Abstract
Imp4p is a component of U3 snoRNP (small nucleolar ribonucleoprotein) involved in the maturation of 18S rRNA. We have shown that Imp4p interacts with Cdc13p, a single-stranded telomere-binding protein involved in telomere maintenance. To understand the role of Imp4p in telomeres, we purified recombinant Imp4p protein and tested its binding activity towards telomeric DNA using electrophoretic mobility-shift assays. Our results showed that Imp4p bound specifically to single-stranded telomeric DNA in vitro. The interaction of Imp4p to telomeres in vivo was also demonstrated by chromatin immunoprecipitation experiments. Significantly, the binding of Imp4p to telomeres was not limited to yeast proteins, since the hImp4 (human Imp4) also bound to vertebrate single-stranded telomeric DNA. Thus we conclude that Imp4p is a novel telomeric DNA-binding protein that, in addition to its role in rRNA processing, might participate in telomere function.
Keywords: DNA binding, Imp4, rRNA, small nucleolar ribonucleoprotein (snoRNP), telomere, yeast
Abbreviations: Brix, biogenesis of ribosomes in Xenopus; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility-shift assay; GST, glutathione transferase; hImp4, human Imp4; hnRNP, heterogeneous nuclear ribonucleoprotein; hPot1, human protection of telomeres1; IPTG, isopropyl β-D-thiogalactoside; Ni-NTA, Ni2+-nitrilotriacetate; snoRNP, small nucleolar ribonucleoprotein
INTRODUCTION
Telomeres are the specialized structures found at the ends of eukaryotic chromosomes. They are essential for maintaining chromosome integrity by protecting chromosomes from end-to-end fusion and nuclease degradation [1]. In most cloned telomeric DNA sequences, telomeres are composed of guanine (G)-rich DNA repeats [2]. For example, the repeated sequences in Saccharomyces cerevisiae are TG1–3 and TTAGGG in vertebrates. In addition to the duplex repeats, single-stranded telomeric G-rich DNA tail is also detected in telomeres. The presence of single-stranded G-tails could serve as a substrate for telomerase extension [3], a damage signal to trigger cell-cycle arrest [4,5], and was postulated to be an intermediate for telomere replication [6]. Protein factors that interact with telomeres have been identified. In S. cerevisiae, Rap1p binds to the double-stranded telomeric DNA and Cdc13p binds specifically to the single-stranded portion of the telomeres [7–10]. These two proteins serve as a core for other proteins to form a multiple protein–DNA complex in telomeres [11–15]. Together they participate in multiple functions on telomeres, such as end protection and replication.
In addition to Cdc13p, several proteins with single-stranded telomeric DNA-binding activities have been identified in yeast [16–19]. Est1p is a telomerase-associated protein that mediates the binding of telomerase to telomeres [13]. Although it is unclear whether the single-stranded binding activity of Est1p is required for its binding to telomeres, it is generally accepted that binding of Est1p to telomeres is through its interaction with Cdc13p [13,14]. Gbp2p also binds to single-stranded telomeric DNA in vitro [17,18]. The telomere-binding activity of Gbp2p is required for its complementation of the temperature-sensitive and telomere length defects of cdc13-1, indicating that it might have a role in telomere function [5]. Interestingly, a single-stranded DNA-binding protein RPA (replication protein A) has also been shown to interact with telomeres at the S-phase of the cell cycle and mediate the binding of Est1p to telomeres [19]. Thus these proteins might affect telomere function through their single-stranded telomeric DNA-binding activities.
Previously, we conducted a two-hybrid screen using the N-terminal 1–252 amino acids of Cdc13p as the bait and identified three Cdc13p-interacting proteins: Zds2p, Sir4p and Imp4p [15]. Zds2p and Sir4p have been shown to affect telomere functions [20,21]; however, the role of Imp4p on telomeres has not been reported. Imp4p is an essential gene. It is a component of U3 snoRNP (small nucleolar ribonucleoprotein), also called SSU processome, that is involved in the maturation of 18S rRNA [22,23]. In both humans and yeast, Imp4p forms a complex with Mpp10p and Imp3p [24,25]. The protein complex then interacts with both U3 snoRNA and 35S rRNA to initiate the maturation of rRNAs [26–28]. Imp4p belongs to a protein superfamily that contains a conserved Brix (biogenesis of ribosomes in Xenopus) domain [29]. In yeast, Imp4p superfamily is composed of Imp4p, Rpf1p, Rpf2p, Brx1p and Ssf1/2p. The Brix domains within these proteins are believed to be required for their binding to RNA substrates [28,30]. For example, the Brix domain of Imp4p has been shown to bind U3 snoRNA [28]. Sequence analysis of Imp4p also revealed a σ70-like motif located at amino acids 245–262, within the predicted Brix domain [31]. The 17-amino-acid σ70-like motif of Rpf1p or Rpf2p is sufficient for it to bind to homoribopolymers, suggesting that this motif is indeed an eukaryotic RNA-binding motif [31].
In the present study, we show that Imp4p bound specifically to single-stranded telomeric DNA and associated with telomeres in vivo. Significantly, the binding of Imp4p to single-stranded telomeric DNA was not limited to yeast protein, as hImp4 (human Imp4) also bound to single-stranded telomeric (TTAGGG)n DNA. Thus our results indicate that Imp4p is a novel telomeric DNA-binding protein that may have a role in telomeres.
