Abstract
The structure of the 30-nm chromatin fibre is an important determinant of the regulation of eukaryotic transcription. A fundamental issue is whether the stacking of nucleosomes in this fibre is organized as a one-start or two-start helix. We argue that all recent experimental data are compatible with a two-start helix and propose that the topology of the fibre, but not the mode of stacking the nucleosomes, is dependent on the length of the linker DNA. This arrangement conserves nucleosome stacking and thus the external morphology of the fibre, and also ensures that the fibre adopts the highest available packing density.
Keywords: 30 nm fibre, chromatin structure, DNA topology, linker histone
Introduction
A sine qua non of chromatin function should be to maintain the highest DNA packing density that is compatible with efficient biological processing. The packaging of nucleosomes must therefore both compact DNA and maintain accessibility for regulatory and transcribing proteins. The initial level of compaction of an array of nucleosomes is the 30-nm fibre (Finch & Klug, 1976; Thoma et al, 1979). This fibre itself probably folds into structures with a greater extent of DNA compaction, which might themselves constitute the fundamental functional units of chromatin (reviewed in Woodcock & Dimitrov, 2001; Tremethick, 2007). A simple analogy would be that, just as in bacteria a DNA ‘rope' can coil into higher-order structures, so in the eukaryotic nucleus the 30-nm fibre acts as an equivalent rope.
Properties of the 30-nm fibre
The structure of the 30-nm fibre is a key element in understanding chromatin compaction. It consists of a helical array of nucleosomes, each comprising a core particle wrapping ∼146 or 147 base pairs (bp) of DNA associated with a linker histone. The globular domain of the linker histone constrains an additional ∼20 bp (Allan et al, 1984), and the resulting chromatosomes are connected by stretches of linker DNA with lengths varying between 0 and ∼70 bp. At the core of current and past controversies is the structural problem of accommodating variable linker lengths between neighbouring core particles.
One characteristic of linker lengths is that in vivo they are usually quantized in increments of 10 bp (Strauss & Prunell, 1983; Widom, 1992), such that linker lengths of 20, 30 and 40 bp correspond to nucleosome repeat lengths of 167, 177 and 187 bp, respectively. The 10-bp increment is close to the 10.5-bp helical repeat of DNA in solution, which implies that, in principle, optimization of the total DNA twist between nucleosomes is an important feature of the structural organization of chromatin. This twist constraint further implies that optimal and uniform linker lengths could facilitate the folding of a nucleosome array.
Certain physical properties of the 30-nm fibre are well established. Early studies, which were in good agreement with the dimensions of native fibres, showed that the diameter of refolded chromatin was ∼30 nm and was largely independent of average linker length (McGhee et al, 1983; Widom & Klug, 1985), although the inclination of individual nucleosomes toward the helical axis changed with linker length (Sen et al, 1986). Another important parameter is the packing density. Early studies of native and reconstituted fibres reported values of both ∼6 and ∼12 nucleosomes per 11 nm (Thomas & Butler, 1980; Widom & Klug, 1985; Gerchman & Ramakrishnan, 1987). However, for native fibres from echinoid spermatozoa with an average linker length of ∼70 nm, both the diameter and the packing density were substantially greater (42 nm and ∼13 nucleosomes per 11 nm, respectively; Athey et al, 1990). Importantly, the packing density depends on the DNA sequence, such that for fibres containing centromeric DNA the packing density is higher (∼12 nucleosomes per 11 nm) than for those containing more mixed sequences (Gilbert & Allan, 2001; J. Allan, personal communication). Other relevant observations include X-ray diffraction patterns of oriented fibre samples showing a strong 11-nm axial reflection, which are interpreted as the long dimension (11 nm) of the core particle being approximately parallel to the superhelical axis. Furthermore, in both natural and reconstituted chromatin fibres with average linker lengths of 50–60 bp, the linker histone is located in the interior of the fibre (Dimitrov et al, 1987; Graziano et al, 1994).
