Abstract
Nrf1 (nuclear factor-erythroid 2 p45 subunit-related factor 1) is negatively controlled by its NTD (N-terminal domain) that lies between amino acids 1 and 124. This domain contains a leucine-rich sequence, called NHB1 (N-terminal homology box 1; residues 11–30), which tethers Nrf1 to the ER (endoplasmic reticulum). Electrophoresis resolved Nrf1 into two major bands of approx. 95 and 120 kDa. The 120-kDa Nrf1 form represents a glycosylated protein that was present exclusively in the ER and was converted into a substantially smaller polypeptide upon digestion with either peptide:N-glycosidase F or endoglycosidase H. By contrast, the 95-kDa Nrf1 form did not appear to be glycosylated and was present primarily in the nucleus. NHB1 and its adjacent residues conform to the classic tripartite signal peptide sequence, comprising n-, h- and c-regions. The h-region (residues 11–22), but neither the n-region (residues 1–10) nor the c-region (residues 23–30), is required to direct Nrf1 to the ER. Targeting Nrf1 to the ER is necessary to generate the 120-kDa glycosylated protein. The n-region and c-region are required for correct membrane orientation of Nrf1, as deletion of residues 2–10 or 23–30 greatly increased its association with the ER and the extent to which it was glycosylated. The NHB1 does not contain a signal peptidase cleavage site, indicating that it serves as an ER anchor sequence. Wild-type Nrf1 is glycosylated through its Asn/Ser/Thr-rich domain, between amino acids 296 and 403, and this modification was not observed in an Nrf1Δ299–400 mutant. Glycosylation of Nrf1 was not necessary to retain it in the ER.
Keywords: endoplasmic reticulum, glycosylation, non-alcoholic steatohepatitis (NASH), nuclear factor-erythroid 2-p45 subunit-related factor (Nrf), oxidative stress, signal anchor sequence
Abbreviations: ARE, antioxidant response element; ATF6, activating transcription factor 6; bZIP, basic-region leucine zipper; DMEM, Dulbecco's modified Eagle's medium; ER, endoplasmic reticulum; GFP, green fluorescent protein; GST, glutathione transferase; LDS, lithium dodecyl sulfate; NASH, non-alcoholic steatohepatitis; NHB, N-terminal homology box; NQO1, NAD(P)H:quinone oxidoreductase 1; Nrf, nuclear factor-erythroid 2 p45 subunit-related factor; NTD, N-terminal domain; SPase, signal peptidase; SREBP, sterol-regulatory-element-binding protein
INTRODUCTION
The Nrf1 (nuclear factor-erythroid 2 p45 subunit-related factor 1) cap ‘n’ collar bZIP (basic-region leucine zipper) transcription factor and its isoforms TCF11 (transcription factor 11) and LCR-F1 (locus control region-factor 1) regulate genes that contain an ARE (antioxidant response element) in their promoter [1–5]. Such genes include those encoding NQO1 [NAD(P)H:quinone oxidoreductase 1], both the catalytic and modifier subunits that form glutamate cysteine ligase, haem oxygenase 1, the ferritin heavy subunit and metallothionein 1 [6]. Transcription factor Nrf2 also controls expression of the ARE-gene battery [7–9].
The relative contribution that Nrf1 and Nrf2 make to expression of ARE-driven genes is unclear. Targeted disruption of nrf1 and nrf2 has revealed marked differences in their biological roles. Global knockout of Nrf1 is embryonically lethal to the mouse [10,11]. The survival of hepatocytes appears to be particularly dependent on Nrf1 as none of the livers in seven chimaeric mice, each generated from injection of nrf1−/− JM-1 ES cells into C57BL/6 blastocysts, contained Nrf1 nulled JM-1 cells [12]. Further evidence that this bZIP factor is essential for normal hepatic function comes from the finding that postnatal liver-specific disruption of nrf1 in mice results by 4 weeks in the accumulation of lipid in the liver that is associated with endogenous oxidative stress, increased apoptosis, inflammation and fibrosis [13], a combination of pathological features that is referred to as NASH (non-alcoholic steatohepatitis) [14]. Furthermore, in the mutant mice, NASH progresses spontaneously to hepatocarcinogenesis with preneoplastic lesions being observed at 4 months, and hepatocellular adenomas and carcinomas evident at 10–12 months of age [13]. In contrast with knockout of Nrf1, global knockout of Nrf2 results in mice that are developmentally normal and under non-stressed laboratory conditions have a full life expectancy, but are more sensitive to environmental and chemical insults than wild-type mice [15]. Livers from nrf2−/− mice show a substantial reduction in the expression of ARE-driven genes [16], but they do not develop fatty liver, NASH or hepatoma.
As Nrf1 and Nrf2 are widely expressed in mouse tissues [17], and regulate common genes [6], it is not immediately obvious why knockout of either factor produces different phenotypes. One possible explanation is that they mediate adaptation to redox stress in different subcellular compartments. Support for this hypothesis comes from the recent finding that Nrf1 is targeted to the ER (endoplasmic reticulum) [18,19], whereas Nrf2 is found primarily in the nucleus [18,20].
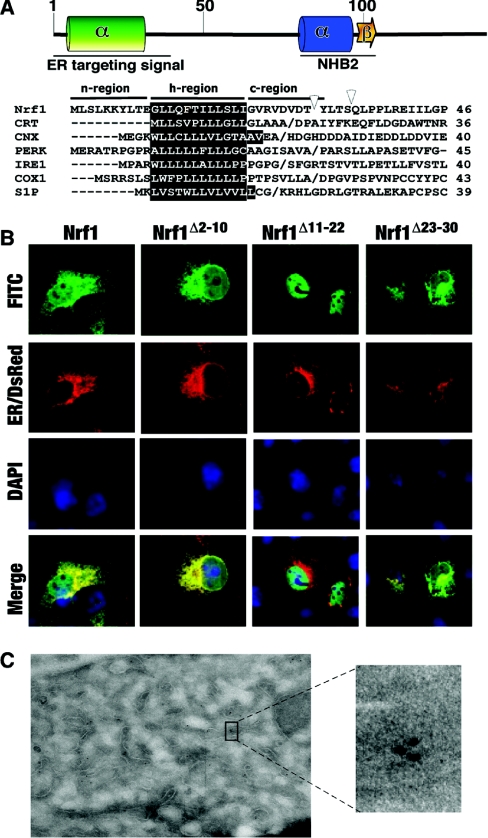
We have found that Nrf1 is directed to the ER through its NTD (N-terminal domain; amino acids 1–124), a region that is not represented in Nrf2 [18]. Two discrete conserved subdomains exist within the NTD of Nrf1, referred to as NHB1 (N-terminal homology box 1; residues 11–30) and NHB2 (residues 81–106) because they share significant sequence identity with equivalent portions of Nrf3 [18] (see Figure 1). Targeting of Nrf1 to the ER has been postulated to involve NHB1 and its neighbouring amino acids [18,19]. It is however unclear whether NHB1 in Nrf1 represents part of a cleavable signal sequence or part of an uncleavable signal anchor sequence. In either event, it would be predicted that the transmembrane peptide requires flanking amino acids at both its N- and C-terminal ends in order to function as a signal sequence. As described in reviews by von Heijne [21] and Martoglio and Dobberstein [22], such sequences are not well conserved but contain the following tripartite features: (i) a core α-helical hydrophobic region (h-region) of 7–15 residues that spans the membrane and is critical for translocation; (ii) a relatively variable hydrophobic or hydrophilic sequence containing some basic amino acids located at the N-terminal end of the transmembrane peptide, called the n-region, that dictates the orientation of insertion of a protein into the ER; and (iii) a hydrophilic sequence of 2–9 residues situated at the C-terminal end of the transmembrane peptide, called the c-region, that determines the cleavage site. Residues in the n- and c-regions influence the function of signal sequences [23]. In the case of a cleavable signal sequence, the c-region frequently contains a recognition site for a SPase (signal peptidase) [24], where proteolysis usually occurs at positions where small residues such as alanine, glycine and serine are found at the −1 position from the cleavage site, and small uncharged residues such as alanine, glycine, leucine, serine and valine are found at the −3 position [25].
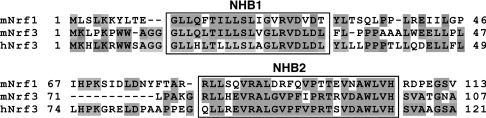
Figure 1. Alignment of N-terminal amino acid sequences in Nrf1 and Nrf3.
Amino acids 1–46 and 67–113 from mouse Nrf1 (mNrf1) are shown aligned with similar regions of mouse Nrf3 (mNrf3) and human Nrf3 (hNrf3). Residues placed on a shaded background represent those that share identity. The positions of NHB1 and NHB2 are indicated.
