Abstract
Centrioles are surrounded by pericentriolar material (PCM), which is proposed to promote new centriole assembly by concentrating γ-tubulin. Here, we quantitatively monitor new centriole assembly in living Caenorhabditis elegans embryos, focusing on the conserved components SAS-4 and SAS-6. We show that SAS-4 and SAS-6 are coordinately recruited to the site of new centriole assembly and reach their maximum levels during S phase. Centriolar SAS-6 is subsequently reduced by a mechanism intrinsic to the early assembly pathway that does not require progression into mitosis. Centriolar SAS-4 remains in dynamic equilibrium with the cytoplasmic pool until late prophase, when it is stably incorporated in a step that requires γ-tubulin and microtubule assembly. These results indicate that γ-tubulin in the PCM stabilizes the nascent daughter centriole by promoting microtubule addition to its outer wall. Such a mechanism may help restrict new centriole assembly to the vicinity of preexisting parent centrioles that recruit PCM.
Introduction
Centrioles are cylindrical structures 100–250 nm in diameter and 100–500 nm in length whose distinguishing structural feature is an outer wall typically containing nine singlet, doublet, or triplet microtubules arranged in a radially symmetric pattern. Centrioles are thought to have originated in the ancestral eukaryote (Richards and Cavalier-Smith, 2005) and have been conserved in the majority of extant lineages (with some exceptions, such as fungi and flowering plants; Azimzadeh and Bornens, 2004). Centrioles perform two distinct functions: (1) they recruit pericentriolar material (PCM) to form centrosomes that nucleate and organize cellular microtubule arrays; and (2) they template the formation of cilia, microtubule-based projections that serve a variety of motile and sensory functions (for reviews see Bettencourt-Dias and Glover, 2007; Marshall, 2007).
A major topic of study in centriole biology is the assembly mechanism for this intricately structured organelle. Electron microscopic studies have outlined a multistep assembly pathway for centrioles that serve as dedicated basal bodies in multiciliated vertebrate epithelial cells (Anderson and Brenner, 1971). Centriole assembly begins with the formation of a cylindrical intermediate termed the annulus (∼125 nm long and 85 nm in diameter), which contains an internal system of radially symmetric filaments. After annulus formation is complete, microtubules are sequentially added to the outer surface to form the procentriole. Finally, the procentriole elongates past the original length of the annulus to form the mature basal body (∼500 nm in length and 250 nm in outer diameter).
Centrioles in the Caenorhabditis elegans embryo are dedicated to organizing centrosomes that function in cell division and are simpler in structure than vertebrate basal bodies. Mature daughter centrioles are only ∼75 nm in diameter and ∼110 nm in length and possess a ninefold symmetric array of singlet rather than triplet microtubules. Nevertheless, the steps of their assembly parallel those reported for basal body formation (Pelletier et al., 2006). Centriole assembly initiates in S phase with the formation, adjacent to the parent centriole, of a cylindrical intermediate termed the “central tube” (∼100 nm long and 70 nm in diameter) that is analogous to the annulus in basal body formation. In late prophase, an array of nine singlet microtubules is added to the outer centriole wall. In contrast to basal body centrioles, where the procentriole subsequently elongates to form a structure approximately four times the original length of the annulus, C. elegans centrioles do not elongate significantly past the central tube. It is therefore likely that mature centrioles in the C. elegans embryo are analogous to the unelongated procentrioles found in vertebrates. Collectively, these studies suggest that centrioles assemble via a conserved morphological pathway that operates both in the context of cilia formation and in dividing cells.
The C. elegans embryo has proven to be a powerful system for dissecting the molecular mechanism of centriole assembly. Mutational analysis and comprehensive RNAi-based screens have identified four C. elegans proteins specifically required for centriole assembly: SAS-4, SAS-5, SAS-6, and the kinase ZYG-1 (O'Connell et al., 2001; Kirkham et al., 2003; Leidel and Gonczy, 2003; Dammermann et al., 2004; Delattre et al., 2004; Leidel et al., 2005). Based on reciprocal depletions, a molecular hierarchy for centriole assembly has been established in which ZYG-1 recruits SAS-5 and SAS-6, which are in turn required to recruit SAS-4 (Dammermann et al., 2004; Delattre et al., 2004, 2006; Leidel et al., 2005; Pelletier et al., 2006). Consistent with this hierarchy, analysis of centriole ultrastructure revealed that central tube assembly requires ZYG-1, SAS-5, and SAS-6, whereas SAS-4 is required for the subsequent addition of microtubules to the outer centriole wall (Pelletier et al., 2006). Drosophila melanogaster and vertebrate orthologues of SAS-4 (CPAP/CENPJ), SAS-6, and ZYG-1 (PLK4/SAK) are also required for centriole formation and can drive ectopic centriole assembly when overexpressed (Dammermann et al., 2004; Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Leidel et al., 2005; Basto et al., 2006; Kleylein-Sohn et al., 2007; Peel et al., 2007; Rodrigues-Martins et al., 2007a,b; Vladar and Stearns, 2007), which suggests that these proteins are part of the core centriole assembly machinery in all eukaryotes.
Centriolar microtubules assemble from α/β-tubulin heterodimers and are extremely stable (Kochanski and Borisy, 1990). Other tubulin isoforms, including γ-, δ-, and ε-tubulin, have also been implicated in centriole assembly (Dutcher, 2003). ε- and δ-tubulin promote the formation of doublet and triplet microtubules. However, these isoforms are not found in D. melanogaster or C. elegans, which suggests that they are not universally required for centriole formation. In contrast, γ-tubulin, which concentrates in the PCM and is best known for its role in the nucleation of centrosomal microtubules (Moritz and Agard, 2001), has been implicated in centriole assembly in ciliated protozoa (Ruiz et al., 1999; Shang et al., 2002), C. elegans (Dammermann et al., 2004), D. melanogaster (Raynaud-Messina et al., 2004), and vertebrate cells (Haren et al., 2006; Kleylein-Sohn et al., 2007). Within the centrosome, centrioles have a symbiotic relationship with the surrounding PCM. Centrioles recruit PCM and organize it into a focal body (Bobinnec et al., 1998), thereby determining centrosome number. In turn, recent work in C. elegans suggests that the PCM promotes centriole duplication by concentrating γ-tubulin around the centrioles (Dammermann et al., 2004). However, the mechanism by which γ-tubulin contributes to centriole assembly and how it interfaces with the core duplication machinery remains unknown.
Here, we develop a method to quantitatively monitor the recruitment of components to the site of new centriole assembly in living embryos. We show that SAS-4 and SAS-6 are coordinately recruited and reach their maximal levels during S phase, concurrent with assembly of the central tube. The amount of SAS-6 is subsequently reduced by half in a process intrinsic to the early steps of the centriole duplication mechanism that does not require cell cycle progression into mitosis, SAS-4, or the assembly of centriolar microtubules. Newly recruited centriolar SAS-4 remains in dynamic equilibrium with the cytoplasmic pool until late prophase, when it is stably incorporated concurrent with the assembly of centriolar microtubules. SAS-4 stabilization requires cell cycle progression into mitosis, γ-tubulin, and microtubule assembly. Our results suggest that γ-tubulin in the PCM organized by the parent stabilizes the nascent daughter centriole by promoting the addition of centriolar microtubules to its outer wall. Such a mechanism would help restrict the formation of new centrioles to the vicinity of existing parent centrioles that have the ability to recruit PCM.
Results
A method to monitor the recruitment of components during new centriole assembly in vivo
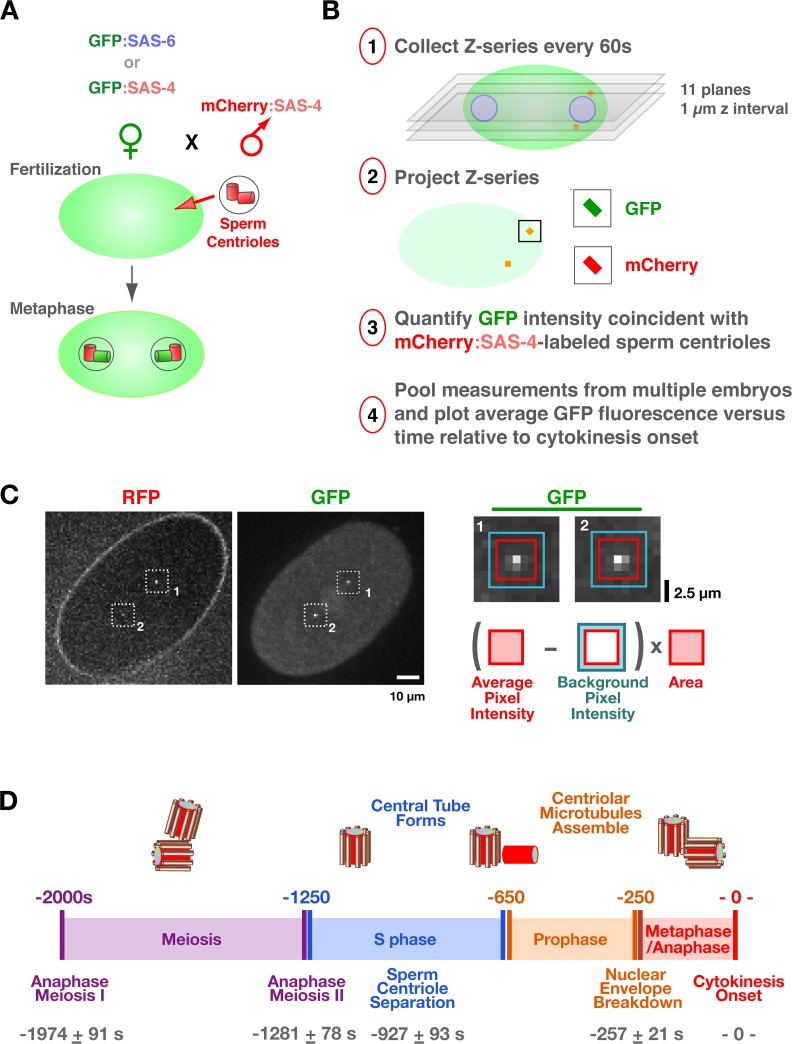
Because of their small size, centriole assembly has typically been monitored by serial section electron microscopy, with its inherent limitations of small sample size and the inability to inform on dynamic behavior. To progress toward a dynamic view of centriole assembly, we developed a fluorescence microscopy–based method to monitor the recruitment of GFP fusions with centriole components during the first division of living C. elegans embryos. The reproducible timing of events during this division (Oegema and Hyman, 2006) makes it possible to generate recruitment curves by pooling measurements from multiple embryos. C. elegans oocytes lack centrioles, which are introduced during fertilization, when the sperm brings in a centriole pair. The sperm-derived centrioles separate, and between S phase and metaphase of the first mitotic division of the resulting embryo, a new centriole assembles adjacent to each sperm-derived parent (Fig. 1 D; (Pelletier et al., 2006). Light microscopic analysis of centriole assembly is limited by the proximity of the newly forming daughter centriole to its parent such that each parent/daughter pair appears as a single diffraction-limited spot. To selectively monitor the recruitment of GFP-tagged centriole components to the newly forming centriole without the complication of a signal caused by labeled protein stably incorporated into the parent, we used mating to independently control the genetic background of the oocyte and sperm. Feminized hermaphrodites producing oocytes loaded with a GFP fusion to a centriole component were mated with males whose sperm centrioles were stably labeled with RFP via expression of an RFP:SAS-4 fusion but which lack a GFP signal (Fig. 1 A). Recruitment of the GFP-labeled protein to the site of new centriole assembly was monitored by measuring the GFP signal in proximity to the RFP-labeled sperm centrioles (Fig. 1, B and C).
