-8) | YPH250 | Hanna et al. (1995) |
| YDH13 | MATa,ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 cka1- Δ1∷HIS3 cka2-Δ1∷TRP1 (pCEN6/ARSH4 LEU2 cka2-13) | YPH250 | Hanna et al. (1995) |
University of California, Santa Cruz, Santa Cruz, CA.
University of California, San Francisco, San Francisco, CA.
University of Ottowa, Ottowa, Canada.
Louisiana State University Medical Center, Shreveport, LA.
California Institute of Technology, Pasadena, CA.
Plasmid construction and antibody generation
To generate an anti-Mih1 antibody, full-length MIH1 was cloned into pGEX4T-3 to create pGP1 (oligos: GCGGGATCCATGAACAATATATTTCAT and GCGGAATTCGCGGGCCTGGGTAAATCT). The GST-Mih1 fusion was expressed in Escherichia coli and purified as previously described (Kellogg and Alberts, 1992).
Cell cycle time courses
For most cell cycle time courses, log-phase culture cells were grown overnight at room temperature to OD600 = 0.65. Cells were arrested in G1 by the addition of either 15 μg/ml α-factor for 2.5 h (for BAR1 strains) or 0.5 μg/ml α-factor for 3 h (for bar1Δ strains) at room temperature. The cells were released from the arrest at 30°C, 1.6-ml samples were collected at 10-min intervals, and the cells were rapidly pelleted in screw-top tubes. The supernatant was removed and ∼250 μl of acid-washed glass beads were added before freezing on liquid nitrogen. To lyse the cells, 150 μl of protein sample buffer (65 mM Tris-HCl, pH 6.8, 3% SDS, 10% glycerol, and 5% β-mercaptoethanol) supplemented with 50 mM NaF, 50 mM β-glycerophosphate, and 2 mM PMSF was added and the tubes were placed in a Multibeater-8 (BioSpec Products, Inc.) and mixed at top speed for 90 s. The PMSF was added to the sample buffer immediately before use from a 100-mM stock made in 100% ethanol and stored at −20°C. The tubes were immediately removed, centrifuged for 10 s in a microfuge, and placed in a boiling water bath for 5 min. The tubes were centrifuged in a tabletop microfuge for 5 min and 20 μl was loaded on a gel. To test for essential kinases or phosphatases that act on Mih1, log-phase cultures of temperature-sensitive alleles were shifted to the restrictive temperature and 1.6-ml samples were collected at 1-h time intervals. For the cell cycle time course shown in Fig. 5 B, cells were arrested in G1 at 22°C and released from the arrest at 30°C, which is a semirestrictive temperature for the yck2-2 allele. For Fig. 6 A, cells were released from an α-factor arrest into fresh YPD at 30°C. At 50 min, the culture was split in half, 25 μM 1NM-PP1 (a gift from C. Zang and K. Shokat, University of California, San Francisco, San Francisco, CA) was added to one half, and an equivalent amount of DMSO was added to the other half (mock treated). 1.6-ml samples were collected from each set at 10-min intervals until 120 min. For Fig. 6 B, log-phase cells were arrested in G1 and released from an α-factor arrest into fresh YPD at 30°C. At 90 min, the culture was split in half, 25 μM 1NM-PP1 was added to one half, and an equivalent amount of DMSO was added to the other half. 1.6-ml samples were collected from each set at 0, 2.5, 5, and 10 min after the addition of 1NM-PP1. The time courses for Fig. 1 (B and C) and Fig. 5 C were repeated a minimum of three times, and in each case the data shown represent the result of a timecourse performed in parallel with an isogenic control and the mutant strains.
Western blotting, microscopy, and cell size analysis
Polyacrylamide gel electrophoresis was done as previously described (Anderson et al., 1973). Gel electrophoresis for analysis of Mih1 phosphorylation by Western blotting was performed with a 10% SDS-PAGE gel of dimensions 17 × 8.5 × 1 mm. The gels were run at 20 mA constant current until a 58-kD prestained molecular mass marker was at the bottom of the gel. Western blot transfers were performed for 75 min at 800 mA at 4°C in a Hoeffer transfer tank in a buffer containing 20 mM Tris base, 150 mM glycine, and 20% methanol. Mitotic spindles were fixed and stained as previously described (Pringle et al., 1991). Cell size analysis was performed as previously described, except that cultures were grown at room temperature in YPD to OD600 = 0.65 (Harvey et al., 2005). All microscopy was done using an Axioskop 2 plus (Carl Zeiss, Inc.) with a camera (HRm; AxioCam) and 63× objective (numerical aperture 1.4). Images were acquired using Axiovision software (Carl Zeiss, Inc).
