Abstract
To elucidate the human colonization process of northern Asia and human dispersals to the Americas, a diverse subset of 71 mitochondrial DNA (mtDNA) lineages was chosen for complete genome sequencing from the collection of 1,432 control-region sequences sampled from 18 autochthonous populations of northern, central, eastern, and southwestern Asia. On the basis of complete mtDNA sequencing, we have revised the classification of haplogroups A, D2, G1, M7, and I; identified six new subhaplogroups (I4, N1e, G1c, M7d, M7e, and J1b2a); and fully characterized haplogroups N1a and G1b, which were previously described only by the first hypervariable segment (HVS1) sequencing and coding-region restriction-fragment–length polymorphism analysis. Our findings indicate that the southern Siberian mtDNA pool harbors several lineages associated with the Late Upper Paleolithic and/or early Neolithic dispersals from both eastern Asia and southwestern Asia/southern Caucasus. Moreover, the phylogeography of the D2 lineages suggests that southern Siberia is likely to be a geographical source for the last postglacial maximum spread of this subhaplogroup to northern Siberia and that the expansion of the D2b branch occurred in Beringia ∼7,000 years ago. In general, a detailed analysis of mtDNA gene pools of northern Asians provides the additional evidence to rule out the existence of a northern Asian route for the initial human colonization of Asia.
The territory of northern Asia is of crucial importance for the study of early human dispersal and the peopling of the Americas. Recent findings about the peopling of northern Asia reconstructed by paleoanthropologists and archaeologists suggest that modern humans colonized the southern part of Siberia 45,000–35,000 years ago. It seems that almost all of northern Asia, including extreme northeastern Siberia, had been colonized by modern humans by ∼15,000 years ago.1–3 Over the past few years, a number of genetic studies about populations from different parts of Siberia were conducted.4–15 Molecular evidence suggests that ancestral Native American populations may have emerged from this region of northern Asia, since several maternally and paternally inherited genetic lineages present in both Siberia and the Americas appear to have evolved in that region of northern Asia.4,16–18 Recent studies have also revealed the presence of both eastern and western Eurasian lineages in gene pools of modern populations of southern Siberia, a pattern that probably reflects a complex history of population movements and interactions since the Paleolithic period.8,11,13,15 However, such issues as timing, origin, and routes of founding migrations to Siberia and the Americas remain ambiguous and controversial.
Because of a new phase of development of genetic studies that is based on complete mitochondrial genome analyses, our chances to improve the phylogenetic resolution of the mtDNA tree and, consequently, to define the timing and direction of human dispersions more precisely are occurring repeatedly. The emergence of complete mtDNA sequences made it possible to reconstruct the phylogenies of African, European, Oceanian, eastern Asian, southeastern Asian, and Indian lineages and to gain detailed insight into human evolution and pioneer settlement processes.19–34 Meanwhile, such systematic analyses have not yet been available for northern Asian populations. Recent analyses of a large data set of eastern Asian complete mtDNA sequences have provided a significant refinement of the eastern Asian mtDNA phylogeny,27,32 which is undoubtedly useful for northern Asian mtDNA phylogeny reconstruction. Nevertheless, to date, only two studies dealing with complete mtDNA variation in northern Asian populations have been published.9,14 The first study reported the analysis of complete mtDNA sequences of haplogroup D2 that are fixed in the gene pool of the Aleuts of the Commander Islands.9 The second study presented the set of different mtDNA lineages characterizing the gene pools of aboriginal populations from the Altai-Sayan Upland and the Lower Amur/Sea of Okhotsk regions.14 Both studies focused primarily on the peopling of the Americas, whereas the problems of initial human colonization of northern Asia fell beyond the scope of those studies.
Recent models based on mtDNA evidence suggest a single human dispersal out of Africa by a southern coastal route to India and farther, to East Asia and Australasia.26,29,30,33,35 However, the early arrival in southern Siberia of Upper Palaeolithic technology from the Middle East (∼40,000 years ago) has often been interpreted as support for the existence of another migration route from Africa toward East Asia through the Levant and farther along the northern Asian route through central Asia and southern Siberia.36 This scenario suggests that unique mtDNA lineages that cannot be derived from southern and southeastern Asian variation should be found in northern and central Asian populations.37 It is worth noting, however, that mtDNA-variation studies of modern Siberian populations have shown a lack of basal M, N, and R lineages in the mtDNA tree of northern Asians.6,11,38 This should be considered a strong argument against the northern Asian–route model. Meanwhile, we should mention that both population coverage of Siberian ethnic groups and a range of phylogenetic resolution of mtDNA data obtained so far were insufficient for a definitive conclusion. To elucidate the human colonization process of northern Asia, as well as human dispersals to the Americas, we use here the complete genome–sequencing approach, in addition to control-region sequencing combined with restriction analysis of the mtDNA coding region, to study mtDNA variation in a large number of populations representing northern, central, eastern, and southwestern Asia.
Material and Methods
Subjects
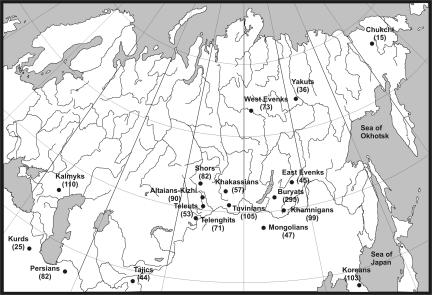
Sampling locations are shown in figure 1. A total of 1,432 DNA samples were analyzed and comprised 107 individuals from southwestern Asia (82 Persians from eastern Iran and 25 Kurds from northwestern Iran), 44 individuals from central Asia (Tajiks from Tajikistan), 150 individuals from East Asia (47 Mongolians from Ulaanbaatar and 103 Koreans from South Korea), and 1,131 individuals from northern Asia (295 Buryats and 99 Khamnigans from Buryat Republic, 105 Tuvinians from Tuva Republic, 118 Evenks [45 East Evenks from Buryat Republic and 73 West Evenks from the Krasnoyarsk region], 36 Yakuts from Sakha [Yakutia] Republic, 82 Shors from the Kemerovo region, 57 Khakassians from Khakassian Republic, 214 South Altaians [90 Altaian-Kizhi, 71 Telenghits from Altai Republic, and 53 Teleuts from the Kemerovo region], 110 Kalmyks from Kalmyk Republic, and 15 Chukchi from Anadyr, Chukotka Autonomous Okrug). Each sample comprises unrelated healthy donors from whom appropriate informed consent was obtained.
Figure 1. .
Population sample sites, with sample sizes given in parentheses
Sequencing and RFLP Typing
The first hypervariable segment (HVS1) (from positions 15999 to 16400) was sequenced in all samples, and HVS2 (from positions 30 to 407) was also sequenced in Chukchi, Telenghits, East Evenks, Tuvinians, Buryats, Khamnigans, Kalmyks, Koreans, Mongolians, Persians, Kurds, and Tajiks, as described elsewhere.11 Sequences were aligned to the revised Cambridge Reference Sequence (rCRS).39 Any unusual mutations (transversions or transitions at sites with a low relative mutation rate) were rechecked. The sequences with uncertain phylogenetic status were selected for complete mtDNA sequencing. RFLP screening was used to resolve haplogroup status in a hierarchical scheme as follows: haplogroups M (+10397 AluI and +10394 DdeI), N (−10397 AluI and −10394 DdeI), R (+12704 MboII), A (+663 HaeIII), C (−13259 HincII), D (−5176 AluI), D1 (−2093 Bst4CI), D4 (−3010 FnuDII), D2 (−7493 Eco130I), E (−7598 HhaI), G (+4831 HhaI), F1 (−12406 HpaI), M7 (+9820 HinfI), M9 (+3391 HaeIII), M10 (+10646 RsaI), X (+14465 AccI), HV (−14766 MseI), H (−7025 AluI), U (+12308 HinfI), U4 (+4646 RsaI), K (−9052 HaeII), T (+13366 BamHI and +15606 AluI), T1 (−12629 AvaII), W (−8994 HaeIII), I (−4529 HaeII, −8250 HaeIII, and +10032 AluI), N1 (−12498 NlaIII), V (+4577 NlaIII), and HV0a (+15904 MseI). Haplogroup B affiliation was checked by screening for the 9-bp deletion in the COII/tRNALys region, and haplogroup J1b2a affiliation was checked by sequencing position 10410 within the fragment 8910–10649. Sequence classification into mtDNA haplogroups was based on generally accepted nomenclatures,26,32,40 with several improvements.
Complete mtDNA Sequencing
A total of 71 mtDNAs representing 36 subhaplogroups were selected for complete sequencing (table 1). These samples include several mtDNAs (6 Koryaks, 1 Evenk, 3 Russians, 1 Polish, and 3 Czechs) from our collection, added for comparative purposes. The mtDNA genomes were amplified and sequenced by means of the procedures described by Torroni et al.21 Sequencing reactions were run on Applied Biosystems 3130 Genetic Analyzer. Sequences were edited and aligned by SeqScape software, version 2.5 (Applied Biosystems), and mutations were scored relative to the rCRS.39 The 71 complete sequences have been submitted to GenBank (accession numbers EF153771–EF153833, EF397558–EF397562, and EF486517–EF486519).
Table 1. .