Figure 1.
A method to monitor the recruitment of centriole components in vivo. (A) Schematic of the mating scheme used to monitor centriolar recruitment of GFP fusions. (B) Imaging and quantification flowchart. (C) A pair of representative images and schematic of the method used to measure the GFP intensity coincident with the RFP-labeled sperm centrioles. The regions corresponding to the two sperm centrioles in the low magnification images are indicated (white dashed boxes). A 5 × 5 pixel box (red) and a larger 7 × 7 pixel box (blue) were drawn around the peak RFP signal for each sperm centriole and GFP intensity was quantified as outlined. (D) Timeline of events between fertilization and onset of the first embryonic cytokinesis. Times for each event (n > 5 embryos) are in seconds relative to cytokinesis onset (t = 0) ± standard deviation. Schematics illustrate intermediates in centriole assembly based on ultrastructural work (Pelletier et al., 2006). After fertilization, the sperm-derived centrioles separate and by early S phase (∼−950 s), a small central tube ∼60 nm in length and 40 nm in diameter is present adjacent and perpendicular to each sperm-derived centriole. By early prophase, the central tube is ∼110 nm in length and 65 nm in diameter. Centriolar microtubules assemble during the second half of mitotic prophase (−450 to −250 s), and their assembly is complete by metaphase (−150 s).
We concentrated our analysis on SAS-6 and SAS-4, two conserved centriolar proteins required for assembly of the central tube and addition of centriolar microtubules to the outer wall, respectively (Pelletier et al., 2006). Photobleaching and mating experiments indicate that both proteins are stably incorporated into new centrioles during their formation (Kirkham et al., 2003; Leidel and Gonczy, 2003; Dammermann et al., 2004; Leidel et al., 2005). Before initiating detailed analysis, we used an RNAi-based approach to confirm that the GFP fusions with SAS-4 and SAS-6 were functional (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1), indicating that their dynamics likely reflect those of the endogenous proteins. The GFP signal coincident with the RFP-labeled sperm centrioles was measured in projections of z series collected at 60-s intervals (Fig. 1 B). A reference differential interference contrast (DIC) image was also acquired at each time point. By using a sensitive electron-multiplying charge-coupled device camera mounted on a spinning disk confocal microscope and imaging conditions that sacrifice resolution to increase the signal (60× with 2 × 2 binning; see sample images in Fig. 1 C), we were able to collect up to 20 z series per embryo without detectable photobleaching (Fig. S2). Higher resolution z series (90× without binning) were also acquired to qualitatively confirm the quantitative results; single planes from these image stacks are presented together with the quantitative data. To generate kinetic curves, measurements from multiple embryos were pooled and the average GFP fluorescence was plotted relative to time of cytokinesis onset. We chose cytokinesis onset as a temporal landmark because it is easily scored in the paired DIC images (Fig. S3). We did not characterize the recruitment of SAS-5, which is also required for central tube formation (Pelletier et al., 2006), because, unlike SAS-4 and SAS-6, it exchanges into preexisting centrioles (Delattre et al., 2004), complicating analysis of its recruitment. However, depletion of SAS-5 was used as a means for preventing central tube assembly.
Interpreting the recruitment curves requires knowledge of the timing of events between fertilization and the first embryonic cytokinesis (Oegema and Hyman, 2006). After fertilization, the oocyte completes two rounds of meiotic segregation, generating the oocyte pronucleus and two polar bodies. Oocyte and sperm pronuclear chromatin subsequently decondenses and is replicated. After S phase, the pronuclei migrate toward each other concurrent with chromosome condensation during mitotic prophase. The pronuclei meet, the nuclear envelopes break down, and the first mitotic spindle assembles, followed by chromosome segregation and cytokinesis. To establish a timeline for these events, we filmed embryos coexpressing GFP:γ-tubulin and GFP:histone H2B to visualize centrosomes and chromosomes, respectively, and measured the timing of anaphase of meioses I and II, sperm centriole separation, nuclear envelope breakdown, and anaphase of the first mitosis relative to cytokinesis onset (Fig. S3 and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1). A timeline constructed from these data incorporating previous information on the timing of S phase (−1,250 to −650 s; Edgar and McGhee, 1988), chromosome condensation (−650 to −250 s; Maddox et al., 2006), and ultrastructural events in the centriole duplication cycle (Pelletier et al., 2006), is shown in Fig. 1 D. Our analysis of centriolar protein recruitment and dynamics is interpreted in the context of this timeline.
The amount of SAS-6 at the site of new centriole assembly oscillates during the duplication cycle
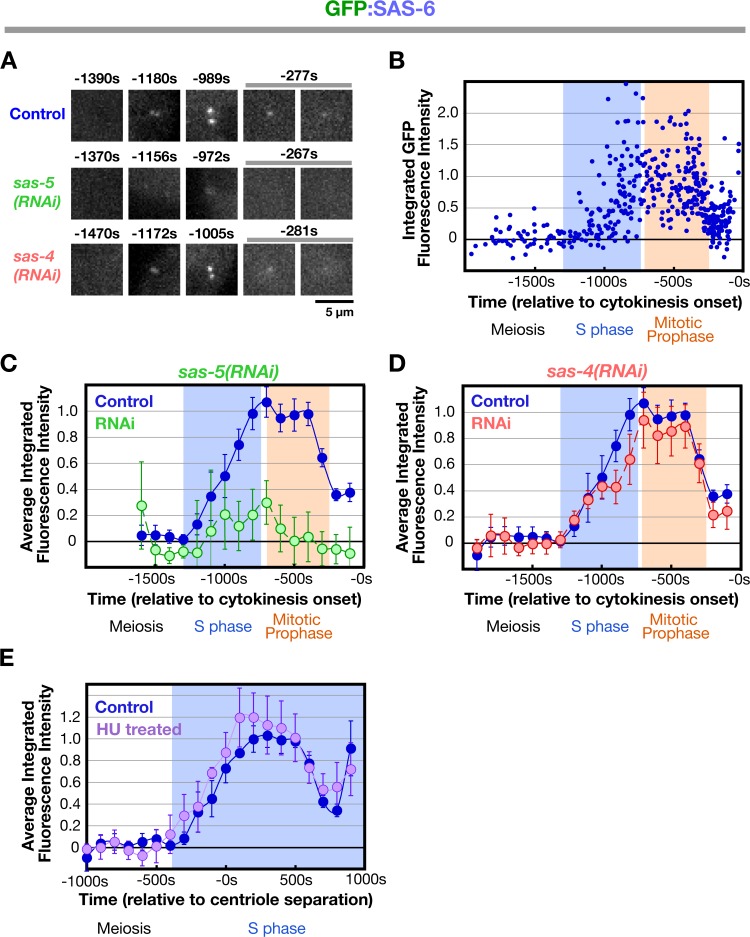
We began by analyzing the recruitment of SAS-6, which is required for central tube assembly (Pelletier et al., 2006). During meiosis, the sperm centrioles do not recruit detectable GFP:SAS-6 from the embryo cytoplasm, confirming that SAS-6 on sperm centrioles does not exchange with the cytoplasmic pool (Leidel et al., 2005). GFP:SAS-6 is first detected at the site of new centriole assembly at S phase onset and its levels steadily increase throughout S phase (Fig. 2, A–C; −1,200 to −800 s) coincident with the time previously reported for the appearance and expansion of the central tube (Fig. 1 D; Pelletier et al., 2006). As embryos transition into prophase, levels of centriolar GFP:SAS-6 plateau for ∼400 s (Fig. 2, A–C; −800 to −400s), which is concurrent with compaction of the nuclear chromatin into discrete linear chromosomes (Maddox et al., 2006). During the second half of mitotic prophase, the amount of centriolar GFP:SAS-6 declines (Fig. 2 A–C; −400 to −200s), reaching a low point ∼50 s after nuclear envelope breakdown, when the amount of centriolar SAS-6 is ∼40% of that observed during the plateau. SAS-6 tagged with GFP at the C terminus also exhibited this loss (unpublished data), indicating that it is not caused by a site-specific cleavage event that liberates the GFP and leaves behind a fragment of SAS-6.
Figure 2.