Immunoaffinity purifications
Immunoaffinity purification of 3XHA-Mih1 was performed in the presence of 1 M KCl using the same protocol used for purification of 3XHA-Swe1 with some modifications (Harvey et al., 2005). For the purification of phosphorylated Mih1 used in Figs. 2 B and 4 B, immunoaffinity beads were prepared by binding 400 μg of affinity-purified polyclonal anti-HA antibodies to 400 μl of protein A beads (Bio-Rad Laboratories). Binding was performed overnight at 4°C on a rotator (Thermo Fisher Scientific). Beads were equilibrated by washing three times in extract buffer (50 mM Hepes-KOH, pH 7.6, 1 M KCl, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, and 0.25% Tween-20) before the addition of extract. Cells containing a 3XHA-tagged copy of Mih1 under the control of the GAL1 promoter (GP16) were grown overnight at 30°C in yeast extract peptone media containing 2% glycerol and 2% ethanol to OD600 = 0.8. Galactose was added to 1% and cells were incubated at 30°C for 3.5 h. The cells were pelleted and ground under liquid nitrogen and 12 g of cell powder was resuspended in 25 ml of extract buffer containing 2 mM PMSF. All subsequent steps were performed at 4°C. The extract was centrifuged for 5 min at 10,000 g, followed by an additional centrifugation step at 45,000 g for 45 min. After the final spin, 15 ml of the extract was added to the anti-HA antibody beads.
The extract and anti-HA beads were gently rotated end over end at 4°C for 1.5 h. The beads were pelleted by centrifugation, the supernatant was removed, and the remaining extract was incubated with the beads for an additional 1.5 h at 4°C. The beads were pelleted by centrifugation and washed three times with 15 ml of ice-cold extract buffer without PMSF. After the final wash, the beads were washed twice with ice-cold elution buffer (50 mM Hepes-KOH, pH 7.6, 250 mM KCl, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, and 0.1% Tween-20). The beads were then transferred to a 1.5-ml Biospin column (Bio-Rad Laboratories) and washed three times by pipetting 1 ml of elution buffer onto the top of the column. To elute the column, 250 μl of elution buffer containing 0.5 mg/ml HA dipeptide was added to the column and the flow-through fraction was collected. After a 30-min incubation, another aliquot was added. This was repeated for a total of six fractions. Elution fractions two through five were aliquoted in 10-μl vol and flash frozen on liquid nitrogen.
For the purification of the trimeric PP2ACdc55 core complex, we utilized a CDC55-3XHA strain (DK354) that lacks the Zds1 and 2 proteins, which associate with the PP2ACdc55 core complex (Gavin et al., 2002). A similar protocol was followed, except that cells were grown in YPD and 10 g of powder was resuspended in 25 ml of extract buffer. For the purification of dephosphorylated Mih1 for Fig. 4 (B and D), the experiment was performed similarly with the following modifications: 500 μg of polyclonal anti-HA antibodies were added to 400 μl of protein A beads. For treatment with λ phosphatase, the beads were washed twice with λ phosphatase buffer (50 mM Tris-HCl, pH 7.5, 5 mM DTT, 2 mM MnCl2, and 100 μg/ml BSA). After the final wash in phosphatase buffer, the beads were resuspended in phosphatase buffer and transferred to a 1.6-ml tube. The beads were centrifuged and the supernatant was aspirated. 400 μl of phosphatase buffer and 30 μl of λ phosphatase were added and the tube was incubated at 30°C for 1 h with gentle mixing every 15 min. The column was washed and the protein was eluted and aliquoted as described in the previous paragraph.
