Abstract
The molecular mechanisms that coordinate cell morphogenesis with the cell cycle remain largely unknown. We have investigated this process in fission yeast where changes in polarized cell growth are coupled with cell cycle progression. The orb6 gene is required during interphase to maintain cell polarity and encodes a serine/threonine protein kinase, belonging to the myotonic dystrophy kinase/cot1/warts family. A decrease in Orb6 protein levels leads to loss of polarized cell shape and to mitotic advance, whereas an increase in Orb6 levels maintains polarized growth and delays mitosis by affecting the p34cdc2 mitotic kinase. Thus the Orb6 protein kinase coordinates maintenance of cell polarity during interphase with the onset of mitosis. orb6 interacts genetically with orb2, which encodes the Pak1/Shk1 protein kinase, a component of the Ras1 and Cdc42-dependent signaling pathway. Our results suggest that Orb6 may act downstream of Pak1/Shk1, forming part of a pathway coordinating cell morphogenesis with progression through the cell cycle.
Eukaryotic cells establish and maintain polarized cellular domains that are essential for intracellular transport, cell differentiation, cell shape, and cell locomotion (1). Changes in cell shape and cell polarity occur during the cell cycle and in response to exogenous signals, but little is known about how cell morphogenesis is regulated or how polarized cell growth is integrated with the cell cycle. Fission yeast Schizosaccharomyces pombe cells grow in a polarized fashion that undergoes distinct changes during the cell cycle. Cells grow from one tip during G1 and S-phase, and activate the second tip during early G2, becoming bipolar in growth (2). Actin dots are located at one end of the cell when cells are growing with one tip and are found at both ends when cells grow in a bipolar manner (3). At the onset of mitosis, polarized cell growth ceases, and actin relocates from the cell tips to the middle of the cell where an actin ring forms. Both activation of bipolar growth and onset of mitosis require the attainment of a minimal cell size (2, 4).
In fission yeast, cell polarity and morphology is thought to be regulated by the product of the ras homologue, ras1, acting upstream of the Cdc42 GTPase (5, 6). Ras1 and Cdc42 are part of a complex including Scd1, the putative Cdc42 guanine nucleotide exchange factor, and Scd2, a Src homology 3 (SH3) domain-containing protein (6). Cdc42 also has been shown to be associated with the serine/threonine kinase Pak1/Shk1, a homologue of mammalian PAKs kinases and the budding yeast Ste20 protein kinase (7). In fission yeast, pak1/shk1 has been shown to be essential for cell viability and to participate in the Ras1-dependent mating response pathway (8, 9). Although overexpression studies suggest a role for the Pak1/Shk1 kinase in the control of cell morphology (9), the molecular details of Pak1/Shk1-dependent regulation of the cytoskeleton have yet to be fully clarified.
We previously identified 19 genes that are required for various aspects of cell morphogenesis (10). The orb6 gene is required to maintain cell polarity throughout the interphase period of the cell cycle. Mutants in orb6 lose growth polarity and become spherical with disorganized microtubule arrays and delocalized actin dots (10). In this paper, we report the cloning of Orb6 and its identification as a fission yeast kinase closely related to a number of higher eukaryotic kinases, including Drosophila warts kinase, mammalian Rho-associated kinase, and the human myotonic dystrophy kinase (DMPK). We show that Orb6 is required for maintenance of cell polarity during interphase and to promote actin reorganization during morphological transitions. Moreover, we demonstrate that Orb6 also has another role during the cell cycle, specifically to delay onset of mitosis, suggesting that it functions to coordinate cell morphogenesis with the cell cycle. Finally, we show that orb6 interacts genetically with Pak1/Shk1 protein kinase and that pak1/shk1 is required for proper Orb6 intracellular localization. We propose that the Orb6 kinase acts downstream of a morphogenetic control pathway involving Cdc42 and Pak1/Shk1, which maintains the cell in a polarized state during interphase while simultaneously delaying the onset of mitosis.
MATERIALS AND METHODS
Strains and Growth Conditions.
The strains used in this paper were orb6–25 ade6-M210 leu1–32h−; orb2–34 ade6-M210 leu1–32h−; wee1–50 leu1–32h−; and leu1–32h−. Cells were cultivated in rich medium (yeast extract liquid or agar), or Edinburgh minimal medium liquid with the appropriate supplements (11), as indicated. Cells in liquid cultures had been growing exponentially for at least eight generations, at densities below 107 cells/ml, before observation.
Libraries and Transformation Procedures.
The libraries used for cloning orb6 and orb2 were a cDNA library cloned in the plasmid Rep3X (29) and a genomic library in plasmid pDB248′ (30). Cells were transformed by the lithium acetate procedure (11).