MATERIALS AND METHODS
Purification of His6-tagged Imp4p, GST (glutathione transferase)–Imp4p-(88–275) (GST–Brix) and hImp4
To purify His6-tagged Imp4p or hImp4, a 2 litre culture of Escherichia coli BL21 (DE3) pLysS cells harbouring the expression plasmids was grown at 37 °C to an attenuance (D600) of 0.6 and was induced with 1 mM IPTG (isopropyl β-D-thiogalactoside). The cells were grown at 25 °C for another 4 h before harvesting by centrifugation at 2560 g for 10 min. The cells were resuspended in 20 ml of sonication buffer [50 mM sodium phosphate, pH 7.8, 300 mM NaCl, 1 mM PMSF and 1× protease inhibitors (Calbiochem)], and sonicated to release the cell contents. The sonicated cells were centrifuged at 20000 g for 15 min at 4 °C to obtain total cell free extract. A 0.5 ml volume of Ni-NTA (Ni2+-nitrilotriacetate)–agarose (GE Healthcare) was added to the total cell-free extract before incubation at 4 °C for 1 h. The resin was washed and eluted with 2.5 ml of buffer containing 50 mM sodium phosphate buffer, pH 8.0, 250 mM imidazole and 20% (v/v) glycerol. The eluted protein was dialysed against the dialysis buffer (50 mM sodium phosphate buffer, pH 8.0, and 50% glycerol) at 4 °C for 16 h. Protein was divided into 5 μl aliquots and frozen using a solid-CO2/ethanol bath.
To purify GST-tagged Imp4p-(88–275) (GST–Brix), a 1 litre culture of E. coli BL21 (DE3) cells harbouring pHTPP2-brix were grown and induced with IPTG. Cells was resuspended in 10 ml of sonication buffer and sonicated to release the cell contents. The sonicated cells were centrifuged at 20000 g for 15 min at 4 °C to obtain total cell-free extract. Then, 0.75 ml of glutathione–Sepharose 4B (GE Healthcare) was added to the total cell-free extract before incubation at 4 °C for 1 h. The resin was washed and then eluted with 0.5 ml of buffer containing 50 mM sodium phosphate buffer, pH 7.8, 200 mM glutathione and 10% (v/v) glycerol. The eluted protein was dialysed, divided into 5 μl aliquots and frozen as described above. Polyclonal antibodies against His6-tagged Imp4p were raised in rabbits using the purified protein. Purification of hPot1 (human protection of telomeres1) was performed, following the procedure of Baumann and Cech [32], who provided the hPot1 expression plasmid.
EMSA (electrophoretic mobility-shift assay)
Oligonucleotide TG15, TG20, TG25, TG30, TG35, TG40 (see Table 1) or (T2AG3)6 was first 5′-end-labelled with [γ-32P]ATP (3000 mCi/mM) (NEN) using T4 polynucleotide kinase (New England Biolabs) and subsequently purified from a 10% sequencing gel after electrophoresis. To perform the assays, purified Imp4p, GST–Brix, GST or hImp4 was mixed with 5 nM 32P-labelled oligonucleotides in a total volume of 15 μl in buffer containing 50 mM Tris/HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl, 1 mM dithiothreitol and 1 μg of heat-denatured poly(dI-dC)·(dI-dC). The reaction mixtures were incubated at room temperature (23 °C) for 10 min and separated on a 6% non-denaturing polyacrylamide gel. Electrophoresis was carried out in TBE (89 mM Tris/borate and 2 mM EDTA) at 125 V for 105 min. The gels were dried and autoradiographed, and the oligonucleotides bound to the proteins were quantified using a PhosphorImager (Molecular Dynamics). The apparent binding constant of Imp4p to telomeric DNA was determined using EMSA and quantified using a PhosphorImager. Values presented in Table 1 were determined from interpolation on a Hill plot. Each value was the average for two or three experiments. For competition analysis, 10 nM 32P-labelled TG30 was mixed with various amounts of unlabelled competitors before addition of the cell extracts.
Table 1. Sequences and Imp4p-binding affinity of yeast telomeric DNA substrates.
Oligonucleotide TG15, TG20, TG25, TG30, TG35 or TG40 was 5′-end-labelled with [γ-32P]ATP and subsequently purified from a 10% sequencing gel after electrophoresis. To perform the assays, purified Imp4p was mixed with 5 nM 32P-labelled oligonucleotides in a total volume of 15 μl in buffer containing 50 mM Tris/HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl, 1 mM dithiothreitol and 1 μg of heat-denatured poly(dI-dC)·(dI-dC). The reaction mixtures were incubated at room temperature for 10 min and separated on a 6% non-denaturing polyacrylamide gel. The apparent binding constant of Imp4p to telomeric DNA was determined from an average of three experiments using EMSA and quantified using a PhosphorImager.