Sample and Ethnic Origin of 71 Complete mtDNA Sequences
| Sample | Haplogroup | Origin |
| Ch 1 | A2a | Chukchi |
| Ch 9 | A2a | Chukchi |
| Ch 5 | A2b | Chukchi |
| Ch 6 | A2b | Chukchi |
| Ch 11 | A2b | Chukchi |
| Krk 39 | A2b | Koryak |
| Br 334 | A4 | Buryat |
| Br 393 | A4 | Buryat |
| Br 523 | A4 | Buryat |
| Br 568 | A4 | Buryat |
| Cz_25_IV | A4 | Czech |
| Khm 16 | A4 | Khamnigan |
| VN 65 | A4a | Russian |
| Alt 163 | A4a1 | Altaian-Kizhi |
| Br 390 | A4a1 | Buryat |
| Br 552 | A4a1 | Buryat |
| Br 627 | A4a1 | Buryat |
| Br 442 | A4b | Buryat |
| Evk 42 | A4b | Evenk |
| Br 406 | A4c | Buryat |
| Br 575 | A4c | Buryat |
| Khm 66 | A4c | Khamnigan |
| Alt 178 | A4d | Altaian-Kizhi |
| Kor 97 | A5a | Korean |
| Br 643 | A5c | Buryat |
| Khm 43 | A5c | Khamnigan |
| Br 618 | A8 | Buryat |
| Br 324 | C1 | Buryat |
| Br 412 | D2a | Buryat |
| Br 608 | D2a | Buryat |
| Ch 2 | D2b | Chukchi |
| Khm 27 | D2a | Khamnigan |
| Yak 44 | D2a | Yakut |
| Br 636 | D3 | Buryat |
| Kor 38 | G1a1a | Korean |
| Alt 145 | G1a1b | Altaian-Kizhi |
| Ch 3 | G1b | Chukchi |
| Ev 23 | G1b | Evenk |
| Krk 115 | G1b | Koryak |
| Krk 120 | G1b | Koryak |
| Krk 35 | G1b | Koryak |
| Krk 41 | G1b | Koryak |
| Kor 33 | G1c | Korean |
| Br 405 | I4 | Buryat |
| Alt 194 | J1b2a | Altaian-Kizhi |
| Br 363 | J1b2a | Buryat |
| Br 345 | M7a2 | Buryat |
| Br 423 | M7b1 | Buryat |
| Khm 83 | M7b1 | Khamnigan |
| Kor 19 | M7c1 | Korean |
| Kor 53 | M7c1 | Korean |
| Khm 51 | M7c2 | Khamnigan |
| Br 365 | M7c2a | Buryat |
| Br 646 | M7c2a | Buryat |
| Br 311 | M7d | Buryat |
| Khm 47 | M7d | Khamnigan |
| Cz_32_III | M7e | Czech |
| Vo 20 | N1a | Russian |
| Br 312 | N1a1 | Buryat |
| Cz_61_III | N1a1 | Czech |
| Bg_1_I | N1a1 | Russian |
| Pl 348 | N1c | Polish |
| Br 397 | N1e | Buryat |
| Alt 16 | X2e | Altaian-Kizhi |
| Alt 208 | X2e | Altaian-Kizhi |
| Br 561 | X2e | Buryat |
| Tl 715 | X2e | Teleut |
| Khm 6 | Y1 | Khamnigan |
| Krk 28 | Y1a1 | Koryak |
| Br 621 | Y2 | Buryat |
| Khm 2 | Y2 | Khamnigan |
Data Analysis
Descriptive statistical indexes, the Tajima’s D41 and Fu’s FS42 neutrality tests (for HVS1 and HVS2 sequence data) and the analysis of molecular variance (AMOVA)43 (for HVS1 sequence data) were calculated using Arlequin software, version 3.01.44 Principal-component (PC) analysis was performed using STATISTICA software, version 6.0, with mtDNA haplogroup frequencies as input vectors.
The complete mtDNA phylogeny was reconstructed manually and was verified by use of the median-joining algorithm with Network, version 4.1.0.9 (Free Phylogenetic Network Software).45 For phylogeny construction, the highly variable site 16519 and the length variation in the poly-C stretches at nucleotides 16180–16193 and 309–315 were not used. Haplogroup divergence estimates ρ and their SEs were calculated as the average number of substitutions in the mtDNA-coding region from the ancestral sequence type.46,47 To estimate the time to the most recent common ancestor of each cluster, the evolutionary rate corresponding to 5,140 years per substitution in the coding region25 was used. In addition to our 71 newly collected mtDNAs, 21 additional complete genomes from the literature9,14,19,22,24,26,27,48,49 were employed for the tree reconstruction.
Results
mtDNA Haplogroup Profile
The complete RFLP haplotypes and HVS1 and HVS1/HVS2 sequence data of the 1,432 individuals from northern, central, eastern, and southwestern Asia and the detailed haplogroup classification are reported in a tab-delimited txt file, which can be imported into a Microsoft Excel spreadsheet. The samples fall into 91 haplogroups or paragroups (unclassified lineages within a clade) within the three major non-African haplogroups M, N, and R; the single Persian mtDNA belongs to the sub-Saharan haplogroup L2a. Table 2 shows the haplogroup distribution within the populations studied. The eastern Eurasian component is represented by haplogroups A, N9a, and Y, which belong to the major haplogroup N; by haplogroups B, F, R9, R11, and R*, which belong to macrohaplogroup R; and by different branches of macrohaplogroup M, such as C, D, G, M*, M3, M7–M11, M13, and Z haplogroups. The latter lineages were shown to be widespread among northern and eastern Asians and, to a lesser extent, central Asians.11,38,50–53 All populations of northern and eastern Asia exhibit the prevalence of eastern Eurasian lineages, with frequencies ranging from 69% in Telenghits to 100% in Koreans and East Evenks. Haplogroups C and D are the most common in northern and eastern Asia, accounting together for 24%–87% of lineages in populations studied. The only exception is the Chukchi sample, which shows the highest frequency of haplogroup A (73%). One of the most common haplogroups in northern and eastern Asian populations is haplogroup B, which falls into two main clades, B4 and B5. The majority of B lineages in northern Asia fall within haplogroup B4, whereas haplogroup B5 is more frequent among Koreans and Mongolians. Haplogroup G and its subgroup G2 occur with the highest frequencies (>8%) in Mongolic-speaking populations, such as the Mongolians, Buryats, Khamnigans, and Kalmyks. It is also worth noting that haplogroup F exhibits very high frequency only in two populations: Shors (41%) and Khakassians (25%).
Table 2. .
Haplogroup Frequencies in Siberia, Southwestern Asia, and Central Asia
| Frequency (%) of Haplogroup in Study Subjects |
||||||||||||||||||
| Haplogroup | Persians (n=82) |
Kurds (n=25) |
Tajiks (n=44) |
Koreans (n=103) |
Mongolians (n=47) |
Kalmyks (n=110) |
Buryats (n=295) |
Khamnigans (n=99) |
Tuvinians (n=105) |
East Evenks (n=45) |
West Evenks (n=73) |
Yakuts (n=36) |
Shors (n=82) |
Khakassians (n=57) |
Altaians-Kizhi (n=90) |
Teleuts (n=53) |
Telenghits (n=71) |
Chukchi (n=15) |
| A2 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 73.0 |
| A4 | 2.4 | … | 2.3 | 2.9 | 13.0 | 2.7 | 4.4 | 4.0 | 1.0 | 2.2 | 4.1 | … | 1.2 | 3.5 | 3.3 | … | 5.6 | … |
| A5 | … | … | … | 3.9 | … | … | .3 | 1.0 | … | … | … | … | … | … | … | … | … | … |
| A8 | … | … | … | … | … | .9 | .3 | … | … | … | … | … | … | … | … | … | … | … |
| B4 | 4.9 | … | … | 13.0 | 11.0 | 2.7 | 3.1 | 5.1 | 1.9 | … | 4.1 | 2.8 | 4.9 | 8.8 | 3.3 | 3.8 | 11.0 | … |
| B5 | 1.2 | … | … | 7.8 | 4.3 | .9 | .3 | 2.0 | … | … | … | … | … | … | 1.1 | … | … | … |
| C* | … | … | 9.1 | 1 | 6.4 | 1.8 | 5.1 | 4.0 | 21.0 | 11.0 | 9.6 | 17.0 | 2.4 | 5.3 | 10.0 | 15.0 | 11.0 | … |
| C1 | … | … | … | … | … | .9 | 1.0 | … | … | … | … | … | … | … | … | … | … | … |
| C4 | 1.2 | … | 2.3 | … | 8.5 | 5.5 | 5.8 | 7.1 | 18.0 | 27.0 | 32.0 | 36.0 | 8.5 | 14.0 | 13.0 | 5.7 | 2.8 | … |
| C5 | … | … | … | … | 2.1 | 2.7 | 4.7 | 5.1 | 11.0 | 24.0 | 6.8 | 11.0 | 1.2 | … | 8.9 | 7.5 | 2.8 | … |
| D2 | … | … | … | … | … | 1.8 | .7 | 1.0 | 2.9 | 2.2 | … | 2.8 | … | … | … | … | … | 13.0 |
| D3 | … | … | … | … | … | … | 2.4 | 5.1 | 1.0 | 13.0 | 5.5 | 2.8 | … | … | 2.2 | … | … | … |
| D4 | 1.2 | 12 | 4.5 | 32.0 | 11.0 | 22.0 | 29.0 | 25.0 | 8.6 | 8.9 | 18.0 | 14.0 | 11.0 | 16.0 | 6.7 | 23.0 | 21.0 | … |
| D5 | 1.2 | … | 2.3 | 7.8 | … | 5.5 | 2.7 | 2.0 | 2.9 | … | 6.8 | 2.8 | 1.2 | … | … | 1.9 | … | … |
| F1 | … | … | … | 4.9 | 6.4 | 5.5 | 2.4 | 4.0 | 7.6 | … | 1.4 | … | 40.0 | 19.0 | 3.3 | 5.7 | 1.4 | … |
| F2a | … | … | … | … | … | … | .7 | … | 1.0 | … | … | … | 1.2 | 5.3 | 2.2 | … | … | … |
| G* | … | … | … | … | … | .9 | … | … | … | … | … | … | … | … | … | … | … | … |
| G1 | … | … | … | 1.9 | 2.1 | .9 | … | … | 1.9 | … | 1.4 | … | … | … | 4.4 | … | … | 6.7 |
| G2a | 1.2 | … | 2.3 | 2.9 | 8.5 | 6.4 | 11.0 | 9.1 | 3.8 | … | 2.7 | 2.8 | … | … | 1.1 | … | 2.8 | … |
| G3 | … | … | 2.3 | 1.9 | … | … | .3 | 1 | 1 | 2.2 | … | … | … | 1.8 | … | … | … | … |
| M* | 7.3 | … | 2.3 | … | 2.1 | … | … | … | … | … | … | … | … | … | … | … | 1.4 | … |
| M3a | 2.4 | … | … | … | … | .9 | … | … | … | … | … | … | … | … | … | … | … | … |
| M7 | … | … | … | 9.7 | 2.1 | 1.8 | 3.4 | 3 | … | … | … | … | … | … | … | … | 1.4 | … |
| M8a2 | … | … | … | 4.9 | 4.3 | … | … | … | … | … | … | … | … | 3.5 | 4.4 | … | … | … |
| M9a | … | … | … | 1.9 | 2.1 | 2.7 | .3 | 1 | 1 | … | … | … | … | … | … | … | … | … |
