Abstract
InsP6 is an intracellular signal with several proposed functions that is synthesized by IP5K [Ins(1,3,4,5,6)P5 2-kinase]. In the present study, we overexpressed EGFP (enhanced green fluorescent protein)–IP5K fusion proteins in NRK (normal rat kidney), COS7 and H1299 cells. The results indicate that there is spatial microheterogeneity in the intracellular localization of IP5K that could also be confirmed for the endogenous enzyme. This may facilitate changes in InsP6 levels at its sites of action. For example, overexpressed IP5K showed a structured organization within the nucleus. The kinase was preferentially localized in euchromatin and nucleoli, and co-localized with mRNA. In the cytoplasm, the overexpressed IP5K showed locally high concentrations in discrete foci. The latter were attributed to stress granules by using mRNA, PABP [poly(A)-binding protein] and TIAR (TIA-1-related protein) as markers. The incidence of stress granules, in which IP5K remained highly concentrated, was further increased by puromycin treatment. Using FRAP (fluorescence recovery after photobleaching) we established that IP5K was actively transported into the nucleus. By site-directed mutagenesis we identified a nuclear import signal and a peptide segment mediating the nuclear export of IP5K.
Keywords: euchromatin; Ins(1,3,4,5,6)P5 2-kinase (IP5K); in situ hybridization; nuclear localization; stress granule
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DTT, dithiothreitol; EGFP, enhanced green fluorescent protein; FISH, fluorescence in situ hybridization; FRAP, fluorescence recover after photobleaching; IPMK, inositol phosphate multikinase; IPK, inositol phosphate kinase; IP3K, Ins(1,4,5)P3 3-kinase; IP5K, Ins(1,3,4,5,6)P5 2-kinase; LMB, leptomycin B; MDD-HPLC, metal-dye-detection-HPLC; mRNP, messenger ribonucleoprotein; NLS, nuclear localization sequence; NRK, normal rat kidney; ORF, open reading frame; PABP, poly(A)-binding protein; RNAi, RNA interference; ROI, region of interest; SG, stress granule; siRNA, small-interfering RNA; TIAR, TIA-1-related protein
INTRODUCTION
There is a considerable literature describing the signalling functions and metabolic pathways of inositol phosphates [1–3]. Since inositol phosphates are both soluble (rather than membrane-bound) and also rapidly diffusible molecules [4], these signals have generally been considered to be freely dispersed throughout the cell. However, there is a growing realization that this is not the case.
For example, the nuclear membrane has emerged to be a diffusional barrier for inositol phosphates [5]. Moreover, the importance of nuclear localization of inositol phosphates and their kinases is becoming increasingly evident [6]. Nuclear localization has been demonstrated for both endogenous and overexpressed IP3K [Ins(1,4,5)P3 3-kinase] isoforms B [7] and C. Overexpressed rat IP3K-C actively shuttles between cytoplasm and nucleus [8,9]. Human inositol phosphate multikinase (termed IPMK or Ipk2) is predominantly localized in the nucleus [10]. The Drosophila melanogaster homologue of IPMK is targeted to the nucleoplasm of salivary glands where it appears in the ‘non-DAPI (4′,6-diamidino-2-phenylindole) stained nucleosol’ [11]. Two of the three isoforms of IP6K (InsP6 kinase) also exhibit nuclear localization although to different extents [12]. Thus vertebrate IPKs (inositol phosphate kinases) may support an inositol phosphate metabolic profile within the nucleus that is at least partly independent of that in the cytosol [2,6,7,10,13–15].
In the present study we provide new information on the intracellular distribution of IP5K [Ins(1,3,4,5,6)P5 2-kinase; also known as Ipk1]. This 2-kinase synthesizes InsP6 from Ins(1,3,4,5,6)P5 and was first observed by Stephens et al. [15a] and later in soybean seeds by Phillippy et al. [15b]. The enzyme was first cloned from Saccharomyces cerevisiae [16]. Human IP5K was identified by its homology with small conserved domains in the fungal sequences [17]. Substrate specificity of IP5K for Ins(1,3,4,5,6)P5 in human and rat cells was confirmed by RNAi (RNA interference) techniques. A significant decrease in cellular InsP6 was measured after knockdown of IP5K [18,19]. Overexpression of IP5K enhanced conversion of Ins(1,3,4,5,6)P5 into InsP6 in HEK-293 (human embryonic kidney 293) cells [20]. This enzyme is considered to have an important role in signal transduction because its product, InsP6, has been reported to have a number of biological functions. For example, InsP6 is a cofactor for DNA-dependent protein kinase activity in non-homologous end-joining [21] and an essential folding factor for an adenosine deaminase that participates in editing of mRNA and tRNA [22]. InsP6 may also have critical roles in chromatin remodelling [23–25].