Cloning of orb6+.
orb6 mutants were transformed with a cDNA library controlled by the thiamine-repressible nmt1 promoter (29); 145,000 colonies were screened for transformants showing reversion of lethality and morphological phenotype. The screen yielded 14 rescuing plasmids. Nine plasmids (pFV 10, 44, 39, 6, 75, 42, 23, 271, and 622) rescued both lethality and shape defects under low level of expression (in thiamine-containing medium). They all identified the same gene sequence. One plasmid, pFV10, was chosen and used for all successive experiments. The coding sequence was cloned in a REP6X plasmid (containing the sup3–5 sequence) and integrated in an orb6–25 ade6–704 strain. Thirty-four integrants were isolated and crossed to a wild-type strain: on average one mutant colony appeared for 500 colonies screened, indicating integration at the orb6 locus and consistent with a 0.5–1 cM distance between the mutant gene and its wild-type integrated copy. As a control, one of these integrants (number 27) was crossed to an orb6–25 ade6–704 strain: orb6+ colonies were found to be adenine prototrophs, indicating that the rescuing sequence was linked to the sup3–5 marker and excluding the possibility of reversion. Plasmid pFV10 contained a single continuous ORF of 1,410 bp.
Deletion of orb6+.
We identified a genomic clone containing the orb6 sequence again by complementation of orb6 mutants. The genomic sequence, ≈10 kilobases long, was shown by PCR and Southern blotting analysis to contain the orb6+ gene, with a 267-bp intron. The deletion construct, substituting the whole catalytic domain of orb6 with the ura4+ gene sequence, was built leaving a 2.9-kilobase 5′ flanking sequence and 0.530-kilobase 3′ flanking sequence.
Cloning of orb2+.
To clone the orb2 gene, we identified a 12-kilobase suppressing genomic fragment that was shown to integrate very closely to the orb2–34 mutation. We identified the suppressing gene by transposon knockout (12) and sequenced outwardly from the transposon insertion site. The orb2–34 mutation also was suppressed by transformation with pak1/shk1 cDNA, which was a kind gift of J. Chernoff (Fox Chase Cancer Center, Philadelphia) (9).
Cytology.
Cells were grown exponentially at 25°C for at least eight generations, then incubated at the indicated temperature for the indicated time. Immunofluorescence was performed as described (13). Cells were fixed in methanol and stained with mouse anti-actin antibody (Sigma) or anti-hemagglutinin (HA) primary antibody (Babco, Richmond, CA) and a CY3-conjugated anti-mouse secondary antibody. Cells then were immobilized on coverslips and observed using a Zeiss Axioskop microscope. Cells were photographed using a Bio-Rad 600 confocal microscope, with a 0.2-μm interval between successive pictures and analyzed using Bio-Rad software. For quantification of actin delocalization, the two-dimensional projections of the confocal microscope pictures were analyzed by using National Institutes of Health image software, measuring the intensity of immunofluorescence signal in different areas of the cell (% of intensity throughout the cell and at the cell tips). Ten cells were measured for each time point. Finally, the penetrance of the corresponding phenotype was quantified in the original sample, by counting 200 cells for each time point.
RESULTS
orb6 Encodes a Serine Threonine Protein Kinase.
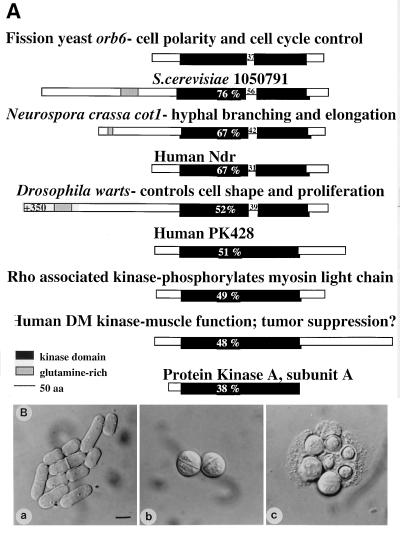
Mutant orb6 cells are round at restrictive temperature (Fig. 1b). Moreover, the number of cells undergoing the process of cell division increases from 14% to 60% after 5 hr at 36°C: 15% of these septa are aberrant (see Fig. 5b). The orb6 gene was cloned by complementation. orb6 mutants were transformed with a cDNA library controlled by the thiamine-repressible nmt1 promoter (29); 145,000 colonies were screened for transformants showing a rescue of the morphological phenotype. The screen yielded 14 rescuing plasmids. Nine plasmids contained the same gene sequence that was shown by integration to encode the orb6+ gene (see Materials and Methods). These plasmids complemented the shape defects of the orb6–25 mutant with low level of expression (in thiamine-containing medium). They contained a single continuous ORF predicted to encode a protein of 469 amino acids; its amino acid sequence was found to encode a member of a serine/threonine protein kinase family, including Neurospora cot1 that affects hyphal morphology (14), Drosophila warts that controls cell shape and cell proliferation (15, 16), Rho-associated kinase (17–19), and the human DMPK (20) (Fig. 2A).