| Name | Sequence | Kd (app) (μM) |
|---|---|---|
| TG15 | 5′-TGTGTGGGTGTGGTG-3′ | 8.7±1.5 |
| TG20 | 5′-TGGTGTGTGTGGGTGTGGTG-3′ | 3.5±1.6 |
| TG25 | 5′-GGGTGTGGTGTGTGTGGGTGTGGTG-3′ | 1.3±0.6 |
| TG30 | 5′-TGTGTGGGTGTGGTGTGTGTGGGTGTGGTG-3′ | 1.2±0.1 |
| TG35 | 5′-TGGTGTGTGTGGGTGTGGTGTGTGTGGGTGTGGTG-3′ | 1.0±0.2 |
| TG40 | 5′-GGGTGTGGTGTGTGTGGGTGTGGTGTGTGTGGGTGTGGTG-3′ | 0.6±0.3 |
ChIP (chromatin immunoprecipitation) analysis
Yeast strain YPH499 (MATa ura3-52 lys2-801amber ade2-101ochre trp1-δ63 his3-δ200 leu2-δ1 [33]) with URA3 placed near the telomere of chromosome VII-L (YPH499UT) was used to immunoprecipitate telomeric DNA by Cdc13p polyclonal antibodies. The isogenic strain with a Myc9 tag fused to the C-terminus of Imp4p was used to immunoprecipitate telomeric DNA by anti-Myc antibody (9E10; Santa Cruz Biotechnology). Cells were grown, fixed with 1% (w/v) formaldehyde at room temperature for 15 min, and growth was arrested using 0.125 M glycine. The cells were then harvested, resuspended in TBS (Tris-buffered saline: 20 mM Tris/HCl, pH 7.4, and 150 mM NaCl) and broken apart by vortex-mixing with glass beads. Sonication was then used to shear chromatin to ∼500 bp in length. The chromatin-sheared cells were centrifuged at 15000 g for 15 min to yield supernatants for immunoprecipitation by antibodies against Cdc13p or Imp4p. The immunoprecipitates were then incubated at 65 °C overnight to reverse cross-links followed by proteinase K treatment. Three DNA sequences were analysed representing the telomeric (TEL), 5 kb from the telomere (ADH, ADH4 gene) and internal (ARO, ARO1 gene) DNA. The PCR primers were designed by following the sequences reported by Taggart et al. [34]. The sizes for the PCR products of ARO, ADH and TEL are 372, 301 and 252 bp respectively. The PCR products were separated on a 2% agarose gel and visualized by staining with ethidium bromide.
RESULTS
Specific binding of Imp4p to single-stranded telomeric DNA
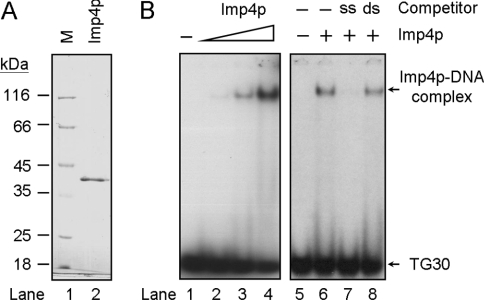
Imp4p is capable of binding to U3 snoRNA [28]. Because the single-stranded character of RNA and single-stranded DNA is similar, RNA-binding proteins tend to bind single-stranded DNA. For example, several human hnRNPs (heterogeneous nuclear ribonucleoproteins) were shown to bind both RNA and single-stranded telomeric DNA [35,36]. We then tested whether Imp4p could also bind to single-stranded telomeric DNA. An E. coli expression system was used to prepare the recombinant Imp4p. In the present study, Imp4p with a His6 tag at the N-terminus was expressed, and the protein was purified to homogeneity using an Ni-NTA–agarose resin (Figure 1A). The purified protein has an apparent molecular mass of 42 kDa, which is higher than the predicted value of 34 kDa. MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) analysis of the purified protein indicated that it was indeed Imp4p (results not shown). Thus Imp4p appears to migrate aberrantly on SDS/PAGE gels.
Figure 1. Imp4p binds to single-stranded telomeric DNA.
(A) Purification of Imp4p. Imp4p with a His6 tag was purified from E. coli using Ni-NTA–agarose (see the Materials and methods section). A Coomassie Blue-stained SDS/10% polyacrylamide gel is shown. Lane 1, molecular-mass markers (M; sizes indicated in kDa). Lane 2, 1 μg of purified Imp4p. (B) Specific binding of Imp4p to single-stranded TG1–3 DNA. 32P-labelled TG30 (5 nM) was mixed with 0, 0.43, 1.3 and 3.9 μM of purified Imp4p respectively, before the gel-shift assay (lanes 1–4). In lanes 5–8, 10 nM 32P-labelled TG30 was first mixed without (lane 6) or with 1 μM unlabelled competitors, TG30 (ss; lane 7) or 270 bp double-stranded telomeric DNA (ds; lane 8). Imp4p (1 μM) was then added to the DNA mixtures.
The binding of Imp4p to telomeric DNA was determined using EMSA analysis. Purified protein was mixed with 32P-labelled single-stranded TG1–3 with various amounts of unlabelled nucleic acid competitors before subjection to the EMSA. As shown in Figure 1(B), a gel-shifted band was apparent in the mixture containing Imp4p and TG30 (a 30-mer TG1–3 repetitive DNA, Table 1). The binding appeared to be specific to single-stranded DNA, as it was competed away by single-stranded TG30, but not by double-stranded telomeric DNA (Figure 1B, lanes 7 and 8). Thus these results provided the first indication that Imp4p was capable of binding to single-stranded telomeric DNA.