In yeast, IP5K/Ipk1 is localized in the nucleus and at the nuclear pore complex [26]. However, the only previous study of the localization of IP5K in mammalian cells found the enzyme to be distributed equally in the cytosol and within the nucleus [18]. In the present study, we confirm that IP5K is distributed throughout the cell, but we make the novel observation that the kinase is especially concentrated in both nuclear and cytoplasmic foci. We show further that endogenous and overexpressed human IP5K are both targeted to euchromatin regions and nucleoli. Amino acid clusters required for the observed nuclear import and export activity of IP5K were identified by mutagenesis of overexpressed IP5K–EGFP (enhanced green fluorescent protein) fusion protein. FISH (fluorescence in situ hybridization) of poly(A)-RNA revealed co-localization with both nuclear mRNA and cytosolic mRNA in so called SGs (stress granules). SGs are large cytoplasmic mRNP (messenger ribonucleoprotein) complexes formed in response to different types of cellular stress, such as that caused by toxic agents, or by oxidative conditions and also stress caused by artificial gene overexpression [27]. We propose that the IP5K has the ability to promote spatial microheterogeneity in the synthesis of InsP6.
EXPERIMENTAL
Construction of fusion genes
Sequencing of cDNA clone IMAGE:4839268 (GenBank® accession no. BG772243; GenBank® gene info identifier: 14082896; ResGen Invitrogen Corporation) revealed total agreement with the cDNA described in [17]. The ORF (open reading frame) of human IP5K was amplified using the following primer pairs: 5′-AGCTAGCATGGAAGAGGGGAAGATGGA-3′ and 5′-GAGATCTGAGACCTTGTGGAGAACTAATG-3′; 5′-CAAGCTTCGATGGAAGAGGGGAAGATGGA-3′ and 5′-TGGATCCTTAGACCTTGTGGAGAACTA-3′. PCR products were cloned into pGEM T-Easy vector (Promega) by TA-cloning. After re-sequencing, the fragment was subcloned by introduced restriction sites (BglII/NheI and BamHI/HindIII) into expression vectors pEGFP-N1 and pEGFP-C1 (Clontech) respectively. IP5K mutants were generated by modified QuikChange™ site-directed mutagenesis as previously described [28] using the following primers: C162Y, 5′-AAGCATAAGGTCTGTCGATACTACATGCACCAGCACCTCAAGGTA; R41A/K42A/K43A, 5′-CTGAAGTTTCCTCCAAATGCGGCGGCGACCTCGGAAGAGATATTT; K36A, 5′-TGCGTCGTGCTGCGGTTTCTGGCGTTTCCTCCAAATGCGGCGGCG; R28A/R33A, 5′-GTGGCCCACGCGCAGGCGTGCGTCGTGCTGGCGTTTCTGGCGTTTCCT; H152S/H156S, 5′-TTCTCGAGTGATGTCACGTCTGAGATGAAGTCTAAGGTCTGTCGATAC; C159S/C162S, 5′-GAGATGAAGCATAAGGTCAGTCGATACAGCATGCACCAGCACCTCAAG; K173A/K175A, 5′-CTCAAGGTAGCAACTGGGGCGTGGGCGCAGATCAGCAAATACTGTC; K179A, 5′-CAGATCAGCGCGTACTGTCCC; K155A/K157A, 5′-CGAGTGATGTCACGCATGAGATGGCGCATGCGGTCTGTCGATACTGCATG; and Del1, 5′-CACTTTGCCTTGAAGAGTTTGCTGTTCAGTAGGAGCCTTCGCTGCCAAGG-3′ (only forward primers are shown). The plasmids encoding the EGFP trimer and tetramer were created using a previously described method [29] with pEGFP-N1 (BD Biosciences Clontech) as a template.
Bacterial expression and purification of recombinant Strep–IP5K fusion proteins
The ORF of human IP5K was amplified using the primer pair 5′-AGCCATGGCATGGAAGAGGGGAAGATGGA-3′ and 5′-AGCTCGAGTCTAGACCTTGTGGAGAACTA-3′, introducing an NcoI restriction site at the 5′-end and an XhoI restriction site at the 3′-end. The amplified DNA fragment was cloned into the pGEM T-Easy vector (Promega). An XhoI restriction site within the coding region interfering with our cloning strategy was removed by introducing a silent mutation using the QuikChange™ site-directed mutagenesis kit from Stratagene according to the manufacturer's instructions using the primer pair 5′-TAGTGGGTTTATTCCTTTTTCGAGTGATGTCACGCAT-3′ and 3′-primer. The fragment was sub-cloned by the introduced restriction sites into a pET17b-based expression vector encoding an N-terminal Strep-tag fusion protein (Novagen). The ORF was completely re-sequenced. Mutations were introduced by primer-based mutagenesis as described above.