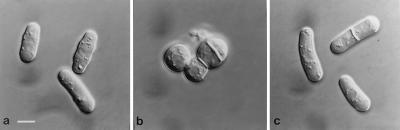
Figure 1.
Cloning of orb6+. (a) leu1–32, pRep3X. (b) orb6–25 leu1–32, pRep3X. (c) orb6–25 leu1–32, pRep3X orb6+. Cells were exponentially grown at 25°C for at least eight generations, then shifted at 36°C for 5 hr.
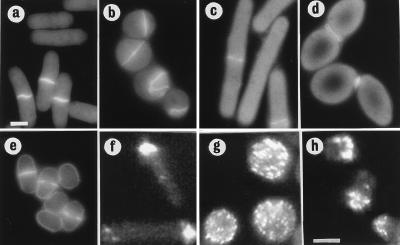
Figure 5.
Overexpression of Wee1 in orb6 mutants and actin localization in orb6–25 and wee1–50 mutants. (a–d) Overexpression of Wee1 in orb6 mutants. (a) leu1–32h− pRep3x. (b) orb6–25 leu1–32h− pRep3x. (c) leu1–32h− pRep3x wee1+. (d) orb6–25 leu1–32h− pRep3x wee1+. (e) wee1–50h−. Cells were grown exponentially at 25°C in the presence of thiamine for at least eight generations; thiamine was washed away, and cells were grown in the absence of thiamine at 25°C for 16 hr, then shifted at 36°C for 5 hr. (f–h) Actin localization in orb6–25 and wee1–50 mutants. (f) leu1–32h−. (g) orb6–25 leu1–32h−. (h) wee1–50h−. (Bar = 5 μm.)
Figure 2.
The orb6+ gene. (A) Sequence comparison of Orb6 and homologous proteins. An homology search using EMBL and GenBank databases identified S. cerevisiae 1050791, Neurospora crassa cot1 (14), human Ndr (23), Drosophila warts (15, 16), human PK428, Rho-associated kinase (17–19), and human DMPK (20). (B) orb6+ gene deletion. Spores were plated on yeast extract plates and allowed to germinate for 16 hr at 32°C. (a) ade- leu1–32 ura4-D18 microcolony. (b) orb6∷ura4+ ade- leu1–32 ura4-D18 microcolony. (c) orb6∷ura4+ ade- leu1–32 ura4-D18 microcolony after 24 hr at 32°C. (Bar = 5 μm.)
The Function of the orb6 Gene Is Essential.
A genomic clone containing the orb6+ gene was isolated from a fission yeast genomic library (30), again by complementation of the orb6 mutation. A deletion mutation was built, which removed most of the orb6+ gene and replaced it with the ura4+ gene. This construct deleted all 11 kinase domains in the orb6 gene, leaving 106 bp at the 5′ end and 224 bp at the 3′ end. The ura4+ gene was substituted for the whole orb6 kinase domain by the one-step gene replacement method in diploids cells using this deletion construct (11). Deletion of the orb6 gene led to cell lethality, establishing that orb6 is essential. Spores deleted for the orb6 gene germinate to form spherical cells that undergo 3–4 cell divisions before lysing (Fig. 2B). Lysis is not prevented by addition of 1 M sorbitol as an osmotic stabilizer.
orb6 Is Necessary for Actin Reorganization After Mitosis and During Activation of Bipolar Growth.
To investigate Orb6 function more fully we constructed a triple HA-tagged orb6 gene under control of the thiamine repressible nmt1 promoter. This construct was integrated into the genome of a strain in which the endogenous orb6+ gene was replaced by the ura4+ marker (orb6∷ura4+). When orb6-HA was expressed in the absence of thiamine, this strain was perfectly viable, showing a generation time of 2 hr and 50 min (at 32°C) as compared with the generation time of 2 hr and 40 min of the ade6-M210 control (determined by monitoring the optical density and the cell number increase of the cell culture). Cells displayed a normal cell morphology with a correctly localized actin cytoskeleton (Fig. 3 a and d). In contrast, when thiamine was added to repress orb6-HA expression, we observed changes in cell morphology and a reorganization of the actin cytoskeleton. Early in the time course, between 4 and 8 hr after thiamine addition, cell length shortened (Fig. 3b) and actin became less localized at the cell tips. Delocalization of 61.5% of actin was found in 67% of cells, as compared with time zero where 91% of actin was found localized at the cell tips (see Materials and Methods) (Fig. 3e). Most cells showed a monopolar pattern of growth (see below). Actin dots also were found in a ring pattern at the new end after septation in 10% of cells, suggesting a delay in actin reorganization after mitosis (Fig. 3e, arrow). Later in the time course, 12 hr after thiamine addition, cells became spherical and actin was found dispersed throughout the cell cortex in 81% of cells (Fig. 3 c and f), with the exception of actin clusters, which persisted in the area where the actin ring had formed during the previous cell division (in 12% of cells) (Fig. 3f, arrow). These observations indicate that when Orb6 levels decrease, the actin cytoskeleton becomes disorganized. Moreover, early in the time course, the reorganization of actin during the cell cycle transitions between monopolar and bipolar growth and between mitosis and interphase is defective, as indicated by an increase in the number of cells growing with one tip only (68% compared with 28% in orb6+ cells) or undergoing the process of cell division (24% compared with 14% in orb6+ cells) (Fig. 3b). We conclude that the Orb6 protein kinase is required to maintain polarity of the actin cytoskeleton during interphase and to promote actin reorganization both after mitosis and during activation of bipolar growth.