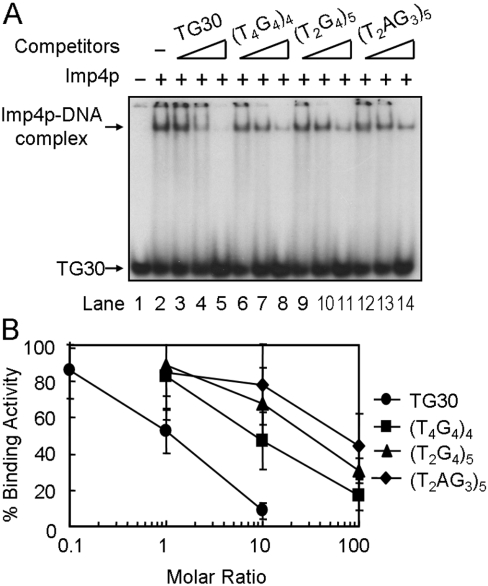
Although the binding of Imp4p to telomeric DNA was not affected by the presence of single-stranded poly(dI-dC)·(dI-dC) (results not shown), 1 μg of single-stranded poly(dI-dC)·(dI-dC) was included to suppress potential non-specific single-stranded DNA binding in the EMSA. Thus the protein–DNA complexes observed in our binding assay should be very specific. To determine further the selectivity of the Imp4p-binding activity, a competition assay was employed in this analysis. Imp4p was mixed with 32P-labelled single-stranded TG1–3 and various amounts of unlabelled competitor nucleic acids, and was assayed using the EMSA. The competitors were ∼30-mer single-stranded G-strand DNA from Saccharomyces (TG30), vertebrate (T2AG3)5, Oxytricha (T4G4)4, and Tetrahymena (T2G4)5 telomeric DNAs. The autoradiograph of the reaction products after gel electrophoresis (Figure 2A) was quantified (Figure 2B) using the binding activity to TG30 in the absence of a specific competitor as 100% (Figure 2A, lane 2). As shown in Figure 2(A), unlabelled TG30 competed efficiently with 32P-labelled TG30 (lanes 3–5). Telomeric DNA from Oxytricha (T4G4), Tetrahymena (T2G4) and vertebrate (T2AG3) also competed for Imp4p binding (Figure 2A, lanes 6–14), although a 10–100-fold molar excess of these competitors was needed to obtain the same level of competition as that from TG30 (Figure 2B). These results indicate that Imp4p prefers to bind to single-stranded TG1–3 DNA.
Figure 2. Imp4p prefers yeast telomeric DNA for binding.
Competition analyses with various telomeric DNAs were used to determine the binding specificity. (A) 32P-labelled TG30 (10 nM) was first mixed with 1, 10 or 100 nM unlabelled yeast TG30 or 10, 100 or 1000 nM of unlabelled vertebrate (T2AG3)5, Oxytricha (T4G4)4 and Tetrahymena (T2G4)5 competitors. The DNA mixtures were then incubated with 0.75 μM purified Imp4p and analysed by gel-shift assay. (B) Quantification of the Imp4p-binding activity. The amount of 32P-labelled TG30 bound to the protein was quantified using a PhosphorImager, and binding without any competitor was taken as 100% (A, lane 2).
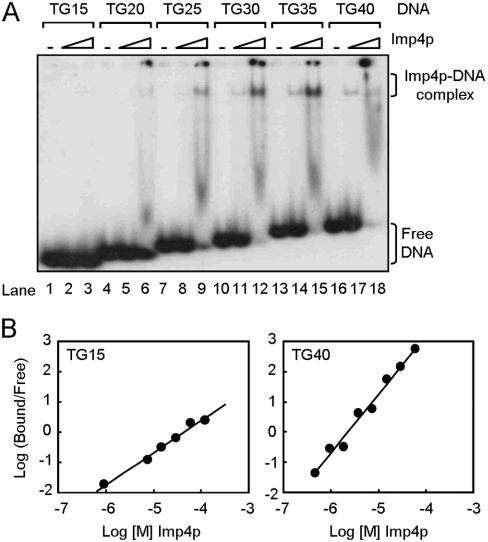
In the organisms that have been analysed, telomeres are double-stranded duplex telomeric DNA with a single-stranded telomeric tail. In yeast, a single-stranded tail with 12–14 nucleotides was detected in most stages of the cell cycle [37]. The length of single-stranded tail is transiently increased to >30 nucleotides at late S-phase of the cell cycle [38]. To determine the binding preference of Imp4p to different lengths of telomeres, the single-stranded TG1–3 substrates with various lengths were tested for their binding by Imp4p. It is apparent from the results of Figure 3(A) that Imp4p preferred long DNA substrates for binding. The binding affinity [Kd (app)] of Imp4p to these various lengths of telomeric DNA substrates also supports this observation (Table 1). Interestingly, while the binding of Imp4p to TG15 did not appear to be co-operative (Hill coefficient=∼1.1), the binding of Imp4p to long telomeric DNA, TG40, was co-operative, with a Hill coefficient of ∼2.1 (Figure 3B).
Figure 3. Imp4p prefers long single-stranded TG1–3 DNA for binding.
(A) 32P-labelled TG15, TG20, TG25, TG30, TG35 or TG40 (5 nM of each) was mixed with 0, 0.15 or 1.5 μM purified Imp4p before subjection to gel-shift assay. (B) Quantitative data from PhosphorImager analysis of gel mobility shifts were used to determine the equilibrium binding constant. Concentrations of Imp4p from 0 to 120 μM were used in these analyses. The fraction of DNA bound/free DNA is plotted against the Imp4p concentration. Representative plots for TG15 (left-hand panel) and TG40 (right-hand panel) are shown.