In early studies, the distribution of linker lengths in a single fibre was unknown and, indeed, in reconstituted fibres it is likely to be irregular. With the identification of strong in vitro nucleosome-positioning signals, arrays of nucleosome-binding sites with different uniform linker lengths were constructed. These revealed that for lengths corresponding to the quantized—and assumed optimal—values, fibres of high packing density, up to and exceeding that for echinoid spermatozoa chromatin, could be assembled in vitro (Robinson et al, 2006). The diameter of these fibres, as visualized by electron microscopy, was dependent on the length of the linker DNA, but in an unusual stepwise fashion, remaining constant at ∼33 nm over a range of 30–60 bp and then increasing abruptly to ∼42 nm from 70–90 bp.
Models for the 30-nm fibre structure
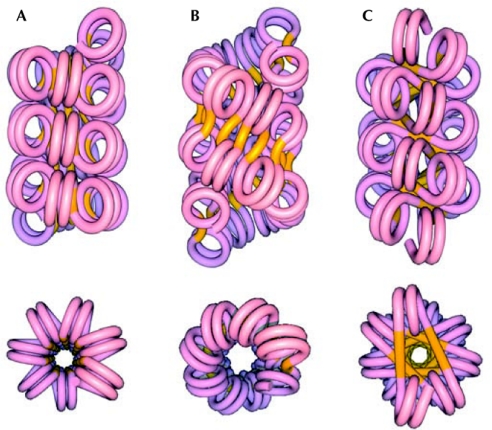
A major unresolved question about the structure of the 30-nm fibre is whether it is organized as a one-start stack of nucleosomes known as a solenoid, which is a single continuous helix, or as a two-start helix known as a zigzag with two continuous stacks, analogous to the structure of DNA (Fig 1B,C).
Figure 1.
Classes of models for the 30-nm chromatin fibre. The image shows longitudinal views above and axial views below. (A) Solenoid model. (B) Helical ribbon model. (C) Crossed-linker model. Nucleosome arrays reveal the two-start organization of the chromatin fibre. Reproduced from Dorigo et al (2004) with permission from the American Association for the Advancement of Science (AAAS).
All current models of the structure of the 30-nm fibre assume, either implicitly or explicitly, that all experimental observations pertain to the same structure. The models divide easily into two classes: the solenoid class in which a one-start helical stack of nucleosomes is linked together by bent DNA between adjacent octamers (Finch & Klug, 1976; Thoma et al, 1979; Felsenfeld & McGhee, 1986; Fig 1A); and the two-start zigzag class in which, in the simplest examples, two helical stacks of nucleosomes are connected by a relatively straight DNA linker (Fig 1B,C). The latter class further divides into the helical/twisted-ribbon model in which the linker DNA is oriented at angles varying from 0° to 50° to the fibre axis (that is, along the length of the fibre; Worcel et al, 1981; Woodcock et al, 1984), and the crossed-linker model in which the linker DNA is oriented approximately perpendicular to the fibre axis (that is, across the fibre; Williams et al, 1986; Smith et al, 1990).
The helical character of the 30-nm fibre was recognized from the earliest studies and the question of whether it comprised a one-start or two-start helix was immediately apparent (Thoma et al, 1979). Opting for simplicity, the one-start model was initially preferred. However, the earlier observation of two parallel stacks of nucleosomes in a single fibre—clearly distinguishable from the nucleosome zigzag of the unfolded fibre (Oudet et al, 1975)—anticipated a two-start structure (Fig 2). This was subsequently formalized into two alternative two-start models that could also explain the high packing density of some natural fibres. To explain this feature, a variant of the one-start model was proposed in which successive turns of a one-start helix partly interdigitate with each other (Daban & Bermudéz, 1998). Subsequently, this model was invoked to explain why the dimensions of highly compacted arrays of nucleosomes, formed with 177–237 bp repeats that increased incrementally by 10 bp, were inconsistent with either the crossed-linker two-start model or the helical-ribbon two-start model individually (Robinson et al, 2006).
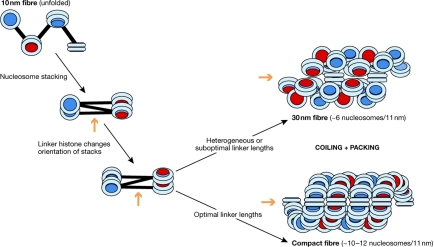
Figure 2.