Figure 3.
Orb6+ gene dosage effects. orb6∷ura4+ ade- leu1–32 ura4-D18 cells containing integrated HA-tagged orb6+ were grown exponentially for at least eight generations at 32°C, then thiamine was added to the culture. (a and d) Cells grown in the absence of thiamine. (b and e) 7.5 hr after thiamine addition. (c and f) 12 hr after thiamine addition. Cells were photographed immediately (a–c) or fixed and stained for actin (d–f). (Bar, a–c = 5 μm.; d–f = 10 μm.)
This experiment also revealed that Orb6 delays the onset of mitosis (Table 1). Five hours after switching off the orb6 gene, cell length was reduced to 84% of orb6+ cells, and at 7.5 hr cell length was reduced to 65%. This reduction in average cell length was found to be highly significant (P < 0.05). Cell width was unchanged, indicating that these cells were advanced into mitosis at a reduced cell size. Shortening of cell length was not caused by a reduction in cell growth rate, which was unchanged until 8.5 hr after thiamine addition (data not shown).
Table 1.
orb6 gene dosage effects
| Strain | Plasmid | Cell length at division* | % wt length |
|---|---|---|---|
| (1) leu1-32 | Multicopy plasmid (Rep3X) | 14.6 ± 1.0 | 100% |
| (1) leu1-32 | Integrant nmt1 orb6HA | 17.8 ± 1.9 | 121% |
| (1) leu1-32 | Multicopy nmt1 orb6HA | 26.6 ± 4.9 | 181% |
| (1) leu1-32 | Multicopy nmt1 orb6HA ala122 | 13.7 ± 2.7 | 93% |
| (2) leu1-32 | Multicopy plasmid (Rep3X) | 14.2 ± 1.0 | 100% |
| (2) leu1-32 | Multicopy nmt1 orb6HA | 28.9 ± 5.0 | 203% |
| Δorb6 | Integrant nmt1 orb6HA | 18.0 ± 1.3 | 122% |
| Δorb6 | Integrant nmt1 orb6HA (+th., 5h.) | 12.4 ± 0.7 | 84% |
| Δorb6 | Integrant nmt1 orb6HA (+th., 7.5h.) | 9.5 ± 0.6 | 65% |
| leu1-32 | Multicopy nmt1 orb6HA | 20.0 ± 5.6 | 137% |
| wee1-50 leu1-32 | Multicopy plasmid (Rep3X) | 7.4 ± 0.9 | 50% |
| wee1-50 leu1-32 | Multicopy nmt1 orb6HA | 7.5 ± 0.9 | 51% |
Thirty cells at division were measured for each sample. The mean values ± SD are shown.
Orb6 Acts as a Dose-Dependent Inhibitor of Mitosis.
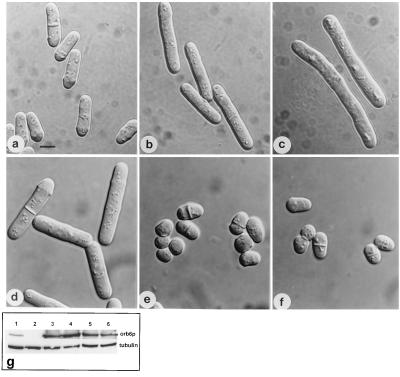
To confirm the function of Orb6 in the regulation of mitosis, we overexpressed orb6-HA in wild-type cells (Fig. 4 and Table 1). Expressing orb6-HA from a single-copy integrant caused cells to divide at a larger cell size (Fig. 4b). Expression of orb6-HA on a multicopy plasmid using the nmt1 promoter led to an increase of cell length at division, indicating that onset of mitosis was substantially delayed (Fig. 4c). FACS analysis established that these cells were delayed in the G2 phase of the cell cycle (data not shown). This delay of mitotic onset required Orb6 protein kinase activity because an orb6 kinase inactive mutant (K122A) did not induce cell elongation when overexpressed (Table 1). orb6-HA overexpression in a wee1–50 strain, which is defective in the Wee1 tyrosine protein kinase that phosphorylates and inhibits p34cdc2, failed to increase cell length at division (Fig. 4 d–f and Table 1). This experiment demonstrated that the Orb6-dependent delay over mitosis acts through the p34cdc2 mitotic kinase. Overexpression of wee1+ in orb6–25 mutant cells did not suppress their morphological defects (Fig. 5 a–d). Moreover, wee1–50 and orb6–25 mutants show a different morphological phenotype at restrictive temperature: orb6–25 cells are round and their actin cytoskeleton is dispersed (Fig. 5 b and g) whereas wee1–50 cells are very short, but still grow in a polarized fashion (Fig. 5e) and show a polarized actin cytoskeleton (Fig. 5h). These experiments indicate that Orb6 kinase has a specific function in the control of cell morphology, maintaining a polarized actin cytoskeleton and promoting polarized cell growth, while delaying the onset of mitosis.