Only a few transcription factors are targeted to the ER. The best-characterized examples include SREBP-1 (sterol-regulatory-element-binding protein-1), a factor through which insulin promotes hepatic lipid synthesis [26,27], and also ATF6 (activating transcription factor 6), a factor that mediates adaptation to ER stress [28]. Both SREBP-1 and ATF6 are negatively controlled through association with the ER. They are retained in this subcellular compartment under normal metabolic conditions and undergo ER-to-Golgi transport when the levels of cholesterol are low, in the case of SREBP-1, or when misfolded proteins accumulate, in the case of ATF6. Once in the Golgi, SREBP-1 and ATF6 are subject to regulated intramembrane proteolysis, a two-step sequential cleavage process that releases them into the cytoplasm [29]. The first of these steps is catalysed by Site-1 protease and cleaves the factors in the transmembrane domain of the protein. The second proteolysis step is catalysed by Site-2 protease approx. 20 amino acids from the first cleavage site. Following intramembrane proteolysis, SREBP-1 and ATF6 translocate to the nucleus and activate their appropriate target genes. However, it is not known whether Nrf1 is processed by the same enzymes that are responsible for release of SREBP-1 and ATF6 from the ER.
In the present study the hypothesis that the NHB1 in the NTD of Nrf1 is part of a signal sequence has been examined. We have also examined whether cleavage occurs within the NTD as this would indicate whether SPase is involved in the activation of Nrf1.
EXPERIMENTAL
Chemicals and enzymes
Chemicals and enzymes were all of the highest quality available and were readily available commercially. All oligonucleotide primers were synthesized by MWG Biotech and are listed in Supplementary Table 1 at http://www.BiochemJ.org/bj/408/bj4080161add.htm.
Expression plasmids
Complementary DNA encoding mouse wild-type Nrf1 was cloned into the pcDNA3.1/V5His B plasmid (Invitrogen) following KpnI/XbaI digestion [18]. PCR amplification and site-directed mutagenesis were performed to create cDNA for various Nrf1 mutants [30], all of which were cloned into pcDNA3.1/V5His B.
Sequences encoding the NTD of Nrf1, along with several internal deletion mutants, were generated by PCR using suitable primers (see Supplementary Table 1) along with the appropriate Nrf1 expression constructs as templates. The cDNA for the N-terminal 156 residues (N156) of Nrf1 was similarly amplified. The PCR products for the NTD and N156 were ligated to the 5′-end of the cDNA for Nrf2 in which Met1 and Met2 had been mutated into valine residues, before being inserted into pcDNA3.1/V5His B to allow production of various Nrf2-containing fusion proteins. An essentially identical strategy was adopted to produce GFP (green fluorescent protein) fusions. Constructs expressed NTD/GFP×2 or N65/GFP×2 in which the cDNAs encoding the NTD or the N-terminal 65 residues of Nrf1 (N65) were amplified using primer pairs listed in Supplementary Table 1 and ligated to the 5′-end of a cDNA encoding two fused copies of GFP; in these constructs the first Kozak's consensus translation initiation site (GCCACCATGG) in each of the two GFP were eliminated, to ensure that free GFP was not produced [31]. The fidelity of all DNA products was confirmed by automated sequencing.
The pRL-TK plasmid (Promega) encoding renilla, as well as pcDNA4/HisMax/lacZ (Invitrogen) encoding β-galactosidase (β-gal), was used to control for transfection efficiency. The pDsRed2-ER plasmid (Clontech), used in confocal microscopy experiments, encoded ER/DsRed, an ER marker protein.
Cell culture, transfection and luciferase reporter assays
Monkey kidney COS-1 cells (5×105) were seeded in 6-well plates and grown for 24 h in DMEM (Dulbecco's modified Eagle's medium). After the cells reached 70% confluence, they were transfected with Nrf1, Nrf2 or NTD/Nrf2 expression constructs and the PTKnqo1-ARE-Luc reporter plasmid using Lipofectin® reagent or Lipofectamine™ 2000 (Invitrogen). Reporter gene activity was determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The activity was normalized to that of the Renilla reporter and was calculated, with reference to the value obtained using the empty pcDNA3.1/V5His B, as a fold change [18]. The significance of differences between the transactivation activity of Nrf1 and its mutants was determined using the Student's t test.
Immunocytochemistry, confocal and electron microscopy
Immunocytochemistry and confocal microscopy for Nrf1, GFP and DsRed were performed in COS-1 cells as described previously [18]. For electron microscopy of V5 epitope-tagged Nrf1, cells were fixed in 0.1% (v/v) glutaraldehyde and 4% (v/v) para-formaldehyde in 200 mM Hepes/NaOH (pH 7.4) for 30 min at room temperature (22 °C). Once fixed, the cells were scraped from culture dishes, centrifuged (at 500 g for 5 min at 4 °C) and snap-frozen in liquid nitrogen before ultrathin frozen sections were prepared and gold labelling was performed [32].
Subcellular fractionation
Nuclear fractions were prepared as described previously [20]. Membrane fractions containing intact ER were obtained by sucrose density-gradient purification [33]. Briefly, COS-1 cells were scraped in serum-free DMEM and centrifuged at 600 g for 5 min at 4 °C. The cell pellets were suspended in 4 pellet-volumes of 1×isotonic extraction buffer [10 mM Hepes (pH 7.8), containing 250 mM sucrose, 1 mM EGTA, 1 mM EDTA and 25 mM KCl] supplemented with 1% (v/v) Complete protease inhibitor cocktail (Roche Applied Science). The resuspended cells were gently homogenized by repetitively passing the mixture through a 23-gauge needle with 20 strokes. The resulting homogenate was centrifuged at 1300 g for 10 min at 4 °C to sediment nuclei, along with the remaining intact cells, and the supernatant was re-centrifuged further at 17000 g for 30 min at 4 °C. The pellet obtained from the 17000 g centrifugation was resuspended in 0.5 ml of MS buffer [5 mM Tris/HCl (pH 7.5), containing 210 mM mannitol, 70 mM sucrose, 1 mM EDTA and 1% Complete protease inhibitor cocktail] and overlaid on to a three-part discontinuous gradient comprising 0.5 ml aliquots of buffered solutions [10 mM Tris/HCl (pH 7.5) and 1 mM EDTA] containing, from top to bottom, 1.0, 1.2 and 1.5 M sucrose [33]. This resuspended pellet was then centrifuged at 39000 g for 30 min at 4 °C. Intact ER-enriched and mitochondria-containing fractions were collected as sedimented material that formed at the interphases between 1.0/1.2 M sucrose buffer and 1.2/1.5 M sucrose buffer respectively. The sediment that formed at each of these two interfaces was transferred to separate tubes and washed with 1.5 ml of MS buffer, centrifuged at 12000 g for 10 min at 4 °C before being dissolved in RIPA buffer [50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1% Ipegal, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM dithiothreito1 and a protease inhibitor cocktail]. The supernatant from the first 17000 g centrifugation step was further ultracentrifuged at 100000 g for 60 min at 4 °C to pellet the relatively lighter membrane components that represented microsomes. The protein in the 100000 g supernatant fraction was concentrated by acetone precipitation, and kept as the cytosolic fraction.
In vitro deglycosylation reactions
Portions (30 μg protein) of either total lysates or subcellular fractions from COS-1 cells transfected with expression constructs for wild-type or mutant Nrf1 protein were heated at 100 °C for 10 min in 15 μl of denaturing buffer consisting of 0.5% SDS, 1% 2-mercaptoethanol and 1% (v/v) Complete protease inhibitor cocktail. The heat-denatured samples were incubated at 37 °C for 1 h with 500 units of PNGase (peptide:N-glycosidase) F or Endo H (endoglycosidase H; both from New England Biolabs) in a final volume of 20 μl containing 1×reactive buffer G7 [50 mM sodium phosphate (pH 7.5) and 1% NP40 (Nonidet P40)] or G5 (50 mM sodium citrate, pH 5.5) respectively. These reactions were stopped by being diluted with 4×NuPAGE LDS (lithium dodecyl sulfate) sample buffer and heated at 100 °C for 10 min. A 10 μg aliquot of ribonuclease B was used as a control protein for the deglycosylation reaction.
Western blotting
Cell lysates were prepared in 1×RIPA buffer and were then clarified by centrifugation at 16000 g for 10 min. If appropriate, supernatant fractions were normalized to β-gal activity [18]. Samples for electrophoresis were diluted in 4×NuPAGE LDS sample buffer (pH 8.4, Invitrogen) and then resolved using LDS/NuPAGE with either the 7% Tris/acetate or 4–12% gradient Bis/Tris gel systems. Normally, the Nrf1 proteins were C-terminally V5 tagged and were visualized by Western blotting using an antibody against the V5 epitope (Invitrogen) [18]. For some experiments an antibody was used that was raised in rabbits against residues 292–741 of Nrf1. This polypeptide was generated by ligating the cDNA for Nrf1Δ1–291 to the 3′ end of the coding sequence of GST (glutathione transferase) in the pGEX-6PA expression vector through an NcoI/XhoI site; the resulting GST/Nrf1Δ1–291 fusion protein was subject to PreScission™ protease (Amersham) digestion to allow removal of the GST prior to immunization.
Bioinformatic searches
Multiple amino acid sequence alignments were carried out using the ClustalW from EMBL-EBI (http://www.ebi.ac.uk/clustalw/). The ER signal sequence in Nrf1 was predicted using both the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) and the PolyPhobius software program (http://phobius.cgb.ki.se/cgi-bin/predict.pl).