Kinetic profiles for SAS-6 recruitment in control, sas-5(RNAi), sas-4(RNAi), and HU-treated embryos. (A) High-resolution images of GFP:SAS-6 recruited to RFP-labeled sperm centrioles in embryos generated using the mating scheme in Fig. 1 A. Times (in seconds relative to cytokinesis onset) correspond to meiosis (−1,470 to −1,370 s), early (−1,180 to −1,156 s) and mid (−1,005 to −972 s) S phase, and late prophase (−281 to −267 s). (B) Normalized individual measurements of the integrated GFP:SAS-6 intensity coincident with sperm centriolar RFP signal (see Materials and methods for details on normalization). (C) Kinetic profiles for the recruitment of GFP:SAS-6 in control and sas-5(RNAi) embryos. Data points are the mean of the normalized GFP intensity measurements collected during the 200-s interval centered on that point. (D) Depletion of SAS-4 does not affect the kinetics of centriolar GFP:SAS-6 recruitment or loss. (E) Recruitment profile of centriolar GFP:SAS-6 in embryos treated with 75 mM HU. The profile was generated as in C and D except that sequences were time-aligned with respect to centriole separation, which occurs at the same time after meiosis II anaphase in control and HU-treated embryos (Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1). The control dataset plotted here is the same as in B–D except that the means were recalculated for the indicated 200-s intervals after converting seconds before cytokinesis onset to seconds after centriole separation. All error bars indicate the 90% confidence interval.
Consistent with prior work (Leidel et al., 2005), GFP:SAS-6 was not recruited to sperm centrioles in embryos depleted of SAS-5 (Fig. 2, A and C) in which the central tube fails to form (Pelletier et al., 2006). SAS-4 is required for the addition of microtubules to the outer centriole wall during the second half of mitotic prophase (Kirkham et al., 2003; Pelletier et al., 2006). In SAS-4–depleted embryos, the kinetics of centriolar GFP:SAS-6 recruitment were essentially identical to those in controls (Fig. 2, A and D). We conclude that the recruitment and subsequent loss of centriolar SAS-6 do not require either SAS-4 or the assembly of centriolar microtubules.
Reduction in centriolar SAS-6 levels does not require cell cycle progression into mitosis
To determine whether the reduction in SAS-6 levels requires cell cycle progression into mitosis, we analyzed SAS-6 recruitment in the presence of the DNA replication inhibitor hydroxyurea (HU), which arrests C. elegans embryos in S phase (Holway et al., 2006). Filming of HU-treated embryos expressing GFP:histone H2B and GFP:γ-tubulin indicated that treated embryos arrest with small centrosomes, no chromosome condensation, and a persistent pseudocleavage furrow (Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1). Because later events, including cytokinesis onset, do not occur in these embryos, we used centriole separation as a temporal landmark, an early event not perturbed by HU treatment that is easily scored in embryos expressing centriolar GFP fusions. Arresting embryos in S phase by HU treatment did not affect the kinetics of recruitment or loss of centriolar SAS-6 (Fig. 2 E). Thus, the reduction in the amount of SAS-6 at the site of new centriole assembly does not require cell cycle progression out of S phase but is intrinsic to the early steps in the assembly mechanism.
SAS-4 is coordinately recruited to centrioles with SAS-6 and remains at constant levels until onset of the subsequent duplication cycle
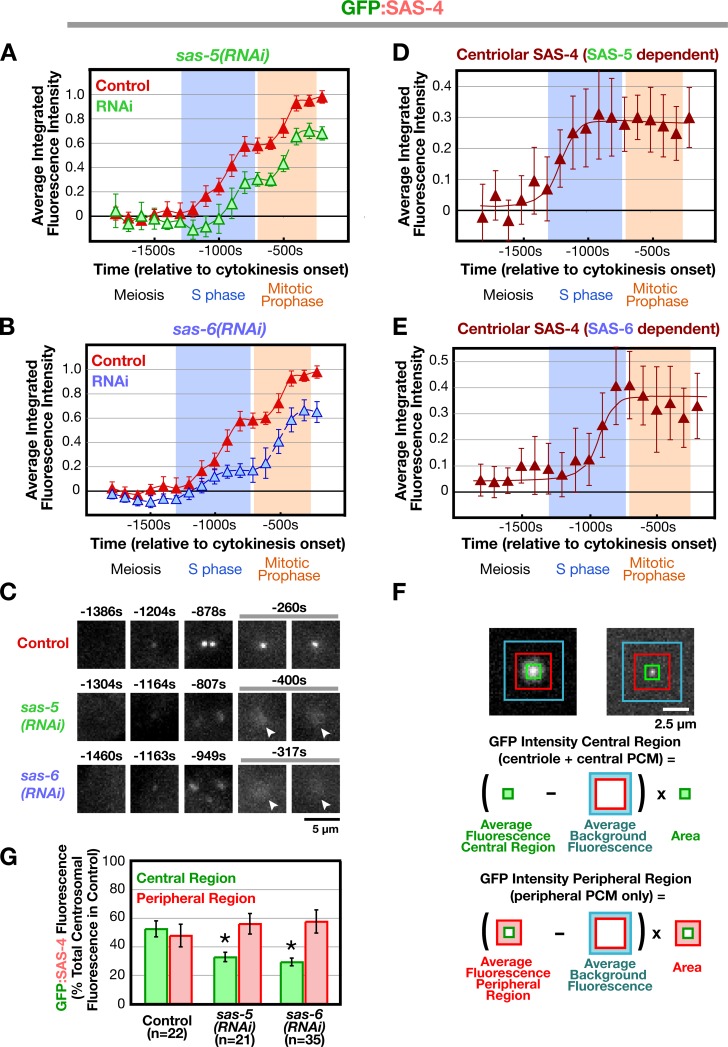
Next, we examined the recruitment of SAS-4, which is required for the addition of microtubules to the outer centriole wall (Pelletier et al., 2006). As for SAS-6, sperm centrioles did not recruit detectable GFP:SAS-4 between fertilization and onset of S phase, which is consistent with prior work (Kirkham et al., 2003; Leidel and Gonczy, 2003; Dammermann et al., 2004). The accumulation of GFP:SAS-4 at the site of new centriole assembly exhibited a biphasic pattern (Fig. 3 A). Levels increased during S phase (between −1,100s and −800 s) followed by a short plateau (−800 to −600 s) and a second period of increase during prophase (−600 to −400 s). Surprisingly, sperm centriole-associated GFP:SAS-4 in sas-5 and sas-6(RNAi) embryos, in which recruitment of centriolar SAS-4 has been reported to be prevented (Dammermann et al., 2004; Delattre et al., 2004; Leidel et al., 2005), exhibited a similar biphasic pattern, although in both cases, the curves were offset from the control curves reflecting lower overall levels (Fig. 3, A and B). This apparently contradictory result was explained by closer examination of high-resolution image sequences (Fig. 3 C). In addition to localizing prominently to centrioles, GFP:SAS-4 localizes weakly to the PCM. Although the PCM signal is dim compared with the brighter centriolar signal, it makes a substantial quantitative contribution because the PCM is much larger than the centrioles (∼100 times greater in cross-sectional area in metaphase stage embryos). The biphasic nature of the curve therefore largely reflects the dynamics of the PCM, which increases in amount during S phase and further during centrosome maturation in prophase (Video 1; Hannak et al., 2001).
Figure 3.
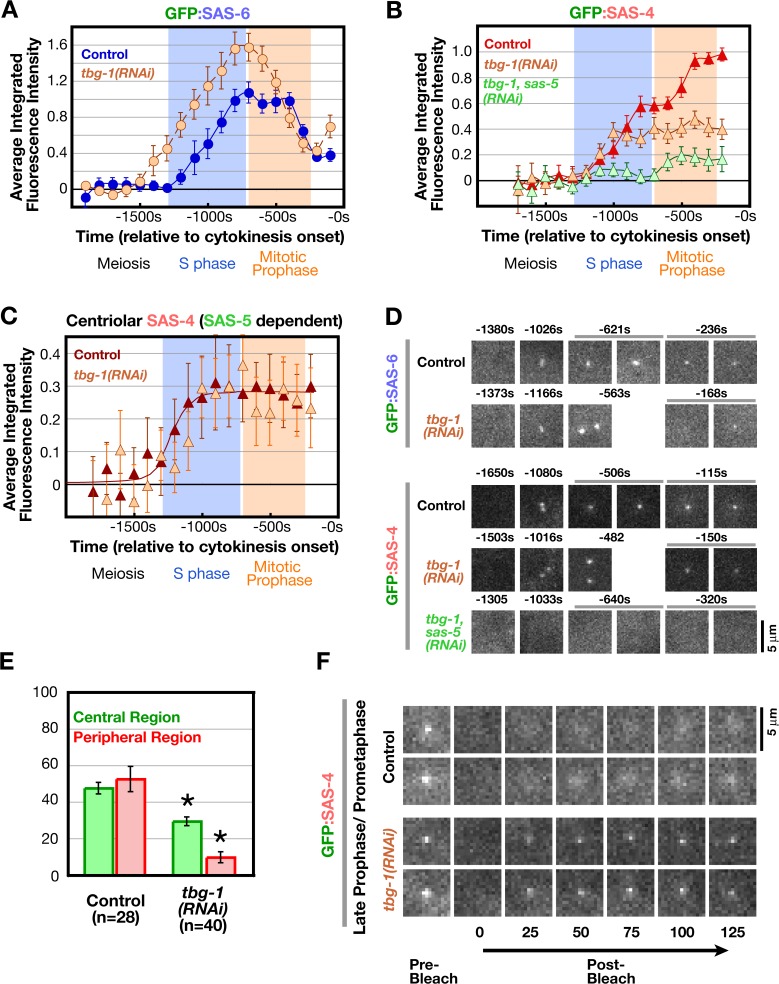
Deconvolving centriolar- and pericentriolar material–associated SAS-4 recruitment by comparison of control and sas-5/sas-6(RNAi) embryos. (A and B) Kinetic profiles for the recruitment of GFP:SAS-4 in control, sas-5(RNAi) (A), and sas-6(RNAi) (B) embryos. Data points are the mean of the normalized GFP intensity of measurements collected during the 200-s interval centered on that point. (C) High-resolution images of GFP:SAS-4 recruited to RFP-labeled sperm centrioles under the indicated conditions. Times (in seconds relative to cytokinesis onset) correspond to meiosis (−1,460 to −1,304 s), early (−1,204 to −1,163 s) and mid (−949 to −878 s) S phase, and late prophase (−400 to −260 s). Note that, although GFP:SAS-4 is not enriched at centrioles in SAS-5– or SAS-6–depleted embryos, it still accumulates in the PCM (arrowheads). Levels of centriolar GFP:SAS-4 were calculated by subtracting the PCM signal, measured in either sas-5(RNAi) (D) or sas-6(RNAi) (E) embryos, from the combined centriole and PCM signal measured in control embryos. (F) Outline of the method used to measure the distribution of GFP:SAS-4 within the centrosome. A pair of high-resolution images of prometaphase/metaphase centrosomes from embryos expressing GFP:γ-tubulin (left) or a GFP centriole marker (right) illustrate how the centrosome was partitioned into central and peripheral regions by concentric boxes. A 7 × 7 pixel central box (green) includes the signal from the centrioles as well as the central PCM, a larger 18 × 18 pixel box (red) includes the peripheral PCM, and the largest 30 × 30 pixel box (blue) was used to measure the background. (G) Depletion of SAS-5 or SAS-6 specifically affects centriolar recruitment of SAS-4. The distribution of GFP:SAS-4 within the centrosome was quantified in prometaphase/metaphase embryos, when PCM recruitment is maximal. Integrated GFP:SAS-4 fluorescence in the central and peripheral regions, expressed as a percentage of the mean total centrosomal fluorescence (central + peripheral) in control embryos, is plotted for the indicated conditions. Asterisks denote statistically significant differences relative to control (P < 0.05 by t test). All error bars indicate the 90% confidence interval.