| M10 | … | … | 2.3 | … | 2.1 | .9 | … | … | … | … | … | … | 3.7 | … | 1.1 | … | … | … |
| M11 | … | … | … | … | … | … | .3 | … | … | … | … | … | … | … | 2.2 | 3.8 | 1.4 | … |
| M13a | … | … | … | … | … | … | 1.0 | 1.0 | … | … | … | … | … | … | … | … | … | … |
| Y | … | … | 2.3 | 1.0 | … | 1.8 | 1.4 | 3.0 | … | 8.9 | 2.7 | … | … | … | … | … | … | … |
| Z | … | … | 2.3 | … | 2.1 | 1.8 | 1.4 | … | 1.0 | … | 1.4 | … | 1.2 | … | 2.2 | 5.7 | … | … |
| N9a | … | … | … | 2.9 | 2.1 | 4.5 | 1.7 | 1.0 | 1.9 | … | … | … | … | … | … | … | 1.4 | … |
| R* | … | … | … | … | … | .9 | … | … | … | … | … | … | … | … | … | … | … | … |
| R9 | … | … | … | … | … | … | .3 | 1.0 | … | … | … | … | … | … | … | … | 4.2 | … |
| R11 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 1.1 | … | … | … |
| L2a | 1.2 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| H | 26.0 | 12 | 25.0 | … | … | 3.6 | 6.8 | 6.1 | 3.8 | … | 1.4 | 2.8 | 11.0 | 7.0 | 5.6 | 7.5 | 11.0 | … |
| HV | 3.7 | 8 | … | … | … | 5.5 | 1.0 | 2.0 | 1.0 | … | … | … | … | … | … | … | 1.4 | … |
| HV0a | … | … | 2.3 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| V | … | … | … | … | … | … | … | … | … | … | … | … | … | 1.8 | … | 5.7 | … | … |
| R0a | 2.4 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| R2 | 1.2 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| J1 | 3.7 | 12 | 2.3 | … | … | .9 | .7 | 1.0 | … | … | … | 5.6 | 6.1 | 3.5 | 5.6 | … | 2.8 | … |
| J2 | 1.2 | 8 | 2.3 | … | … | 2.7 | … | … | … | … | … | … | … | 1.8 | … | … | … | … |
| T* | 4.9 | 4 | … | … | … | .9 | .3 | 1.0 | … | … | … | … | … | … | … | … | 1.4 | … |
| T1 | 8.5 | 8 | 2.3 | … | … | .9 | .7 | … | … | … | … | … | … | 5.3 | … | 5.7 | 4.2 | 6.7 |
| N1 | 1.2 | … | 2.3 | … | 2.1 | … | .7 | … | … | … | … | … | … | … | … | … | 1.4 | … |
| I | 3.7 | … | 2.3 | … | … | .9 | .3 | … | 2.9 | … | … | … | 2.4 | … | 1.1 | … | 1.4 | … |
| W | 2.4 | … | 4.5 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| N2 | 1.2 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| U1 | 1.2 | 12 | … | … | … | … | .3 | … | … | … | … | … | … | … | … | … | … | … |
| U2 | 1.2 | … | 2.3 | … | … | 1.8 | … | … | … | … | … | … | … | … | … | 3.8 | 2.8 | … |
| U3 | 2.4 | … | 2.3 | … | … | … | … | … | 1.9 | … | … | … | … | … | 2.2 | … | … | … |
| U4 | … | … | 6.8 | … | 6.4 | 1.8 | 1.0 | … | 1.0 | … | … | … | 2.4 | 1.8 | 2.2 | 1.9 | 1.4 | … |
| U5 | 4.9 | … | … | … | … | .9 | 2.0 | 2.0 | 1.9 | … | … | … | … | 1.8 | 1.1 | … | 1.4 | … |
| U7a | 1.2 | 16 | 4.5 | … | … | .9 | … | … | … | … | … | … | … | … | … | … | … | … |
| U8 | 1.2 | 4 | … | … | … | … | .7 | … | … | … | … | … | … | … | … | … | 1.4 | … |
| K | 3.7 | … | 6.8 | … | 2.1 | 2.7 | 1.4 | 2.0 | … | … | 2.7 | … | 1.2 | … | 6.7 | 1.9 | … | … |
| X2e | … | 4 | … | … | … | … | .3 | … | … | … | … | … | … | … | 4.4 | 1.9 | … | … |
Most northern and eastern Asian populations (with the exception of Koreans and East Evenks) indicate a significant contribution of the western Eurasian mtDNA component. The proportion of these mitochondrial lineages is considerably higher (>22%) in the western part of southern Siberia (in populations of Shors, Khakassians, Tuvinians, and Telenghits) than in the eastern part. In northern Asia, the western Eurasian mtDNA component displays numerous lineages belonging to major haplogroups H, HV, V, J, T, U, I, N1, and X. Among them, haplogroups H, J, and U are the most frequent. Populations from southwestern (Persians and Kurds) and central (Tajiks) Asia have a number of other western Eurasian mtDNA haplogroups not found (or very rare) in northern and eastern Asian populations (W, N2, R0a, R2, HV0a, HV2, U1, U2b, and U7a) but characteristic of the Indo-Pakistani region.53,54 Some western Eurasian haplogroups observed at low frequencies in northern Asian populations are also present in central and southwestern Asian populations. For instance, haplogroup U4 is absent in Persians and Kurds and is present in Tajiks (at a frequency of 6.8%), whereas haplogroup U5a is found (at a frequency of 4.9%) only in Persians. In general, the western Eurasian mtDNA component clearly predominates in central and southwestern Asian populations, found at a frequency of >65% in each population studied. The eastern Eurasian lineages in central and southwestern Asian populations are represented by haplogroups A4, B4, B5, C, D4, D5, G2a, G3, M10, Y, and Z. Their proportion is much higher in Tajiks (in total, 31.8%) than in Kurds and Persians (12% and 13.5%, respectively). It is noteworthy that subgroup B4b1 (defined by mutations at positions 16086 and 16136, in addition to the B4b general motif), which is characteristic of populations of southern Siberia and Mongolia, was also found in northeastern Iran among Persians (at a frequency of 2.4%). Conversely, subgroup J1b2—which is relatively frequent in Indo-Iranian populations,53,54—was found, for instance, at frequencies of 12% and 3.7% in our Kurd and Persian samples, respectively, and is present at a marked frequency (∼3%) in Altaian populations (both in Telenghits and Altaians-Kizhi).
Population Summary Statistics
HVS1 and HVS2 sequences have been used to gain information about internal population diversity (tables 3 and 4). Overall, HVS1 presented slightly higher values of sequence diversity and a higher number of different haplotypes, segregating sites, and average number of nucleotide differences than did HVS2. Most populations demonstrate similar sequence-diversity values, with the Chukchi showing the lowest (0.781 for HVS1/0.467 for HVS2) and the Kalmyks, Kurds, and Tajiks showing the highest (0.997/0.959, 0.993/0.972, and 0.995/0.983, respectively). The low diversity exhibited by the Chukchi is also evident in the low mean number of pairwise differences observed both in HVS1 and HVS2 (4.381 and 1.657, respectively). These are among the lowest values of all the populations, which otherwise ranged from 5.970 in Kurds to 7.002 in Telenghits (for HVS1) and from 1.618 in East Evenks to 3.840 in Kurds (for HVS2).
Table 3. .
Diversity Indexes and Neutrality Tests for the Studied Populations, Based on HVS1-Variability Data
| Population | na | Hb (SE) | K (%)c | Sd | Pie (SE) | θk (95% CI) | Tajima’s D (P)f | Fu’s FS (P)f |
| Persians | 82 | .987 (.005) | 62 (76) | 79 | 6.192 (2.972) | 114.10 (7.73–187.89) | −2.03 | −25.18 |
| Kurds | 25 | .993 (.013) | 23 (88) | 36 | 5.970 (2.946) | 133.89 (44.79–45.042) | −1.41 (.076) | −18.02 |
| Tajiks | 44 | .995 (.006) | 39 (89) | 67 | 6.314 (3.052) | 16.77 (73.29–382.2) | −2.10 | −25.21 |
| Koreans | 103 | .984 (.007) | 77 (75) | 80 | 6.511 (3.104) | 149.81 (96.80–236.12) | −1.90 | −25.04 |
| Mongolians | 47 | .994 (.006) | 40 (85) | 65 | 7.709 (3.656) | 157.02 (75.30–351.29) | −1.72 | −24.94 |
| Kalmyks | 110 | .997 (.002) | 92 (84) | 81 | 6.605 (3.143) | 287.42 (178.33–477.81) | −1.88 | −24.99 |
| Buryats | 295 | .990 (.002) | 154 (52) | 92 | 6.014 (2.874) | 129.31 (101.93–164.00) | −2.00 | −24.71 |
| Khamnigans | 99 | .990 (.004) | 71 (72) | 81 | 6.169 (2.957) | 11.96 (72.63–171.79) | −1.98 | −25.13 |
| Tuvinians | 105 | .960 (.008) | 43 (41) | 58 | 6.248 (2.990) | 27.43 (18.37–4.69) | −1.48 (.061) | −22.70 |
| East Evenks | 45 | .902 (.022) | 14 (31) | 24 | 5.414 (2.658) | 6.58 (3.43–12.29) | −.04 (.499) | −.61 (.429) |
| West Evenks | 73 | .952 (.013) | 31 (42) | 47 | 5.982 (2.885) | 2.11 (12.45–32.22) | −1.24 (.106) | −11.66 |
| Yakuts | 36 | .898 (.034) | 18 (50) | 27 | 4.835 (2.415) | 13.66 (7.16–25.92) | −.89 (.199) | −5.46 (.026) |
| Shors | 82 | .839 (.035) | 29 (35) | 50 | 6.359 (3.045) | 15.57 (9.70–24.66) | −1.19 (.117) | −7.40 (.034) |
| Khakassians | 57 | .980 (.008) | 38 (67) | 52 | 6.683 (3.2) | 48.66 (28.63–83.84) | −1.39 (.788) | −24.45 |
| Altaians-Kizhi | 90 | .982 (.004) | 48 (53) | 59 | 6.364 (3.044) | 41.06 (26.99–62.46) | −1.47 (.063) | −25.11 |
| Teleuts | 53 | .980 (.007) | 33 (62) | 49 | 6.361 (3.063) | 36.38 (21.19–63.03) | −1.49 (.062) | −18.62 |
| Telenghits | 71 | .986 (.005) | 49 (69) | 76 | 7.002 (3.329) | 68.63 (42.24–113.09) | −1.87 | −25.02 |
| Chukchi | 15 | .781 (.102) | 7 (47) | 18 | 4.381 (2.292) | 4.50 (1.72–11.56) | −.85 (2.219) | .36 (.604) |
Sample size.