Proposal for a folding pathway for chromatin fibres. With constant and optimal short linker lengths, the ‘10-nm' zigzag folds to a twisted crossed-linker structure corresponding to the tetranucleosome crystallized by Schalch et al (2005). In the presence of linker histone, a more coiled structure is formed that, for shorter linker lengths, compacts into a helical ribbon corresponding to the condensed structures visualized by Robinson et al (2006). For clarity, the nucleosomes in the condensed fibres are shown parallel to the superhelical axis, although in actual fibres they are substantially tilted with respect to the axis. With suboptimal or heterogeneous linker lengths, a less-compact fibre—corresponding to the classic 30-nm fibre—will be formed (top right). The orange arrows indicate the approximate direction of the long axis of the fibres.
There is now compelling experimental evidence indicating that the 30-nm fibre can adopt a two-start organization. The crystal structure of a tetranucleosome with a zero-length linker and without a linker histone revealed two stacks of nucleosomes connected by ‘straight' linkers (Schalch et al, 2005). To reconcile the apparent dichotomy between this structure and the compact fibres with a linker histone, Robinson & Rhodes (2006) suggested that the former represents ‘active' unfolded chromatin, whereas the latter represents the canonical folded state of the 30-nm fibre. However, cross-linking studies on longer reconstituted arrays with uniform optimal linkers indicated that nucleosome i was immediately adjacent to nucleosomes i ± 2, as predicted by a two-start helix (Dorigo et al, 2004). Crucially, the same geometry was observed for regular arrays with linker lengths of 20, 40 and 60 bp when compacted either with divalent cations or with linker histone. These direct demonstrations of a two-start connectivity are consistent with other observations—the DNA-cleavage patterns induced by ionizing radiation in intact cells (Rydberg et al, 1998), the straight trajectory of linkers in a slightly extended fibre (Bednar et al, 1998) and their relative resistance to ultraviolet-induced pyrimidine-dimer formation (Pehrson, 1995).
Fibre dimensions and linker length
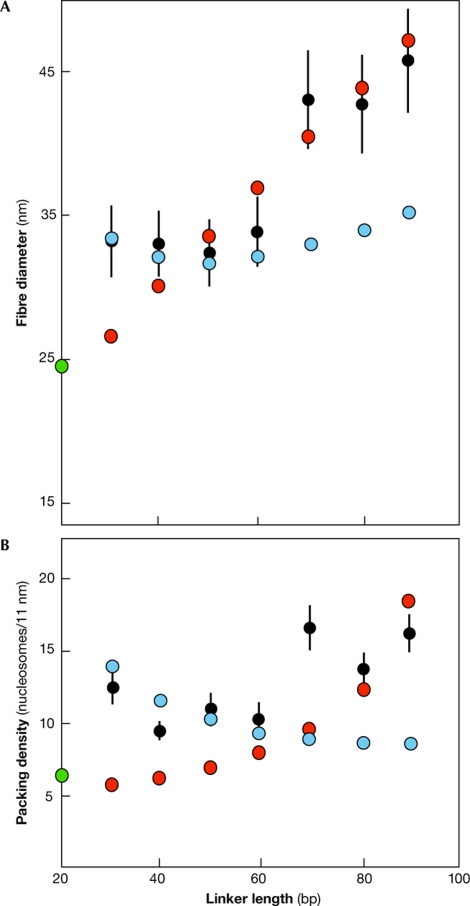
Any model of the structure of the 30-nm fibre must be compatible with the variation of fibre diameter with linker length. Assuming the simple geometry of both of the two-start models, do the predicted diameters and packing densities of regularly packed fibres with repeat lengths of 167 + 10 n bp (where n = 0–8) fit the available experimental data? For every 10-bp increment, the linker DNA increases by 3.4 nm in length, and the twist between adjacent nucleosomes changes by 17° (as 10 bp comprises only 343° of a full helical turn of 10.5 bp). With these constraints, the fibre diameter of a helical ribbon remains relatively constant, whereas the packing density decreases as the linker length increases (Fig 3; see supplementary Fig 1 online). By contrast, for a crossed-linker structure, both the diameter and packing density increase as the linker length increases. For all linker lengths tested, the closest calculated diameter is within one standard deviation of the experimental value. Notably, at short linker lengths, the packing density of the helical-ribbon form is higher than that of the crossed-linker model, whereas the converse is true at the longest linker lengths.
Figure 3.