It is not possible to spatially separate the centriolar GFP:SAS-4 signal from that derived from the surrounding PCM by light microscopy. However, measurement of the peripheral PCM signal alone (performed by excluding the central bright region containing the centrioles) in high-resolution images indicated that GFP:SAS-4 accumulates to essentially identical levels in the PCM of control, sas-5(RNAi), and sas-6(RNAi) embryos (Fig. 3, F and G). This result, together with the fact that no daughter centriole structures are detected in sas-5 or sas-6 RNAi embryos, suggested a convenient means to exclusively measure the centriolar signal of GFP:SAS-4 subtraction of the GFP:SAS-4 kinetic curves obtained in sas-5 (Fig. 3 A) or sas-6 (Fig. 3 B) RNAi embryos (PCM signal only) from the curve obtained in control embryos (PCM plus centriolar signal). Such a subtraction analysis revealed that the amount of GFP:SAS-4 at newly forming centrioles increases during S phase coincident with the recruitment of SAS-6, after which it remains constant until the onset of the subsequent duplication cycle (Fig. 3, D and E).
The results above indicate that SAS-6 and SAS-4 are coordinately recruited to the site of new centriole assembly during S phase. The amount of SAS-6 but not SAS-4 at the site of centriole assembly is subsequently reduced by half in a step that does not require SAS-4, centriolar microtubule assembly, or cell cycle progression. In addition to being present at centrioles, SAS-4 is also found in the PCM, which could contribute to its role in centriole duplication.
SAS-4 at newly forming centrioles exchanges with the cytoplasmic pool until it is stably incorporated in late prophase
The in vivo analysis indicated that SAS-4 and SAS-6 are coordinately recruited to the site of centriole assembly during S phase. This conclusion appears to contradict results from previous fixation-based analyses of GFP:SAS-4 recruitment that used the same mating-based scheme described here. In fixed embryos, GFP:SAS-4 became gradually detectable concurrent with chromosome condensation during prophase, and clear foci of GFP:SAS-4 were reliably detected only after late prophase (Kirkham et al., 2003; Dammermann et al., 2004; Delattre et al., 2006). These results have been central to the current sequential recruitment model of centriole assembly in which SAS-4 is recruited and incorporated after execution of the SAS-6– and (SAS-5–) dependent step of central tube formation. Because the live imaging data instead suggest that SAS-4 is coordinately recruited to centrioles along with SAS-6 during S phase, it was imperative to provide an explanation for the discrepancy between the fixation-based analysis and the live imaging assay.
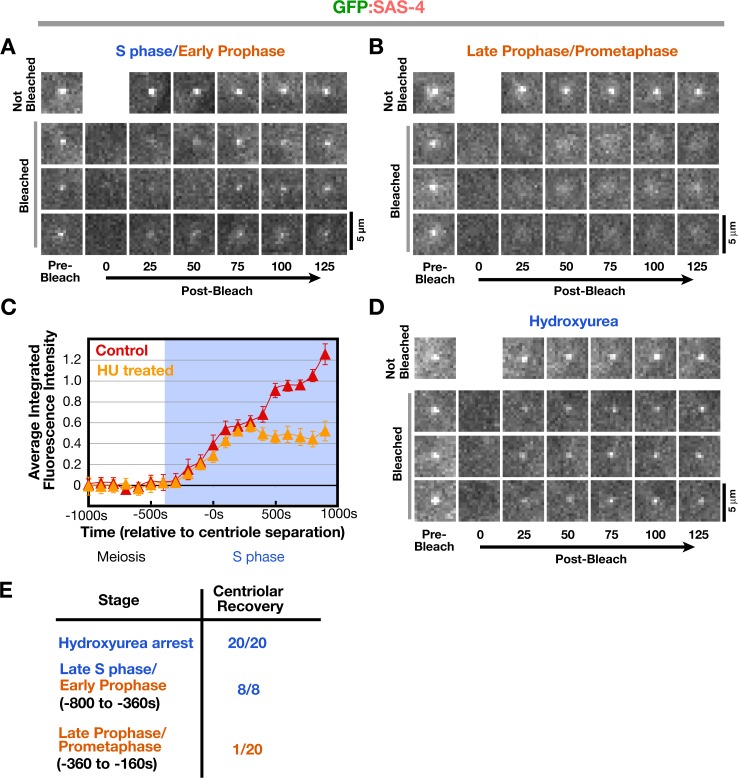
We hypothesized that the discrepancy is caused by a change in the state of GFP:SAS-4 between early and late prophase that renders it stable to fixation. To test this, we used FRAP to reexamine the dynamics of centriolar GFP:SAS-4 during different cell cycle stages. Initially, we compared the recovery of the centriolar signal during late S phase/early prophase to that during late prophase/prometaphase in embryos generated using the mating scheme outlined in Fig. 1 A. Recovery of the centriolar GFP:SAS-4 signal is straightforward to assess by visual inspection and was qualitatively scored; the complications arising from the PCM pool of SAS-4 (Fig. 3) precluded quantitative analysis. As previously reported (Leidel and Gonczy, 2003), recovery of newly recruited centriolar GFP:SAS-4 was not observed in late prophase/prometaphase (Fig. 4, B and E). However, when centrioles were bleached in late S phase and early prophase, recovery of the centriolar signal was observed in all cases (Fig. 4, A and E).
Figure 4.
Centriolar SAS-4 is in dynamic equilibrium with the cytoplasmic pool during late S phase and early prophase but becomes stable to exchange in late prophase. (A and B) Three examples of centrosomes in GFP:SAS-4 embryos, generated as in Fig. 1 A, photobleached in S phase/early prophase (A; bleached at 693, 756, and 424 s before cytokinesis onset, from top to bottom, respectively) and late prophase/prometaphase (B; bleached at 120, 336, and 170 s before cytokinesis onset, from top to bottom, respectively). Times are in seconds after photobleaching. (C) Recruitment profile of centrosomal GFP:SAS-4 after HU treatment. Recruitment curves were generated after time-aligning the data points with respect to centriole separation as described in Fig. 2 E. Error bars indicate the 90% confidence interval. (D) Three examples of photobleached centrosomes in embryos arrested in S phase by HU treatment. Times are given in seconds after photobleaching. (E) Summary of fluorescence recovery after photobleaching analysis. Bars, 5 μm.
Centriolar GFP:SAS-4 does not change in levels after late S phase (Fig. 3, D and E), which suggests that the recovery observed was not caused by recruitment of additional SAS-4. To conclusively show that this was the case, we bleached centrioles in HU-treated embryos expressing GFP:SAS-4. In these embryos, GFP:SAS-4 is recruited with normal kinetics until it reaches the maximum level normally seen during S phase in controls (Fig. 4 C, 300 s after centriole separation). After this point, SAS-4 levels remain constant throughout the duration of the S phase arrest (Fig. 4 C). Recovery of the centriolar GFP:SAS-4 signal was observed for all centrioles bleached in S phase arrested embryos (Fig. 4, D and E).
We conclude that although centriolar GFP:SAS-4 reaches its maximal levels by late S phase, it remains in dynamic equilibrium with the cytoplasmic pool during late S phase and early prophase. During late prophase, concurrent with the addition of microtubules to the outer centriole wall, centriolar GFP:SAS-4 is stabilized and can no longer exchange. This stabilization appears to be required to render SAS-4 stable to fixation using the standard methanol-based protocol used in previous studies. Thus, investigation of the discrepancy between the live and fixed analysis revealed that centriolar SAS-4 exhibits distinct behaviors at different stages of the duplication cycle. The change in behavior of SAS-4 is linked to cell cycle progression into mitosis.