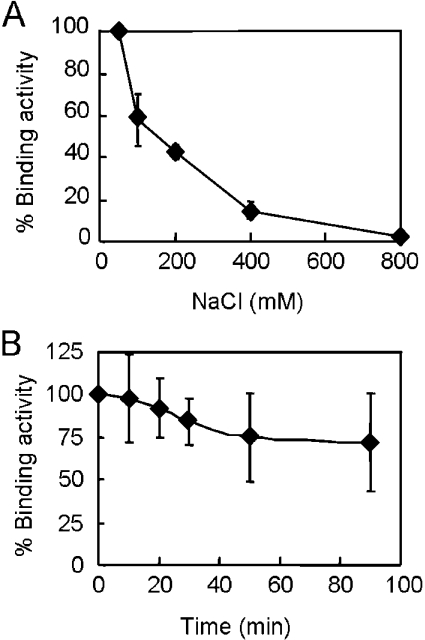
Binding of Oxytricha α and β proteins to TTTTGGGG DNA is salt-stable [39]. Oxytricha α and β proteins remain bound to single-stranded TTTTGGGG when the NaCl concentration was as high as 2 M. To analyse the salt-stability of Imp4p, binding of the purified protein to TG30 was conducted at various levels of NaCl (Figure 4A). To avoid re-association of the proteins to TG30 after loading the sample, an excess amount of unlabelled TG30 was added to the reaction mixtures before loading. It is apparent that Imp4p preferred low salt for binding. At 200 mM NaCl, the binding activity of Imp4p fell to less than 50% relative to 50 mM NaCl (Figure 4A). To evaluate the stability of the protein–DNA complex, the dissociation rate of the protein–DNA complex was measured. Imp4p was first bound to 32P-labelled TG30 and then an excess amount of unlabelled TG30 was added to prevent re-association of protein to labelled DNA. The dissociation rate was estimated as the time required for half of the protein–DNA complex to dissociate. As shown in Figure 4(B), Imp4p had a half-life of ∼90 min on DNA, suggesting that the binding of Imp4p to single-stranded telomeric DNA is stable.
Figure 4. Binding properties of Imp4p.
(A) Imp4p prefers low salt for binding. 32P-labelled TG30 (5 nM) was incubated with 1 μM Imp4p and 0.05, 0.1, 0.2, 0.4 or 0.8 M NaCl at room temperature for 10 min. Unlabelled TG30 (1 μM) was added to the reaction mixtures and a gel-shift assay was then performed. The binding was quantified using a PhosphorImager, and the activity with 0.05 M NaCl was taken as 100%. (B) Slow dissociation of Imp4p from single-stranded TG1–3 DNA. 32P-labelled TG30 (15 nM) was incubated with 1 μM Imp4p at room temperature for 10 min. Unlabelled TG30 (1 μM) was then added at zero time. Aliquots of the mixtures were withdrawn at the indicated time points and were loaded on to a running gel. After electrophoresis, the amount of 32P-labelled TG30 remaining bound was quantified using a PhosphorImager and the binding at zero time was taken as 100%.
The Brix domain of Imp4p binds to single-stranded telomeric DNA
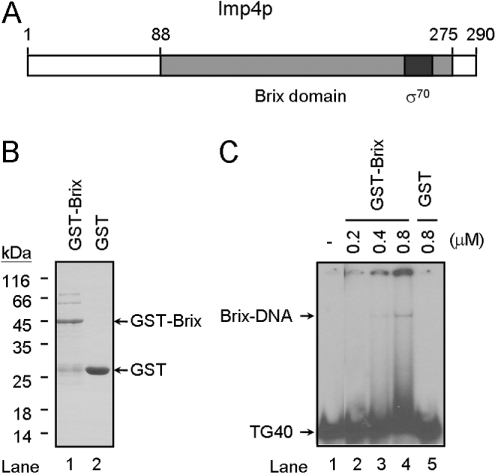
Sequence analysis of Imp4p identified a Brix domain and a σ70-like motif within the protein (Figure 5A). It has been shown that the Brix domain of Imp4p is sufficient for binding to U3 snoRNA [28]. To define the TG1–3-binding domains of Imp4p, the E. coli expression system was used to purify the Brix domain of Imp4p. In the present study, a GST-fusion protein was used to purify the Brix domain of Imp4p (residues 88–275; Figure 5A). The fusion protein with an apparent molecular mass of 45 kDa was purified using a glutathione–Sepharose resin (Figure 5B, lane 1). Western blotting indicated that the 45 kDa protein was recognized by antibodies against GST (results not shown). As a control, GST protein was also purified and had an apparent mass of 26 kDa (Figure 5B, lane 2). The binding ability of the Imp4p Brix domain to telomeric DNA TG40 was determined using EMSA. As shown in Figure 5(C), a faint gel-shift band was detected with GST–Brix fusion protein (Figure 5C, lanes 2–4). The apparent gel-shift band, although weak, was due to the binding activity of the Brix domain, because the GST protein did not yield any detectable gel-shift band with a similar amount of protein added to the reaction mixture (Figure 5C, lane 5). Thus our results indicate that the Brix domain of Imp4p is capable of binding to single-stranded telomeric DNA. It also appears that the other portion of Imp4p might be required for its proper binding to telomeric DNA.
Figure 5. The Brix domain is important for binding to single-stranded telomeric DNA.
(A) Schematic representation of the Imp4p domain. The Imp4p is 290 amino acids in length. Sequence analysis reveals a Brix domain located at amino acids 88–275 and a σ70-like motif is located at amino acids 245–263. The relative locations of the Brix domain and σ70-like motif are indicated. (B) Purification of Imp4p Brix domain and GST. The GST–Brix domain fusion protein and GST protein were purified from E. coli using a glutathione–Sepharose resin (see the Materials and methods section). A 2 μg sample of each of purified Imp4p, GST–Brix and GST was analysed by SDS/12% PAGE and stained with Coomassie Blue. Molecular masses are indicated in kDa. (C) The Brix domain of Imp4p binds to single-stranded TG1–3 DNA. 32P-labelled TG40 (5 nM) was mixed with various amounts of GST–Brix or GST and was then analysed by EMSA.