Dependence of fibre diameter and packing density on linker length. Comparison of (A) fibre diameter and (B) packing density calculated assuming helical-ribbon (blue circles) and crossed-linker (red circles) models, with values obtained from compact fibres with linker histone (black circles with error bars indicating one standard deviation; Robinson et al, 2006), and from the fibre inferred from the crystal structure of a tetranucleosome without linker histone and 20 base pair linker length (green circle; Dorigo et al, 2004). At short linker lengths the transition from the relaxed to the supercoiled form increases packing density, whereas at longer linker lengths the opposite transition has the same effect. The linker length is defined as the number of base pairs between adjacent core particles.
Therefore, the diameters of the 177–207 bp repeat fibres visualized by electron microscopy are well fitted by the helical-ribbon model, whereas those of the 217–237 bp repeat fibres are better fitted by the crossed-linker model, especially at higher repeat lengths (Fig 3A). The predictions of the crossed-linker model also extrapolate well to the fibre diameter calculated from the crystal structure of a zero-linker tetranucleosome lacking linker histone. The predictions of the helical-ribbon model reproduce the observed reduction in packing density when the repeat length is increased from 177 to 187 bp, whereas those for the crossed-linker model reproduce the subsequent increase (Fig 3B). In the latter case, the fit is less good, especially for the 217-bp repeat fibre. However, because of end effects, the experimental determination of diameter is likely to be more robust that that of packing density (for further details see supplementary Fig 2 online).
The properties of the helical-ribbon model and the crossed-linker model potentially resolve the divergent interpretations of the crystal structure of a ‘tetranucleosome', and the electron-microscopy images of compact fibres. Both are, in principle, consistent with a two-start straight-linker model. As the predictions of the helical-ribbon and crossed-linker models intersect, the form of the two-start helix could depend on linker length. With short linkers and linker histone, the preferred structure is a more coiled fibre, such as a helical ribbon, whereas with long linkers, the preferred structure is a crossed-linker form. Additional coiling could similarly be achieved by an intermediate structural change akin to the B-to-A-form transition in DNA. The transition between different forms probably occurs between 187 and 217 bp. Within this range it is possible that, depending on the nature of the linker histone, either form can be adopted by the fibre. A transition between 207 and 217 bp would be consistent with the stepped increase in the diameter of compact fibres in this region. Such an intrinsic dimorphism of the 30-nm chromatin fibre could explain why a unique structural solution has been elusive.
Topology of the 30-nm fibre
A variable topology explains three important features of the morphology of the 30-nm fibre: the relative invariance of its diameter at shorter linker lengths, the abrupt increase in diameter of compact fibres at linker lengths greater than 60 bp (corresponding to a nucleosome repeat of 207 bp) and an apparent linker-histone requirement for the higher density fibres at short linker lengths. However, why should a topological switch occur and how might the stacking of nucleosomes be conserved during a switch?
A change in fibre topology with increasing optimal linker length depends on the properties of DNA. If the incremental increase in linker length differs from the helical repeat of relaxed DNA, then the difference in twist will be reflected in the structure of the fibre itself. In vivo measurements of linker length pointed to an approximate quantization in increments of 10 bp (Widom, 1992), whereas in vitro the tested increments of linker length were exactly 10 bp (Robinson et al, 2006). For each 10-bp increment, the linker DNA incurs a twist deficit of ∼17° (see above). This change is an intrinsic property of the quantization of linker length and can account for the topological changes proposed. In the tetranucleosome crystal structure the two stacks of core nucleosomes are twisted relative to each other by −71.5° (Schalch et al. 2005). Given that a two-start helical ribbon is a supercoiled form of a two-start crossed linker (Schalch et al, 2005), we suggest that the globular domain of a conventional linker histone with an extended basic carboxy-terminal tail alters the angles of the entering and exiting DNA of the core particle, and imparts a positive twist. With a short DNA linker, such a twist would place the two nucleosome stacks in a different configuration and concomitantly drive their concerted cooperative coiling into a fibre with the helical-ribbon form (Fig 2), which would result in an additional compaction. Therefore, altering the angle of entering and exiting DNA might represent a fundamental role of the linker histone. In vitro the extent of the coiling would be dependent on exogenous factors, such as the ionic strength, implying that—consistent with electron-microscope and linear-dichroism studies (Thoma et al, 1979; Makarov et al, 1983)—the inclination of the nucleosomes to the superhelical axis would change little with varying degrees of compact folding. However, as the linker length incrementally increases, the added positive twist is reduced by the cumulative negative twist deficit until the fibre is no longer supercoiled—that is, the net twist is ∼0°—and assumes a relaxed crossed-linker form. Up to and beyond this point, the cumulative changes could be accommodated by changes in the pitch of the nucleosome stacks.