RESULTS
Identification of regions within the NTD that mediate negative regulation of Nrf1
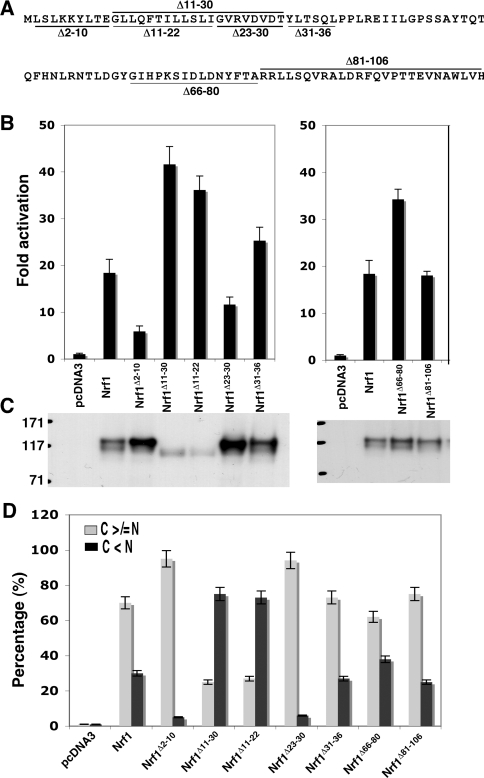
Expression constructs encoding Nrf1 mutants lacking sequentially larger portions of the N-terminus were generated (Figure 2A) to help evaluate whether both NHB1 (residues 11–30) and NHB2 (residues 82–106) negatively regulated the bZIP factor. These were each transfected into COS-1 cells along with the PTKnqo1-ARE-Luc reporter plasmid. Transactivation of ARE-driven gene expression by Nrf1 decreased significantly when residues 2–10, flanking NHB1, were omitted from its N-terminus (Figure 2A). Progressive deletion of increasing portions of the NTD from Nrf1 led to a discrete step-wise increase in luciferase reporter gene activity following removal of residues 11–22 (i.e. the leucine-rich core of NHB1), and again following deletion of residues 66–80 (i.e. the region flanking the N-terminal boundary of NHB2).
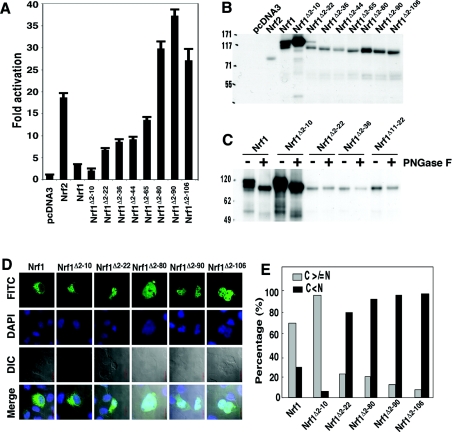
Figure 2. Discrete regions within the NTD regulate Nrf1 activity and its localization.
(A) Portions (1.2 μg DNA) of expression constructs for wild-type Nrf1 and its mutants possessing increasing N-terminal deletions were each transfected into COS-1 cells together with 0.6 μg of PTKnqo1-ARE-Luc and also 0.2 μg of both the β-gal and pRL-TK reporter plasmids. Luciferase reporter gene activity is shown as a fold-change (mean±S.D.) from three independent experiments, each in triplicate, relative to the activity observed following transfection with the empty pcDNA3.1 expression vector. (B) The wild-type and mutant V5-tagged Nrf1 proteins were examined by Western blotting following electrophoresis in 7% LDS/NuPAGE containing a Tris/acetate buffer system. The amount of cell lysates added to each sample well was normalized to ensure equal loading of β-gal activity. (C) Heat-denatured samples (30 μg protein of each) from total lysates of COS-1 cells transfected with Nrf1 expression constructs were digested for 1 h at 37 °C by PNGase F (500 units). The reaction products were separated on 4–12% LDS/NuPAGE Bis/Tris gels and visualized by Western blotting. (D) COS-1 cells were transfected with expression constructs for V5-tagged wild-type Nrf1 or its truncation mutants (1.3 μg DNA of each) and allowed to recover for 24 h before the location of the ectopic proteins was examined by immuocytochemistry followed by confocal imaging. A FITC-labelled secondary antibody was used to locate V5-tagged proteins. Nuclear DNA was stained by DAPI (4′-6-diamidino-2-phenylindole). Images labelled as DIC (differential interference contrast) are those from normal light microscopy. The merge signal represents the results obtained when the three images were superimposed. Scale bar=20 μm. (E) The cytoplasmic–nuclear distribution of the FITC signals was quantified by calculating the percentage of cells (100 cells were counted) with predominantly extra-nuclear (cytoplasmic, C) and nuclear (N) staining.
Production of the 120-kDa glycosylated Nrf1 isoform is dependent on NHB1
To examine whether differences in transactivation of ARE-driven transcription affected by the various Nrf1 mutants might be due to either modification of the factor or its relative abundance, Western blotting experiments were performed. Wild-type Nrf1 was resolved by electrophoresis using the NuPAGE Novex® Tris/acetate gel system into several discrete bands. The smallest of these had an apparent molecular mass of approx. 95 kDa, whereas the largest had an apparent mass of approx. 120 kDa (Figure 2B). In addition, an electrophoretic band with an intermediate mobility was frequently observed that was estimated to have a molecular mass of between 105 and 110 kDa. This additional band was not always clearly discernible, possibly because of its inherent biological variability, or because of incomplete resolution of the band in the NuPAGE electrophoresis system. The finding that wild-type Nrf1 yielded multiple electrophoretic bands suggests that it is subject to glycosylation and/or proteolytic processing. In contrast with the wild-type protein, the Nrf1Δ2–10 mutant yielded essentially a single major polypeptide band that migrated in NuPAGE Tris/acetate gel with a mass of approx. 120 kDa (Figure 2B). The abundance of Nrf1Δ2–10 protein and its high molecular mass were intriguing, given the fact that it was particularly poor at transactivating ARE-driven luciferase activity. In comparison with ectopic Nrf1Δ2–10, the Nrf1Δ2–22 mutant translated into two relatively minor protein products. These had molecular masses of 95 kDa and 115 kDa. Other expression constructs for Nrf1 deletion mutants, in which residues 2–106 were progressively and cumulatively deleted, each yielded a unique single polypeptide of between 75 and 90 kDa (Figure 2B). Comparison of the luciferase reporter gene data (Figure 2A) with the Western blot analysis (Figure 2B) suggested that the presence of the 120-kDa form of Nrf1 was associated with lower ARE-driven gene activity, whereas the presence of the 95-kDa form of Nrf1 was associated with higher transactivation of ARE-driven genes. We subjected the wild-type and Nrf1 mutants to enzymatic deglycosylation to test whether the Nrf1 electrophoretic 120-kDa band represents a glycosylated form of the bZIP protein, and whether the increase in the 120-kDa band produced by Nrf1Δ2–10 is because it is extensively glycosylated. As shown in Figure 2(C), digestion of COS-1 cell lysates expressing wild-type Nrf1 with PNGase F resulted in loss of the two high-molecular-mass forms of the protein (i.e. the bands of approx. 105 kDa and 120 kDa) and a corresponding increase in the faster migrating 95-kDa protein. As postulated, cell lysates expressing Nrf1Δ2–10 contained almost exclusively the 120-kDa protein, but following digestion with PNGase F this band disappeared to be replaced with a 95-kDa band. Similar experiments using COS-1 lysates expressing Nrf1Δ2–22, Nrf1Δ2–36 or Nrf1Δ11–22 showed that the single electrophoretic band they each yielded, of between 90 and 95 kDa, did not change following digestion with PNGase F (Figure 2C). These results suggest that a major fraction of Nrf1 is glycosylated, and that the Nrf1Δ2–10 mutant is more extensively glycosylated than the wild-type protein. In contrast, the Nrf1 mutants lacking residues 11–36, which include NHB1, appear to escape glycosylation.
Sequences associated with NHB1 control the subcellular distribution of Nrf1
We performed immunocytochemistry to relate the ability of the Nrf1 mutants to transactivate ARE-driven gene expression with their subcellular distribution (Figure 2D). This revealed that approx. 30% of COS-1 cells expressing ectopic wild-type Nrf1 showed predominantly nuclear staining, with approx. 70% of cells showing primarily extra-nuclear staining (Figure 2E). By contrast, following transfection with an expression construct for Nrf1Δ2–10, only 5% of the cells exhibited principally nuclear staining. Ectopic Nrf1Δ2–22, Nrf1Δ2–80, Nrf1Δ2–90 and Nrf1Δ2–106 proteins demonstrated an increasingly greater propensity to accumulate in the nucleus rather than in an extra-nuclear compartment (Figure 2E). These results also suggest that the low reporter gene activity observed with Nrf1Δ2–10, which is associated with the 120-kDa polypeptide, correlates with it being targeted to an extra-nuclear cellular compartment. Further, the high activity of Nrf1 mutants lacking NHB1, which is associated with the electrophoretically fast migrating band, correlates with greater nuclear accumulation of the protein.