γ-Tubulin is required for the stable incorporation of SAS-4 during late prophase
In previous work using fixation-based analysis, we demonstrated a requirement for γ-tubulin in formation of GFP:SAS-4 foci at sites of new centriole assembly (Dammermann et al., 2004). Although we had originally interpreted this result to mean that γ-tubulin is required for the recruitment of SAS-4, the FRAP results raised the possibility that SAS-4 is recruited normally to centrioles in γ-tubulin–depleted embryos but is not subsequently stabilized. We therefore performed a series of experiments to examine the basis for the centriole assembly defect resulting from γ-tubulin depletion. First, we analyzed the recruitment of GFP:SAS-6 in γ-tubulin–depleted embryos. Because γ-tubulin depletion inhibits cytokinesis, sequences were time-aligned with respect to the onset of cortical contractility, which occurs at approximately the time of cytokinesis onset in control embryos. Although this results in some uncertainty, the recruitment kinetics of GFP:SAS-6 were similar between control and γ-tubulin–depleted embryos (Fig. 5, A and D). We do not know if the slightly earlier onset of recruitment and increased maximal amount are significant; however, we can conclude that γ-tubulin is not required for SAS-6 recruitment or subsequent reduction.
Figure 5.
γ-Tubulin is not required to recruit SAS-6 or SAS-4 to centrioles but is required for stable incorporation of SAS-4 during late prophase. (A) Kinetic profiles for recruitment of GFP:SAS-6 in γ-tubulin–depleted embryos (tbg-1[RNAi]) generated as described in Fig. 2 (B and C). (B) Kinetic profiles for the recruitment of GFP:SAS-4 in embryos depleted of γ-tubulin (tbg-1[RNAi]) or simultaneously depleted of γ-tubulin and SAS-5 (tbg-1,sas-5[RNAi]), generated as described in Fig. 3 A. (C) Levels of centriolar GFP:SAS-4 in γ-tubulin–depleted embryos, calculated by subtracting the GFP:SAS-4 signal in tbg-1,sas-5(RNAi) embryos from the combined centriole and PCM signal in tbg-1(RNAi) embryos, compared with centriolar GFP:SAS-4 in control embryos (control data are from Fig. 3 D). (D) High-resolution images of control and tbg-1(RNAi) embryos, generated by mating as in Fig. 1 A, expressing GFP:SAS-4 or GFP:SAS-6. Times (in seconds relative to cytokinesis onset) correspond to meiosis (−1,305 to −1,650 s), early (−1,166 to −1,002s) and mid (−698 to −482 s) S phase, and late prophase (−320 to −115 s). (E) Analysis of the central and peripheral GFP:SAS-4 signal in prometaphase/metaphase tbg-1(RNAi) embryos performed as in Fig. 3 F. Asterisks denote statistically significant differences relative to control (P < 0.05 by t test). (F) Two representative examples of photobleached centrosomes in late prophase/prometaphase control (top and bottom centrioles were bleached at 215 and 332 s before cytokinesis onset, respectively) or tbg-1(RNAi) (top and bottom centrioles were bleached at 321 and 32 5s before the onset of cortical contractility, respectively) embryos. Times are in seconds after photobleaching. All error bars indicate the 90% confidence interval. Bars, 5 μm.
Next, we analyzed recruitment of GFP:SAS-4 in γ-tubulin–depleted embryos. The recruitment of GFP:SAS-4 in control embryos is biphasic and largely reflects the dynamics of the PCM-associated population (Fig. 3). Surprisingly, GFP:SAS-4 recruitment in γ-tubulin–depleted embryos (Fig. 5 B) was not biphasic and resembled the recruitment curve for centriolar GFP:SAS-4, which is calculated by subtracting the PCM signal measured in either sas-5 or sas-6(RNAi) embryos from the combined centriole and PCM signal measured in control embryos (Fig. 3, D and E). Consistent with the idea that the residual GFP:SAS-4 signal in γ-tubulin–depleted embryos consists primarily of centriolar SAS-4, it was abolished by codepletion of SAS-5 (Fig. 5, B and C). We additionally analyzed the spatial distribution of the GFP:SAS-4 signal in high-resolution images (Fig. 5, D and E). Consistent with specific loss of the PCM signal, the GFP:SAS-4 signal in the peripheral centrosomal region was almost completely absent in γ-tubulin–depleted embryos and the GFP:SAS-4 signal in the central region was reduced by an amount corresponding to the contribution that we estimate to be due to the PCM (based on analysis of sas-5 and sas-6[RNAi] embryos in Fig. 3, F and G). Thus, depletion of γ-tubulin results in a specific loss of PCM-associated GFP:SAS-4, whereas its recruitment to centrioles appears unaffected.
Because normal levels of SAS-6 and SAS-4 were recruited to centrioles in γ-tubulin–depleted embryos, we reasoned that the effect observed on SAS-4 levels in γ-tubulin–depleted embryos in the fixation-based analysis reflects a contribution of γ-tubulin to the change in SAS-4 dynamics between early and late prophase (Fig. 4). To test this possibility, we analyzed the effect of γ-tubulin depletion on the turnover of centriolar GFP:SAS-4 by photobleaching. We observed recovery of centriolar GFP:SAS-4 signal for 11/15 bleached centrioles in late prophase/prometaphase γ-tubulin–depleted embryos (Fig. 5 F), by which time centriolar GFP:SAS-4 in all control embryos is stable to exchange (Fig. 4). The fact that a reduction of γ-tubulin levels prevents the stabilization of GFP:SAS-4 and the detection of GFP:SAS-4 foci in fixed embryos also provides further evidence that dynamic centriolar SAS-4 is lost during fixation. We conclude that γ-tubulin is not required for the initial SAS-6/5–dependent recruitment of SAS-4 to newly forming centrioles but is required for its stable incorporation into the new centriole during late prophase.
Microtubule assembly is required for stable incorporation of SAS-4 during late prophase
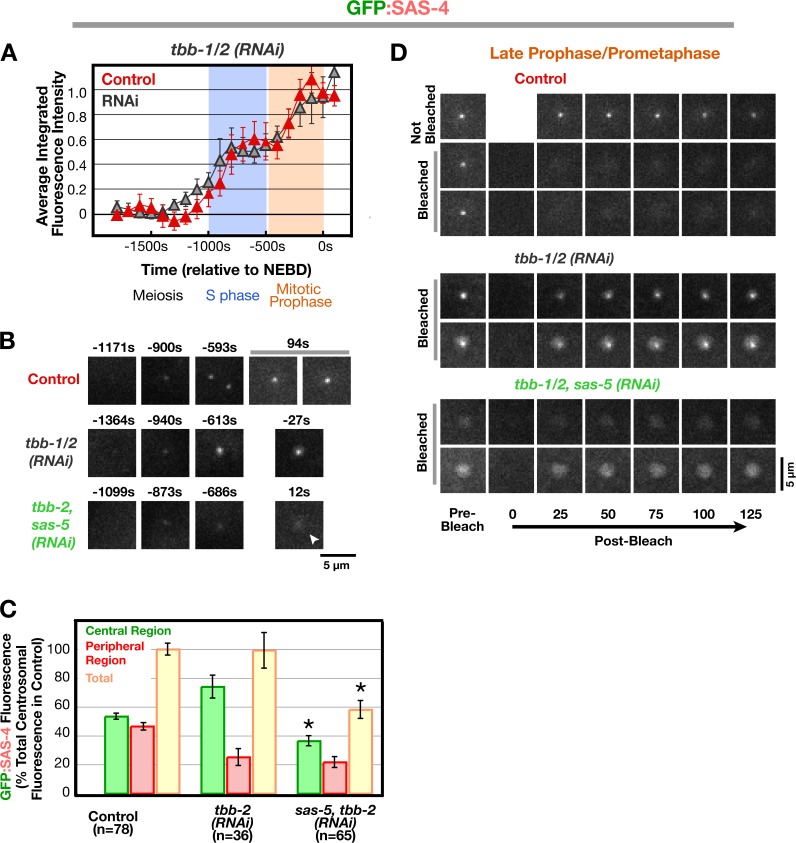
Our data indicate that SAS-4 is recruited to centrioles and reaches normal levels in γ-tubulin–depleted embryos but fails to become stable to cytoplasmic exchange during late prophase. Because late prophase is when centriolar microtubules form (Pelletier et al., 2006), centriolar SAS-4 may not be stabilized because the assembly of centriolar microtubules is compromised by γ-tubulin depletion. To test this idea, we wanted to inhibit microtubule assembly by other means and determine the consequences on SAS-4 recruitment and stabilization. To do this, we examined embryos depleted of β-tubulin by RNAi. β-Tubulin depletion was found to be more effective than nocodazole in inhibiting microtubule assembly because of the low permeability of embryos to drug access. In β-tubulin–depleted embryos, GFP:SAS-4 was recruited to centrosomes with biphasic kinetics essentially identical to those in control embryos (Fig. 6 A), which indicates that both the PCM and centriole populations are recruited normally. This finding also suggests that the effect of γ-tubulin depletion on the PCM population of SAS-4 is not an indirect consequence of inhibiting centrosomal microtubule assembly. Centrosome separation frequently fails in β-tubulin–depleted embryos. In addition, the PCM was more compact. Consequently, although the total centrosomal GFP:SAS-4 fluorescence was the same in the control and β-tubulin–depleted embryos, more signal was present in the central region and less in the peripheral region when GFP:SAS-4 distribution was analyzed (Fig. 6 C). Codepletion of SAS-5 resulted in the expected reduction of signal in the central region, confirming that β-tubulin–depleted embryos recruit GFP:SAS-4 to newly forming centrioles in a fashion similar to controls. To determine whether inhibition of microtubule assembly results in a defect in the stable incorporation of SAS-4 similar to that after γ-tubulin depletion, we performed FRAP on the centrosomes in the depleted embryos (Fig. 6 D). Because of the compaction of the PCM, higher resolution imaging conditions were used than for the experiments in Figs. 4 and Figs.5. Recovery of centriolar GFP:SAS-4 signal was observed for 12/12 bleached centrioles in late prophase/prometaphase β-tubulin–depleted embryos. Centrosomes in embryos codepleted of SAS-5 and β-tubulin were also bleached as a control. As expected, codepleted embryos exhibited recovery of the PCM signal but no centriolar signal either before or after bleaching. We conclude that microtubule assembly, like γ-tubulin, is required for stable incorporation of centriolar SAS-4 during late prophase.
Figure 6.