Nucleosome stacking
Another important consideration is the ability of nucleosome core particles to stack on top of each other to form extended structures (Finch et al, 1977). Interestingly, these stacks can adopt a gentle helical configuration (Livolant & Leforestier, 2000) that itself would favour coiling in a more compact helix. Also, the wedge shape of the core particle (Finch et al, 1977; Luger et al, 1997) is well suited to wrapping in the tight helices of the 30-nm fibre where the narrow end of the wedge occupies the inner surface of the superhelix (Bednar et al, 1998). However, if the shape of the particle is adapted to packing into helices, this raises the important question of how the packing can be maintained with the narrow ends of the wedge pointing inwards if the orientation of the linker DNA relative to the fibre axis changes towards that required for the canonical helical-ribbon form. For a helical ribbon, but less so for an ‘A-type' chromatin fibre, the linker orientation would have to be uncoupled from the histone octamer orientation while still being compatible with the length of the linker. In the case of short linkers, such as 30 bp, this is not a trivial problem, as some rotation would probably still be required to accommodate the 30-bp length of DNA (equivalent to 10.5 nm) between octamers and the ∼11-nm repeat between successive helical stacks. The exact nature of the interaction between the linker histone and the octamer is unknown at present, but one possibility would be that this interaction could flex and change orientation with topology. Indeed, the basic C-terminal domain of the linker histone stabilizes a ‘stem' projecting from the particle both in isolation and in the 30-nm fibre itself (Hamiche et al, 1996; Bednar et al, 1998). This stem potentially binds to two duplexes on opposing faces of a bivalent α-helix (Hill et al, 1989) and could, in principle, be part of a flexible hinge (Sivolob & Prunell, 2003); however, for short linkers such an effect is unlikely to be sufficient.
We have argued that tight stacking of adjacent nucleosomes is a prerequisite for forming fibres with high packing density. This implies that when a nucleosome array with a constant, albeit suboptimal, linker length is condensed in the presence of a linker histone, a compact structure with nucleosome stacks will be formed. These stacks will be twisted relative to each other as in a crossed-linker structure, although the resulting packing density will be less than that in the most compact structures. In essence, this configuration would be similar to those proposed by Schalch et al (2005) for an optimal repeat length without linker histone, and by Athey et al (1990) for natural chromatin. Similarly, a nucleosome array with varying linker lengths that average to an optimal value could fold to form the less compact ‘classic' 30-nm fibre with the same fundamental two-start helical organization. Such a structure is consistent with the observed and inferred lower packing densities of ∼6 nucleosomes per 11 nm (Thomas & Butler, 1980; Widom & Klug, 1985; Gerchman & Ramakrishnan, 1987) and the irregular fibre structures visualized by atomic-force microscopy (Leuba et al, 1994).
Any model of the 30-nm fibre would have to explain the intrinsic topological properties of linker DNA and be consistent with nucleosome packing, in addition to explaining the dimensions and packing density of the fibre. The two-start variable topology model accounts for the dimensions and topology of the 30-nm fibre structure, but it remains to be demonstrated whether it can adequately accommodate nucleosome packing. The one-start interdigitated model is consistent with the external dimensions of the fibre, but proposes a different mode of packing. However, these two models also differ in their predictions for the number of nucleosomes in each helical turn of the fibre, especially in the most stringent case with a 30-bp linker (a 177-bp nucleosome repeat length). Whereas the one-start model for this fibre predicts 10 or 11 nucleosomes per turn—being limited by the internal width of the nucleosome (Robinson & Rhodes, 2006)—a two-start compact fibre predicts 13 or 14 nucleosomes per turn, or 26–28 nucleosomes per double-helical turn and, consequently, a higher packing density (Fig 3).