Sequence diversity.
Number of different haplotypes and percentage of sample size.
Number of segregating sites.
Average number of pairwise differences.
All P values are <.05 (for Tajima’s D) and <.02 for Fu’s FS), except where noted.
Table 4. .
Diversity Indexes and Neutrality Tests for the Studied Populations, Based on HVS2-Variability Data
| Population | na | Hb (SE) | K (%)c | Sd | Pie (SE) | θk (95% CI) | Tajima’s D (P)f | Fu’s FS (P)f |
| Persians | 85 | .951 (.014) | 47 (55) | 37 | 3.156 (1.650) | 42.44 (27.59–65.36) | −1.71 | −26.38 |
| Kurds | 26 | .972 (.021) | 20 (77) | 21 | 3.840 ( 1.994) | 37.88 (16.96–88.94) | −1.09 (.147) | −14.84 |
| Tajiks | 44 | .983 (.009) | 33 (75) | 30 | 3.680 (1.897) | 58.31 (31.10–112.88) | −1.64 | −26.00 |
| Koreans | 105 | .956 (.012) | 50 (48) | 41 | 2.775 (1.480) | 36.78 (24.80–54.37) | −1.98 | −26.66 |
| Mongolians | 50 | .951 (.016) | 27 (54) | 18 | 2.676 (1.449) | 23.19 (13.35–4.29) | −1.21 (.115) | −24.14 |
| Kalmyks | 109 | .959 (.009) | 50 (46) | 40 | 2.841 (1.509) | 35.17 (23.83–51.71) | −1.92 | −26.58 |
| Buryats | 296 | .924 (.010) | 70 (24) | 59 | 2.504 (1.354) | 28.64 (21.43–37.97) | −2.14 | −26.50 |
| Khamnigans | 99 | .941 (.016) | 42 (42) | 29 | 2.595 (1.401) | 27.02 (17.87–4.59) | −1.76 | −26.79 |
| Tuvinians | 104 | .935 (.010) | 28 (27) | 23 | 2.595 (1.401) | 12.24 (7.73–19.04) | −1.47 (.062) | −17.86 |
| East Evenks | 45 | .695 (.069) | 9 (20) | 9 | 1.618 ( .976) | 3.10 (1.45–6.33) | −1.01 (.164) | −2.18 (.149) |
| Telenghits | 76 | .917 (.021) | 33 (43) | 33 | 2.485 (1.356) | 21.64 (13.57–34.25) | −2.13 | −26.85 |
| Chukchi | 15 | .467 (.148) | 4 (27) | 6 | 1.657 (1.031) | 1.43 (.46–4.12) | −.36 (.384) | .83 (.675) |
Sample size.
Sequence diversity.
Number of different haplotypes and percentage of sample size.
Number of segregating sites.
Average number of pairwise differences.
All P values are <.05 (for Tajima’s D) and <.02 for Fu’s FS), except where noted.
As shown in tables 3 and 4, the studied populations exhibited similar neutrality test results for both HVS1 and HVS2 data. The southwestern Asian Persians, central Asian Tajiks, eastern Asian Koreans, and northern Asian populations of Kalmyks, Buryats, Khamnigans, and Telenghits yielded significantly negative values for both Tajima’s D and Fu’s FS neutrality tests. The Kurds, Tuvinians, West Evenks, Khakassians, Altaians-Kizhi, and Teleuts exhibited significantly negative Fu’s FS values and unimodal mismatch distributions (not shown), but the Tajima’s D statistic was not significantly different from 0. This contrasting pattern may be the result of mutation-rate heterogeneity along the HVS1 and HVS2 regions; this effect has been shown to confuse the signature of population expansion in Tajima’s test, leading to higher D values.55 For the East Evenks, Yakuts, Shors, and Chukchi, both neutrality tests gave nonsignificantly negative values (tables 3 and 4), and the mismatch distribution was unequivocally multimodal (data not shown), thus rejecting the hypothesis of population growth and pointing to the strong effects of drift and/or the small sample sizes for these groups.
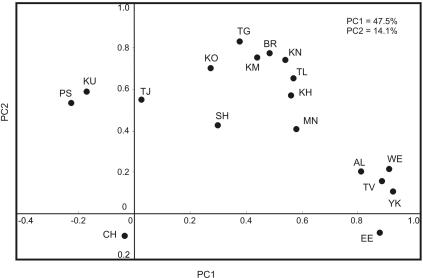
The basal mtDNA haplogroup frequencies of the 18 populations were used as input vectors to perform a PC analysis. Figure 2 shows the PC plot for the first two PCs, which account for 47.5% and 14.1% of the total variation, respectively. When the one outlier, the Chukchi, is ignored, geographic grouping of populations is apparent in the diagram. The first PC (PC1) reveals mainly a southwest-to-northeast cline by separating a group of Indo-European–speaking populations of southwestern and central Asia (Persians, Kurds, and Tajiks) from the Altaic-speaking populations of northern and eastern Asia. The latter, in turn, constitute two distinct clusters with Kalmyks, Mongolians, Buryats, Khamnigans, Shors, Telenghits, Teleuts, and Koreans in the upper right and Altaians-Kizhi, Tuvinians, Yakuts, and two groups of Evenks in the lower right parts of the plot. PC2 essentially displays the outlier genetic position of the Chukchi, who are clearly separated from the other populations studied.
Figure 2. .
PC plot based on haplogroup frequencies for the 18 population samples. Populations are coded as follows: PS = Persians, KU = Kurds, TJ = Tajiks, Ko = Koreans, MN = Mongolians, KM = Kalmyks, BR = Buryats, KN = Khamnigans, TV = Tuvinians, EE = East Evenks, WE = West Evenks, YK = Yakuts, SH = Shors, KH = Khakassians, AL = Altaians-Kizhi, TL = Teleuts, TG = Telenghits, and CH = Chukchi.
AMOVA confirmed that the southwest-to-northeast pattern seen in the PC analysis is highly significant. When the populations were split into three groups relative to their geographic location (Persians, Kurds, and Tajiks as the southwestern Asian/central Asian group; Chukchi as the northeastern Asian group; and the remaining populations as the northern Asian/eastern Asian group), the difference among the three groups was found to be 5.96% of the total variation (P<.0001). This difference was still significant (3.19%; P<.0001) when a major northern Asian/eastern Asian group was further subdivided into four other regional groups (Tuvinians, Shors, Khakassians, Altaians-Kizhi, Teleuts, and Telenghits as southern Siberians; Yakuts and western Evenks as central Siberians; Buryats, Khamnigans, Kalmyks, and eastern Evenks as eastern Siberians; and Mongolians and Koreans as eastern Asians). However, a significant difference was also found when populations were separated, according to language, into Indo-European– and Altaic-speaking groups (3.6%; P<.0001). Therefore, the division by linguistic and geographic affiliations is reflected in mtDNA variation of the population studied.
Phylogeography of Western Eurasian Haplogroups
The average frequency of mtDNAs from western Eurasia among northern Asian populations analyzed (with consideration of our previously published data11) is 16.3%. Meanwhile, in the most-western Altaian (Altaians-Kizhi, Teleuts, and Telenghits) and western Sayan (Shors and Khakassians) populations, their frequency is greater (>25%) than that in populations of eastern Sayan (Tuvinians, Todjins, and Tofalars) and, especially, Baikal (Buryats, Khamnigans, Sojots, and eastern Evenks) regions. Western Eurasian contribution to the northern Asian mitochondrial gene pool can be broadly divided into three different components. The first is represented by haplogroups H1a, H1b, H11a, U4, U5a, U5b, U8a, V, T1, and K, which are, in general, characteristic of eastern European populations.56–58 It is likely that these mtDNA lineages were carried to southern Siberia from the Volga-Ural region, where they occur at relatively high frequencies. Some of these maternal lineages are also present at high frequencies in western Siberia (for instance, haplogroups U4, H1b, T1, and U5a).8 The second component comprises haplogroups HV, U3, J1b2, X, H8, and H20, which are typical of populations of western Asia and the Caucasus region.52,53,59 The remaining component, found in southern Siberian populations, is represented by haplogroups H2a1, H6a, J1b1, N1a, and U2e, which are present at relatively low frequencies both in eastern Europe and western Asia. It should be noted, however, that haplogroups H6a and H2a1, as well as H8 and U3, may have been involved in the Late Upper Paleolithic population expansion in the southern Caucasus and the Near East.54,58,59 Therefore, one can suggest that populations from this area had an impact on the mitochondrial gene pool of southern Siberians. In summary, the composition of mtDNA lineages of western Eurasian origin revealed in southern Siberian populations allows us to suggest that there were at least two migrations into southern Siberia, one from eastern Europe and the other from western Asia/the Caucasus. Traces of both migrations associated with different mtDNA haplogroups were detected in all southern Siberian regional groups, with minor influence on the most northeastern of the eastern Sayan populations.