Screening for kinases that phosphorylate Mih1
To screen for nonessential kinases that phosphorylate Mih1, kinase deletion strains were grown to log phase at 30°C in 2-ml 96-well plates. Table S1 provides a list of the nonessential kinase deletion strains that were tested. Cells were pelleted by centrifugation, resuspended in 50 mM Hepes-KOH, pH 7.6, aliquoted to 1.6-ml screw-top tubes, and pelleted again. The supernatant was removed and glass beads were added to each tube before freezing on liquid nitrogen. For Western blotting of the crude extracts, 100 μl of protein sample buffer was added to each tube, cells were lysed by bead beating followed by incubation at 100°C for 5 min, and 20 μl of each sample was used for SDS-PAGE.
To screen essential kinases, TAP-tagged kinases were purified and tested for their ability to phosphorylate Mih1. Table S1 provides a list of TAP-tagged proteins that were tested. For each purification, 15 μl of IgG Sepharose beads (GE Healthcare) was transferred into a 500-μl microfuge tube. The beads were washed three times with 500 μl of extract buffer (50 mM Tris-HCl, pH 7.5, 700 mM NaCl, 75 mM NaF, 75 mM BGP, 2 mM EGTA, 5% glycerol, and 0.25% Tween 20). To prepare cells for purification experiments, 50 ml of cells at OD600 = 0.65 were pelleted, resuspended in 1 ml of 50 mM Hepes-KOH, pH 7.6, aliquoted to 1.6-ml screw-top tubes, and pelleted again, and 250 μl of glass beads was added to each tube before freezing on liquid nitrogen. Extracts for purifications were made by adding 300 μl of room-temperature extract buffer with 2 mM PMSF. The tubes were placed immediately into a Multibeater-8 and beaten at top speed for 30 s. The tubes were then placed in an ice-water bath for 30 s before a 5-min spin in a microfuge at 13,000 rpm. 300 μl of the supernatant was removed and replaced with 200 μl of extract buffer with 2 mM PMSF and the tubes were beaten again for 30 s. The supernatants were pooled, spun in a microfuge at top speed for 5 min, and then added to the IgG Sepharose beads equilibrated in lysis buffer. The tubes were rotated gently end over end at 4°C for 1.5 h and the beads were then washed three times with 500 μl of extract buffer without PMSF, followed by three more washes with 500 μl TEV cleavage buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 0.1% Tween-20, and 5% glycerol). At the end of these washes, the beads were resuspended in 70 μl TEV cleavage buffer and 1 μl of 1.6-mM TEV protease (a gift from J. Little, University of California, Santa Cruz, Santa Cruz, CA) was added to each tube. The tubes were rotated gently end over end at room temperature for 2 h. The tubes were spun in a microfuge at top speed for 2 min and 65 μl of the supernatant was transferred to a fresh tube. The composition of the supernatant was adjusted to 2 mM MgCl2 and 0.5 mM ATP. Kinase assays were performed by mixing 15 μl of purified kinase with 5 μl of dephosphorylated Mih1, followed by incubation at 30°C for 45 min with gentle mixing every 10 min. For control reactions, 15 μl of purified kinase was incubated with 5 μl of the elution buffer used to purify dephosphorylated Mih1. This control ensured that bands detected in the reactions that included Mih1 were not caused by background bands from the added kinase. Each sample was mixed with 5 μl of 4× protein sample buffer and incubated at 100°C for 5 min, and 25 μl was loaded onto a 10% SDS polyacrylamide gel for Western blot analysis. This protocol was modified to scale up the purification of Yck1-TAP kinase in the in vitro kinase assay shown in Fig. 4 D. In brief, 4 liters of Yck1-TAP cells were grown and 8 g of cell powder was resuspended in 20 ml of extract buffer. The cell lysate was centrifuged at 10,000 g for 5 min followed by 40,000 g for 45 min and the supernatant was bound to 200 μl IgG beads for 2 h. The beads were washed three times with extract buffer without PMSF and three times with TEV cleavage buffer with 0.25% Tween-20. The beads were suspended in 400 μl TEV cleavage buffer and 7 μl of 1.6-mM TEV protease was added. The beads were rotated at room temperature for 2.5 h and 350 μl of the supernatant was removed and frozen in 25-μl aliquots on liquid nitrogen.