The recombinant fusion proteins with an N-terminal Strep-tag were overexpressed in Escherichia coli BL21(DE3)-pLysS[pREP4] and Strep-tag purified. Bacterial cell growth at 37 °C was monitored by measuring the attenuance at 600 nm (D600). At an D600 of 0.5, IPTG (isopropyl β-D-thiogalactoside) was added to a final concentration of 0.25 mM. After 2 h expression at 37 °C cells were harvested (4000 g, 5 min, 4 °C) and resuspended in 1/20 of the culture volume in buffer containing 50 mM Hepes (pH 7.5), 1 mM EDTA, 1 mM DTT (dithiothreitol), 1% (v/v) Triton X-100, 0.5 mM benzamidine and 1 mM PMSF. After sonification (3×45 s) the lysate was centrifuged at 27000 g for 10 min (at 4 °C). The NaCl concentration in the supernatant was adjusted to 500 mM and then the protein was purified by application to a 1 ml Strep-tactin column (IBA) and washing using a buffer containing 500 mM NaCl, 100 mM Hepes (pH 8.0), 1 mM EDTA, 1 mM DTT and 0.1% Triton X-100. The protein was eluted using washing buffer supplemented with 2.5 mM desthiobiotin. The purified protein was analysed by SDS/PAGE and staining with Coomassie Blue. Additionally, after separation by SDS/PAGE and transfer on to a PVDF membrane, recombinant Strep–IP5K was detected using an avidin-alkaline phosphatase conjugate (Bio-Rad) according to the manufacturer's instructions. Immediately after preparation glycerol was added to a final concentration of 50% and the material was stored at −20 °C.
Immunobead purification of EGFP–IP5K fusion protein
COS7 cells (8×106) overexpressing EGFP fusion proteins for 24 h were lysed in MPER® (Pierce) for 10 min at 24 °C, homogenized and centrifuged (16000 g, 15 min, 4 °C). Supernatant was mixed with a Protein-G–agarose/anti-GFP complex (Roche) and immunobead purification of EGFP–IP5K fusion proteins was performed as described previously [30].
Assay of the kinetic properties of purified IP5K enzymes
Kinetic assays of immunobead-adsorbed EGFP–IP5K fusion proteins were performed in a 100 μl or 200 μl assay volume for 2 h at 37 °C. For each assay 20–40 ng of IP5K (determined by densitometry of the specific fusion polypeptide after SDS/PAGE) and between 0.05 and 25 μM Ins(1,3,4,5,6)P5 were added to the assay buffer [50 mM Hepes (pH 7.0), 100 mM KCl, 0.5 mM ATP, 5 mM MgCl2 and 1 mM DTT].
The activity of the purified bacterial-expressed Strep–IP5K and its mutants was assayed for various times at 30 °C in 100 μl of a reaction mixture containing 50 mM Hepes (pH 7.0), 63.5 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1.1 mM DTT, 1 mM ATP, 0.13 mM EDTA, 0.32 mM desthiobiotin, 0.013% Triton X-100, 15% glycerol and 10 μM Ins(1,3,4,5,6)P5. The reaction was started with approx. 0.1 μg of either wild-type enzyme or mutant. Assays were quenched and inositol phosphates were extracted [10] and analysed by MDD-HPLC (metal-dye-detection-HPLC) as described below. Kinetic parameters were determined using PRISM. Owing to a sigmoidal dependence of specific activity on substrate concentration, the v compared with [S] dependence was fitted to a Hill-type function [31].
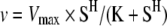 |
where v is the initial velocity, S is the substrate concentration, K is the apparent Km and H is the Hill coefficient.
Analysis of inositol phosphates by MDD-HPLC
Ins(1,3,4,5,6)P5 and InsP6 were measured by MDD-HPLC as previously described [32–34]. A gradient from 0.2 mM HCl, 10 μM YCl3 to 0.5 M HCl, 10 μM YCl3 within 20 min was employed to separate Ins(1,3,4,5,6)P5 and InsP6. Inositol phosphates were quantified by external standardization. The data provided in the text which describe the effects of overexpressed IP5K upon cellular levels of Ins(1,3,4,5,6)P5 and InsP6 are corrected for the transfection efficiency of the two fusion proteins (IP5K–EGFP=40%; EGFP–IP5K=30%).
Cell culture, transfection and drug incubation
H1299 human lung carcinomal cells (CRL-5803) and COS7 cells were cultured as described for NRK (normal rat kidney) cells [7] at 37 °C and 5% CO2. Cell transfection was performed using either FuGENE™6 (Roche) or metafectene (Biontex Laboratories), and 500 ng of cDNA per cm2 culture surface, according to the manufacturer's instructions. For all experiments cells were cultured for 24 h after transfection. Leptomycin B was purchased from A.G. Scientific and was added to a final concentration of 20 nM. Before analysis, cells were incubated with or without leptomycin B for an additional 5 h. To induce SGs, cells were incubated with 50 μg/ml puromycin for 1 h [35].
Immunofluorescence and fluorescence in situ hybridization
Immunofluorescence was carried out as previously described [7] employing the following antibody dilutions: rabbit anti-IP5K peptide antibody (1:100; Invitrogen; see Results), rabbit anti-PABP [poly(A)-binding protein; 1:1750; Dr Evita Mohr, UKE, Hamburg, Germany], mouse anti-TIAR (TIA-1-related protein; 1:1750; BD Biosciences), goat anti-rabbit (1:1750) and goat anti-mouse (1:1500; Invitrogen) and rabbit anti-nucleolin (1:10000; BD Biosciences).
FISH was performed as previously described [36]. Results were evaluated by epi-fluorescence microscopy as described in [10].