Figure 4.
Effects of orb6 overexpression. (a–c) Overexpression of HA-tagged orb6+ in a leu1–32 strain (experiment 1 in Table 1). (a) leu1–23 pRep3X. (b) leu1–23 cells, carrying integrated HA-tagged orb6+, were grown exponentially for at least eight generations at 32°C in the absence of thiamine. (c) leu1–32 cells, carrying HA-tagged orb6+ on the multicopy plasmid pRep3X, were grown in the absence of thiamine for 18 hr at 32°C. (d–f) Overexpression of HA-tagged orb6+ in a wee1–50 mutant strain. Cells were grown exponentially at 25°C in the presence of thiamine for at least eight generations; thiamine was washed away and cells were grown in the absence of thiamine at 25°C for 16 hr, then shifted at 36°C for 5 hr. (d) leu1–32, pRep3X orb6HA. (e) wee1–50 leu1–32 pRep3X. (f) wee1–50 leu1–32 pRep3X orb6HA. (g) Western blot comparing the levels of expression of orb6-HA in the experiments described above, as compared with the control tubulin: lane 1, leu1–32 with an integrated copy of orb6-HA; lane 2, leu1–23 pRep3X; lane3, leu1–32 pRep3x orb6 HA; lane 4, leu1–32 pRep3x orb6 K122A HA; lane 5, wee1–50 leu1–32 pRep3x; lane 6, wee1–50 leu1–32 pRep3x orb6-HA. (Bar = 5 μm.)
Orb6 Localizes to Sites of Cell Growth.
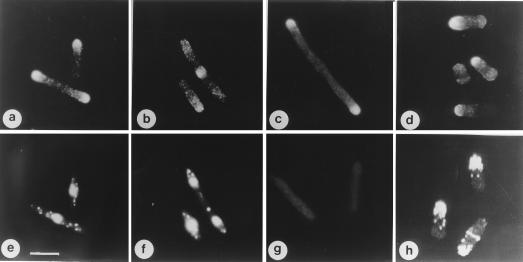
The intracellular location of Orb6-HA protein was determined by using the Orb6-HA integrant strain (Fig. 6). Immunofluorescence showed that Orb6-HA protein is enriched at the cell tips during interphase (Fig. 6 a and e). During mitosis and cytokinesis Orb6-HA protein disappears from the tips and is found in the region of the developing septum (Fig. 6 b and f), indicating that Orb6-HA protein localizes to areas of cellular growth. This observation was confirmed by staining Orb6-HA in cdc25ts and orb2ts mutant cells. Both tips were stained in the cdc25–22ts mutant cells, which were arrested in G2 with both ends growing (Fig. 6c), whereas in the orb2–34 mutant, which cannot activate bipolar growth and only grows at one end (10) (Fig. 6h), the Orb6-HA protein was found only at one growing tip (Fig. 6d).
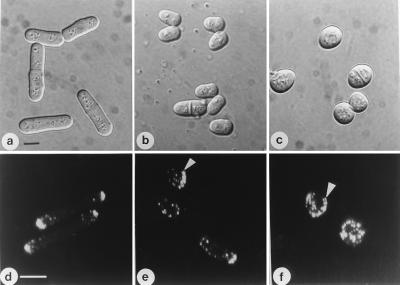
Figure 6.
orb6+ gene product intracellular localization in the cell cycle. (a, b, e, and f) orb6∷ura4+ ade- leu1–32 ura4-D18 cells containing integrated HA-tagged orb6+ were grown exponentially at 32°C in the absence of thiamine, then fixed and stained with an antibody recognizing the HA tag (a and b) and with 4′,6-diamidino-2-phenylindole (e and f). (g) Control staining of orb6∷ura4+ ade- leu1–32 ura4-D18 cells containing integrated untagged orb6+. (c) cdc25–22 leu1–32 Rep3X orb6 HA cells were grown exponentially at 25°C in the presence of thiamine for at least eight generations; thiamine was washed away and cells were grown in the absence of thiamine at 25°C for 16 hr, then shifted at 36°C for 4 hr. (d) orb2–34 leu1–32 pRep3X orb6 HA at 32°C; cells were stained 14 hr after thiamine depletion, which is about 2 hr after derepression of the nmt1 promoter. (h) orb2–34 leu1–32 stained for actin at 32°C. (Bar = 10 μm.)