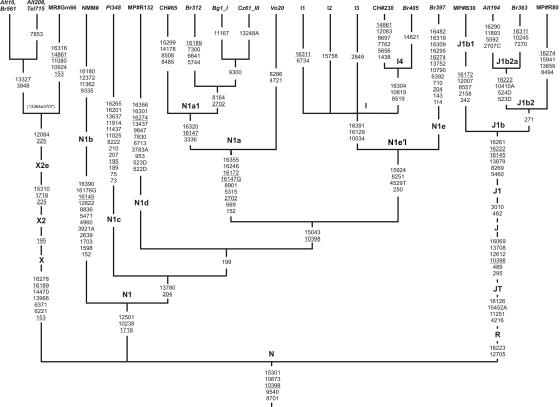
To further elucidate the origin of western Eurasian lineages found in mitochondrial gene pools of northern Asians, complete genome sequencing of particular mtDNAs was performed. Haplogroup X is found in both western Eurasians60 and some northern groups of Native Americans,61 but it is absent in northern Siberian and eastern Asian populations,4,5,61 which are genetically and geographically closest to Native Americans. Among Siberians, haplogroup X mtDNAs were detected in Altaians of southern Siberia7 and Evenks from central Siberia.48 All Siberian haplogroup X sequences, along with the majority of southern Caucasian X mtDNAs, belong to the X2e clade, whereas Native American X sequences constitute the distinctive X2a clade.48 On the basis of extensive phylogeographic analysis of the haplogroup X subclades, Reidla et al.48 suggested that the Near East was likely to be a geographical source for the spread of subhaplogroup X2, with the associated population dispersal that occurred around or after the last postglacial maximum (LGM). They also suggested that Altaians had relatively recently acquired haplogroup X2 (<6,700 years ago). In the present study, we have considerably extended the population screening of haplogroup X mtDNAs and have detected single X2e lineages in Buryats and Teleuts (in addition to Altaians-Kizhi7) from southern Siberia and in Kurds from northwestern Iran (see our txt file and table 2). To obtain further information about the extent of haplogroup X diversity, four mtDNAs (from two Altaians-Kizhi, one Teleut, and one Buryat) were completely sequenced and were compared with the only published X2e sequence from the southern Caucasus.48 A maximum-parsimony tree (fig. 3) reveals that southern Siberian X2e mtDNAs form a separate cluster defined by two coding-region mutations (3948 and 13327). In addition, two samples share one coding-region mutation at position 7853, although all four southern Siberian genomes have identical HVS1/HVS2 sequences. It should be considered, however, that a mutation at position 3948 was also identified in one Georgian individual of haplotype X (GEO0553), who was characterized by the −3944TaqI variant. At the same time, the southern Caucasian X2e subhaplogroup, represented by a single Georgian mtDNA in figure 3, differs from the root of haplogroup X2e by three coding- and two control-region transitions. Thus, taking into account the high level of divergence within the entire X2e branch, it is likely that X2e haplotypes could have been present in southern Siberia for 13,364±3,707 years.
Figure 3. .
The phylogenetic tree of complete mtDNA sequences of haplogroups N1, X2e, and J1b. The tree is rooted in haplogroup N. Mutations are scored relative to the rCRS.39 The information of the reported samples is presented in table 1. Seven additional complete sequences were taken from the literature,22,26,48,49 and particular sequences from these articles are referred to as CH, MP, MR, and NMM, respectively, followed by a number sign (#) and the original sample code. For subhaplogroups I1, I2, and I3, only diagnostic mutations are shown, according to the classification.20,26,28 For sample CH#230,22 only coding-region information is available. Mutations are shown on the branches and are transitions unless the base change is explicitly indicated. Deletions are indicated by a “D” after the deleted nucleotide position. Underlined nucleotide positions occur at least twice in the tree.
In southern Siberian populations, haplogroup N1 consists mainly of two related groups: N1a and I. Haplogroup I is present at low frequencies in different western Eurasian26 and central Asian35 populations, and it is also detected in populations of Altai-Sayan and Baikal regions of southern Siberia (at 0.5%–1.2%). Haplogroup N1a is rare but widespread in Eurasia and North Africa,62 but, in Siberia, it is observed only among Altaians (1.2%) and Buryats (0.2%). It is noteworthy that N1a haplotypes revealed in southern Siberia belong to the central Asian N1a subcluster, which comprises the mtDNA lineages found also in central Asia and in the South Ural and Volga regions.62 This subcluster probably originated in the central Asia/southern Siberia region, because it was found in a 2,500-year-old Scytho-Siberian burial in the Altai region.62,63 Meanwhile, N1a is very rare in modern populations of central Asia (in Turkmens, Karakalpaks, and Uzbeks), as well as in southwestern Asia (in Iranians and Indians).53,54,64
To reconstruct the phylogeny of haplogroup N1a, we sequenced mitochondrial genomes of four individuals from populations of northern Asia and Europe (fig. 3). The N1a tree shows two major phylogenetic branches. The most ancient eastern African/southern Asian branch contains mtDNA characterized by the 16147G variant revealed in Russian individual Vo20, whereas the European/central Asian branch (designated “N1a1”) is characterized by transitions at positions 3336, 16147 (giving the 16147A variant), and 16320. It is worth emphasizing that almost all N1a1 sequences analyzed (with an exception of sample CH#65 studied by Herrnstadt et al.22) belong to the subcluster defined by the transition at position 8164 and the back mutation at position 2702. This subcluster consists of the central Asian branch represented by the Buryat sample (Br312) and European samples (Russian Bg1_I and Czech Cz61_III, joined by mutation at position 9300) and shows a coalescence time of 11,993±4,533 years. This finding suggests that the dispersal of subhaplogroup N1a1 could be associated with interactions of European and southern Siberian/central Asian populations from Neolithic times.
It seems important that southwestern Asian populations (according to data reported elsewhere53,54) also share the other members of haplogroup N1–N1b (in Iran and Pakistan), N1c (in Iran), and N1d (in India and Pakistan), pointing to southwestern Asia as the source of lineage diversification within haplogroup N1. In this context, our finding of a previously unobserved ancestral node of haplogroup I phylogeny in the Baikal region is very intriguing. This lineage, named here as “N1e,” was revealed in the Buryat population. Complete genome sequencing has demonstrated that haplogroup N1e appears to be a sister branch of haplogroup I and has allowed us to identify four mutations (at positions 250, 4529, 8251, and 15924) representing an N1e′I trunk. Therefore, the only coding-region mutation at 10034 remains diagnostic for haplogroup I. Figure 3 also demonstrates the other examples of completely sequenced mtDNAs that allowed resolution of some questions regarding the N1-family evolution. Subgroup I4 coding- and noncoding-region markers are now available. Additionally, for the first time to our knowledge, we give here an example of the complete mitochondrial-genome sequence of haplogroup N1c (fig. 3).
In southern Siberian populations, haplogroup J is represented in all but one subject by subhaplogroup J1 haplotypes. Among them, subhaplogroup J1b, defined by mutations at positions 5460, 8269, 13879, 16145, 16222, and 16261, prevails. Subcluster J1b1, bearing a key mutation at HVS2 position 242, was revealed in different regions of south Siberia—in Altai (0.6%), western Sayan (4.2%), and eastern Sayan (1.7%). Meanwhile, subcluster J1b2, characterized by HVS2 mutation at position 271, is present almost solely in Altaians (2.8%). Until now, there has been only one completely sequenced J1b2 individual of Indian origin.26 Thus, the addition of two Siberian individuals (Altaian-Kizhi and Buryat) allowed us to find the J1b2-diagnostic mutation, which occurs only at position 271 (fig. 3). All studied southern Siberian individuals fall into a separate subcluster defined by transversion T→A at position 10410 and a back mutation at site 16222. To check the possibility of in situ origin of this mtDNA subcluster, the polymorphism at position 10410 was studied in Kurd and Persian individuals carrying HVS1/2 J1b2 haplotypes similar or identical to those revealed in southern Siberians. All of them were found to be characterized by the 10410A marker (txt file), indicating that J1b2 lineages reached southern Siberia, most probably from Iran, where the frequency of this subcluster seems to be relatively high.53,54
Phylogeography of Eastern Eurasian Haplogroups
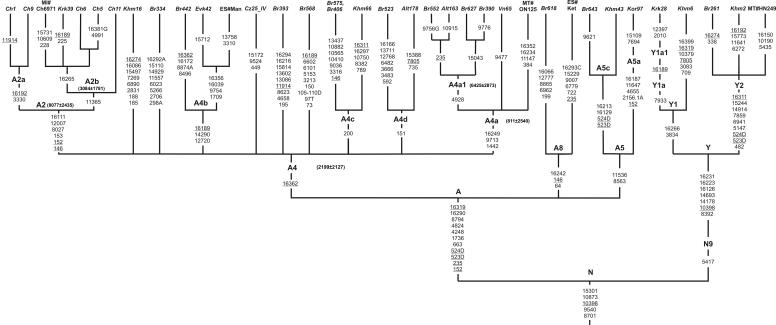
One of the most common haplogroups in northern and eastern Asia is haplogroup A, which falls into two main (A4 and A5) and several minor (A3, A6, and A7) subclades.32,37 The frequency of haplogroup A in eastern Asia is generally between 5% and 10%. Similarly, in central Asia, it accounts for <10% of the mtDNAs of eastern Asian origin.65 Importantly, only one subclade of A, A4, is present in northern Asia, where it is rare even though it was found in the majority of populations studied.5,11,14 Subhaplogroup A4 includes the A2 subcluster, which appears at the highest frequencies (>68%) in the northeastern Siberian populations of Chukchi and Eskimos.4,5 In contrast to A4, the other subclades of haplogroup A are found predominantly in Korea and Japan.27,37 It has been proposed recently37 that haplogroup A can display further region-specific subclades, but only a fraction of A4 can be assigned to subclades with use of the HVS1/HVS2 motifs of the four completely sequenced A4 (excluding the Native American A2) mtDNAs. To verify this possibility, we analyzed a large set of northern and eastern Asian haplogroup A lineages, using complete mtDNA sequencing.