Western blot analysis
For Western blot analysis, 48 h after transfection, cells were harvested, lysis buffer [8 M urea, 15 mM EDTA and 30 mM Tris/HCl (pH 7.4)] was added and, after freezing and thawing in liquid nitrogen and centrifugation (13000 g, 15 min, 4 °C), the supernatant was removed and the protein concentration was determined using a Bradford assay. Next, 50 μg of protein was applied on to SDS/PAGE gels. After gel electrophoresis (150 V), protein was transferred on to PVDF membranes followed by blocking non-specific binding sites with 3% (w/v) BSA in TTBS [10 mM Tris/HCl (pH 8.0), 150 mM NaCl and 0.05% Tween 20] for 20 min. The membranes were washed three times with TTBS for 10 min each and incubated with anti-IP5K overnight at 4 °C. After washing with TTBS under the same conditions, the membranes were incubated with secondary antibody conjugated with horseradish peroxidase for 1 h. Membranes were washed again as above and specific immune complexes were visualized by employing the ECL™ Advance Western Blotting Detection kit (GE Healthcare).
Knockdown of IP5K with Stealth™ siRNA (small-interfering RNA)
H1299 cells were seeded in chamber slides (1×104 cells per chamber), incubated for 24 h and transfected with a mixture of 3 μl of Stealth™ IP5K-siRNA (20 nM; Invitrogen, siRNA #HSS127724) or Stealth™ RNAi negative control and 1 μl of Lipofectamine™ (Invitrogen) according to the manufacturer's instructions. At 48 h after transfection the cells were fixed with 3% (w/v) paraformaldehyde, permeabilized with 0.3% Triton X-100 in PBS and analysed for IP5K expression by immunofluorescent staining with an anti-IP5K antibody and a TRITC-labelled secondary antibody. Nuclei were stained with DAPI (Carl Roth) and visualized by immunofluorescence microscopy (see below).
Fluorescence microscopy and live-cell imaging for FRAP (fluorescence recovery after photobleaching) experiments
Epi-fluorescence microscopy [10] was performed using an Axiovert 25 CFL (Zeiss). Confocal images were taken on a Zeiss LSM 510 Meta (Zeiss) using a Plan-Apochromat 63X/1.4 Oil DIC objective. A Coherent Mira titanium-sapphire ultrafast pulsed laser tuned to 750 nm was used for multiphoton excitation of DAPI. A 390–465 bandpass filter with IR blocking was used to collect images with an open pinhole. For excitation of EGFP a 488 nm laser line from a krypton/argon laser was used (500–500 nm bandpass emission filter). For excitation of Alexa Fluor® 546 a 543 nm laser line from a helium/neon laser was used (560 nm long pass emission filter), pinhole setting for both was 1 airy unit.
FRAP experiments were performed with an object carrier heated to 37 °C using the Zeiss LSM 510 confocal microscope with a 100× oil-immersion objective. For imaging, cells grown on coverslips were mounted in CO2-independent Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% (w/v) BSA (Sigma). EGFP fluorescence was excited at 488 nm using 25% transmission and was emission-collected using a 505 nm filter provided by the manufacturer. An image of the whole cell was obtained before and after the experiment. Photobleaching was performed at 100% transmission by scanning the bleached ROI (region of interest) fitted to the nucleus for 100 iterations. Recovery data were collected over 16 min at 30 s intervals. Fluorescence intensity data for the bleached ROIs were calculated using the ImageJ software. For each FRAP data set, three to five separate experiments were performed, typically including up to four cells investigated per condition on a given day.
Quantitative image analysis
For each experiment, the EGFP fluorescence intensities in the nucleus and cytoplasm were quantified for at least 25 images of representative cells. In detail, EGFP fluorescence intensities of six ROIs in both the nuclear and cytoplasmic areas per cell were averaged to calculate the ratio of nuclear over cytoplasmic intensity using the Image J program (W. Rasband, NIH, Washington, DC, U.S.A.) as previously described [10]. For evaluation of data an unpaired Student's t test was performed using GraphPad InStat version 3.06 (GraphPad Software). A value of P<0.05 was considered statistically significant.
The distribution pattern of IP5K was quantified with Metamorph (Molecular Devices). The fluorescence intensity of EGFP–IP5K in the whole cells was measured by fitting the ROIs to 50 cells with and without puromycin treatment. Nuclei and SGs were identified by FISH of poly(A)-RNA. In the EGFP picture ROIs were fitted to these compartments and their fluorescence intensities were subtracted separately from the total fluorescence intensity.
RESULTS
Intracellular localization of IP5K and targeting to nuclear substructures
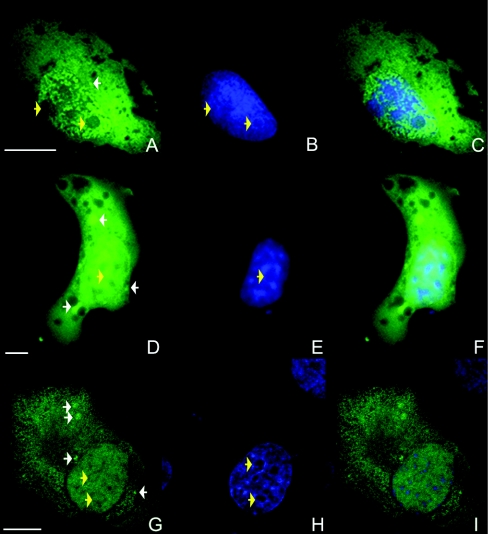
To investigate the intracellular localization of human IP5K, the full-length enzyme, either N-terminally (EGFP–IP5K) or C-ter-minally (IP5K–EGFP) fused to EGFP was transiently overexpressed. Figure 1 shows intracellular distribution of the C-terminally EGFP-fused construct in NRK cells (Figures 1A–1C), COS7 cells (Figures 1D–1F) and H1299 cells (Figures 1G–1I).