In vitro assays with λ phosphatase, PP2ACdc55, and Yck1-TAP
To demonstrate that Mih1 modification is caused by phosphorylation (Fig. 2 B), 10 μl of purified phosphorylated Mih1 was treated with 2 μl λ phosphatase in phosphatase assay buffer (50 mM Hepes-KOH, pH 7.6, 0.05% Tween 20, 5% glycerol, 1 mM DTT, 1 mM MnCl2, and 50 μg/ml BSA) in a total volume of 40 μl. The reactions were incubated at 30°C for 30 min and terminated by adding 12.5 μl of 4× sample buffer. The samples were boiled at 100°C for 5 min and 20 μl of each reaction was analyzed by Western blotting. To show that Mih1 is a direct substrate of PP2ACdc55, a similar protocol was followed, where 10 μl of purified Cdc55-3XHA was added to the 5 μl of purified dephosphorylated 3XHA-Mih1 in phosphatase assay buffer in a total volume of 40 μl. To demonstrate that Cdk1-Clb2-3XHA and Yck1-TAP can directly phosphorylate Mih1, 5 μl of the purified dephosphorylated 3XHA-Mih1 was incubated with 10 μl of purified Cdk1-Clb2-3XHA or Yck1-TAP in the presence of kinase assay buffer (50 mM Hepes-KOH, pH 7.6, 2 mM MgCl2,1 mM DTT,10% glycerol, and 2 mM ATP) in a total volume of 40 μl. The reaction was performed at 30°C for 1 h and was terminated by adding 12.5 μl of 4× protein sample buffer.
Online supplemental material
Tables S1 provides a list of gene deletion strains tested for effects on Mih1 phosphorylation. Table S2 provides a list of essential kinases tested for their ability to phosphorylate Mih1 in vitro. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200711014/DC1.
Supplemental Material
Acknowledgments
We thank Lucy Robinson, Stacy Harvey, Hendri Tjandra, Martha Cyert, Ray Deshaies, David Morgan, Adam Rudner, Jeff Ubersax, Claiborne Glover, and Michael Stark for strains and members of the Kellogg Laboratory for critical reading of the manuscript. We also thank John Little for providing TEV protease, Kevan Shokat and Chao Zhang for providing 1NM-PP1, and Derek McCusker for helpful suggestions regarding identification of kinases that phosphorylate Mih1.
This work was supported by a grant from the National Institutes of Health (GM69602).
Abbreviation used in this paper: PP, protein phosphatase.
References
- Altman, R., and D.R. Kellogg. 1997. Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon, A., U. Surana, I. Muroff, and K. Nasmyth. 1992. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae.Nature. 355:368–371. [DOI] [PubMed] [Google Scholar]
- Amon, A., M. Tyers, B. Futcher, and K. Nasmyth. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 74:993–1007. [DOI] [PubMed] [Google Scholar]
- Anderson, C.W., P.R. Baum, and R.F. Gesteland. 1973. Processing of adenovirus 2-induced proteins. J. Virol. 12:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P.D., and M.J. Stark. 2000. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae.J. Cell Sci. 113:507–520. [DOI] [PubMed] [Google Scholar]
- Babu, P., J.D. Bryan, H.R. Panek, S.L. Jordan, B.M. Forbrich, S.C. Kelley, R.T. Colvin, and L.C. Robinson. 2002. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 115:4957–4968. [DOI] [PubMed] [Google Scholar]
- Bishop, A.C., J.A. Ubersax, D.T. Petsch, D.P. Matheos, N.S. Gray, J. Blethrow, E. Shimizu, J.Z. Tsien, P.G. Schultz, M.D. Rose, et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 407:395–401. [DOI] [PubMed] [Google Scholar]
- Booher, R.N., R.J. Deshaies, and M.W. Kirschner. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12:3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros, R., C. Dozier, and B. Ducommun. 2006. The when and wheres of CDC25 phosphatases. Curr. Opin. Cell Biol. 18:185–191. [DOI] [PubMed] [Google Scholar]
- Clarke, P.R., I. Hoffmann, G. Draetta, and E. Karsenti. 1993. Dephosphorylation of cdc25-C by a type-2A protein phosphatase: specific regulation during the cell cycle in Xenopus egg extracts. Mol. Biol. Cell. 4:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy, W.G. 1994. The decision to enter mitosis. Trends Cell Biol. 4:202–207. [DOI] [PubMed] [Google Scholar]