orb6 Interacts Genetically with orb2.
orb6 mutants were found to be synthetically lethal with orb2 mutants (10). Synthetic lethality interactions were determined by tetrad analysis, and phenotypes were observed at 25°C, 32°C, and 36°C. At 32°C mutants in either orb6 or orb2 are viable, but the double-mutant orb6–25 orb2–34 is lethal, with the cells rapidly undergoing lysis (Fig. 7a). orb2 mutants are unable to activate bipolar growth and grow with only one tip (10). Analysis of the actin cytoskeleton in orb2 mutants revealed that actin is localized only at one pole of growth (Fig. 6h) and that its reorganization after mitosis is delayed (data not shown). This phenotype is reminiscent of the one observed when expression levels of Orb6 decrease, suggesting that Orb2 and Orb6 might fulfill a similar function.
Figure 7.
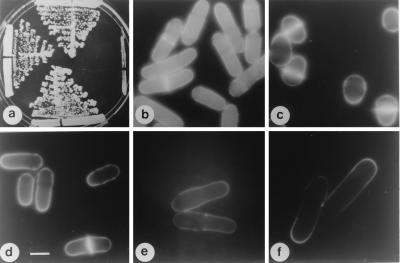
Cloning of the orb2+ gene. (a) top, ade6-M210 leu1–32h−; left, orb6–25 ade6-M210 leu1–32h−; bottom, orb2–34 ade6-M210 leu1–32h−; right, orb2–34 orb6–25 ade6-M210 leu1–32 h−. Cells were grown at 32°C. (b) ade6-M210 leu1–32h−, Rep3X. (c) orb2–34 ade6-M210 leu1–32h−, pRep3X. (d) orb2–34 ade6-M210 leu1–32h− pDB248′-8 (genomic fragment). (e) orb2–34 ade6-M210 leu1–32h− pRep3X pak1/shk1+. (f) orb2–34 ade6-M210 leu1–32h− pRep3X orb6+. Cells were grown exponentially at 25°C in the presence of thiamine for at least eight generations; thiamine was washed away and cells were grown in the absence of thiamine at 25°C for 16 hr, then shifted at 36°C for 6 hr. (Bar = 5 μm.)
orb2 Encodes the Protein Kinase Pak1/Shk1.
Given this interaction with orb6, orb2 was cloned by complementation (Fig. 7 c and d), and its sequence confirmed that orb2 is identical to the previously identified pak1/shk1 gene (8, 9). The orb2–34 mutant was also suppressed by pak1/shk1 cDNA (Fig. 7e). The pak1/shk1 gene encodes a protein kinase that interacts with Cdc42 and is similar to the CDC42 and RAC1-interacting PAK1 protein kinase identified in mammalian cells and STE20 found in budding yeast cells (7). Overexpression of orb6+ partially suppressed the pak1/shk1/orb2ts mutant shape defect (Fig. 7f). Overexpression of pak1/shk1+ did not rescue the orb6ts mutant phenotype (data not shown), but did induce cell elongation in wild-type cells (data not shown). These results suggest that the Orb6 kinase might function downstream of the Pak1/Shk1 protein kinase. This finding also is supported by the observation that in pak1/shk1/orb2ts mutants, which are defective in the activation of the second tip, the Orb6-HA protein was found only at the one growing tip, suggesting that pak1/shk1 is required to bring about bipolar growth and to localize Orb6 protein correctly (Fig. 6d).
DISCUSSION
Orb6 Belongs to a Family of Kinases Essential for Morphological Control.
In the present study, we report the cloning and characterization of a fission yeast gene, orb6, which is required for maintenance of cell polarity during interphase and to promote actin reorganization during morphological transitions. Orb6 encodes a serine/threonine kinase and belongs to a kinase family that includes several serine/threonine kinases whose catalytic domains are highly conserved. These kinases are more closely related to each other then to their closest relatives, the cyclic nucleotide-dependent protein kinases, and, at least in the case of the DMPK and Rho-associated kinase, show distinct biochemical properties (17–19, 21). Warts, orb6, and cot1 have been identified by mutagenesis screens and are required for maintenance of cell shape and polarity. Rho-associated kinase is regulated and translocated to the plasma membrane by rho-GTPase (17–19), which is involved in the control of various aspects of the actin cytoskeleton. Rho-associated kinase is the only member of this kinase family with known substrates and has been shown to have a role in the regulation of actin-myosin interaction by modulating myosin phosphorylation (22). Only one related kinase has been identified in the budding yeast, Saccharomyces cerevisiae, suggesting that this kinase family might have diverged in higher Eukaryotes. We presently are investigating to determine whether S. pombe orb6 and S. cerevisiae 1050791 are functionally homologous to mammalian Rho-associated kinase and whether the modality of regulation by rho-GTPase is conserved.