Figure 1. Intracellular localization of overexpressed IP5K in mammalian cells.
Wild-type IP5K–EGFP fusion protein (A, D and G) was transiently overexpressed in mammalian cells for 24 h. (A–F) Epi-fluorescence images of a representative NRK cell (A–C) and a COS7 cell (D–F). Confocal fluorescence images of representative H1299 cells are shown in (G–I). Nuclei were stained with DAPI (B, E and H). Overlays of both signals are shown in (C, F and I). Cytoplasmic IP5K–EGFP foci are marked by white arrows, and nucleoli by yellow arrows. Scale bars represent 10 μm.
All transfected cells showed IP5K–EGFP fluorescence in the cytoplasm (Figure 1). As indicated by the white arrows in Figures 1 and 2, additional cytosolic foci were observed in approx. 24% of transfected NRK cells (Figures 1A and 2A), in 59% of COS7 cells (Figures 1D and 2D) and 30% of H1299 cells (Figures 1G and 2G). The nature of these foci was clarified in the experiments described below.
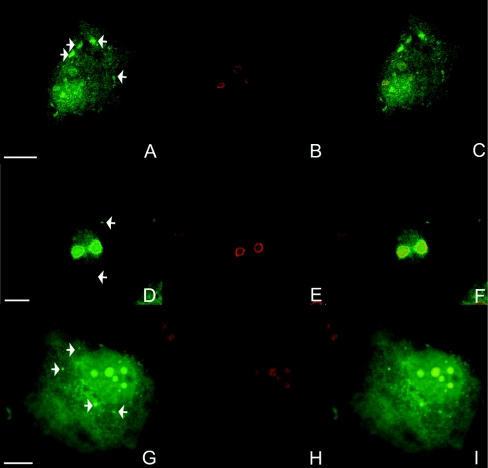
Figure 2. Localization of overexpressed EGFP–IP5K in nucleoli.
EGFP–IP5K fusion protein was overexpressed for 24 h in NRK (A–C), COS7 cells (D–F) and H1299 cells (G–I). All panels show confocal images. Nucleoli were detected by indirect immunofluorescence employing an anti-nucleolin antibody and Alexa Fluor® 546-conjugated secondary antibody (B, E and H). (C, F and I) show overlays of both signals. Cytoplasmic EGFP–IP5K foci are marked by white arrows. Scale bars represent 10 μm.
IP5K was also localized in the nucleus (Figure 1). The IP5K–EGFP fluorescence was heterogeneously distributed, being typically most intense in regions where the DAPI signal was weakest (Figure 1). Regions of the nucleoplasm (i.e. the nuclear area outside the nucleoli) that are stained only weakly by DAPI (Figures 1B, 1E and 1H) are defined as euchromatin [37]. Thus Figure 1 provides evidence of ‘euchromatin targeting’ of IP5K. A similar nuclear localization was observed for IPMK in D. melanogaster [11].
Another region of the nucleus that tends to exclude the DAPI stain is the nucleolus, which can often be revealed as a well-defined circular ‘hole’ in the staining (indicated by yellow arrows in Figure 1). Alternatively, the nucleolus can be delineated by the confocal immunofluorescence signal from anti-nucleolin antibodies (Figure 2). Both C-terminally and N-terminally tagged IP5K were able to enter nucleoli in all three of the cell types that were studied. The N-terminally tagged protein (i.e. EGFP–IP5K) in particular tended to accumulate in the nucleolus to a higher extent than in the nucleoplasm (Figure 2). This enrichment was observed in 50% of H1299 cells 24 h after transfection. It is possible that a C-terminal tag on IP5K reduces the efficiency of targeting to the nucleolus; this was the only significant difference in localization between N-terminally and C-terminally EGFP-tagged IP5K.
To study whether EGFP–IP5K and IP5K–EGFP were expressed functionally, their enzymatic activity was investigated by detailed kinetic analyses in vitro and in vivo. The two fusion proteins were enriched with anti-GFP-conjugated Protein-G–agarose beads [30]. The specific activity of the purified N-terminal fusion protein was 55 nmol/min per mg and that of the C-terminal fusion protein 80 nmol/min per mg. These activities are slightly higher than previously reported for IP5K overexpressed in Schneider cells (31 nmol/min per mg) [17].