Microtubule assembly is not required to recruit SAS-4 to centrioles but is required for its stable incorporation during late prophase. (A) Kinetic profiles for recruitment of GFP:SAS-4 in embryos depleted of β-tubulin (tbb-1/2[RNAi]). Because tubulin-depleted embryos do not undergo cytokinesis, sequences were aligned relative to nuclear envelope breakdown, which occurs ∼257 s before cytokinesis onset in control embryos and with normal timing with respect to earlier events when microtubule assembly is inhibited (Portier et al., 2007). (B) High-resolution images of control, tbb-1/2(RNAi), and sas-5,tbb-1/2 (RNAi) embryos expressing GFP:SAS-4, generated by mating as described in Fig. 1 A. Times (in seconds relative to nuclear envelope breakdown), correspond to meiosis (−1,364 to −1,099s), early (−940 to −873 s) and mid (−686 to −593 s) S phase, and late prophase/prometaphase (−27 to +94 s). Note that, although GFP:SAS-4 is not enriched at centrioles in SAS-5,TBB-1/2–depleted embryos, it still accumulates in the PCM (arrowhead). (C) Analysis of the central and peripheral GFP:SAS-4 signal in control, tbb-1/2(RNAi), and sas-5,tbb-1/2 (RNAi) embryos in prometaphase performed as in Fig. 3 F. Asterisks denote statistically significant differences relative to control (P < 0.05 by t test). (D) Two representative examples each of photobleached centrosomes in late prophase/prometaphase from control, tbb-1/2(RNAi), and sas-5,tbb-1/2(RNAi) embryos. Times are in seconds after photobleaching. All error bars indicate the 90% confidence interval. Bars, 5 μm.
Discussion
Here, we describe a method to quantitatively monitor the recruitment of centriolar proteins to the site of new centriole assembly in living C. elegans embryos. By relating the recruitment and turnover of the conserved centriolar components SAS-6 and SAS-4 to the appearance of previously described ultrastructural intermediates in the duplication cycle, we define two new steps in the centriole assembly pathway: (1) a reduction by ∼50% of SAS-6 levels via a process intrinsic to the early steps of the assembly pathway that does not require cell cycle progression out of S phase; and (2) the stable incorporation of SAS-4 during late prophase in a step that requires progression into mitosis, γ-tubulin, and microtubule assembly. Importantly, our results provide mechanistic insight into the widely conserved role of γ-tubulin and the PCM in centriole assembly, which suggests that γ-tubulin in the PCM organized by the parent stabilizes the nascent daughter centriole by promoting the addition of microtubules to its outer wall.
SAS-4 is recruited during S phase and subsequently stabilized during late prophase
Our first unexpected finding using the live imaging assay was the coordinate recruitment of SAS-4 and SAS-6 to the site of centriole assembly during S phase. Previous work in fixed embryos suggested that SAS-4 was recruited only later during mitotic prophase (Kirkham et al., 2003; Dammermann et al., 2004; Delattre et al., 2006), when it is required for the addition of microtubules to the outer centriole wall (Pelletier et al., 2006). Reinvestigation of the dynamics of centriolar SAS-4 using photobleaching suggested an explanation for this discrepancy as discussed below.
Although SAS-4–dependent centriolar microtubule assembly occurs during late prophase, levels of centriolar SAS-4 plateau much earlier. Thus, centriolar microtubules do not influence the amount of centriolar SAS-4 recruited; instead, an earlier process that is complete by the end of S phase dictates the level of centriolar SAS-4. Because assembly of the central tube is complete by this time (Pelletier et al., 2006), we speculate that SAS-4 associates with this nascent structure and that the dimensions of the central tube determine the amount of SAS-4 recruited to centrioles. Partial depletion experiments provide some support for this idea. Partial depletion of SAS-4 results in assembly of centrioles that recruit less than normal amount of PCM (Kirkham et al., 2003). This phenotype can be mimicked by partial depletion of components required for central tube formation (Delattre et al., 2004; Dammermann et al., 2004; Leidel et al., 2005).
Recruitment of SAS-4 to centrioles during S phase raises the question of whether it functions at this stage. Although SAS-4 is not required for initial formation of the central tube, it may be required for its expansion, i.e., for the increase in its diameter between early S phase (∼40 nm) and early prophase (∼65 nm; Pelletier et al., 2006). However, as this suggestion is based on analysis of a single embryo, additional work will be needed to confirm a role for SAS-4 in central tube expansion.
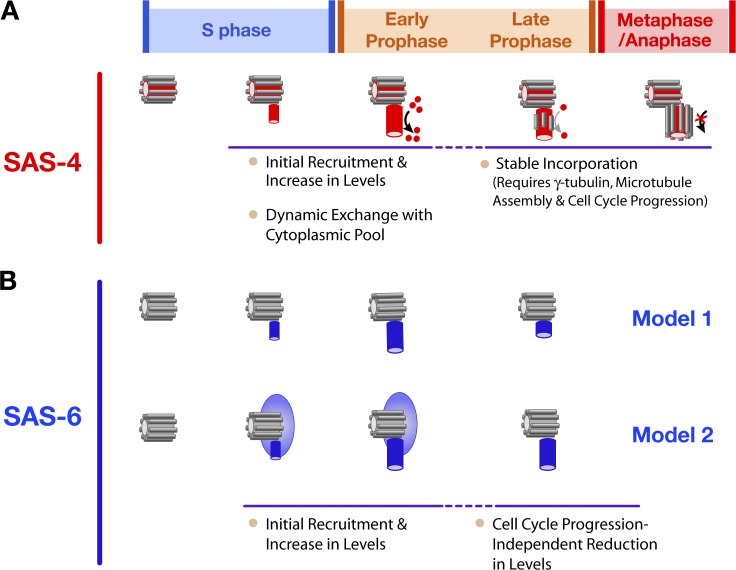
The centriolar SAS-4 recruitment profile provided the necessary framework for using photobleaching to investigate the turnover of centriolar SAS-4 during centriole assembly. When recovery is observed during assembly of a structure, information on the kinetics of protein recruitment is necessary to determine whether recovery is due to continued recruitment of a stably associated component or to component turnover. Because turnover of centriolar SAS-4 was observed during early prophase, after its levels plateaued, we were able to conclude that centriolar SAS-4 is in dynamic equilibrium with the cytoplasmic pool at this stage. SAS-4 is subsequently stably incorporated into the new centriole in late prophase. Arresting embryos in S phase prevents centriolar SAS-4 from transitioning from a dynamic to a stable state. Thus the second step in SAS-4 incorporation, which involves locking it into the stable centriolar structure, is coupled to cell cycle progression. This step also requires γ-tubulin, whose contribution to new centriole formation is discussed below. In summary, the results reveal that SAS-4 incorporation into centrioles is a two-step process, with the transition between the steps being coupled to cell cycle progression.
An intrinsic reduction in SAS-6 levels is programmed into the early steps of centriole assembly
SAS-4 is recruited to nascent daughter centrioles, likely by association with the central tube whose assembly is promoted by components upstream in the duplication pathway. SAS-6 recruitment is more challenging to interpret because it is required to form the central tube, the first detectable structural intermediate in new centriole assembly. SAS-6 may therefore be associated with the mother centriole before its incorporation into the daughter. We consider two models in interpreting SAS-6 recruitment: (1) SAS-6 is recruited from the cytoplasm to the central tube of the daughter centriole coincident with its formation (Fig. 7 B, top); in this model, newly recruited SAS-6 is exclusively daughter centriole–associated. (2) SAS-6 initially associates with the parent centriole in a dynamic manner, and a portion of this localized population is incorporated into the central tube of the daughter centriole (Fig. 7 B, bottom). In model 2, SAS-6 is initially associated with both mother and daughter centrioles. Our analysis indicated that SAS-6 is recruited to the site of new centriole assembly and reaches its maximal levels during S phase. After a brief plateau, ∼60% of SAS-6 is lost during the second half of mitotic prophase. If model 1 is correct, the reduction in SAS-6 levels may represent partial disassembly of an SAS-6–containing structure, likely the central tube, within the newly forming daughter centriole. If model 2 is correct, the reduction in SAS-6 may represent loss of the parent-associated SAS-6 population, leaving behind the SAS-6 that has been stably incorporated into the newly forming daughter centriole.
Figure 7.
Model relating the recruitment and dynamics of SAS-4 and SAS-6 to ultrastructural steps in the duplication cycle. (A) SAS-4 (red) is recruited to the central tube as it forms during S phase. Between late S phase and early prophase, centriolar SAS-4 is in dynamic equilibrium with the cytoplasmic pool but its levels remain constant. During late prophase, centriolar SAS-4 is stably incorporated into the outer centriole wall in a step that requires γ-tubulin and cell cycle progression into mitosis and likely corresponds to assembly of the centriolar microtubules. (B) Two models to explain the recruitment and subsequent reduction in the amount of SAS-6 at the site of new centriole assembly. In the first model (top), newly recruited SAS-6 is strictly associated with the daughter centriole. In this model, an SAS-6–containing structure forms during S phase and is subsequently reduced in size by half in a step that normally occurs in late prophase but does not require cell cycle progression into mitosis. In the second model (bottom), SAS-6 is recruited to the parent centriole before central tube formation. Subsequently, a portion of this SAS-6 is incorporated into the central tube of the daughter centriole. Assembly of the central tube triggers loss of the SAS-6 associated with the parent centriole that was not incorporated into the daughter.