Orb6 Acts as a Dose-Dependent Inhibitor of Mitosis.
Orb6 also acts as a dose-dependent inhibitor of mitosis, a function requiring the activity of wee1 kinase, an inhibitor of p34cdc2 kinase activation. Thus Orb6 kinase has two roles, to control cell shape and to regulate G2/M transition, possibly through the wee1 pathway, or some other mechanism dependent on Cdc2 tyrosine phosphorylation (Fig. 8). Although immunofluorescence microscopy showed that Orb6 is predominantly localized in the cytoplasm, Orb6 has a conserved sequence (KDKMATWKKNRR) that has been shown to contain the nuclear localization signal in Ndr kinase (23), so possibly it may directly interact with Wee1. These findings also suggest an interesting functional similarity with Drosophila warts. The warts gene is one of more than 50 genes in which mutations give an overgrowth phenotype, and which therefore are considered tumor suppressor genes (24). Homozygous loss of warts (wts) in somatic clones results in cell-autonomous formation of epithelial tumors (15, 16), whose cells also show an hypertrophy of the apical ends. Thus, the warts gene product is important for controlling cellular morphogenesis as well as proliferation. Myotonic dystrophy is also sometimes associated with calcifying epitheliomas, neurofibromas, and parathyroid adenomas, suggesting that the DMPK also may have a role in the control of cell proliferation (25).
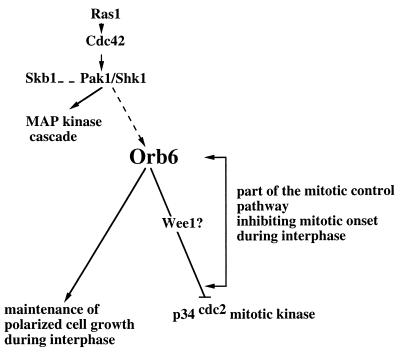
Figure 8.
Orb6 kinase is proposed to be part of a regulatory cascade contributing to the mitotic control and responding to the Ras1 and Cdc42-dependent morphological control pathway via the Pak1/Shk1 kinase. Orb6 kinase regulates independently cell morphogenesis and the cell cycle, mediating its effect on p34cdc2 kinase directly through the Wee1 protein kinase or through some other mechanism dependent on p34cdc2 tyrosine phosphorylation.
What is the role of these kinases in the regulation of the cell cycle and what is its functional significance? One possibility is that the deregulation of cell proliferation is an indirect consequence of cell shape alteration. Our results, though, suggest that a decrease of Orb6 levels leads to mitotic advance before an alteration of cell shape and that Orb6 overexpression can delay onset of mitosis without altering cell morphology. This finding might point to a novel pathway in the control of mitotic onset and in the coordination of cell growth and cell proliferation. Future work will clarify the extent of functional similarity between warts and orb6. If this pathway is conserved, the identification of orb6 in fission yeast, which allows the dissection of the cell cycle machinery by the use of cell cycle (cdc) mutants, may provide the opportunity to easily study the mechanism of cell cycle regulation by these kinases.
Orb6 Protein Kinase Interacts Genetically with pak1/shk1.
To identify other partners in Orb6 function, we cloned orb2, because orb2 mutants showed a strong synthetic lethality interaction with orb6 mutants and their phenotype displayed similarities to the effects of orb6 loss of function. Cloning of orb2 showed that orb2 is identical to the previously identified pak1/shk1 gene (8, 9). The pak1/shk1 gene encodes a protein kinase that interacts with Cdc42, and is similar to the CDC42 and RAC1-interacting PAK1 protein kinase identified in mammalian cells and STE20 found in budding yeast cells (7). Ras1, Cdc42, and Pak1/Shk1 have a regulatory role in fission yeast cell morphogenesis and are thought to function in an hierarchical order whereby ras1 GTPase interacts functionally with Cdc42 GTPase (6) and Pak1/Shk1 kinase acts as an effector of Cdc42 (8, 9).
We identified temperature-sensitive mutants of pak1/shk1, which confirm the morphological role of pak1/shk1 previously indicated by overexpression studies (9). Moreover, our study suggests that Pak1/Shk1 also might have a function in cell cycle control because pak1/shk1/orb2ts cells are short in cell length, and overexpression of pak1/shk1 induces cell elongation in wild type (data not shown). These results are consistent with the observation that Skb1, which interacts with Pak1/Shk1, also causes cell elongation when overexpressed (26). The fact that orb6 overexpression partially suppresses the orb2ts phenotype suggests that the Orb6 kinase might function downstream of the Pak1/Shk1 protein kinase, in a pathway that controls and coordinates maintenance of cell polarity with progression through the cell cycle (Fig. 8). Alternatively, pak1/shk1 and orb6 might represent parallel pathway with similar and partially overlapping functions. The former model, though, is supported also by the observation that in pak1/shk1/orb2ts mutants, which are defective in the activation of the second tip, the Orb6-HA protein is found only at the one growing tip, whereas another marker of cell polarity, Tea1, is found localized at both ends (27). This finding indicates that pak1/shk1 is required to bring about bipolar growth and to localize Orb6 protein correctly. Future work will address the interaction between Pak1/Shk1 and Orb6 and will clarify whether the Orb6 kinase activity or cortical localization is dependent on Pak1/Shk1.