- Dunphy, W.G., and A. Kumagai. 1991. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 67:189–196. [DOI] [PubMed] [Google Scholar]
- Esteban, V., M. Blanco, N. Cueille, V. Simanis, S. Moreno, and A. Bueno. 2004. A role for the Cdc14-family phosphatase Flp1p at the end of the cell cycle in controlling the rapid degradation of the mitotic inducer Cdc25p in fission yeast. J. Cell Sci. 117:2461–2468. [DOI] [PubMed] [Google Scholar]
- Evans, D.R.H., and M.J.R. Stark. 1997. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics. 145:227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes, P., and P. Nurse. 1977. Control of cell size in fission yeast by a growth modulated size control over nuclear division. Exp. Cell Res. 107:377–386. [DOI] [PubMed] [Google Scholar]
- Ferrell, J.E. 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14:140–148. [DOI] [PubMed] [Google Scholar]
- Ferrell, J.E., and W. Xiong. 2001. Bistability in cell signaling: how to make continuous processes discontinuous, and reversible processes irreversible. Chaos. 11:227–236. [DOI] [PubMed] [Google Scholar]
- Fitch, I., C. Dahmann, U. Surana, A. Amon, K. Nasmyth, L. Goetsch, B. Byers, and B. Futcher. 1992. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae.Mol. Biol. Cell. 3:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, J., M.J. Solomon, R.N. Booher, J.F. Bazan, and M.W. Kirschner. 1991. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 67:197–211. [DOI] [PubMed] [Google Scholar]
- Goldbeter, A., and D.E. Koshland Jr. 1981. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. USA. 78:6840–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter, A., and D.E. Koshland Jr. 1984. Ultrasensitivity in biochemical systems controlled by covalent modification. Interplay between zero-order and multistep effects. J. Biol. Chem. 259:14441–14447. [PubMed] [Google Scholar]
- Goris, J., J. Hermann, P. Hendrix, R. Ozon, and W. Merlevede. 1989. Okadaic acid, a specific protein phosphatase inhibitor, induces maturation and MPF formation in Xenopus laevis oocytes. FEBS Lett. 245:91–94. [DOI] [PubMed] [Google Scholar]
- Gould, K.L., and P. Nurse. 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 342:39–45. [DOI] [PubMed] [Google Scholar]
- Hanna, D.E., A. Rethinaswamy, and C.V. Glover. 1995. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae.J. Biol. Chem. 270:25905–25914. [DOI] [PubMed] [Google Scholar]
- Harvey, S.L., and D.R. Kellogg. 2003. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr. Biol. 13:264–275. [DOI] [PubMed] [Google Scholar]
- Harvey, S.L., A. Charlet, W. Haas, S.P. Gygi, and D.R. Kellogg. 2005. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 122:407–420. [DOI] [PubMed] [Google Scholar]
- Healy, A.M., S. Zolnierowicz, A.E. Stapleton, M. Goebl, A.A. DePaoli-Roach, and J.R. Pringle. 1991. Cdc55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, I., P.R. Clarke, M.J. Marcote, E. Karsenti, and G. Draetta. 1993. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self amplification of MPF at mitosis. EMBO J. 12:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, T., and J.L. Maller. 1993. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol. Biol. Cell. 4:1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, T., and J.L. Maller. 1995. Phosphorylation and activation of the Xenopus Cdc25 phosphatase in the absence of Cdc2 and Cdk2 kinase activity. Mol. Biol. Cell. 6:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, T., D.H. Walker, and J.L. Maller. 1992. Periodic changes in the phosphorylation of the Xenopus Cdc25 phosphatase regulate its activity. Mol. Biol. Cell. 3:927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., J.L. Nishikawa, B.J. Breitkreutz, and M. Tyers. 2002. Systematic identification of pathways that couple cell growth and cell division in yeast. Science. 297:395–400. [DOI] [PubMed] [Google Scholar]