Association of Imp4p with telomeres in vivo
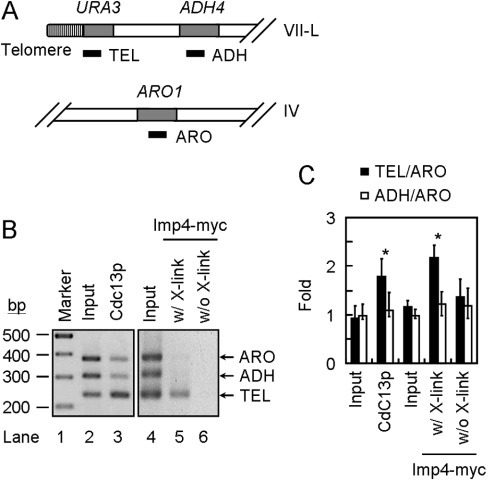
In the present study, we have shown that Imp4p bound to single-stranded telomeric DNA in vitro. To determine whether the binding reflects the association of Imp4p to telomeres in vivo, the ChIP analysis was employed. To conduct the immunoprecipitation experiments, we constructed a yeast strain that expressed the Imp4p with a Myc9 tag fused to the C-terminus of the protein for this analysis. The Myc9-tagged Imp4p did not appear to affect the growth of yeast cells or the telomere length, suggesting that it did not affect the function of Imp4p (results not shown). The yeast cells were treated with formaldehyde to cross-link DNA with binding proteins. Chromatin was isolated, sheared and immunoprecipitated with antibodies against Cdc13p or Myc. The precipitated DNA was then analysed using three sets of primers [34]. Amplification of a telomere-proximal URA3 gene (TEL) is used as an indication for telomere binding. The additional ADH4 gene (ADH) and ARO1 gene (ARO) represent binding to subtelomeric and internal parts of chromosome respectively (Figure 6A). Under our assay conditions, the Cdc13p immunoprecipitates preferentially amplified TEL DNA, consistent with the role of Cdc13p on telomeres (Figures 6B and 6C). Using the anti-Myc antibody, we found that the TEL DNA was preferentially amplified in its immunoprecipitates (Figures 6B and 6C). As a control, pre-immune serum or cells not treated with formaldehyde did not preferentially precipitate TEL DNA (results not shown, and Figure 6B). Because the antibodies used for precipitating Imp4p and Cdc13p were different, the binding selectivity of Imp4p and Cdc13p towards TEL DNA cannot be readily compared. Nevertheless, our results clearly indicate that Imp4p associates with telomeres in vivo.
Figure 6. Association of Imp4p with telomeres in vivo.
(A) Schematic presentation of URA3, ADH4 and ARO1 chromosomal loci monitored by ChIP [34]. The positions of the PCR-amplified DNA fragments are presented by solid bars. (B) Imp4p interacts with telomeres in vivo. Total yeast extract was immunoprecipitated with antibodies against Cdc13p or Myc. The immunoprecipitates were then subjected to multiplex PCR analysis. The PCR products were separated on a 2% agarose gel and stained with ethidium bromide. Sizes are indicated in bp. (C) Quantification of the experiments was conducted using densitometry-scanning of the signals. In each set of experiments, the TEL or ADH signal is normalized with ARO signals (TEL/ARO or ADH/ARO). Results are means±S.D. for four independent experiments. *P<0.05 compared with input control.
hImp4 binds to vertebrate single-stranded telomeric DNA
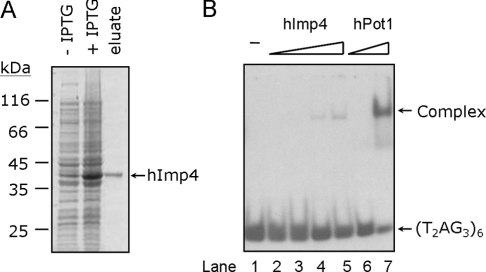
The sequences of different Imp4 proteins are conserved among species. The yeast Imp4p shares 50% sequence identity with and has 65% similarity to the hImp4 protein [22,25]. To evaluate whether hImp4 also binds to single-stranded (TTAGGG)n DNA, we cloned the cDNA of hImp4 and expressed the recombinant protein in E. coli. The His6-tagged recombinant hImp4 was then purified using Ni-NTA resin. Similar to that of yeast Imp4p, the hImp4 has an apparent molecular mass of ∼42 kDa (Figure 7A). The purified protein was mixed with 32P-labelled single-stranded (TTAGGG)6 for the EMSA. As shown in Figure 7(B), a gel-shifted band was apparent in mixtures containing hImp4 and (TTAGGG)6. As a control, the human single-stranded telomeric DNA-binding protein hPot1 also bound to the same DNA with a Kd (app) value of 0.18 μM (Figure 7B, lanes 6 and 7). The binding affinity of hImp4 [Kd (app) of 4.9 μM] to telomeric DNA substrates is similar to that of yeast protein. The results indicate that hImp4 is capable of binding to single-stranded telomeric DNA, although the binding of hImp4 to telomeric DNA appeared to be less efficient than that of hPot1. Thus the binding of Imp4 proteins to telomeric DNA is not limited to the yeast protein, as hImp4 also bound to vertebrate telomeric DNA.
Figure 7. hImp4 binds to vertebrate single-stranded (T2AG3)6 telomeric DNA.
(A) Purification of hImp4. hImp4 with a His6 tag was purified from E. coli using Ni-NTA–agarose (see the Materials and methods section). A 1 μg sample of purified hImp4 was separated by SDS/10% PAGE and stained with Coomassie Blue. Molecular masses are given in kDa. (B) 32P-labelled (T2AG3)6 was mixed with 0.26, 0.52, 1.03 or 2.06 μM of hImp4 (lanes 2–5 respectively) or 0.12 or 0.49 μM of hPot1 (lanes 6 and 7 respectively) before being subjected to gel-shift assay.