A tree of 31 complete haplogroup A mtDNA sequences is illustrated in figure 4, which also incorporates information from four mtDNA genomes published elsewhere.14,19,27 The phylogenetic analysis confirmed a large number of independent basal branches, some giving rise to subclades that have several basal subbranches themselves. Among these subclades, representatives of three (A2, A4, and A5) of six previously proposed subhaplogroups (A2–A737) were present. In our sample, we detected three A5 mtDNAs characterized by two substitutions (8563 and 11536). One of these lineages (Kor97) belongs to the A5a subgroup, with two additional mutations (at positions 7694 and 15109). Two remaining A5 lineages (Br643 and Khm43) have been assigned to the A5c subhaplogroup that is defined by 16129 and 16213 substitutions. It should be noted that transition at position 12816, indicated as “A5c specific,”32,37 is absent in both of our Siberian A5c samples, thus suggesting that the 12816 mutation may be population specific. We have defined one novel subgroup, designated “A8,” by three control-region mutations (64, 146, and 16242) shared with the Ket33 sequence.14 We have also defined several subhaplogroups within the A4 branch. The following novel subhaplogroup names were assigned to these clades: A4a (1442-9713-16249), A4b (12720-14290-16189), A4c (200), and A4d (151). Among these newly defined subgroups, A4a appeared to be the most frequent in our sample, encompassing four Siberian and one Russian mtDNAs, as well as one Japanese sample.27 Interestingly, all four A4a Siberian samples share mutation at position 4928, thus allowing us to define the Siberian-specific A4a1 branch with coalescence age of 6,425±2,873 years. The A2 subgroup is represented in the tree by six Chukchi and one Koryak, all sharing six mutations (146, 152, 153, 8027, 12007, and 16111) in its basal branch. Since two Chukchi samples (Ch1 and Ch9) harbor the 16192 mutation, they have been categorized as an A2a subgroup, whereas three Chukchi (Ch5 and Ch6 from the present study and Ch6971 from the study of Ingman et al.19) and one Koryak (Krk39) who shares the 16265 mutation have been categorized as an A2b subgroup, as proposed by Helgason et al.66 Our phylogenetic analysis reveals that both these subgroups can be defined now by coding-region mutations−A2a has one coding-region substitution at position 3330 in addition to 16192, whereas A2b has a mutation at position 11365, distinctive of this subgroup, in addition to 16265. The coalescence time of the A2b clade is 3,084±1,781 years ago, which is very close to the value obtained from HVS1 data (3,000±1,400 years).47 The coalescence time of the entire subhaplogroup A2 is 8,077±2,435 years ago, but it should be noted that this value corresponds only to the western Beringian component of A2.
Figure 4. .
Phylogenetic tree of complete mtDNA sequences of haplogroups A and Y. The tree is rooted in haplogroup N. The tree includes 36 mtDNAs, of which 31 are novel and 5 were reported elsewhere.14,19,27 Insertions are indicated by a dot followed by the number and type of inserted nucleotide(s). For additional information, see the figure 3 legend.
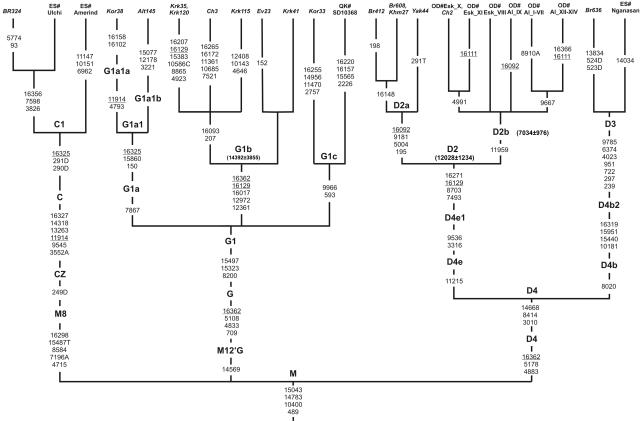
The other Beringian subhaplogroup D2 was previously considered to have a limited distribution, restricted to Na-Dene, Aleuts, Chukchi, and Eskimos.9,14 Geographic specificity and diversity of D2 lineages are thought to support the refugial hypothesis that assumes that the founding population of Eskimo-Aleut originated in Beringian/southwestern Alaskan refugia during the early postglacial period.9,14 Extensive screening of mtDNA variability in northern Asian populations demonstrates that, besides Chukchi, where D2 mtDNA lineages account for 13%, subhaplogroup D2 occurs in populations of southern Siberia with frequencies ranging from 0.7% in Buryats to 2.9% in Tuvinians (table 2). Moreover, our results of complete mtDNA sequencing have significantly changed the topology of D2 and have showed that this subhaplogroup constitutes two different clusters with contrasting geographic distribution (fig. 5). The first cluster, designated here as “D2a,” is represented by two Buryat, one Khamnigan, and one Yakut mtDNAs, all sharing four mutations (195, 5004, 9181, and 16092) in its basal branch. The second cluster, which we designate here as “D2b,” includes at least three subclades, with various mtDNA lineages revealed in Aleuts, Siberian Eskimo, and Chukchi. D2b is distinguished by mutation at position 11959. Two mutations in the coding region (7493 and 8703) and two mutations in the control region (16129 and 16271) make up the root of the entire D2 clade. The coalescent time of the entire D2 haplogroup is estimated as 12,028±1,234 years ago, whereas the age of its D2b subclade dates to 7,034±976 years. Hence, our finding of the southern Siberian–specific D2a subcluster testifies to a southern Siberian rather than Beringian origin of haplogroup D2 lineages.
Figure 5. .
Phylogenetic tree of complete mtDNA sequences of haplogroups C1, G1, and D4. The tree is rooted in haplogroup M. For subhaplogroup D2b, only diagnostic mutations for specific lineages are indicated, according to data reported elsewhere.9 D2 and D2b coalescence-time estimates were obtained with consideration of the whole diversity within D2b subclusters. For additional information, see the figure 3 legend.
We have also completely sequenced one C1 genome that represents the haplogroup C1 characteristic of Native Americans and that rarely occurs in some Asian populations.67 The sequence of Buryat (Br324), together with the only published C1 genome of Ulchi from the Lower Amur region,14 forms a separate cluster, defined by transitions at positions 3826 and 7598 in the coding region and 16356 in the control region (fig. 5). It should be noted that the Buryat C1 genome displays two additional mutations (93 and 5774), pointing to a certain divergence within the Asian-specific branch of C1.
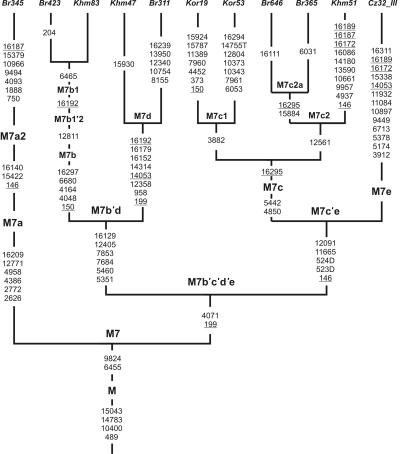
Haplogroup M7, which is characteristic of eastern Asian populations, has not been found in northeastern Asia.4,5,38,68 It is also very rare in central Asians.54,65 This haplogroup has been detected in Island Southeast Asia, China, Vietnam, Korea, and Japan, as well as among Mongols, the western Siberian Mansi, and the southern Siberian Buryats, Todjins, Khamnigans, and Telenghits.8,11,27,50,51,69 M7a, M7b2, and M7c1b are found almost exclusively in Korea and Japan, and M7b1 is characteristic of Chinese populations, whereas M7c1c is specific to Island Southeast Asia.50,69 In the current study, we revised the classification of haplogroup M7 that was defined elsewhere32,50,70 as having two major branches: M7a, characterized by five coding-region (2626, 2772, 4386, 4958, and 12771) and one control-region (16209) transitions, and M7b′c, characterized by 199 and 4071 transitions. In our survey of almost 1,500 samples across southwestern, central, eastern, and northern Asia, we found a specific mtDNA lineage that seemed to be prevalent in southern Siberians. One Khamnigan, one Telenghit, and three Buryat samples harbor an HVS1 motif (16129-16152-16179-16192-16223-16362) that was not present in other studied populations and published data sets. Information from complete mtDNA sequencing reveals that, besides M7 haplogroup-specific mutations, samples Khm47 and Br311 share mutations at sites 958, 12358, 14053, and 14314 and therefore may represent a new subgroup within the M7 haplogroup. We designate this lineage “M7d” (fig. 6). Our phylogenetic tree points to 5351, 5460, 7684, 7853, 12405, and 16129 as the basal mutations for the M7b′d clade, whereas, 150, 4048, 4164, 6680, and 16297 are common only to the M7b subgroup. The M7a classification remains as proposed elsewhere,50 whereas mutations at positions 199 and 4071 now define the large clade M7b′c′d′e. Haplogroup M7c is redefined here as requiring only two coding-region mutations (4850 and 5442). Another three transitions (at positions 146, 11665, and 12091) may characterize the enlarged M7c′e branch, which includes also the highly divergent mtDNA found in one Czech individual and is here designated “M7e.” Unfortunately, the lack of other M7e representatives in our and published data sets does not allow us to describe the M7e-specific coding-region motif. It is noteworthy that, within M7c2, we identified a new Buryat-specific subcluster, M7c2a, defined by transition at position 15884 and a back mutation at position 16295.
Figure 6. .
Phylogenetic tree of complete mtDNA sequences of haplogroup M7. The tree is rooted in haplogroup M. For additional information, see the figure 3 legend.
Haplogroup Y, distributed predominantly in northeastern Asia,5,68,71 where it is found in Itelmens (4.3%), Koryaks (9.7%), Evenks (10.8%), Nivkhs (64.9%), and Ainu (22%), is much less frequent in southern Siberia, where it is found in Buryats, Sojots, Tuvinians, Todjins, Khamnigans, and Evenks with frequencies ranging from 1.4% to 8.9%.11 It has been suggested that haplogroup Y has evolved in the lower Amur River region of southeastern Siberia, where it was found with considerable frequencies in Ulchi (37.9%), Negidals (21.2%), and Udegeys (8.7%).14 The occurrence of haplogroup Y mtDNAs in Japanese and Koreans was explained by gene flow from the Ainu or other Siberian groups who have these mtDNAs.5 Interestingly, the overwhelming majority of Y mtDNAs revealed so far belong to the Y1 subhaplogroup defined by two transitions—at positions 3884 and 16266. The Y2 branch—distinguished by six coding (482, 5147, 6941, 7859, 14914, and 15244) and one HVS1 (16311) mutations—is much less frequent, found in Japanese (0.3%),27 Koreans (0.3%),72 Taiwanese aboriginals (1.4%),30 Buryats (0.3%), and Khamnigans (2%). With relatively high frequencies (1.2%–12.9%), haplogroup Y2 has been recently found in Island Southeast Asia, where it is thought to be linked with the mid-Holocene “Out of Taiwan” dispersal.69 To assess the nature and extent of subhaplogroup Y2 variation, we have sequenced two southern Siberian complete mtDNAs and compared them with the only published Y2 mtDNA from Japan.27 As shown in figure 4, mtDNAs Br261, Khm2, and HN24927 harbored a string of mutations specific for haplogroup Y2 and further characterized by a unique combination of mutations distinctive of each branch. The diversity of Y2 is apparently high, since all three lineages are different. The age of Y2 is dated to 8,567±3,831 years, thereby suggesting the Neolithic expansion of these lineages in eastern Asia and southern Siberia.