CONCLUSION
We conclude that Orb6 has two roles during the cell cycle: to maintain polarized cell growth and to delay onset of mitosis. We propose that the Orb6 kinase acts downstream of a morphogenetic control pathway involving Cdc42 and Pak1/Shk1, which maintains the cell in a polarized state during interphase while delaying the onset of mitosis (Fig. 8). When the cell has reached the appropriate size and is ready to undergo mitosis, a reduction in Orb6 kinase activity could remove an inhibitory signal over the p34cdc2 mitotic kinase and thus contribute to bringing about the onset of mitosis. The similarity of orb6 with Drosophila warts, which is thought to function as a tumor suppressor (15, 16) and other genes in mammalian cells that affect cell shape (22, 28), suggests that its function in coordinating cell morphogenesis with the cell cycle may be conserved in higher eukaryotes. Future work will compare the functional properties of this kinase family in different organisms and elucidate their role in the mechanisms integrating cell morphogenesis and proliferation.
Acknowledgments
We thank Dr. J. Chernoff for kindly providing the pak1/shk1 cDNA. We thank Dr. R. Myers, Dr. R. Assoian, and Z. Chen for critically reading the manuscript, and Dr. P. Salas for help with the confocal microscope. This work has been supported by an European Economic Community and an Imperial Cancer Research Fund fellowship and by the Sylvester Comprehensive Cancer Center (University of Miami).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HA, hemagglutinin; DMPK, myotonic dystrophy kinase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF009512).
References
- 1.Drubin D G, Nelson W J. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison J M, Nurse P. J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- 3.Marks J, Hyams J S. Eur J Cell Biol. 1985;110:417–425. [Google Scholar]
- 4.Nurse P. Nature (London) 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- 5.Miller P J, Johnson D I. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang E C, Barr M, Wang Y, Jung V, Xu H P, Wigler M H. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 7.Sells M A, Chernoff J. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 8.Marcus S, Polverino A, Chang E, Robbins D, Cobb M H, Wigler M H. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ottilie S, Miller P J, Johnson D I, Creasy C L, Sells M A, Bagrodia S, Forsburg S L, Chernoff J. EMBO J. 1995;14:5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verde F, Mata J, Nurse P. J Cell Biol. 1995;131:1–10. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreno S, Klar A, Nurse P. J Gen Microbiol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 12.Morgan B A, Conlon F L, Manzanares M, Millar J B, Kanuga N, Sharpe J, Krumlauf R, Smith J C, Sedgwick S G. Proc Natl Acad Sci USA. 1996;93:2801–2806. doi: 10.1073/pnas.93.7.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagan I M, Hyams J S. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- 14.Yarden O, Plamann M, Ebbole D J, Yanofsky C. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Justice R W, Zilian O, Woods D F, Noll M, Bryant P J. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 16.Xu T, Wang W, Zhang S, Stewart R A, Yu W. Development (Cambridge, UK) 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 17.Leung T, Manser E, Tan L, Lim L. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- 18.Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 19.Ishizaki T M M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N S Y, Kakizuka A, Morii N, Narumiya S. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 20.Brook J D, McCurrach M E, Harley H G, Buckler A J, Church D, Aburatani H, Hunter K, Stanton V P, Thirion J P, Hudson T, et al. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 21.Dunne P W, Walch E T, Epstein H F. Biochemistry. 1994;6:10809–10814. doi: 10.1021/bi00201a031. [DOI] [PubMed] [Google Scholar]
- 22.Kimura K I M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 23.Millward T C P, Hemmings B A. Proc Natl Acad Sci USA. 1995;92:5022–5026. doi: 10.1073/pnas.92.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson, K. L., Justice, R. W. & Bryant, P. J. (1994) J. Cell. Sci. 18, Suppl., 19–33. [DOI] [PubMed]
- 25.Hamshere M G B. Trends Genet. 1996;12:332–334. doi: 10.1016/s0168-9525(96)80002-3. [DOI] [PubMed] [Google Scholar]
- 26.Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb M H, Marcus S. Proc Natl Acad Sci USA. 1996;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mata J, Nurse P. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 28.Ishizaki T N M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- 29.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 30.Hirano T, Hiraoka Y, Yanaghida M. J Cell Biol. 1988;106:1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]