Implications and conclusions
A variable topology model allows the fibre to adopt the form with the highest packing density depending on linker length—either crossed linker or helical ribbon—and at the same time permits facile linker histone-dependent cooperative transitions between folded and unfolded chromatin. Additionally, for short optimal linker lengths (for example, 30–50 bp), the linker would be closer to the exterior of the fibre and, hence, more accessible. With the transition from the supercoiled helical-ribbon form to the relaxed crossed-linker form of the fibre, the packing density would increase as the linker DNA becomes more internal and, presumably, less accessible. Such a variation in accessibility correlates well with the observation that transcriptionally highly active nuclei, such as those of yeast or neurons, often have short linkers, whereas those in less-active or inactive nuclei, as in echinoid spermatozoa, have longer linkers and more tightly packed chromatin (Spadafora et al, 1976; Thomas & Thompson, 1977; Thomas & Furber, 1977).
The available data on the properties and structure of the 30-nm fibre strongly indicate that the fibre has a two-start helical structure with straight linker DNA between nucleosomes. Consequently, the local spatial connectivity between nucleosomes is dependent on linker length, unlike the fundamental organization of the fibre or the nucleosome stacking. The linker length-dependent topological transition between the helical-ribbon and crossed-linker forms should ensure that the fibre adopts the form with the higher packing density, and therefore maintains optimal compaction in the biological milieu. There are two further considerations. First, two other abundant chromosomal proteins, high-mobility group A2 (HMGA2) and HMGB (Funayama et al, 2006; Ner et al, 2001), can compete with linker histones, and therefore the 30-nm fibre might be one of several related structures. Second, even though the 30-nm fibre is the initial level of folding of a nucleosome array, it might constitute the rope component of a higher-order functional structure.
While this manuscript was under revision, Wong et al (2007) proposed a different multi-structure model for the 30-nm fibre in which different linker lengths generate structures of different connectivity but broadly similar topology.
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Modelling of the 30nm fibre
Dependence of the conclusion on modelling parameters.

Andrew Bassett

Chenyi Wu

Andrew Travers
Acknowledgments
A.B. was supported by a Medical Research Council studentship. We are grateful to S. Teichmann and V. Ramakrishnan for encouragement.
References
- Allan J, Rau DC, Harborne N, Gould H (1984) Higher order structure in a short repeat length chromatin. J Cell Biol 98: 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athey BD, Smith MF, Rankert DA, Williams SP, Langmore JP (1990) The diameters of frozen-hydrated chromatin fibers increase with DNA linker length: evidence in support of variable diameter models for chromatin. J Cell Biol 111: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA 93: 14173–14178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daban JR, Bermúdez A (1998) Interdigitated solenoid model for compact chromatin fibers. Biochemistry 37: 4299–4304 [DOI] [PubMed] [Google Scholar]
- Dimitrov SI, Russanova R, Pashev IG (1987) The globular domain of histone H5 is internally located in the 30 nm fibre: an immunochemical study. EMBO J 8: 2387–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306: 1571–1573 [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, McGhee JD (1986) Structure of the 30 nm chromatin fiber. Cell 44: 375–377 [DOI] [PubMed] [Google Scholar]
- Finch JT, Klug A (1976) Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA 73: 1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A (1977) Structure of nucleosome core particles of chromatin. Nature 289: 29–36 [DOI] [PubMed] [Google Scholar]
- Funayama R, Saito M, Tanobe H, Ishikawa F (2006) Loss of linker histone H1 in cellular senescence. J Cell Biol 175: 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerchman SE, Ramakrishnan V (1987) Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc Natl Acad Sci USA 84: 7802–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Allan J (2001) Distinctive higher-order chromatin structure at mammalian centromeres. Proc Natl Acad Sci USA 98: 11949–11954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano V, Gerchman SE, Schneider DK, Ramakrishnan V (1994) Histone H1 is located in the interior of the chromatin 30-nm filament. Nature 368: 351–354 [DOI] [PubMed] [Google Scholar]
- Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A (1996) Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol 257: 30–41 [DOI] [PubMed] [Google Scholar]