With our newly collected sequences, it was also possible to gain more insight into the phylogeny of haplogroup G, which was considerably refined recently32 but is still poorly understood in terms of its subgroup variation. Thus, haplogroup G1b was previously characterized by the HVS1 mutations only, but now we have defined this haplogroup by two coding-region mutations (12361 and 12972) accompanying the transitions at 16017 and 16129. The subhaplogroup G1b dates to 14,392±3,855 years ago, which provides further evidence of post-LGM expansion within northeastern Asia. Inside haplogroup G1, a new subclade named “G1c,” with characteristic mutations at positions 593 and 9966, has been identified on the basis of the sharing of mutations between our data and the data of Kong et al.24 (fig. 5).
Discussion
Our analysis of 1,432 mtDNA sequences from 18 southwestern, central, eastern, and northern Asian populations shows that the highest variation is observed in populations located both in the southwestern Asia and the Altai-Sayan region of southern Siberia, thus highlighting these regions as places where western Eurasian lineages interacted with eastern Eurasian genetic components. The coexistence of different genetic lineages in these areas may have resulted from various migrations from diverse geographical sources at different times, beginning with the early human settlements in the Paleolithic era. In addition, the southern Siberian region is characterized by the traces of recent migration events, such as the northward expansion into subarctic and arctic regions that occurred after the LGM. In this respect, the phylogeographic structure of the D2 subhaplogroup observed so far in the circumarctic populations appears to be only more complicated. Here, we demonstrate that a separate subclade of D2 is present in Buryats, Khamnigans, and Yakuts and consequently points to the southern Siberian rather than Beringian origin of haplogroup D2 lineages. The possible split between southern Siberian D2a and Beringian D2b mtDNA clades may have been ∼12,000 years ago, whereas the Beringian-specific D2b branch, with a coalescent age of ∼7,000 years, appears to be the consequence of population expansion that occurred exclusively in Beringia. It seems that the same expansion was responsible for generating diversity within another Beringian-specific haplogroup, A2. Our results confirm the subdivision of A2 into A2a and A2b clades, which are defined here by coding-region mutations.
The high-resolution analysis of selected complete mtDNAs in Buryats reveals the presence of subhaplogroup C1, which is phylogenetically most related to Native American C1. This conclusion confirms that Native American C1 really has its Asian counterparts and points to the presence of this subhaplogroup in the mtDNA pool of the Siberian progenitors of Paleoindians. In contrast to the haplogroup C1 mtDNAs, haplogroup D1, which is considered to be one of the founder haplogroups for the Americas, is not found in our data set. Although we identified the single D4 lineage with a mutation at 16325, which is diagnostic for Native American haplogroup D1, in Telenghits from the South Altai region, the lack of the second D1-specific mutation at 2092 does not permit us to assign this lineage to D1. This observation is concordant with the previous conclusion67 about the absence of haplogroup D1 mtDNAs in Asia.
As mentioned above, the significant influx of western Eurasian mtDNA lineages was revealed in southern Siberian populations. In the present study, new western Eurasian subhaplogroup N1e, which is most closely related to haplogroup I, was identified in Buryats. The phylogeny of another four western Eurasian haplogroups found in southern Siberia (I4, J1b2, N1a, and X2e) was reconstructed with the use of complete-genome data. Western Eurasian haplogroups found in gene pools of southern Siberians demonstrate an obvious link between populations of Siberia and those of western Asia, the Caucasus, and eastern Europe. It is noteworthy, however, that complete genome–based coalescence times for haplogroups X2e, J1b2, and N1a suggest that their post-LGM flows from the west. Similarly, the possible dispersal from eastern Asia to southern Siberia has been dated on the basis of haplogroups M7 and Y mtDNA phylogenies. Late Upper Paleolithic and/or early Neolithic dispersals may explain the distribution of haplogroups M7c2, M7d, and Y2, which are almost entirely restricted to Baikal-region populations and are dated to ∼14,000, ∼13,000, and ∼9,000 years ago, respectively.
In addition, a detailed analysis of northern Asian mtDNA phylogeography has confirmed the lack of lineages ancestral to major Eurasian haplogroups M, N, and R in southern Siberia, thereby providing supplementary evidence to rule out the existence of a northern Asian route. Further complete genome–based studies will be extremely informative for revealing spatial patterns attributable not only to primary colonization events and late-glacial expansions but also to more-recent events of gene flow.
Supplementary Material
Acknowledgments
We are grateful to all the voluntary donors of DNA samples used in this study, to Tsendsuren Tsedev for Mongolian samples, to Mohamad Reza Mohamad Abadi for Persian samples, and to Jalal Rostamzadeh for Kurd samples. We thank Jaroslaw Bednarek, Jakub Czarny, Ewa Lewandowska, and Aneta Jakubowska for their technical assistance. We also thank Walther Parson for quality checking of some samples by independent sequencing of HVS1/HVS2. This study was supported by Russian Foundation for Basic Research grants 04-04-48746, 07-04-00445 (to M.D.), 05-04-97226 (to I.D.), and 06-04-48136 (to B.M.), by Polish State Committee for Scientific Research grant 3P04C 04823 (to T.G.), by Far-East Branch of the Russian Academy of Sciences grants 06-III-A-06-175 (to M.D.) and 06-I-Π11-032 (to B.M.), and by Program of Basic Research of Russian Academy of Sciences “Biodiversity and Dynamics of Gene Pools” (to B.M.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Arlequin, http://anthropologie.unige.ch/arlequin/ (for software v. 3.01)
- Free Phylogenetic Network Software, http://www.fluxus-engineering.com/sharenet.htm (for the Network 4.1.0.9 software package)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for 71 complete mtDNA sequences [accession numbers EF153771–EF153833, EF397558–EF397562, and EF486517–EF486519])
References
- 1.Vasiliev SA (1993) The Upper Paleolithic of northern Asia. Curr Anthropol 34:82–92 10.1086/204141 [DOI] [Google Scholar]
- 2.Vasil’ev SA, Kuzmin YV, Orlova LA, Dementiev VN (2002) Radiocarbon-based chronology of the Paleolithic in Siberia and its relevance to the peopling of the New World. Radiocarbon 44:503–530 [Google Scholar]
- 3.Goebel T (1999) Pleistocene human colonization of Siberia and peopling of the Americas: an ecological approach. Evol Anthropol 8:208–227 [DOI] [Google Scholar]
- 4.Starikovskaya YB, Sukernik RI, Schurr TG, Kogelnik AM, Wallace DC (1998) mtDNA diversity in Chukchi and Siberian Eskimos: implications for the genetic history of Ancient Beringia and the peopling of the New World. Am J Hum Genet 63:1473–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC (1999) Mitochondrial DNA variation in Koryaks and Itel’men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am J Phys Anthropol 108:1–39 [DOI] [PubMed] [Google Scholar]
- 6.Derenko MV, Malyarchuk BA, Dambueva IK, Shaikhaev GO, Dorzhu CM, Nimaev DD, Zakharov IA (2000) Mitochondrial DNA variation in two South Siberian Aboriginal populations: implications for the genetic history of North Asia. Hum Biol 72:945–973 [PubMed] [Google Scholar]
- 7.Derenko MV, Grzybowski T, Malyarchuk BA, Czarny J, Miścicka-Śliwka D, Zakharov IA (2001) The presence of mitochondrial haplogroup X in Altaians from South Siberia. Am J Hum Genet 69:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derbeneva OA, Starikovskaya EB, Wallace DC, Sukernik RI (2002) Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am J Hum Genet 70:1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derbeneva OA, Sukernik RI, Volodko NV, Hosseini SH, Lott MT, Wallace DC (2002) Analysis of mitochondrial DNA diversity in the Aleuts of the Commander islands and its implications for the genetic history of Beringia. Am J Hum Genet 71:415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF (2002) High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol 74:761–789 10.1353/hub.2003.0006 [DOI] [PubMed] [Google Scholar]
- 11.Derenko MV, Grzybowski T, Malyarchuk BA, Dambueva IK, Denisova GA, Czarny J, Dorzhu CM, Kakpakov VT, Miścicka-Śliwka D, Wozniak M, et al (2003) Diversity of mitochondrial DNA lineages in South Siberia. Ann Hum Genet 67:391–411 10.1046/j.1469-1809.2003.00035.x [DOI] [PubMed] [Google Scholar]
- 12.Fedorova SA, Bermisheva MA, Villems R, Maksimova NR, Khusnutdinova EK (2003) [Analysis of mitochondrial DNA haplotypes in Yakut population.] Mol Biol (Mosk) 37:643–653 [PubMed] [Google Scholar]
- 13.Pakendorf B, Wiebe V, Tarskaia LA, Spitsyn VA, Soodyall H, Rodewald A, Stoneking M (2003) Mitochondrial DNA evidence for admixed origins of central Siberian populations. Am J Phys Anthropol 120:211–224 10.1002/ajpa.10145 [DOI] [PubMed] [Google Scholar]
- 14.Starikovskaya EB, Sukernik RI, Derbeneva OA, Volodko NV, Ruiz-Pesini E, Torroni A, Brown MD, Lott MT, Hosseini SH, Huoponen K, et al (2005) Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann Hum Genet 69:67–89 10.1046/j.1529-8817.2003.00127.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derenko M, Malyarchuk B, Denisova GA, Wozniak M, Dambueva I, Dorzhu C, Luzina F, Miścicka-Śliwka D, Zakharov I (2006) Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum Genet 118:591–604 10.1007/s00439-005-0076-y [DOI] [PubMed] [Google Scholar]
- 16.Sukernik RI, Shur TG, Starikovskaia EB, Uolles DK (1996) [Mitochondrial DNA variation in native inhabitants of Siberia with reconstructions of the evolutional history of the American Indians. Restriction polymorphism.] Genetika 32:432–439 [PubMed] [Google Scholar]