Furthermore, the activities of EGFP–IP5K and IP5K–EGFP in vivo were analysed by measuring the effects of their overexpression upon cellular levels of Ins(1,3,4,5,6)P5 and InsP6 in COS7 cells. Control cells contained 75 pmol of Ins(1,3,4,5,6)P5 per 106 cells; overexpression of EGFP–IP5K or IP5K–EGFP reduced Ins(1,3,4,5,6)P5 levels to 60 and 55 pmol per 106 cells respectively. Control cells contained 125 pmol of InsP6 per 106 cells; overexpression of EGFP–IP5K or IP5K–EGFP increased InsP6 levels to 137 and 145 pmol per 106 cells respectively (means of two independent experiments). After correcting for transfection efficiency of 30–40% (see the Experimental section) we calculated that both fusion proteins decreased Ins(1,3,4,5,6)P5 levels by 66%. The absolute decrease in Ins(1,3,4,5,6)P5 levels is near-quantitatively matched by an increase in InsP6 levels. These results provide strong evidence that the fusion proteins are catalytically active in vivo.
IP5K partially co-localizes with mRNA
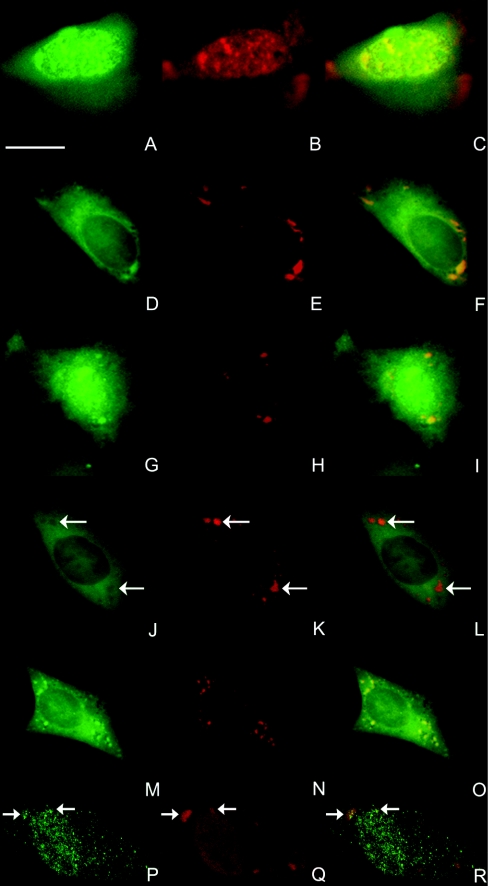
Results described above indicate that IP5K is particularly concentrated in transcriptionally active areas of the nucleus, i.e. euchromatin. Since the InsP6 that is synthesized by IP5K also participates in mRNA export from the nucleus [38], we investigated whether IP5K might co-localize with nuclear RNA in vertebrate cells. In NRK cells in which EGFP–IP5K was overexpressed, we detected mRNA by FISH of poly(A)-RNA. Figure 3 shows a co-localization of IP5K and poly(A)-RNA in the nucleus (Figures 3A–3C).
Figure 3. Co-localization of IP5K with mRNA.
NRK cells (A–O) overexpressed either IP5K–EGFP (A–I, M–O) or a catalytically inactive EGFP–IP5K mutant C162Y (J–L) for 24 h. mRNA was detected by FISH of poly(A)-RNA employing Alexa Fluor® 555-labelled poly(dT)50 oligonucleotide (red signal in B, C, E and F). Two representative sets of images are shown. SGs were detected either by FISH of poly(A)-RNA (N and O), or by indirect immunofluorescence employing anti-PABP (H, I, K and L) or anti-TIAR (Q and R) antibodies and Alexa Fluor® 546-conjugated secondary goat anti-rabbit antibody. For clarity, in some panels the positions of the SGs are highlighted with arrows. In some experiments (M–R) SGs were induced by puromycin treatment (50 μg/ml for 1 h). Endogenous IP5K in H1299 cells (P–R) was detected by indirect immunofluorescence employing an anti-IP5K peptide antibody and Alexa Fluor® 488-conjugated secondary goat anti-rabbit antibody. In epi-fluorescence images, scale bars represent 10 μm.
IP5K is a component of stress granules
Interestingly, FISH analysis of poly(A)-RNA in NRK cells additionally revealed that the cytoplasmic foci of EGFP–IP5K, mentioned above (Figures 1 and 2), also co-localized with high amounts of mRNA (Figures 3D–3F). We suspected that this co-localization might occur in SGs. These are large mRNP complexes that are sites of mRNA storage; they can form in response to artificial protein overexpression [27].
To explore this idea, we employed antibodies against two of the mRNA-binding proteins present in SG, namely PABP and TIAR [39]. Cytosolic EGFP–IP5K foci (Figure 3G) co-localized with PABP (Figure 3H) and TIAR (results not shown) and thus are localized in SGs.
To investigate whether the co-localization of IP5K and SGs depends on catalytic activity of the IP5K we generated a fully inactive IP5K mutant with an N-terminal EGFP tag. On the basis of the earlier demonstration that the yeast homologue Ipk1 is inactivated by conversion of Cys139 into a tyrosine residue [40], we mutated the corresponding Cys162 in IP5K to a tyrosine residue. The immunosorbent-purified mutant enzyme was completely inactive (results not shown). When overexpressed in NRK cells, EGFP–IP5K/C162Y (Figure 3J) no longer exhibited a co-localization with SGs detected by FISH of poly(A)-RNA (Figure 3K) or by anti-PABP (results not shown). Instead, the catalytically dead kinase seemed to be excluded from SGs (Figure 3L).