NHB1 is required for the negative regulation of Nrf1, its ER targeting, and its glycosylation
Having studied N-terminal deletion mutants, we engineered a series of expression constructs for Nrf1 containing internal deletions to help identify more accurately sequences involved in repression of its activity (Figure 3A). In COS-1 cells, a mutant lacking the NHB1, Nrf1Δ11–30, exhibited significantly greater transactivation activity than wild-type Nrf1 (Figure 3B, left-hand panel). The Nrf1Δ11–22 mutant exhibited a similar increase in transactivation activity as Nrf1Δ11–30, suggesting that the N-terminal 60% of NHB1 is essential for negative regulation. This conclusion was supported by the fact that Nrf1Δ23–30 possessed less transactivation activity than wild-type Nrf1. In Nrf1Δ31–36, deletion of residues flanking the C-terminal border of NHB1 did not increase transactivation activity. By contrast, in Nrf1Δ66–80, a moderate enhancement in ARE-driven reporter gene activity was observed when residues immediately adjacent to NHB2 were deleted. The transactivation activity of Nrf1 was not markedly affected by deletion of NHB2 residues 81–106 (Figure 3B, right-hand panel).
Figure 3. NHB1, but not NHB2, regulates Nrf1 by controlling its subcellular distribution.
(A) Diagrammatic representation of sequences deleted from within the NTD of Nrf1. (B) The expression constructs for wild-type Nrf1 or those lacking the sequences shown in (A) (1.2 μg DNA of each) were transfected into COS-1 cells together with 0.6 μg of PTKnqo1-ARE-Luc and 0.2 μg of the pRL-TK and β-gal reporter plasmids. Approx. 36 h after transfection, the cells were harvested and luciferase reporter activity was measured. The transactivation activity was calculated and is presented as a fold change (mean±S.D.) from three independent experiments, each performed in triplicate. (C) Ectopic V5-tagged Nrf1 proteins in the COS-1 cells were resolved by 7% NuPAGE and examined by immunoblotting; samples loaded in each well contained equal amounts of β-gal activity. (D) The Nrf1 expression constructs (1.3 μg DNA of each), together with an ER/DsRed construct, were cotransfected into COS-1 cells. Subsequently, the Nrf1 proteins were analysed using immunocytochemistry with a FITC-coated secondary antibody, followed by confocal microscopy, as described in the legend for Figure 2. The quantitative data shown here were calculated by determining the percentage of cells showing predominantly an extra-nuclear stain (i.e. cytoplasmic plus ER, called simply C) was greater than or equal to the nuclear stain (called N), as opposed to the percentage of cells in which the extranuclear stain was less than the nuclear stain (100 cells were counted); some of the images from which these data were obtained are shown in Supplementary Figure S2 at http://www.BiochemJ.org/bj/408/bj4080161add.htm.
Two Nrf1 mutants which exhibited the highest transactivation activity, namely Nrf1Δ11–30 and Nrf1Δ11–22, both migrated during NuPAGE as a single 95-kDa band (Figure 3C). In the case of Nrf1Δ11–22, this band has been shown by PNGase F digestion to represent a non-glycosylated polypeptide (Figure 2C). Importantly, neither Nrf1Δ11–30 nor Nrf1Δ11–22 yielded the 120-kDa glycosylated electrophoretic band.
Immunocytochemistry showed that these two mutants were present in the nucleus at particularly high levels (Figure 3D and Supplementary Figure S1 at http://www.BiochemJ.org/bj/408/bj4080161add.htm). As was noted for Nrf1Δ2–10 above, Nrf1Δ23–30 was found to exhibit low transactivation activity, it possessed a mass of 120 kDa, and was present almost exclusively in an extra-nuclear compartment. On the other hand, deletion of NHB2 yielded a protein that exhibited a similar subcellular localization to wild-type Nrf1 (Figure 3D).
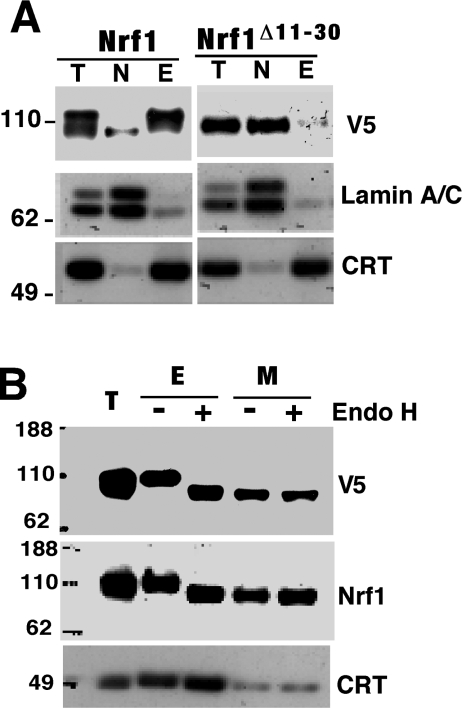
Subcellular fractionation was performed to confirm that NHB1 is required to target Nrf1 to the ER, and that this event is necessary to allow formation of the 120-kDa glycosylated form. Figure 4(A) shows that the wild-type Nrf1 120-kDa polypeptide was almost entirely present in the intact ER fraction (E), and that the wild-type Nrf1 95-kDa subunit was found predominantly in the nuclear fraction (N). Mutant Nrf1Δ11–30 was represented by a 95-kDa band that was only recovered in the nuclear fraction (Figure 4A, right-hand panel). In vitro digestion with Endo H increased substantially the electrophoretic mobility of Nrf1 protein in the ER, resulting in its apparent molecular mass being reduced from 120 kDa to approx. 95 kDa (Figure 4B). Together with data shown in Figure 2(C), these results indicate that the NHB1 sequence is required for both ER targeting of Nrf1 and its glycosylation within this organelle.
Figure 4. Nrf1 is glycosylated in the ER.
(A) COS-1 cells were transfected with expression constructs for wild-type Nrf1. At 36 h after transfection, these cells were subjected to subcellular fractionation as described in the Experimental section. Proteins in the different fractions were resolved by electrophoresis using the 4–12% NuPAGE Bis/Tris gel system and visualized by Western blotting. The abbreviations at the top of the lanes in the gels are as follows: T, total cell lysates; N, nuclei purified from NP40 homogenates; E, intact ER-enriched fraction obtained from sucrose density-gradient purification. Blotting with antibodies against calreticulin (CRT) and Lamin A/C was undertaken to assess the purity of subcellular fractions. (B) Membrane fractions containing microsomes (M) were pelleted by centrifugation at 100000 g for 1 h. The E and M fractions purified from COS-1 cells expressing ectopic Nrf1 were heat-denatured and then digested by Endo H. The reaction products were resolved by electrophoresis and visualized by Western blotting using antibodies against the V5 epitope and Nrf1.
Nrf1 contains an ER targeting signal at its N-terminus
Bioinformatic analyses predict that residues 7–26 in Nrf1 form a transmembrane α-helix, in which residues 11–22 span the hydrocarbon core region of the phospholipid bilayer [34], and residues 7–10 and 23–26 may occupy respectively, the relatively hydrophilic interfaces of the bilayer head-groups at the cytoplamic side and the ER luminal side. According to the classic structural organization of signal peptides [22], residues 1–10, 11–22 and 23–30 in Nrf1 may correspond to n-region, h-region and c-region sequences respectively (Figure 5A). The contribution that these sequences make to the ER targeting of Nrf1 was examined by confocal microscopy. Ectopic wild-type Nrf1 gave a confocal image that was similar, but not identical, to that obtained from ER/DsRed, a fusion protein containing both the ER targeting and retention signals of calreticulin (Figure 5B); wild-type Nrf1 gave greater nuclear staining than ER/DsRed. However, the fluorescence signal obtained from Nrf1Δ2–10 more closely resembled that from ER/DsRed than did wild-type Nrf1. By contrast, Nrf1Δ11–22 gave a strong nuclear stain that was clearly distinct from the ER/DsRed signal (Figure 5B). Like Nrf1Δ2–10, the stain obtained from Nrf1Δ23–30 was similar to that produced by ER/DsRed. COS-1 cells expressing wild-type Nrf1, Nrf1Δ2–10, Nrf1Δ11–22 and Nrf1Δ23–30 were subjected to immuno-electron microscopy to determine more accurately their localization. This revealed that wild-type Nrf1 protein was targeted to the ER, but not the mitochondria (Figure 5C).
Figure 5. Nrf1 contains an ER-targeting signal sequence around the NHB1.
(A) NHB1 and its flanking residues are predicted to form a hydrophobic α-helix, whereas the residues around NHB2 may fold as a basic amphipathic α-helix and β-sheet structure. Multiple sequence alignments are shown of signal sequences from Nrf1 and other ER-resident proteins including CRT (calreticulin), CNX (calnexin), PERK (PKR-related ER kinase), IRE1 (inositol-requiring kinase 1), COX1 (cyclo-oxygenase 1) and S1P (Site-1 protease). (B) Expression constructs for wild-type Nrf1 and its specific deletion mutants around NHB1, along with an ER/DsRed construct encoding an ER marker protein, were cotransfected into COS-1 cells. Subsequent immunocytochemistry and imaging were performed as described in the legend of Figure 2. Scale bar=20 μm. (C) COS-1 cells were transfected with Nrf1/pcDNA3.1/V5His before being subjected to immuno-electron microscopy. The dots in original and enlarged images represent positive signals for ectopic Nrf1 protein.