The reduction in SAS-6 levels at the site of new centriole assembly does not require cell cycle progression out of S phase. SAS-4, and hence the SAS-4–dependent addition of centriolar microtubules to the outer centriole wall, are also not required. These results suggest that the reduction in SAS-6 levels is intrinsic to the early steps of the assembly mechanism. One attractive possibility, consistent with model 2 described above, is that the reduction in SAS-6 levels results from a negative feedback loop in which assembly of the SAS-6–dependent central tube of the daughter centriole inhibits ZYG-1–dependent recruitment of SAS-6 to the site of centriole assembly. This idea is supported by prior work showing that SAS-6 modulates the centriolar levels of ZYG-1, the polo family kinase that recruits SAS-6 to centrioles. Centriolar ZYG-1 fluctuates during the cell cycle, with high levels in early S phase and low levels in prophase (O'Connell et al., 2001; Delattre et al., 2006). In embryos depleted of SAS-6 but not SAS-4, the amount of centriolar ZYG-1 remains high throughout the cell cycle (Delattre et al., 2006). We speculate that a negative feedback loop between SAS-6 at the daughter centriole and ZYG-1 on the parent centriole controls the cell cycle–independent loss of SAS-6. After the SAS-6–containing portion of the daughter centriole has formed, ZYG-1–dependent recruitment of SAS-6 to the parent centriole ceases and SAS-6 that was not incorporated into the daughter centriole is lost. Such a mechanism may contribute to restricting the number of daughter centrioles assembled in dividing cells.
We emphasize that the intrinsically programmed reduction in SAS-6 levels during the early steps of centriole assembly that we describe here is different from the loss of centriolar SAS-6 at the end of the duplication cycle that has recently been described in human cells. In human cells, HsSAS-6 levels are kept in check by the presence of a KEN box that targets the protein for proteosomal degradation via an APCCdh1-dependent pathway in telophase at the end of the duplication cycle (Strnad et al., 2007). HsSAS-6 disappears from centrioles at this time, which suggests that, although HsSAS-6 is essential for new centriole assembly, it is not required for their maintenance (Kleylein-Sohn et al., 2007; Strnad et al., 2007). This proteolytic mechanism likely serves to limit HsSAS-6 levels between telophase of one cell division and the beginning of the subsequent S phase to prevent ectopic centriole assembly (Strnad et al., 2007). In the C. elegans embryo, the period between duplication cycles is extremely brief. As there is no G1 phase and the second duplication cycle begins as the chromosomes decondense after anaphase, there is likely not sufficient time to resynthesize SAS-6 before initiating a second round of centriole assembly. Consistent with this, C. elegans SAS-6 does not contain a KEN box and there is currently no evidence that SAS-6 is completely removed from centrioles at any point in the duplication cycle, which suggests that the proteolytic mechanism of safeguarding against excess SAS-6 levels during G1 does not operate in this system.
γ-Tubulin recruited by the PCM mediates the stable incorporation of SAS-4 into newly forming centrioles
γ-Tubulin has been shown to have a conserved, essential role in centriole assembly (Ruiz et al., 1999; Shang et al., 2002; Dammermann et al., 2004; Raynaud-Messina et al., 2004; Haren et al., 2006; Kleylein-Sohn et al., 2007). However, the mechanism by which γ-tubulin contributes to centriole assembly has remained unclear. We have previously shown that the centriole assembly defect resulting from inhibition of γ-tubulin is essentially identical to that resulting from depletion of SPD-5, a coiled-coil protein that is a critical structural component of the PCM (Dammermann et al., 2004). In SPD-5–depleted embryos, centrioles fail to recruit PCM proteins including γ-tubulin and to organize a centrosome (Hamill et al., 2002). In contrast, depletion of γ-tubulin does not perturb PCM assembly (Hannak et al., 2002; Dammermann et al., 2004). This result suggested that the role of the PCM in centriole assembly is to recruit and provide a localized source of γ-tubulin.
In fixed late prophase/prometaphase-stage embryos depleted of γ-tubulin, GFP:SAS-4 foci were frequently absent (Dammermann et al., 2004). We now know the basis for this defect. SAS-4 as well as SAS-6 are recruited to centrioles and reach normal levels in γ-tubulin–depleted embryos. However, centriolar SAS-4 fails to become stably incorporated during late prophase. Because late prophase is when centriolar microtubules form (Pelletier et al., 2006), this result suggests that γ-tubulin contributes to SAS-4 stabilization by promoting the assembly of centriolar microtubules. Consistent with this idea, direct inhibition of microtubule assembly via depletion of β-tubulin results in a similar defect to the one that follows γ-tubulin depletion. Although highly suggestive, analysis of centrioles in γ-tubulin–depleted embryos by EM tomography is needed to confirm a role for γ-tubulin in centriolar microtubule assembly. Determining whether other components of γ-tubulin–containing complexes required for microtubule nucleation by the PCM are also required for centriolar microtubule assembly, will also be an important future direction.
Depletion of γ-tubulin also prevents the targeting of GFP:SAS-4 to the PCM. Levels of centriolar GFP:SAS-4 are normal in γ-tubulin–depleted embryos, indicating that GFP:SAS-4 does not need to localize to the PCM to be effectively recruited to centrioles. One possibility is that the γ-tubulin–dependent localization of GFP:SAS-4 to the PCM reflects an interaction between GFP:SAS-4 and γ-tubulin that contributes to the nucleation of centriolar microtubules. This would represent a separate pool of SAS-4 from centriolar SAS-4 and both pools may coordinately act, perhaps by homotypic interactions, to drive centriolar microtubule assembly. Because γ-tubulin–dependent stabilization of SAS-4 does not occur in S phase arrested embryos, posttranslational cues are likely required to trigger this event.
The γ-tubulin–dependent stable incorporation of SAS-4 at newly forming centrioles is an interesting addition to the emerging symbiosis between centrioles and their surrounding PCM. Centrioles direct assembly of the PCM and are required to maintain its organization (Bobinnec et al., 1998). In turn, our results indicate that γ-tubulin in the PCM organized by the parent stabilizes the nascent daughter centriole by promoting the addition of microtubules to its outer wall. In dividing cells, this mechanism might serve to prevent the formation of stable SAS-4–containing structures in other locations, ensuring that new centrioles form only adjacent to existing parent centrioles that have the ability to accumulate PCM.
Materials and methods
Worm strains and culture conditions
Strains expressing GFP:SAS-4 (TH26), GFP:histone H2B, and GFP:γ-tubulin (TH32) and the strain for analysis of GFP:SAS-4 recruitment (OD19) have been described previously (Dammermann et al., 2004). Strains expressing mCherry:SAS-4 (OD68) and GFP:SAS-6 (OD45) were generated by cloning the corresponding genomic loci into pAA65 (mCherry fusion; McNally et al., 2006) and pIC26 (GFP fusion; Cheeseman and Desai, 2005) and introducing the constructs into unc-119(ed3) worms by microparticle bombardment. The recruitment strain for GFP:SAS-6 (OD103) was generated as described previously (Dammermann et al., 2004). The genotypes of all strains used are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1). Strains were maintained at 16°C (OD19 and OD103) or 20°C (TH26, OD45, TH32 and OD68). For the recruitment assay, L4 fem-1 mutant larvae (OD19 and OD103) were shifted to the restrictive temperature (25°C) and their progeny at the L4 larval stage were mated to OD68 males. Sperm introduced by mating restore embryogenesis, resulting in viable offspring. The L4 progeny generated at the restrictive temperature remain unable to produce their own sperm even when shifted back to 16°C for RNAi-mediated depletions.
RNA-mediated interference
Double-stranded RNAs (dsRNAs) were prepared as described previously (Oegema et al., 2001) using the primers listed in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1) to amplify regions from N2 genomic or specific cDNAs. For double depletions, RNAs were mixed to obtain equal concentrations of ≥1 mg/ml for each RNA. For β-tubulin depletions, young adults were injected with RNA against tbb-2, which targets both tbb-2 and tbb-1, and incubated at 25°C for 15–18 h before filming. For double depletions of β-tubulin and SAS-5, L4s were injected with sas-5 RNA 48 h before filming and the resulting young adults were reinjected with a 1:1 mixture of sas-5 and tbb-2 RNA and incubated at 25°C for 15–18 h before filming. For all other depletions, L4 hermaphrodites were injected with dsRNA and incubated at 16°C for 48 h (20°C for 36 h for γ-tubulin depletions).
HU treatment
For HU treatment, adult worms were incubated on seeded nematode growth medium plates containing 75mM HU for 5 h before dissection, and embryos were filmed in blastomere culture medium supplemented with 75 mM HU.
Imaging conditions
Embryos were filmed without compression (Monen et al., 2005) using a spinning disk confocal mounted on an inverted microscope (Nikon TE2000-E; Nikon) equipped with a 60× 1.4 NA Plan Apochromat lens (Nikon), a krypton-argon 2.5-W water-cooled laser (Spectra-Physics) and an electron multiplication back-thinned charge-coupled device camera (iXon; Andor Technology). Acquisition parameters, shutters, and focus were controlled by MetaMorph software (MDS Analytical Technologies). For quantitative analysis of the recruitment of GFP:SAS-4 and GFP:SAS-6, an 11 × 1 μm RFP/GFP z series with 2 × 2 binning and a single central reference DIC image with no binning were collected every 60 s (<20 z series per embryo). Exposures were 200 and 1,000 ms for GFP and RFP images, respectively (laser power = 40%). For high resolution centriolar imaging, 11 × 0.5 μm RFP/GFP z series were collected (less than five per embryo) at 90× with no binning. Exposures were 400 and 1,000 ms for GFP and RFP images, respectively (laser power = 60%). For FRAP experiments, the 488-nm laser line was steered into a custom-modified epifluorescence port, creating a single diffraction limited spot (full-width half-max ∼800 nm) in the objective focal plane. The exposure time for the bleach was 500 ms (laser power = 40%). 0.5-μm z series were collected before the bleach at irregular short intervals to locate the centriole and after the bleach at 25-s intervals (with 90×, 2 × 2 binning, and 200-ms GFP exposures). For β-tubulin RNAi embryos, 90× views were used without binning. In embryos not treated with HU, DIC microscopy was used to follow the embryo until onset of cytokinesis.