SGs also form in response to certain cellular stresses, such as puromycin treatment [41]. NRK cells that overexpressed IP5K–EGFP were incubated with puromycin (Figure 3M) and we detected drug-induced SGs by mRNA staining (Figure 3N). A clear co-localization of IP5K with puromycin-induced SGs was visible (overlay Figure 3O).
To determine the percentage of overexpressed IP5K that co-localizes with SGs, the distribution of IP5K localization patterns was quantified. In H1299 cells in which EGFP–IP5K was overexpressed, SGs were detected by FISH of poly(A)-RNA. Images of 100 cells treated with puromycin and 100 non-treated control cells were analysed with Metamorph software. Note that SGs can accumulate as a consequence of the stress caused by protein overexpression [27], so a substantial number of SGs were observed in cells that overexpressed EGFP–IP5K, even in the absence of puromycin. Without puromycin treatment the fusion protein showed an average nuclear localization of 34.0% which included nucleolar localization of 4.8%. When SGs were present, approx. 1.6% of the total cell fluorescence resides in SGs. Nuclear and nucleolar localization was not significantly altered by SG formation induced by protein overexpression. Incubation with puromycin increased the percentage of cells that showed SGs from 58% to 73% and the number of SGs per cell from 2–10 to 4–21. The localization pattern of IP5K in puromycin-treated cells was determined to be 32.1% nuclear localization including 5.2% nucleolar localization and a localization in SGs of 2.1% of the fusion protein. An unpaired Student's t test showed that these average fluorescence intensities per cell compartment were not significantly altered compared with untreated cells.
Intracellular localization of endogenous IP5K
We also investigated whether endogenous IP5K could be detected in SGs. Therefore we used H1299 cells and SGs were induced by puromycin treatment (Figures 3P–3R). The SGs were monitored using an anti-TIAR antibody. To detect IP5K a rabbit anti-IP5K peptide antibody was employed (see below and Supplementary data at http://www.BiochemJ.org/bj/408/bj4080335add.htm for a characterization of its specificity). Our experiments clearly show that endogenous IP5K co-localized with SGs (Figures 3P–3R).
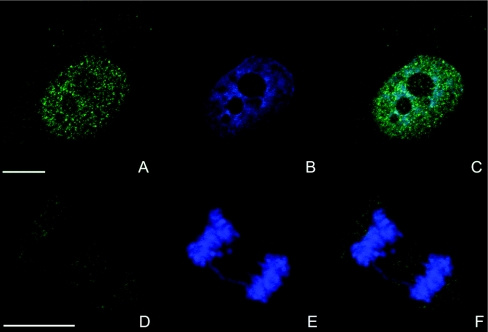
In these experiments, we noted that a greater proportion of total IP5K was located in the nucleus (Figures 3P–R and 4A–4C) compared with cells in which IP5K was overexpressed (Figures 1G–1I and Figures 2G–2I). The signal that was observed in the cytoplasm was punctate, but the foci were too small to be considered SGs (Figure 4). This observation is consistent with previous reports [27] that the SGs form in response to the cellular stress caused by protein overexpression. In dividing metaphase cells, where nuclear membranes are absent, a punctate distribution of IP5K was observed throughout the cytoplasm. Condensed metaphase chromosomes did not show any IP5K (Figures 4D–4F).
Figure 4. Localization of endogenous IP5K in human H1299 cells.
Endogenous IP5K was detected in H1299 cells by indirect immunofluorescence employing an anti-IP5K peptide antibody and Alexa Fluor® 488-conjugated secondary goat anti-rabbit antibody (A and D). Nuclei were stained with DAPI (B and E). Overlays are shown in (C) and (F). In confocal images, scale bars represent 10 μm.
The nuclear distribution of endogenous IP5K mirrored that observed following overexpression of EGFP-tagged IP5K. The nuclear staining of endogenous IP5K in the nucleoplasm again was inversely correlated with DAPI staining (compare Figures 1A–1I with Figures 4A–4C). Punctae of IP5K were visible in nucleoli, but most of them were located in the weakly DAPI-stained euchromatin regions. In the Supplementary data (at http://www.BiochemJ.org/bj/408/bj4080335add.htm) we provide evidence that the anti-IP5K antibody may underestimate the amount of endogenous IP5K in nucleoli. It is possible that the epitope is masked by proteins that might form a complex with IP5K.