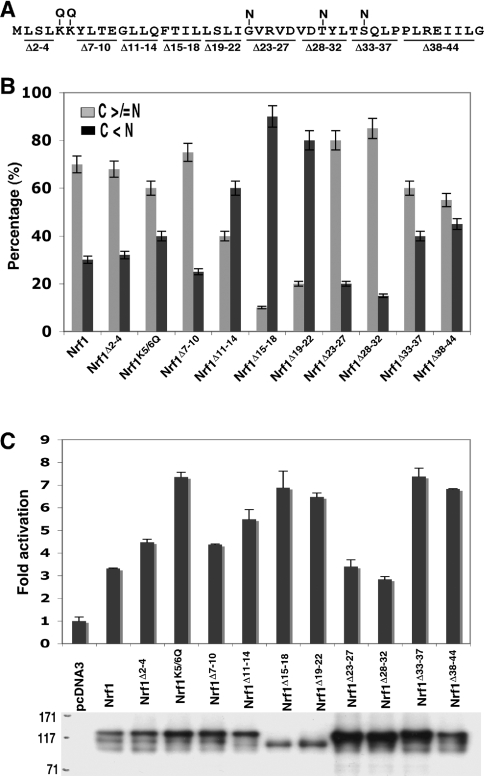
The NHB1 and its flanking residues comprise a signal-anchoring sequence
A series of Nrf1 mutants with small internal deletions and point mutations (Figure 6A) were generated to explore the contribution made by certain N-terminal residues to signal sequence function. In constrast with Nrf1Δ2–10, deletion of residues 2–4 and 7–10 from Nrf1 did not substantially change ER localization, transactivation activity or the electrophoretic banding pattern of the proteins (Figures 6B and 6C, and see Supplementary Figure S2 at http://www.BiochemJ.org/bj/408/bj4080161add.htm), suggesting that the number of residues required in the putative n-region for correct orientation of Nrf1 in the ER membrane is relatively small. It is probable that Nrf1 obeys the positive-inside rule [35–37] and, if so, the di-lysine sequence at positions 5 and 6 would control translocation of the signal peptide into the lumen of the ER. To test this hypothesis we mutated the di-lysine to di-glutamine (Nrf1K5/6Q) and found that this led to a protein with a modest increase in nuclear localization and a more substantial increase in transactivation activity, but there appeared to be no accompanying decrease in the level of the 120-kDa electrophoretic band (Figures 6B and 6C).
Figure 6. Identification of the signal anchor peptide in Nrf1.
(A) Diagrammatic representation depicting amino acids deleted (shown as horizontal lines) or mutated (shown as a vertical lines) in and around NHB1. (B) Expression constructs for these proteins (1.3 μg DNA of each), along with an ER/DsRed construct, were cotransfected into COS-1 cells. Subsequent immunocytochemistry and confocal imaging were performed as described in the legend for Figure 2. Quantification of data was achieved by calculating the percentage of cells in which the cytoplasmic stain, including the ER, was greater than or equal to the nuclear stain (100 cells were counted). (C) Each of the indicated expression constructs, together with PTKnqo1-ARE-Luc, pRL-TK and β-gal plasmids, were transfected into COS-1 cells. Subsequent luciferase reporter assays and immunoblotting were carried out as described in the text.
Among three h-region mutants, Nrf1Δ15–18 and Nrf1Δ19–22 possessed the highest nuclear accumulation and transactivation activity, and during electrophoresis both ran as a high-mobility 95-kDa band (Figures 6B and 6C), indicating that residues 15–22 are required to target Nrf1 to the ER. By contrast, residues 11–14 appeared to be dispensable.
We also examined c-region mutants. Deletion of either residues 23–27 or 28–32 had little effect on transactivation of ARE-driven gene expression, but both appeared to increase the amount of the 120-kDa subunit. Unexpectedly, deletion of either residues 33–37 or 38–44, flanking the c-region increased transactivation activity without increasing the relative amount of the 95-kDa polypeptide.
Based on bioinformatic predictions [24,25], we tested the hypothesis that the peptide bond adjacent to Gly23, Thr30 or Ser34 might be cleaved by SPase. Mutation of all three residues into asparagine, either individually or collectively (Nrf1G23/T30/S34N), had no effect on its ability to locate to the nucleus, its electrophoretic protein pattern or its ability to activate ARE-driven reporter gene (see Supplementary Figure S2). These findings suggest that no SPase cleavage site exists around the c-region, indicating that NHB1 acts as a signal anchor sequence.
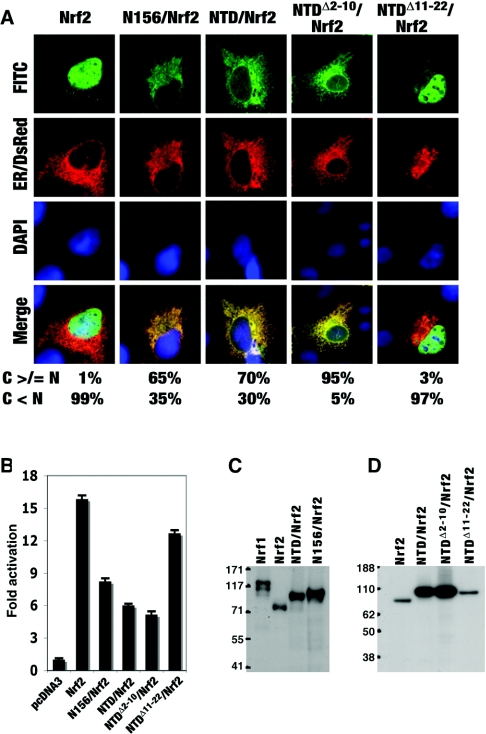
NHB1 is responsible for directing NTD/Nrf2 fusion proteins to the ER
Attachment of the NTD from Nrf1 to the N-terminus of Nrf2 results in the fusion protein being redirected from the nucleus to the ER [18]. We therefore decided to use NTD/Nrf2 as a model to test whether the NHB1 is essential for targeting chimaeric proteins to the ER, and also to examine whether the NTD is itself the object of glycosylation or proteolytic cleavage. Figure 7(A) shows that ectopic Nrf2 is essentially a nuclear protein, but attachment of either residues 1–156 of Nrf1 or residues 1–125 of Nrf1, to its N-terminus, giving N156/Nrf2 or NTD/Nrf2 respectively, creates fusion proteins that locate primarily to an extra-nuclear compartment. Removal of residues 2–10 from NTD/Nrf2 increased targeting of the fusion protein to the ER, whereas removal of residues 11–22 from NTD/Nrf2 largely negated the ability of the NTD to redirect Nrf2 to this organelle. Reporter assays showed that the ability of Nrf2 to transactivate an ARE-driven luciferase gene was inhibited by it being fused at its N-terminus with NTD (Figure 7B). This inhibition was increased modestly when residues 2–10 were deleted from the NTD. However, the inhibition achieved by attaching NTD to the N-terminus of Nrf2 was diminished substantially when residues 11–22 were deleted (Figure 7B). Thus the finding that residues 11–22 in the NTD are required for the negative control of Nrf1 and its targeting to the ER can be recapitulated in an NTD/Nrf2 fusion protein.
Figure 7. The NHB1 of Nrf1 redirects Nrf2 fusion proteins to the ER.
(A) COS-1 cells were transfected with expression constructs for chimaeric proteins containing the NTD and its mutants from Nrf1 fused to the N-terminus of the full-length Nrf2, as described in the Experimental section. Subcellular localization of these fusion proteins was determined by immuocytochemistry and confocal imaging as described in the legends for Figures 2 and 3. Scale bar=20 μm. (B) The transactivation activity of these fusion proteins was examined as described in the text. (C and D) The V5-tagged fusion proteins were separated by NuPAGE in a 7% polyacrylamide Tris/acetate buffer system (C) or 4–12% polyacrylamide gradient Bis/Tris buffer system (D).
Ectopic NTD/Nrf2 and N156/Nrf2 fusion proteins both migrated during LDS/NuPAGE as a single band rather than the multiple polypeptides generated by Nrf1 (Figure 7C). V5-tagged Nrf2 migrated with an apparent molecular mass of 75 kDa, whereas similarly tagged NTD/Nrf2 and N156/Nrf2 migrated with masses of approx. 92 kDa and 96 kDa respectively. The increased size of the fusion proteins is consistent with them containing either 125 or 156 extra amino acids. We also found that both NTDΔ2–10/Nrf2 and NTDΔ11–22/Nrf2 migrated during electrophoresis as a single band, and that they were indistinguishable from NTD/Nrf2 (Figure 7D); in the context of a V5-tagged polypeptide of 722 amino acids, the loss of 9 or 12 residues is unlikely to be detected by NuPAGE. By contrast, Nrf1Δ2–10 and wild-ype Nrf1 are clearly electrophoretically distinct, with Nrf1Δ2–10 yielding more of the 120-kDa glycosylated polypeptide found in the ER (Figure 2B). These results suggest that the 120-kDa Nrf1 electrophoretic band results from modification of a domain other than the NTD. Furthermore, as NTD/Nrf2 and NTDΔ2–10/Nrf2 were still targeted to the ER, it appears that the post-translational modification is not required to direct them to this organelle.