DISCUSSION
Our previous studies have provided evidence for the formation of a Cdc13p-mediated telosomic complex through its N-terminal region that was involved in telomere maintenance, telomere length regulation and cell growth control [15]. The Cdc13p-mediated telosomic complex contains proteins including Stn1p, Ten1p, Est1p, Pol1p, Zds2p, Sir4p and Imp4p [11,12,14,15]. Imp4p was identified from yeast two-hybrid screening experiments using Cdc13p as the bait. Further analysis indicates that Imp4p interacts with Cdc13p directly and co-immunoprecipitates with Cdc13p [15]. The role of Imp4p in this telosomic complex was investigated. In the present study, we show that Imp4p bound specifically to single-stranded telomeric DNA directly in vitro and associated with telomeres in vivo. We have also demonstrated the telomeric DNA-binding activity of hImp4, indicating that the telomeric DNA-binding activity of Imp4 is conserved in both yeast and humans. Thus the effects of Imp4p on telomeres might arise through its direct binding to telomeres. However, the question of how Imp4p affects telomere length is still unclear.
Unlike Cdc13p that binds specifically to telomeres and has an affinity for yeast single-stranded telomeric DNA of ∼50 nM [40], the binding affinity of Imp4p towards single-stranded telomeric DNA is relatively low under similar assay conditions. It has been shown that Imp4p is capable of binding to U3 snoRNA [28] or other RNA species [31], which is important for its function in RNA processing. Since the function of Imp4p is not limited to telomeres, a high binding affinity of Imp4p towards telomeric DNA is not anticipated. Nevertheless, although with lower binding affinity, Imp4p binds to telomeres and with enough selectivity to discriminate telomeric DNA sequences from other DNAs (Figure 6). Moreover, in both yeast and human cells, Imp4p was shown to form a complex with Imp3p and Mpp10p [24,25]. It is likely that the apparent low-affinity binding for Imp4p is due to the lack of Imp3p and Mpp10p in the in vitro assays. Indeed, it was shown that the α subunit of Oxytricha telomere-binding protein is capable of binding to single-stranded telomeric G4T4 DNA. However, high-affinity and terminus-specific binding requires the β subunit which is not directly involved in binding [41,42]. Thus Imp3p and Mpp10p might be required to facilitate tight binding of Imp4p to telomeres. It will be interesting to test the effects of these two proteins on Imp4p-binding activity. It is also noteworthy that the yeast single-stranded telomere DNA-binding protein Cdc13p also forms a heterotrimeric protein complex with Stn1p and Ten1p to protect telomeres and regulate telomere length [43]. Imp4p might interact with telomeres in a form of trimeric complex with Imp3p and Mpp10p.
Several proteins involved in RNA metabolism also participate in telomere functions [44]. These RNA-binding proteins have dual roles in cells that are important for both RNA metabolism and telomere length regulation. They could affect telomere function through their interaction with telomerase RNA. For example, Tlc1 RNA mutations on the predicted binding site of Sm protein, a component of the Sm snRNPs (small nuclear ribonucleoprotein particles), affect the steady-state Tlc1 RNA levels and have short telomeres [45]. The Sm protein appears to be involved in telomerase metabolism. Other groups of RNA-binding proteins are capable of binding directly to single-stranded telomeric DNA. For example, hnRNP A1 was shown to bind single-stranded telomeric DNA and telomerase RNA [35]. Depletion of hnRNP A1 expression causes short telomeres and reduces telomerase activity [46,47]. It was postulated that binding of these hnRNPs to telomeres might help in recruiting telomerase to the telomeres. In the present study, we provide evidence that Imp4 both binds to single-stranded telomeric DNA in vitro and is associated with telomeres in vivo. Thus, in addition to its role in rRNA processing, our findings suggest that Imp4p might affect telomeres through direct association with telomeres. Our results also suggest that it might be a more general property of RNA-processing proteins to have dual functions in the nucleus.
Proteins belonging to the Imp4 family share a high level of sequence homology. A Brix domain is identified in this family of proteins that was postulated to be involved in RNA binding. Indeed, it was shown biochemically that the Brix domain of Imp4p is involved in binding to U3 snoRNA and the amino acid residues Arg220 and Arg253 are important for its binding to RNA [28]. However, the crystal structure of Methanothermobacter thermautotrophicus Mil protein, a member of the Imp4 superfamily, indicated an alternative [30]. The structure and charge distribution of the N-terminal half of the Mil protein suggested that it is capable of binding to its RNA substrate; this raised doubts regarding the role of Brix domain in these proteins. In the present study, we showed that the telomere-binding domain of Imp4p is mapped to its Brix domain, which is consistent with the conclusion made by Gerczei and Correll [28]. However, since the telomeric DNA-binding activity is significantly decreased in the Brix domain, our results also indicated that the Brix domain alone is not sufficient for proper binding to telomeric DNA, suggesting that other portions of Imp4p are also involved in binding to telomeric DNA. We speculate that the N-terminal portion of Imp4p might also participate in binding to telomeric DNA. Interestingly, unlike the binding of single-stranded telomeric DNA, the binding of U3 snoRNA by full-length Imp4p and the Brix domain of Imp4p appears to be similar [28]. Thus the binding of U3 snoRNA and single-stranded telomeric DNA by Imp4p, although similar, might still be different.
Acknowledgments
We thank the members of the Institute of Biopharmaceutical Sciences for the help and support. We are grateful to Dr S. J. Baserga for providing IMP4 reagents and to Dr P. Baumann and Dr T. R. Cech for providing hPot1. This research was supported by National Science Council grants NSC 94-2311-B-010-012 and NSC 94-3112-B-010-020 and National Health Research Institute grant NHRI-EX95-9436SI.