- Kafadar, K.A., H. Zhu, M. Snyder, and M.S. Cyert. 2003. Negative regulation of calcineurin signaling by Hrr25p, a yeast homolog of casein kinase I. Genes Dev. 17:2698–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson-Rosenthal, C., and J.B. Millar. 2006. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 16:285–292. [DOI] [PubMed] [Google Scholar]
- Keaton, M.A., and D.J. Lew. 2006. Eavesdropping on the cytoskeleton: progress and controversy in the yeast morphogenesis checkpoint. Curr. Opin. Microbiol. 9:540–546. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R. 2003. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J. Cell Sci. 116:4883–4890. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R., and B.M. Alberts. 1992. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol. Biol. Cell. 3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, K., T. Nemoto, K. Nabeshima, H. Kondoh, H. Niwa, and M. Yanagida. 1996. The regulatory subunits of fission yeast protein phosphatase 2A (PP2A) affect cell morphogenesis, cell wall synthesis and cytokinesis. Genes Cells. 1:29–45. [DOI] [PubMed] [Google Scholar]
- Knippschild, U., A. Gocht, S. Wolff, N. Huber, J. Lohler, and M. Stoter. 2005. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 17:675–689. [DOI] [PubMed] [Google Scholar]
- Kovelman, R., and P. Russell. 1996. Stockpiling of Cdc25 during a DNA replication checkpoint in Schizosaccharomyces pombe. Mol. Cell. Biol. 16:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, A., and W.G. Dunphy. 1991. The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell. 64:903–914. [DOI] [PubMed] [Google Scholar]
- Kumagai, A., and W.G. Dunphy. 1992. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 70:139–151. [DOI] [PubMed] [Google Scholar]
- Kumagai, A., and W. Dunphy. 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 273:1377–1380. [DOI] [PubMed] [Google Scholar]
- LaPorte, D.C., and D.E. Koshland Jr. 1983. Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature. 305:286–290. [DOI] [PubMed] [Google Scholar]
- Lew, D.J. 2000. Cell cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 10:47–53. [DOI] [PubMed] [Google Scholar]
- Lew, D.J. 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15:648–653. [DOI] [PubMed] [Google Scholar]
- Lin, F.C., and K.T. Arndt. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie, D. DeMarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Ma, X.J., Q. Lu, and M. Grunstein. 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10:1327–1340. [DOI] [PubMed] [Google Scholar]
- Margolis, S.S., and S. Kornbluth. 2004. When the checkpoints have gone: insights into Cdc25 functional activation. Cell Cycle. 3:425–428. [PubMed] [Google Scholar]
- Margolis, S.S., J.A. Perry, C.M. Forester, L.K. Nutt, Y. Guo, M.J. Jardim, M.J. Thomenius, C.D. Freel, R. Darbandi, J.H. Ahn, et al. 2006. a. Role for the PP2A/B56delta phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell. 127:759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis, S.S., J.A. Perry, D.H. Weitzel, C.D. Freel, M. Yoshida, T.A. Haystead, and S. Kornbluth. 2006. b. A role for PP1 in the Cdc2/Cyclin B-mediated positive feedback activation of Cdc25. Mol. Biol. Cell. 17:1779–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker, D., C. Denison, S. Anderson, T.A. Egelhofer, J. Yates III, S.P. Gygi, and D.R. Kellogg. 2007. Cdk1 coordinates cell surface growth with the cell cycle. Nat. Cell Biol. 9:506–515. [DOI] [PubMed] [Google Scholar]
- McNulty, J.J., and D.J. Lew. 2005. Swe1p responds to cytoskeletal perturbation, not bud size, in S. cerevisiae. Curr. Biol. 15:2190–2198. [DOI] [PubMed] [Google Scholar]
- Minshull, J., A. Straight, A.D. Rudner, A.F. Dernburg, A. Belmont, and A.W. Murray. 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6:1609–1620. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O. 2006. The Cell Cycle: Principles of Control. New Science Press Ltd., London, UK. 297 pp.