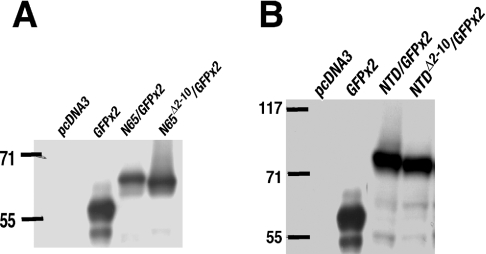
The NTD does not contain a proteolytic cleavage site
In the experiments described above, we found no evidence for the presence of an SPase cleavage site in the ER signal peptide of Nrf1. However, as loss of approx. 30 residues from V5-tagged Nrf1 might be difficult to identify by LDS/NuPAGE, we examined the electrophoretic properties of a series of NTD/GFP×2 fusion proteins because they were considerably smaller than Nrf1. Immunoblotting COS-1 cell lysates expressing these fusion proteins revealed that both N65/GFP×2 and NTD/GFP×2 were clearly resolved from GFP×2 by LDS/NuPAGE (Figures 8A and 8B). During electrophoresis, the GFP×2 protein migrated with an apparent molecular mass of 58 kDa, whereas N65/GFP×2 was estimated to have a mass of 65 kDa (Figure 8A) and NTD/GFP×2 had a mass of 73 kDa (Figure 8B). The relative mobilities of N65/GFP×2 and NTD/GFP×2 are consistent with them containing an additional 65 and 125 amino acids respectively. The NTDΔ2–10/GFP×2 also gave a single polypeptide of approx. 72 kDa.
Figure 8. The NTD/GFP×2 fusion proteins do not contain a SPase cleavage site.
(A and B) COS-1 cells were transfected with an expression construct for GFP×2 (two copies) fused at its N-terminus to various portions of the NTD of Nrf1 that contain just NHB1 (i.e. N65) (A) or the NHB1 and the NHB2 (i.e. NTD) (B). Wild-type or mutant forms of N65 or NTD were fused to GFP×2 as indicated; it should be noted that nucleotides for the GFP translation initiation site in the two proteins were mutated in these fusion constructs to prevent synthesis of GFP×2 alone. The cell lysates were analysed by Western blotting with antibodies against GFP.
Nrf1 is glycosylated within its NST domain
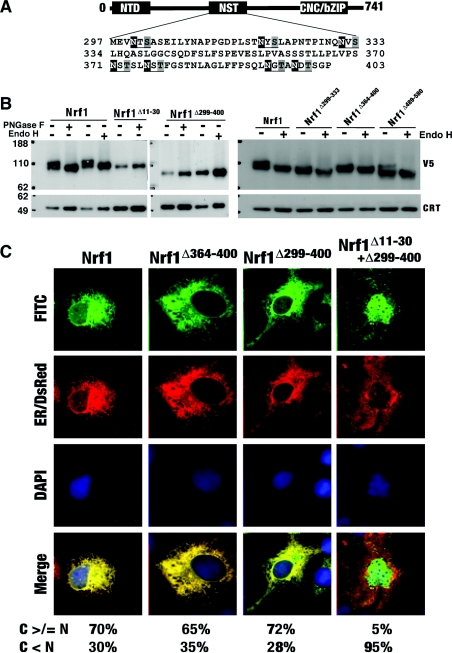
PNGase F and Endo H digestions revealed that Nrf1 is glycosylated through an NHB1-mediated association with the ER (Figures 2 and 4). Examination of various Nrf2- and GFP-containing fusion proteins suggests that the glycosylation of Nrf1 does not occur within its NTD (Figures 7 and 8). Thus the identity of the glycosylation site(s) and its significance requires clarification. Figure 9(A) shows that the NST domain (residues 296–403) of Nrf1 contains seven asparagine-glycosylation consensus sites (-Asn-X-Ser/Thr-, where X is any amino acid except proline [38]). Deletion of residues 299–400 from Nrf1 gave rise to a mutant protein that migrated as a single electrophoretic band of approx. 75 kDa. The mobility of this protein was unaltered by digestion with either PNGase F or Endo H (Figure 9B, middle panel), even though it was localized in the ER (Figure 9C). By contrast, although the Neh6-like domain (residues 489–580) has a single consensus site at Asn543, deletion of this domain in Nrf1Δ489–580 yielded two major electrophoretic protein bands, the slower of which was diminished after digestion with Endo H (Figure 9B, right-hand panel). These data indicate that Nrf1 is asparagine-glycosylated within its NST, but not its Neh6-like, domain. Within the NST domain three asparagine-glycosylation sites are located between residues 299 and 333 and four asparagine-glycosylation sites are located between residues 364 and 400 (see Figure 9A). Deletion of either of these regions from Nrf1 did not prevent its glycosylation (Figure 9B, right-hand panel), suggesting that Nrf1 is probably modified at several sites within its NST domain. Nrf1Δ11–30 migrated during electrophoresis as a single band of approx. 95 kDa and its mobility was unaffected by digestion with either PNGase F or Endo H (Figure 9B). Therefore Nrf1 has to be targeted to the ER in order that asparagine-glycosylation of the factor can occur through its NST domain. In Figure 9(C) we show that Nrf1Δ299–400 is located primarily in the ER, suggesting that glycosylation is not required for the factor to be retained in this organelle.
Figure 9. Nrf1 is asparagine-glycosylated within its NST domain in the ER.
(A) Seven asparagine-glycosylation consensus sites (Asn-X-Ser/Thr) were identified in the NST domain of Nrf1. The +1 position asparagine is shown on a black background, whereas serine or threonine at the +3 position is shown on a grey background. (B) Total lysates of COS-1 cells that had been transfected with expression constructs for wild-type Nrf1 and the mutants indicated were heat-denatured and then digested for 1 h at 37 °C with either PNGase F or Endo H. The reaction products were analysed by a 4–12% NuPAGE Bis/Tris gel system and were visualized by Western blotting using antibodies against the V5 epitope. Calreticulin (CRT) was used as a loading control. (C) The COS-1 cells were cotransfected with an ER/DsRed construct together with each of the indicated expression constructs for Nrf1 and its mutants. Subsequent confocal imaging was performed as described in the legend for Figure 2. Correspondingly quantitative data were shown by calculating the percentage of cells (100 cells were counted). Scale bar=20 μm.
DISCUSSION
We have characterized the NTD in Nrf1 because it appears to be, at least partially, responsible for the unique activity of this cap ‘n’ collar bZIP factor. Database searching has revealed two motifs in the NTD of Nrf1 that are conserved in the N-terminal region of Nrf3. The first of these, between residues 11 and 30, was designated NHB1 and the second, between residues 82 and 106, was designated NHB2 [18]. The purpose of the present study was to determine whether NHB1 and/or NHB2 are responsible for the negative regulation of Nrf1.
NHB1 and its adjacent sequences anchor Nrf1 to the ER
In the present study, we have shown that inhibition of Nrf1 activity by the NTD can be almost entirely attributed to NHB1, rather than NHB2, and that this sub-domain is responsible for targeting Nrf1 to the ER. To gain further insight into the mechanism responsible for directing Nrf1 to the ER we mutated sequences within and around NHB1. Confocal microscopy provided evidence that the peptide GLLQFTILLSLI (single-letter amino acid code) between amino acids 11 and 22 represents the core hydrophobic α-helix (h)-region of a signal sequence in Nrf1 that is required to target it to the ER membrane. In addition, sequences that flank this h-region called, according to established terminology, the n-region [i.e. MLSLKKYLTE (single-letter amino acid code), amino acids 1–10] and the c-region [i.e. GVRVDVDT (single-letter amino acid code), amino acids 23–30] are probably involved in determining how the h-region is orientated within the membrane [22,39].
A striking finding during the present study was that deletion of the n-region increased targeting of Nrf1 to the ER and decreased ARE-driven luciferase reporter activity. This was associated with a substantial increase in the amount of Nrf1Δ2–10 protein recovered in cell lysates and also its molecular mass. Mutation of the di-lysine motif within the n-region to di-glutamine modestly increased nuclear accumulation of Nrf1K5/6Q and robustly increased reporter gene activity. These results suggest that the n-region may quantitatively control ER targeting and might be required for the correct membrane orientation of Nrf1. The reason why replacement of di-lysine with di-glutamine in the n-region substantially augments reporter gene activity without showing a corresponding increase in nuclear accumulation of Nrf1 may be due to the mutation affecting two different phenomena. First, according to the positive-inside rule, that states polypeptide regions of the membrane proteins which are enriched with positively charged amino acids tend to reside in the cytoplasm [35–37], the two lysine residues may contribute an overall positive charge to the n-region and in so doing ensure correct orientation of the protein within the ER membrane. Secondly, as a di-lysine sequence can allow proteins to interact with COPI (coatomer protein complex I) [40–42], it is also possible the di-lysine motif in Nrf1 prevents it trafficking from the ER to the Golgi apparatus, and its ultimate translocation into the nucleus (if indeed this happens). It therefore seems probable that the Nrf1K5/6Q mutant is mis-orientated within the ER membrane but can traffic to the Golgi apparatus more readily than the wild-type protein. Further mutagenesis studies in which the di-lysine sequence is disrupted but overall charge is maintained, should clarify whether the ER-to-Golgi migration of Nrf1 occurs normally.