Quantification: recruitment curves
The GFP signal coincident with the RFP-labeled sperm centrioles was measured in maximum intensity projections of the z series collected at each time point. Images were only quantified if both centriole pairs in the embryo were captured in the z series and neither was moving appreciably during acquisition. A 5 × 5 pixel box and a larger 7 × 7 pixel box were drawn around the peak of the RFP signal. The integrated GFP intensity in the smaller box (which encompassed the PCM as well as the centriole) was calculated by subtracting the mean fluorescence intensity in the area between the two boxes (mean background) from the mean GFP intensity in the smaller box and multiplying by the area of the smaller box. Before separation of the sperm-derived centrioles, both sperm centrioles fell within a single box; after separation, measurements for the two centrioles were summed for each time point. Kinetic curves were generated by pooling measurements from multiple embryos and often multiple datasets for each condition (the number of datasets/measurements for each condition are provided in Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1). Individual measurements from both control and RNAi embryos were normalized by dividing by the mean intensity for the set of measurements made over the interval between −750 and −500 s (SAS-6) or −400 to 0 s (SAS-4) in control embryos imaged on the same day. This allowed us to quantitatively compare the signals in RNAi embryos to those in control embryos and to pool datasets collected on different days. Data points in the graphs are the mean of the normalized GFP intensity measurements collected during the 200 s interval centered on that point. Error bars indicate the 90% confidence interval for the mean, which takes into account the standard deviation and number of data points collected for each interval. For subtraction curves, 90% confidence intervals were propagated using the GraphPad online calculator (http://www.graphpad.com).
Quantification: spatial distribution
Quantification of the spatial distribution of centriolar proteins was performed on single plane high resolution images of prometaphase/metaphase centrioles. A 7 × 7 pixel central box was drawn around the sperm centriole signal in the RFP channel. In the GFP channel, this box included the signal from the centrioles as well as the central PCM; a larger 18 × 18 pixel box included the peripheral PCM and the largest 30 × 30 pixel box was used to measure the background. The integrated GFP intensity in the 7 × 7 pixel central box was calculated by subtracting the mean background fluorescence intensity in the area between the 18 × 18 and 30 × 30 pixel boxes from the mean GFP intensity in the central box and multiplying by the area of the central box. The integrated GFP intensity in the peripheral centrosomal region (between the 7 × 7 and 18 × 18 pixel boxes) was calculated by subtracting the same mean background value from the mean intensity in the region between the 7 × 7 and 18 × 18 pixel boxes and multiplying by the area of the peripheral region.
Online supplemental material
Fig. S1 shows GFP fusions with SAS-4 and SAS-6 can functionally substitute for the corresponding endogenous proteins. Fig. S2 shows that the centriolar GFP:SAS-6 and GFP:SAS-4 signals are not subject to detectable photobleaching under the imaging conditions used. Fig. S3 establishes a timeline of events during the first mitotic division. Table S1 lists worm strains used in this study. Table S2 lists dsRNAs used in this study. Table S3 shows recruitment curve statistics. Video 1 shows a wild-type embryo coexpressing GFP:γ-tubulin and GFP:histone. Video 2 shows an HU-treated embryo coexpressing GFP:γ-tubulin and GFP:histone. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200709102/DC1.
Supplementary Material
Acknowledgments
We thank members of the Oegema and Desai laboratories for their support throughout this project; Laurence Pelletier, Thomas Müller-Reichert, and Tony Hyman for communicating unpublished results; Andrew Fire for the synthetic C. elegans intron sequences; Yuji Kohara for γ-tubulin cDNA; and the Caenorhabditis Genetics Center for strains.
This work was supported by funding from the Ludwig Institute for Cancer Research to K. Oegema and A. Desai and a National Institutes of Health grant (R01-GM074207) to K. Oegema. K. Oegema is a Pew Scholar in the Biomedical Sciences.
P.S. Maddox's present address is Department of Pathology and Cell Biology, Institute for Research in Immunology and Cancer, Faculty of Medicine, Université de Montréal, Pavillon Marcelle-Coutu, Quai 20, Montreal QC H3T 1J4, Canada
Abbreviations used in this paper: DIC, differential interference contrast; dsRNA, double-stranded RNA; HU, hydroxyurea; PCM, pericentriolar material.
References
- Anderson, R.G.W., and R.M. Brenner. 1971. The formation of basal bodies (centrioles) in the rhesus monkey oviduct. J. Cell Biol. 50:10–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh, J., and M. Bornens. 2004. The Centrosome in Evolution. In Centrosomes in Development and Disease. E.A. Nigg, editor. Wiley-VCH, Weinheim, Germany. 93–122.
- Basto, R., J. Lau, T. Vinogradova, A. Gardiol, C.G. Woods, A. Khodjakov, and J.W. Raff. 2006. Flies without centrioles. Cell. 125:1375–1386. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., and D.M. Glover. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., A. Rodrigues-Martins, L. Carpenter, M. Riparbelli, L. Lehmann, M.K. Gatt, N. Carmo, F. Balloux, G. Callaini, and D.M. Glover. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. [DOI] [PubMed] [Google Scholar]
- Bobinnec, Y., A. Khodjakov, L.M. Mir, C.L. Rieder, B. Edde, and M. Bornens. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., and A. Desai. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., T. Muller-Reichert, L. Pelletier, B. Habermann, A. Desai, and K. Oegema. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829. [DOI] [PubMed] [Google Scholar]
- Delattre, M., S. Leidel, K. Wani, K. Baumer, J. Bamat, H. Schnabel, R. Feichtinger, R. Schnabel, and P. Gonczy. 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6:656–664. [DOI] [PubMed] [Google Scholar]
- Delattre, M., C. Canard, and P. Gonczy. 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16:1844–1849. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K. 2003. Long-lost relatives reappear: identification of new members of the tubulin superfamily. Curr. Opin. Microbiol. 6:634–640. [DOI] [PubMed] [Google Scholar]
- Edgar, L.G., and J.D. McGhee. 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell. 53:589–599. [DOI] [PubMed] [Google Scholar]
- Habedanck, R., Y.D. Stierhof, C.J. Wilkinson, and E.A. Nigg. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. [DOI] [PubMed] [Google Scholar]
- Hamill, D.R., A.F. Severson, J.C. Carter, and B. Bowerman. 2002. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell. 3:673–684. [DOI] [PubMed] [Google Scholar]
- Hannak, E., M. Kirkham, A.A. Hyman, and K. Oegema. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak, E., K. Oegema, M. Kirkham, P. Gonczy, B. Habermann, and A.A. Hyman. 2002. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is γ-tubulin dependent. J. Cell Biol. 157:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., M.H. Remy, I. Bazin, I. Callebaut, M. Wright, and A. Merdes. 2006. NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holway, A.H., S.H. Kim, A. La Volpe, and W.M. Michael. 2006. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J. Cell Biol. 172:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham, M., T. Muller-Reichert, K. Oegema, S. Grill, and A.A. Hyman. 2003. SAS-4 Is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn, J., J. Westendorf, M. Le Clech, R. Habedanck, Y.D. Stierhof, and E.A. Nigg. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202. [DOI] [PubMed] [Google Scholar]
- Kochanski, R.S., and G.G. Borisy. 1990. Mode of centriole duplication and distribution. J. Cell Biol. 110:1599–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel, S., and P. Gonczy. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 4:431–439. [DOI] [PubMed] [Google Scholar]
- Leidel, S., M. Delattre, L. Cerutti, K. Baumer, and P. Gonczy. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125. [DOI] [PubMed] [Google Scholar]
- Maddox, P.S., N. Portier, A. Desai, and K. Oegema. 2006. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc. Natl. Acad. Sci. USA. 103:15097–15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W.F. 2007. What is the function of centrioles? J. Cell. Biochem. 100:916–922. [DOI] [PubMed] [Google Scholar]
- McNally, K., A. Audhya, K. Oegema, and F.J. McNally. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen, J., P.S. Maddox, F. Hyndman, K. Oegema, and A. Desai. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 7:1248–1255. [DOI] [PubMed] [Google Scholar]
- Moritz, M., and D.A. Agard. 2001. γ-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11:174–181. [DOI] [PubMed] [Google Scholar]
- O'Connell, K.F., C. Caron, K.R. Kopish, D.D. Hurd, K.J. Kemphues, Y. Li, and J.G. White. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558. [DOI] [PubMed] [Google Scholar]
- Oegema, K., and A.A. Hyman. 2006. Cell division. WormBook, editor. The C. elegans Research Community, Wormbook. doi/ 10.1895/wormbook.1.72.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel, N., N.R. Stevens, R. Basto, and J.W. Raff. 2007. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17:834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, L., E. O'Toole, A. Schwager, A.A. Hyman, and T. Muller-Reichert. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. [DOI] [PubMed] [Google Scholar]
- Portier, N., A. Audhya, P.S. Maddox, R.A. Green, A. Dammermann, A. Desai, and K. Oegema. 2007. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev. Cell. 12:515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina, B., L. Mazzolini, A. Moisand, A.M. Cirinesi, and M. Wright. 2004. Elongation of centriolar microtubule triplets contributes to the formation of the mitotic spindle in γ-tubulin-depleted cells. J. Cell Sci. 117:5497–5507. [DOI] [PubMed] [Google Scholar]
- Richards, T.A., and T. Cavalier-Smith. 2005. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 436:1113–1118. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins, A., M. Bettencourt-Dias, M. Riparbelli, C. Ferreira, I. Ferreira, G. Callaini, and D.M. Glover. 2007. a. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17:1465–1472. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins, A., M. Riparbelli, G. Callaini, D.M. Glover, and M. Bettencourt-Dias. 2007. b. Revisiting the role of the mother centriole in centriole biogenesis. Science. 316:1046–1050. [DOI] [PubMed] [Google Scholar]
- Ruiz, F., J. Beisson, J. Rossier, and P. Dupuis-Williams. 1999. Basal body duplication in Paramecium requires γ-tubulin. Curr. Biol. 9:43–46. [DOI] [PubMed] [Google Scholar]
- Shang, Y., B. Li, and M.A. Gorovsky. 2002. Tetrahymena thermophila contains a conventional γ-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 158:1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad, P., S. Leidel, T. Vinogradova, U. Euteneuer, A. Khodjakov, and P. Gonczy. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar, E.K., and T. Stearns. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.