- 17.Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, et al (1999) Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zegura SL, Karafet TM, Zhivotovsky LA, Hammer MF (2004) High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol Biol Evol 21:164–175 10.1093/molbev/msh009 [DOI] [PubMed] [Google Scholar]
- 19.Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 10.1038/35047064 [DOI] [PubMed] [Google Scholar]
- 20.Finnila S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, et al (2001) Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69:1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, et al (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingman M, Gyllensten U (2003) Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines. Genome Res 13:1600–1606 10.1101/gr.686603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong Q-P, Yao Y-G, Sun C, Bandelt H-J, Zhu C-L, Zhang Y-P (2003) Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet 73:671–676 (erratum 75:157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA 100:171–176 10.1073/pnas.0136972100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palanichamy Mg, Sun C, Agrawal S, Bandelt H-J, Kong Q-P, Khan F, Wang C-Y, Chaudhuri TK, Palla V, Zhang Y-P (2004) Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet 75:966–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka M, Cabrera VM, González AM, Larruga JM, Takeyasu T, Fuku N, Guo L-J, Hirose R, Fujita Y, Kurata M, et al (2004) Mitochondrial genome variation in Eastern Asia and the peopling of Japan. Genome Res 14:1832–1850 10.1101/gr.2286304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandelt HJ, Achilli A, Kong QP, Salas A, Lutz-Bonengel S, Sun C, Zhang YP, Torroni A, Yao YG (2005) Low “penetrance” of phylogenetic knowledge in mitochondrial disease studies. Biochem Biophys Res Commun 333:122–130 10.1016/j.bbrc.2005.04.055 [DOI] [PubMed] [Google Scholar]
- 29.Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, Blackburn J, Semino O, Scozzari R, Cruciani F, et al (2005) Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308:1034–1036 10.1126/science.1109792 [DOI] [PubMed] [Google Scholar]
- 30.Trejaut JA, Kivisild T, Loo JH, Lee CL, He CL, Hsu CJ, Li ZY, Lin M (2005) Traces of archaic mitochondrial lineages persist in Austronesian speaking Formosan populations. PLoS Biol 3:e247 10.1371/journal.pbio.0030247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, Passarino G, Underhill PA, Scharfe C, Torroni A, et al (2006) The role of selection in the evolution of human mitochondrial genomes. Genetics 172:373–387 10.1534/genetics.105.043901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF, et al (2006) Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet 15:2076–2086 10.1093/hmg/ddl130 [DOI] [PubMed] [Google Scholar]
- 33.Sun C, Kong QP, Palanichamy MG, Agrawal S, Bandelt HJ, Yao YG, Khan F, Zhu CL, Chaudhuri TK, Zhang YP (2006) The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol Biol Evol 23:683–690 10.1093/molbev/msj078 [DOI] [PubMed] [Google Scholar]
- 34.Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ (2006) Harvesting the fruit of the human mtDNA tree. Trends Genet 22:339–345 10.1016/j.tig.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 35.Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 10.1038/70550 [DOI] [PubMed] [Google Scholar]
- 36.Lahr M, Foley R (1994) Multiple dispersals and modern human origins. Evol Anthropol 3:48–60 10.1002/evan.1360030206 [DOI] [Google Scholar]
- 37.Metspalu M, Kivisild T, Bandelt HJ, Richards M, Villems R (2006) The pioneer settlement of modern humans in Asia. In: Bandelt HJ, Macaulay V, Richards M (eds) Human mitochondrial DNA and the evolution of Homo sapiens. Springer-Verlag, Berlin, pp 181–199 [Google Scholar]
- 38.Torroni A, Sukernik RI, Schurr TG, Starikorskaya YB, Cabell MF, Crawford MH, Comuzzie AG, Wallace DC (1993) mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet 53:591–608 [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews RM, Kubacka I, Chinnery PF, Lightowlers R, Turnbull D, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- 40.Achilli A, Rengo C, Magri C, Battaglia V, Olivieri A, Scozzari R, Cruciani F, Zeviani M, Briem E, Carelli V, et al (2004) The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet 75:910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider S, Roessli D, Excoffier L (2000) Arlequin version 2.0: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- 45.Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- 46.Forster P, Harding R, Torroni A, Bandelt HJ (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- 47.Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby SS (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reidla M, Kivisild T, Metspalu E, Kaldma K, Tambets K, Tolk H-V, Parik J, Loogvali E-L, Derenko M, Malyarchuk B, et al (2003) Origin and diffusion of mtDNA haplogroup X. Am J Hum Genet 73:1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genet 2:13 10.1186/1471-2156-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R (2002) The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 19:1737–1751 [DOI] [PubMed] [Google Scholar]
- 51.Yao Y-G, Kong Q-P, Bandelt H-J, Kivisild T, Zhang Y-P (2002) Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 70:635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong QP, Yao YG, Liu M, Shen SP, Chen C, Zhu CL, Palanichamy MG, Zhang YP (2003) Mitochondrial DNA sequence polymorphisms of five ethnic populations from northern China. Hum Genet 113:391–405 10.1007/s00439-003-1004-7 [DOI] [PubMed] [Google Scholar]
- 53.Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, et al (2004) Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74:827–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, Serk P, Karmin M, Behar DM, Gilbert MT, et al (2004) Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet 5:26 10.1186/1471-2156-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aris-Brosou S, Excoffier L (1996) The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol Biol Evol 13:494–504 [DOI] [PubMed] [Google Scholar]
- 56.Bermisheva M, Tambets K, Villems R, Khusnutdinova E (2002) [Diversity of mitochondrial DNA haplotypes in ethnic populations of the Volga-Ural region of Russia.] Mol Biol (Mosk) 36:990–1001 [PubMed] [Google Scholar]
- 57.Malyarchuk BA, Grzybowski T, Derenko MV, Czarny J, Wozniak M, Miscicka-Sliwka D (2002) Mitochondrial DNA variability in Poles and Russians. Ann Hum Genet 66:261–283 10.1046/j.1469-1809.2002.00116.x [DOI] [PubMed] [Google Scholar]
- 58.Loogvali EL, Roostalu U, Malyarchuk BA, Derenko MV, Kivisild T, Metspalu E, Tambets K, Reidla M, Tolk HV, Parik J, et al (2004) Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol Biol Evol 21:2012–2021 10.1093/molbev/msh209 [DOI] [PubMed] [Google Scholar]
- 59.Roostalu U, Kutuev I, Loogvali EL, Metspalu E, Tambets K, Reidla M, Khusnutdinova EK, Usanga E, Kivisild T, Villems R (2007) Origin and expansion of haplogroup H, the dominant human mitochondrial DNA lineage in West Eurasia: the Near Eastern and Caucasian perspective. Mol Biol Evol 24:436–448 10.1093/molbev/msl173 [DOI] [PubMed] [Google Scholar]
- 60.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- 61.Brown MD, Hosseini SH, Torroni A, Bandelt H-J, Allen JC, Schurr TG, Scozzari R, Cruciani F, Wallace DC (1998) mtDNA haplogroup X: an ancient link between Europe/Western Asia and North America? Am J Hum Genet 63:1852–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tanzer M, Villems R, Renfrew C, Gronenborn D, Alt KW, et al (2005) Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science 310:1016–1018 [DOI] [PubMed] [Google Scholar]
- 63.Ricaut FX, Keyser-Tracqui C, Bourgeois J, Crubezy E, Ludes B (2004) Genetic analysis of a Scytho-Siberian skeleton and its implications for ancient Central Asian migrations. Hum Biol 76:109–125 10.1353/hub.2004.0025 [DOI] [PubMed] [Google Scholar]
- 64.Chaix R, Quintana-Murci L, Hegay T, Hammer MF, Mobasher Z, Austerlitz F, Heyer E (2007) From social to genetic structures in central Asia. Curr Biol 17:43–48 10.1016/j.cub.2006.10.058 [DOI] [PubMed] [Google Scholar]
- 65.Comas D, Plaza S, Wells RS, Yuldaseva N, Lao O, Calafell F, Bertranpetit J (2004) Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages. Eur J Hum Genet 12:495–504 10.1038/sj.ejhg.5201160 [DOI] [PubMed] [Google Scholar]
- 66.Helgason A, Palsson G, Pedersen HS, Angulalik E, Gunnarsdottir ED, Yngvadottir B, Stefansson K (2006) mtDNA variation in Inuit populations of Greenland and Canada: migration history and population structure. Am J Phys Anthropol 130:123–134 10.1002/ajpa.20313 [DOI] [PubMed] [Google Scholar]
- 67.Bandelt HJ, Herrnstadt C, Yao YG, Kong QP, Kivisild T, Rengo C, Scozzari R, Richards M, Villems R, Macaulay V, et al (2003) Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet 67:512–524 10.1046/j.1469-1809.2003.00049.x [DOI] [PubMed] [Google Scholar]
- 68.Derenko MV, Shields GF (1998) [Diversity of mitochondrial DNA nucleotide sequences in three groups of aboriginal inhabitants of Northern Asia.] Mol Biol (Mosk) 31:784–789 [PubMed] [Google Scholar]
- 69.Hill C, Soares P, Mormina M, Macaulay V, Clarke D, Blumbach PB, Vizuete-Forster M, Forster P, Bulbeck D, Oppenheimer S, et al (2007) A mitochondrial stratigraphy for Island Southeast Asia. Am J Hum Genet 80:29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, et al LB (2001) Genetic evidence on the origins of Indian caste populations. Genome Res 11:994–1004 10.1101/gr.GR-1733RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horai S, Murayama K, Hayasaka K, Matsubayashi S, Hattori Y, Fucharoen G, Harihara S, Park KS, Omoto K, Pan IH (1996) mtDNA polymorphism in East Asian Populations, with special reference to the peopling of Japan. Am J Hum Genet 59:579–590 [PMC free article] [PubMed] [Google Scholar]
- 72.Lee HY, Yoo JE, Park MJ, Chung U, Shin KJ (2006) Mitochondrial DNA control region sequences in Koreans: identification of useful variable sites and phylogenetic analysis for mtDNA data quality control. Int J Legal Med 120:5–14 10.1007/s00414-005-0005-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.