We failed to obtain evidence that the c-region contains a SPase cleavage site, despite addressing this question using a number of experimental strategies. These included comparing the electrophoretic mobilities of wild-type Nrf1 with mutants that lack either ER targeting sequences (i.e. Nrf1Δ11–22) or candidate SPase cleavage sites (i.e. Nrf1G23/T30/S34N). Also, we compared the electrophoretic mobility of a GFP×2 fusion protein with N65/GFP×2, a chimearic protein containing amino acids 1–65 of Nrf1 attached to the N-terminus of GFP×2; the difference in size of these two proteins (58 and 65 kDa respectively) was sufficiently large to suggest there is no cleavage site in or around the c-region. In the absence of an SPase site in the c-region, we presume that the NHB1 will function as a signal to anchor Nrf1 in the ER membrane.
NHB2 is not required to target Nrf1 to the ER
Examination of internal deletion mutants within the NTD by reporter gene assay indicated that NHB2 does not contribute to the negative regulation of Nrf1. Furthermore, confocal microscopy and subcellular fractionation showed that Nrf1Δ81–106 displayed the same subcellular distribution as wild-type protein. We considered the hypothesis that the RRLL84 tetrapeptide within NHB2 represents a Site-1 protease recognition consensus sequence (RxxL, where x is any amino acid) that is processed in a similar fashion to ATF6 [29], but obtained no evidence that this is the case (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/408/bj4080161add.htm). Indeed, C-terminally V5-tagged Nrf2 and N156/Nrf2 had apparent molecular masses in LDS/NuPAGE of 75 and 96 kDa respectively, a result which indicates that the N-terminal 156 amino acids of Nrf1 lack a cleavage site. Similar results were obtained by comparing Nrf2 with NTD/Nrf2 and GFP×2 with NTD/GFP×2.
As NHB2 does not appear to regulate Nrf1 activity, and does not contain a functional Site-1 protease cleavage site, the biological function of this subdomain remains unclear. Bioinformatic analysis suggested that the residues around NHB2 may form a basic amphipathic α-helix, which could lie associated with the cytoplasmic surface of the phospholipid membrane bilayer, but this possibility remains to be tested.
Curiously, deletion of residues immediately adjacent to the N-terminal boundary of NHB2 increased the activity of Nrf1 (see Figures 2A and 3B), but we can offer no explanation for this finding.
Control of the subcellular localization of Nrf1
Our subcellular fractionation experiments revealed that under normal homoeostatic conditions Nrf1 is distributed between the cytoplasm, ER and nucleus. The relative amounts of Nrf1 in these three compartments may be controlled through residues 1–30 representing a signal sequence that can be regulated during stress in a fashion that ensures it is only incorporated into the ER under normal homoeostatic conditions [43]. Alternatively, residues 1–30 of Nrf1 may represent a constitutive signal sequence that it is continuously incorporated into the ER under all circumstances, but this bZIP factor can be released subsequently from the membrane in response to stress [43]. Thus it is not known whether during our transfection experiments a fraction of newly translated Nrf1 failed to be inserted into the ER membrane and escaped to the nucleus, or whether all Nrf1 was inserted into the ER and only a portion was subsequently released to translocate into the nucleus.
As Nrf1 associates with the ER through sequences in its N-terminal region, it is likely to be co-translationally inserted into the ER membrane. According to this hypothesis, the signal sequence around NHB1 could constitutively target the newly synthesized Nrf1 protein to the translocon Sec61 complex in the ER membrane through the cotranslational pathway. Residues 7–26 are postulated to form a hydrophobic amphipathic α-helix, and this region may be integrated laterally into the ER membrane upon synthesis [37]. Based on the aforementioned positive-inside rule [35–37], we propose that the first transmembrane (TM1) region is likely to be orientated in a Ncyt/Clum fashion with its N- and C-terminally flanking regions in the cytoplasm and the ER lumen respectively. The TM1 in Nrf1 is not as hydrophobic as the single transmembrane region found in calnexin (see Figure 5A). As a consequence, the insertion of Nrf1 into the ER via residues 7–26 may not be particularly efficient and therefore other regions of the NTD may be required to interact with TM1 in order to stabilize it within the membrane. Secondary structure predictions suggest that residues 31–50 of Nrf1 can form an amphipathic α-helix [44]. We propose that the stability of the h-region of Nrf1 within the ER membrane could be increased through an interaction with this second putative α-helical sequence. Thus these two α-helices, with a short connecting loop, may form a hairpin structure that fixes Nrf1 in the membrane, as described previously for the anion exchanger Band 3 [37,45]. The orientation of TM1 or its interaction with residues 31–50 are events that could be controlled in a way that enables the factor to dissociate from the ER following appropriate stimulation. An alternative to a route that entails avoidance of capture of Nrf1 by the ER, is a pathway involving controlled release of the bZIP factor from the ER, presumably in response to stress. We have found that several Nrf1 forms of distinct size exist in different organelles. The largest form of Nrf1 is the glycosylated 120-kDa subunit which is present in the ER, whereas the smallest is the essentially non-glycosylated 95-kDa polypeptide found in the nucleus. An important unanswered question is whether a product–precursor relationship exists between the 95-kDa and 120-kDa forms. It is not known whether the 120-kDa Nrf1 subunit can be converted into the 95-kDa form through a regulated deglycosylation process. This could represent part of a retro-translocation event [46] that controls nuclear accumulation of Nrf1. It will be important to establish the relationship between the various Nrf1 forms because glycosylation may be required for activity. Alternatively, it could represent a means of retaining Nrf1 in an extra-nuclear subcellular compartment and/or a means of eliminating it from the cell.
Post-translational modification of Nrf1 and its activation
Regulated intramembrane proteolysis is a well-described mechanism by which transcription factors located in the ER are activated [47,48]. This process requires stress-stimulated escape of the transcription factors from the ER to the Golgi apparatus, whereupon they are cleaved sequentially by Site-1 and Site-2 proteases. Following cleavage, the transcription factor is free to translocate to the nucleus. Although our experiments failed to demonstrate proteolytic cleavage sites in Nrf1 they were carried out under normal homoeostatic conditions and therefore do not exclude regulated intramembrane proteolysis as a mechanism by which it is activated during stress.
Another possibility is that Nrf1 is cleaved in a domain other than the NTD. Short forms of Nrf1 have been described previously which were presumed to have arisen by translation from internal methionine residues [1,49,50], but could have equally arisen from proteolytic cleavage. The data in Figure 9 show that Nrf1 is glycosylated through several sites within the NST domain, indicating that this domain lies in the lumen of the ER. If this conclusion is correct it is possible that cleavage of Nrf1 may occur within or close to the sites of asparagine-glycosylation. This hypothesis remains to be tested.
A less conventional route by which Nrf1 may activate gene expression, is one in which the ER-associated factor might travel directly through the ER membrane to the outer nuclear membrane. In this case, a non-glycosylated or deglycosylated form of Nrf1 may migrate to the inner nuclear membrane either by passive diffusion through the nuclear pore membrane [51,52] or by a flip-flop mechanism [53]. According to this membrane-routing model, once Nrf1 enters the inner nuclear membrane it must somehow be able to interact with chromatin and gain access to ARE-containing gene promoters. The mechanism by which this might occur is unclear.
Concluding comments
In the present study a tripartite signal peptide in Nrf1, between residues 1 and 30, that is responsible for targeting the bZIP protein to the ER has been characterized. No evidence has been found for SPase, Site-1 or Site-2 protease cleavage sites in the NTD of Nrf1 suggesting that it is anchored to the ER through its N-terminus. Also, we have found that Nrf1 is asparagine-glycosylated via its NST domain, an observation that indicates this portion of the bZIP factor is situated in the lumen of the ER.
The biochemical processes that are dependent on Nrf1 being targeted to the ER are unknown but presumably identification of such events may help explain why knockout of Nrf1 in the liver causes steatosis that develops into NASH, and ultimately leads to hepatoma, whereas knockout of Nrf2 does not produce such phenotypes. Day and James [54] have proposed that NASH arises through a ‘two-hit’ mechanism: (i) lipid accumulation, and (ii) oxidative stress. The reason why knockout of Nrf1 should result in fatty liver is not clear, though as this bZIP protein controls expression of glutamate cysteine ligase it is readily understood why knockout of this factor produces oxidative stress. At least three hypotheses can be advanced to explain why loss of Nrf1 might result in fatty liver disease. First, Nrf1 may uniquely control the redox status within the ER and failure to maintain the correct antioxidant to oxidant balance in this compartment may cause incorrect folding of proteins that control lipid export, transport or catabolism. In particular, the ER contains relatively high levels of GSSG and if the GSH to GSSG ratio becomes drastically abnormal it is probable that proteins containing a significant number of cysteine residues will mis-fold. If such proteins control lipid homoeostasis then steatosis may result. Secondly, through its association with the ER, Nrf1 may negatively regulate SREBP-1 and SREBP-2, or other transcription factors targeted to the ER that are involved in either the synthesis or uptake of lipids, through putative interactions that interfere with either trafficking or regulated intramembrane proteolysis. Although protein–protein interactions between Nrf1 and other membrane-bound factors have not been studied, it is noteworthy that a transgenic mouse expressing a nuclear SREBP-1c mutant protein (nSREBP) develops NASH [55], which is consistent with the idea that Nrf1 might inhibit nuclear translocation of SREBP-1. Thirdly, Nrf1 may regulate directly the expression of genes involved in either lipid export from the liver or lipid oxidation. If activation of these genes involves Nrf1 gaining access to chromatin in close proximity to the inner nuclear membrane, through its trafficking via the ER, it is conceivable that Nrf1 regulates a different subset of ARE-driven genes than Nrf2.