- Hill CS, Martin SR, Thomas JO (1989) A stable α-helical element in the carboxy-terminal domain of free and chromatin-bound H1 from sea urchin sperm. EMBO J 8: 2591–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, Bustamante C (1994) Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc Natl Acad Sci USA 91: 11621–11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livolant F, Leforestier A (2000) Chiral discotic columnar germs of nucleosome core particles. Biophys J 78: 2716–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Makarov VL, Dimitrov SI, Petrov PT (1983) Salt-induced chromatin transitions. A flow linear dichroism study. Eur J Biochem 133: 491–497 [DOI] [PubMed] [Google Scholar]
- McGhee JD, Nickol JM, Felsenfeld G, Rau DC (1983) Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell 33: 831–841 [DOI] [PubMed] [Google Scholar]
- Ner SS, Blank T, Perez-Parallé ML, Grigliatti TA, Becker PB, Travers AA (2001) HMG-D and histone H1 interplay during chromatin assembly and early embryogenesis. J Biol Chem 276: 37569–37576 [DOI] [PubMed] [Google Scholar]
- Oudet P, Gross-Bellard M, Chambon P (1975) Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4: 281–300 [DOI] [PubMed] [Google Scholar]
- Pehrson J (1995) Probing the conformation of nucleosome linker DNA in situ with pyrimidine dimer formation. J Biol Chem 270: 22440–22444 [PubMed] [Google Scholar]
- Robinson PJJ, Rhodes D (2006) Structure of the ‘30 nm' chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol 16: 336–343 [DOI] [PubMed] [Google Scholar]
- Robinson PJJ, Fairall L, Huynh VAT, Rhodes D (2006) EM measurements define the dimensions of the “30 nm” chromatin fiber. Evidence for a compact interdigitated structure. Proc Natl Acad Sci USA 103: 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Holley WR, Mian IS, Chatterje A (1998) Chromatin conformation in living cells: support for a zig-zag model of the 30 nm chromatin fiber. J Mol Biol 284: 71–84 [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent D, Richmond TJ (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436: 138–141 [DOI] [PubMed] [Google Scholar]
- Sen D, Mitra S, Crothers DM (1986) Higher order structure of chromatin: evidence from photochemically detected linear dichroism. Biochemistry 25: 3441–3447 [DOI] [PubMed] [Google Scholar]
- Sivolob A, Prunell A (2003) Linker histone-dependent organization and dynamics of nucleosome entry/exit DNAs. J Mol Biol 331: 1025–1040 [DOI] [PubMed] [Google Scholar]
- Smith MF, Athey BD, Williams SP, Langmore JP (1990) Radial density distribution of chromatin: evidence that chromatin fibers have solid centers. J Cell Biol 110: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C, Bellard M, Compton JL, Chambon P (1976) The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett 69: 281–285 [DOI] [PubMed] [Google Scholar]
- Strauss F, Prunell A (1983) Organization of internucleosomal DNA in rat liver chromatin. EMBO J 2: 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A (1979) Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol 83: 403–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Butler PJG (1980) Size-dependence of a stable higher-order structure of chromatin. J Mol Biol 144: 89–93 [DOI] [PubMed] [Google Scholar]
- Thomas JO, Furber V (1977) Yeast chromatin structure. FEBS Lett 66: 274–280 [DOI] [PubMed] [Google Scholar]
- Thomas JO, Thompson RJ (1977) Variation in chromatin structure in two types of cell from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell 10: 633–640 [DOI] [PubMed] [Google Scholar]
- Tremethick DJ (2007) Higher-order structures of chromatin: the elusive 30 nm fiber. Cell 128: 651–654 [DOI] [PubMed] [Google Scholar]
- Widom J (1992) A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci USA 89: 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J, Klug A (1985) Structure of the 300 Å chromatin filament: X-ray diffraction from oriented samples. Cell 43: 207–213 [DOI] [PubMed] [Google Scholar]
- Williams SP, Athey BD, Muglia LJ, Schappe RS, Gough AH, Langmore JP (1986) Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J 49: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H, Victor JM, Mozziconacci J (2007) An all-atom model of the chromatin fibre containing linker histones reveals a versatile structure tuned by nucleosomal repeat length. PLoS ONE 2: e877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Dimitrov S (2001) Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev 11: 130–135 [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Frado LL, Rattner JB (1984) The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol 99: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A, Strogatz S, Riley D (1981) Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci USA 78: 1461–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modelling of the 30nm fibre
Dependence of the conclusion on modelling parameters.