References
- 1.Cech T. R. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 2.Zakian V. A. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 3.Greider C. W., Blackburn E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira M. G., Miller K. M., Cooper J. P. Indecent exposure: when telomeres become uncapped. Mol. Cell. 2004;13:7–16. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- 5.Pang T.-L., Wang C. Y., Hsu C.-L., Chen M. Y., Lin J.-J. Exposure of single-stranded telomeric DNA causes G2/M cell cycle arrest in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:9318–9321. doi: 10.1074/jbc.M208347200. [DOI] [PubMed] [Google Scholar]
- 6.Wellinger R. J., Ethier K., Labrecque P., Zakian V. A. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 7.Conrad M. N., Wright J. H., Wolf A. J., Zakian V. A. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 8.Lustig A. J., Kurtz S., Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 9.Nugent C. I., Hughes T. R., Lue N. F., Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 10.Lin J.-J., Zakian V. A. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandin N., Reed S. I., Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 12.Grandin N., Damon C., Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans S. K., Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 14.Qi H., Zakian V. A. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu C.-L., Chen Y.-S., Tsai S.-Y., Tu P.-J., Wang M.-J., Lin J.-J. Interaction of Saccharomyces Cdc13p with Pol1p, Imp4p, Sir4p, and Zds2p is involved in telomere replication, telomere maintenance and cell growth control. Nucleic Acids Res. 2004;32:511–521. doi: 10.1093/nar/gkh203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virta-Pearlman V., Morris D. K., Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 17.Lin J.-J., Zakian V. A. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1–3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994;22:4906–4913. doi: 10.1093/nar/22.23.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konkel L. M. C., Enomoto S., Chamberlain E. M., McCune-Zierath P., Iyadurai S. J. P., Berman J. A class of single-stranded telomeric DNA-binding proteins required for Rap1p localization in yeast nuclei. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5558–5562. doi: 10.1073/pnas.92.12.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schramke V., Luciano P., Brevet V., Guillot S., Corda Y., Longhese M. P., Gilson E., Geli V. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- 20.Roy N., Runge K. W. The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma. 1999;108:146–161. doi: 10.1007/s004120050364. [DOI] [PubMed] [Google Scholar]
- 21.Aparicio O. M., Billington B. L., Gottschling D. E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 22.Dragon F., Gallagher J. E. G., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Beyer A. L., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 24.Lee S. J., Baserga S. J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granneman S., Gallagher J. E. G., Vogelzangs J., Horstman W., van Venrooij W. J., Baserga S. J., Pruijn G. J. M. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60–80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wormsley S., Samarsky D. A., Fournier M. J., Baserga S. J. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA. 2001;7:904–919. doi: 10.1017/s1355838201010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wehner K. A., Gallagher J. E. G., Baserga S. J. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 2002;22:7258–7267. doi: 10.1128/MCB.22.20.7258-7267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerczei T., Correll C. C. Imp3p and Imp4p mediate formation of essential U3-precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15301–15306. doi: 10.1073/pnas.0406819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenhaber F., Wechsellberger C., Kreil G. The Brix domain protein family: a key to the ribosomal biogenesis pathway? Trends Biochem. Sci. 2001;26:345–347. doi: 10.1016/s0968-0004(01)01851-5. [DOI] [PubMed] [Google Scholar]
- 30.Ng C. L., Waterman D., Koonin E. V., Antson A. A., Ortiz-Lombardia M. Crystal structure of Mil (Mth680): internal duplication and similarity between the Imp4/Brix domain and the anticodon-binding domain of class IIa aminoacyl-tRNA synthetase. EMBO Rep. 2005;6:140–146. doi: 10.1038/sj.embor.7400328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wehner K. A., Baserga S. J. The σ70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell. 2002;9:329–339. doi: 10.1016/s1097-2765(02)00438-0. [DOI] [PubMed] [Google Scholar]
- 32.Baumann P., Cech T. R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taggart A. K. P., Teng S.-C., Zakian V. A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 35.McKay S. J., Cooke H. hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res. 1992;20:6461–6464. doi: 10.1093/nar/20.24.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa F., Matunis M. J., Dreyfuss G., Cech T. R. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larrivee M., LeBel C., Wellinger R. J. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellinger R. J., Wolf A. J., Zakian V. A. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 39.Gottschling D. E., Zakian V. A. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 40.Lin Y.-C., Hsu C.-L., Shih J.-W., Lin J.-J. Specific binding of single-stranded telomeric DNA by Cdc13p of Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:24588–24593. doi: 10.1074/jbc.M101642200. [DOI] [PubMed] [Google Scholar]
- 41.Fang G., Gray J. T., Cech T. R. Oxytricha telomere-binding protein: separable DNA-binding and dimerization domains of the α-subunit. Genes Dev. 1993;7:870–882. doi: 10.1101/gad.7.5.870. [DOI] [PubMed] [Google Scholar]
- 42.Fang G., Cech T. R. Oxytricha telomere-binding protein: DNA-dependent dimerization of the α and β subunits. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6056–6060. doi: 10.1073/pnas.90.13.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao H., Cervantes R. B., Mandell E. K., Otero J. H., Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 44.Ford L. P., Wright W. E., Shay J. W. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21:580–583. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]
- 45.Seto A. G., Zaug A. J., Sobel S. G., Wolin S. L., Cech T. R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 46.LaBranche H., Dupuis S., Ben-Davis Y., Bani M.-R., Wellinger R. J., Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 2001;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q.-S., Manche L., Xu R.-M., Krainer A. R. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA. 2006;12:1116–1128. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]