It is apparent that a considerable amount of work needs to be undertaken in order to provide a satisfactory explanation of why liver-specific Nrf1 knockout mice develop NASH but Nrf2 knockout mice do not. It does however seem probable that differences in the subcellular localization of the two factors and the types of agonists involved in their activation will underpin the explanation for the distinct liver phenotypes of the Nrf1 and Nrf2 knockout mice.
Online data
Acknowledgments
This work was supported by the Association for International Cancer Research (grants 03-074 and 06-015). We thank Dr M. McMahon and Dr L.G. Higgins (both at the Biomedical Research Centre, University of Dundee, Dundee, Scotland, U.K.) for providing expression constructs and for critical advice.
References
- 1.Chan J. Y., Han X. L., Kan Y. W. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luna L., Johnsen O., Skartlien A. H., Pedeutour F., Turc-Carel C., Prydz H., Kolsto A. B. Molecular cloning of a putative novel human bZIP transcription factor on chromosome 17q22. Genomics. 1994;22:553–562. doi: 10.1006/geno.1994.1428. [DOI] [PubMed] [Google Scholar]
- 3.Caterina J. J., Donze D., Sun C. W., Ciavatta D. J., Townes T. M. Cloning and functional characterization of LCR-F1: a bZIP transcription factor that activates erythroid-specific, human globin gene expression. Nucleic Acids Res. 1994;22:2383–2391. doi: 10.1093/nar/22.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnsen O., Murphy P., Prydz H., Kolsto A. B. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes J. D., McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 6.Leung L., Kwong M., Hou S., Lee C., Chan J. Y. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 7.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J. D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwak M. K., Wakabayashi N., Itoh K., Motohashi H., Yamamoto M., Kensler T. W. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 9.Devling T. W., Lindsay C. D., McLellan L. I., McMahon M., Hayes J. D. Utility of siRNA against Keap1 as a strategy to stimulate a cancer chemopreventive phenotype. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7280A–7285A. doi: 10.1073/pnas.0501475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer S. C., Sun C. W., Winnier G. E., Hogan B. L., Townes T. M. The bZIP transcription factor LCR-F1 is essential for mesoderm formation in mouse development. Genes Dev. 1997;11:786–798. doi: 10.1101/gad.11.6.786. [DOI] [PubMed] [Google Scholar]
- 11.Chan J. Y., Kwong M., Lu R., Chang J., Wang B., Yen T. S., Kan Y. W. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Kwong M., Lu R., Ginzinger D., Lee C., Leung L., Chan J. Y. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell. Biol. 2003;23:4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z., Chen L., Leung L., Yen T. S., Lee C., Chan J. Y. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anstee Q. M., Goldin R. D. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int. J. Exp. Pathol. 2006;87:1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan K., Lu R., Chang J. C., Kan Y. W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chanas S. A., Jiang Q., McMahon M., McWalter G. K., McLellan L. I., Elcombe C. R., Henderson C. J., Wolf C. R., Moffat G. J., Itoh K., et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon M., Itoh K., Yamamoto M., Chanas S. A., Henderson C. J., McLellan L. I., Wolf C. R., Cavin C., Hayes J. D. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 18.Zhang Y., Crouch D. H., Yamamoto M., Hayes J. D. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J. 2006;399:373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Chan J. Y. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J. Biol. Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 20.McMahon M., Itoh K., Yamamoto M., Hayes J. D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 21.von Heijne G. Protein targeting signals. Curr. Opin. Cell Biol. 1990;2:604–608. doi: 10.1016/0955-0674(90)90100-s. [DOI] [PubMed] [Google Scholar]
- 22.Martoglio B., Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 23.von Heijne G. Signals for protein targeting into and across membranes. Subcell. Biochem. 1994;22:1–19. doi: 10.1007/978-1-4615-2401-4_1. [DOI] [PubMed] [Google Scholar]
- 24.Paetzel M., Karla A., Strynadka N. C., Dalbey R. E. Signal peptidases. Chem. Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 25.Liu H., Yang J., Ling J. G., Chou K. C. Prediction of protein signal sequences and their cleavage sites by statistical rulers. Biochem. Biophys. Res. Commun. 2005;338:1005–1011. doi: 10.1016/j.bbrc.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 26.Sato R., Yang J., Wang X., Evans M. J., Ho Y. K., Goldstein J. L., Brown M. S. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1) J. Biol. Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- 27.Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 28.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye J., Rawson R. B., Komuro R., Chen X., Dave U. P., Prywes R., Brown M. S., Goldstein J. L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Cho Y. Y., Petersen B. L., Bode A. M., Zhu F., Dong Z. Ataxia telangiectasia mutated proteins, MAPKs, and RSK2 are involved in the phosphorylation of STAT3. J. Biol. Chem. 2003;278:12650–12659. doi: 10.1074/jbc.M210368200. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson M., Mathers J., Dickinson R. J., Mandl M., Keyse S. M. Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase MKP-3 and its ability to anchor MAP kinase in the cytoplasm are mediated by a conserved nuclear export signal. J. Biol. Chem. 2004;279:41882–41891. doi: 10.1074/jbc.M406720200. [DOI] [PubMed] [Google Scholar]
- 32.Lucocq J. M., Habermann A., Watt S., Backer J. M., Mayhew T. M., Griffiths G. A rapid method for assessing the distribution of gold labeling on thin sections. J. Histochem. Cytochem. 2004;52:991–1000. doi: 10.1369/jhc.3A6178.2004. [DOI] [PubMed] [Google Scholar]
- 33.Chua B. T., Volbracht C., Tan K. O., Li R., Yu V. C., Li P. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 2003;5:1083–1089. doi: 10.1038/ncb1070. [DOI] [PubMed] [Google Scholar]
- 34.Bowie J. U. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 35.Martoglio B. Intramembrane proteolysis and post-targeting functions of signal peptides. Biochem. Soc. Trans. 2003;31:1243–1247. doi: 10.1042/bst0311243. [DOI] [PubMed] [Google Scholar]
- 36.Goder V., Spiess M. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 2001;504:87–93. doi: 10.1016/s0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- 37.van Geest M., Lolkema J. S. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helenius A., Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 39.Lyko F., Martoglio B., Jungnickel B., Rapoport T. A., Dobberstein B. Signal sequence processing in rough microsomes. J. Biol. Chem. 1995;270:19873–19878. doi: 10.1074/jbc.270.34.19873. [DOI] [PubMed] [Google Scholar]
- 40.Eugster A., Frigerio G., Dale M., Duden R. The α- and β'-COP WD40 domains mediate cargo-selective interactions with distinct di-lysine motifs. Mol. Biol. Cell. 2004;15:1011–1023. doi: 10.1091/mbc.E03-10-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroder-Kohne S., Letourneur F., Riezman H. α-COP can discriminate between distinct, functional di-lysine signals in vitro and regulates access into retrograde transport. J. Cell Sci. 1998;111:3459–3470. doi: 10.1242/jcs.111.23.3459. [DOI] [PubMed] [Google Scholar]
- 42.Cosson P., Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 43.Swanton E., High S. ER targeting signals: more than meets the eye? Cell. 2006;127:877–879. doi: 10.1016/j.cell.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 44.Heinrich S. U., Rapoport T. A. Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. EMBO J. 2003;22:3654–3663. doi: 10.1093/emboj/cdg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Heijne G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 46.Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 47.Brown M. S., Ye J., Rawson R. B., Goldstein J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 48.Rawson R. B. Regulated intramembrane proteolysis: from the endoplasmic reticulum to the nucleus. Essays Biochem. 2002;38:155–168. doi: 10.1042/bse0380155. [DOI] [PubMed] [Google Scholar]
- 49.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Husberg C., Murphy P., Martin E., Kolsto A. B. Two domains of the human bZIP transcription factor TCF11 are necessary for transactivation. J. Biol. Chem. 2001;276:17641–17652. doi: 10.1074/jbc.M007951200. [DOI] [PubMed] [Google Scholar]
- 51.Voeltz G. K., Rolls M. M., Rapoport T. A. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mattaj I. W. Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2004;5:65–69. doi: 10.1038/nrm1263. [DOI] [PubMed] [Google Scholar]
- 53.Bowie J. U. Flip-flopping membrane proteins. Nat. Struct. Mol. Biol. 2006;13:94–96. doi: 10.1038/nsmb0206-94. [DOI] [PubMed] [Google Scholar]
- 54.Day C. P., James O. F. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 55.Shimomura I., Shimano H., Korn B. S., Bashmakov Y., Horton J. D. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J. Biol. Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.