- Mortensen, E.M., W. Haas, M. Gygi, S.P. Gygi, and D.R. Kellogg. 2005. Cdc28-dependent regulation of the Cdc5/Polo kinase. Curr. Biol. 15:2033–2037. [DOI] [PubMed] [Google Scholar]
- Nurse, P. 1975. Genetic control of cell size at cell division in yeast. Nature. 256:547–551. [DOI] [PubMed] [Google Scholar]
- Nurse, P., P. Thuriaux, and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe.Mol. Gen. Genet. 146:167–178. [DOI] [PubMed] [Google Scholar]
- Panek, H.R., J.D. Stepp, H.M. Engle, K.M. Marks, P.K. Tan, S.K. Lemmon, and L.C. Robinson. 1997. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 16:4194–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerening, J.R., E.D. Sontag, and J.E. Ferrell Jr. 2003. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat. Cell Biol. 5:346–351. [DOI] [PubMed] [Google Scholar]
- Pomerening, J.R., S.Y. Kim, and J.E. Ferrell Jr. 2005. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 122:565–578. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., A.E.M. Adams, D.G. Drubin, and B.K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565–602. [DOI] [PubMed] [Google Scholar]
- Qian, Y.W., E. Erikson, C. Li, and J. Maller. 1998. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis.Mol. Cell. Biol. 18:4262–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Y.W., E. Erikson, and J.L. Maller. 1999. Mitotic effects of a constitutively active mutant of the Xenopus polo-like kinase Plx1. Mol. Cell. Biol. 19:8625–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Y.W., E. Erikson, F.E. Taieb, and J. Maller. 2001. The polo-like kinase Plx1 is required for activation of the phosphatase Cdc25c and CyclinB-Cdc2 in Xenopus Oocytes. Mol. Biol. Cell. 12:1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, L.C., M.M. Menold, S. Garrett, and M.R. Culbertson. 1993. Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol. Cell. Biol. 13:2870–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, L.C., C. Bradley, J.D. Bryan, A. Jerome, Y. Kweon, and H.R. Panek. 1999. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol. Biol. Cell. 10:1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner, A.D., K.G. Hardwick, and A.W. Murray. 2000. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 149:1361–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupes, I. 2002. Checking cell size in yeast. Trends Genet. 18:479–485. [DOI] [PubMed] [Google Scholar]
- Russell, P., and P. Nurse. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 45:145–153. [DOI] [PubMed] [Google Scholar]
- Russell, P., and P. Nurse. 1987. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 49:559–567. [DOI] [PubMed] [Google Scholar]
- Russell, P., S. Moreno, and S.I. Reed. 1989. Conservation of mitotic controls in fission and budding yeasts. Cell. 57:295–303. [DOI] [PubMed] [Google Scholar]
- Shu, Y., H. Yang, E. Hallberg, and R. Hallberg. 1997. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, R.A., H.A. Herald, and D.J. Lew. 1996. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell. 7:1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, R.A.L., E.S.G. Bardes, and D.J. Lew. 1998. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 17:6678–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, P.K., and A.W. Murray. 1992. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature. 355:365–368. [DOI] [PubMed] [Google Scholar]
- Stark, M.J.R. 1996. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast. 12:1647–1675. [DOI] [PubMed] [Google Scholar]
- Strausfeld, U., J.C. Labbé, D. Fesquet, J.C. Cavadore, A. Picard, K. Sadhu, P. Russell, and M. Dorée. 1991. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human CDC25 protein. Nature. 351:242–245. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., J. Petersen, F. MacIver, D.P. Mulvihill, D. Glover, and I.M. Hagan. 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 20:1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., and D.J. Burke. 1997. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for kinetochore/spindle checkpoint in Saccharomyces cerevisiae.Mol. Cell. Biol. 17:620–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B.A., and K.L. Gould. 2004. a. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 23:919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B.A., and K.L. Gould. 2004. b. Inactivating Cdc25, mitotic style. Cell Cycle. 3:601–603. [PubMed] [Google Scholar]
- Yang, H., W. Jiang, M. Gentru, and R.L. Hallberg. 2000. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p hyperphosphorylation. Mol. Cell. Biol. 20:8143–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.