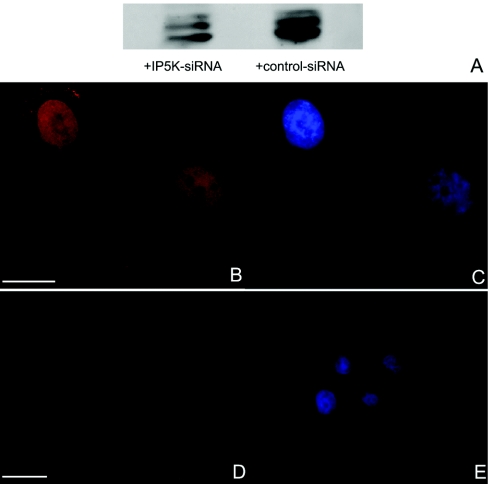
We used siRNA to study the specificity of our antibody against endogenous IP5K. H1299 cells were transfected either with Stealth IP5K-siRNA or Stealth™ RNAi negative control. After 48 h, cells were analysed for IP5K expression by Western blot analysis (Figure 5A) and by indirect immunofluorescence employing an anti-IP5K antibody and a TRITC-labelled secondary antibody (Figures 5B–5E). Western blot analysis showed that the IP5K protein level throughout the cell population was reduced by approx. 65% after RNAi (Figure 5A, left-hand lane) compared with the control cells (Figure 5A, right-hand lane). In the transfected cells RNAi eliminated the IP5K signal (Figure 5D). Control siRNA did not alter intracellular localization of endogenous IP5K (Figure 5B).
Figure 5. Knockdown of IP5K by RNAi.
H1299 cells were transfected with Stealth RNAi negative control (A, right-hand lane; B and C) or Stealth™ IP5K-siRNA (A, left-hand lane; D and E). At 48 h after transfection, cells were lysed and 50 μg of protein was loaded on to an SDS/PAGE, transferred on to a PVDF membrane and IP5K was detected with an anti-IP5K antibody and HRP-conjugated secondary antibody. Signal was developed by ECL. In corresponding cells IP5K was detected by indirect immunofluorescence employing an anti-IP5K peptide antibody and Alexa Fluor® 546-conjugated secondary goat anti-rabbit antibody (B and D). Nuclei were stained with DAPI (C and E). In epi-fluorescence images, scale bars represent 10 μm.
Investigation of IP5K nucleocytoplasmic shuttling by FRAP experiments and identification of IP5K-targeting domains
To investigate whether nuclear IP5K localization is caused by active transport or by passive diffusion, FRAP experiments with EGFP-tagged IP5K were performed. EGFP–IP5K was overexpressed in H1299 cells, nuclei were selectively bleached and nuclear fluorescence recovery was monitored.
For these experiments, we divided the cells into two groups. In one group (‘high’ nuclear signal) the intensity of nuclear fluorescence was equal or greater to the intensity of cytoplasmic fluorescence. In the other group of cells (‘low’ nuclear signal), nuclear fluorescence was significantly lower than that in the cytoplasm (Figure 6).
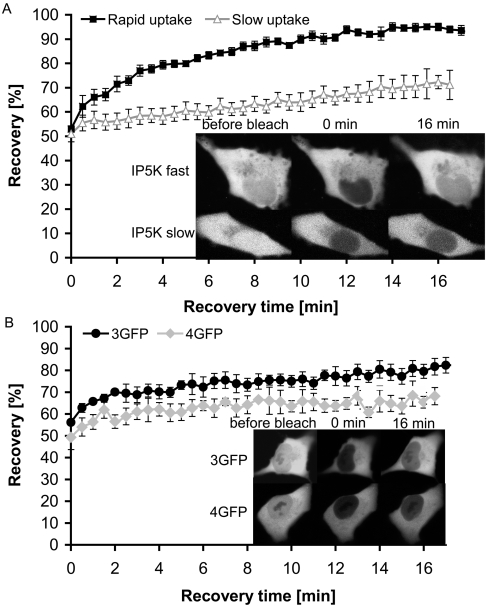
Figure 6. FRAP experiments to investigate IP5K nuclear import.
H1299 cells in which EGFP–IP5K was overexpressed for 24 h were mounted in CO2-independent medium on a 37 °C heated carrier of a Zeiss LSM 510 microscope. EGFP fluorescence was excited at 488 nm using 25% transmission. Emission was measured at 505 nm. Photobleaching was performed at 100% transmission by scanning the bleached ROI fitted to the nucleus for 100 iterations. Recovery data were collected over 16 min at 30 s intervals. (A) Mean recovery curves from EGFP–IP5K-expressing H1299 cells. The ratio of the fluorescence in the bleached compartment to that of the whole cell prior to photobleaching was set at 100%. Approx. 50% of the bleached cells showed rapid half-time of recovery (3 min, black squares, n=8 cells). The other fraction of the investigated cells showed an infinite half-time (grey triangles, n=8 cells). Images of a representative cell ‘before bleach’, directly post bleach (0 min) and after 16 min recovery are shown. These particular cells show no SGs. (B) Mean recovery curves from GFP-multimer-expressing cells. The ratio of the fluorescence in the bleached compartment to that of the whole cell prior to photobleaching was set at 100%. Both curves show infinite recovery half-times (3GFP, black circles, n=7 cells; 4GFP, grey rhombuses, n=3 cells). Representative images ‘before bleach’, post bleach (0 min) and after 16 min recovery are shown.
In the group of cells which originally showed a ‘high’ nuclear signal, there was a relatively slow recovery of nuclear fluorescence after bleaching, such that fluorescence returned from 50% to 65% of the initial level within 16 min (Figure 6A; labelled IP5K slow). It is possible that nuclei of these cells were already saturated with IP5K and there was little or no further import of IP5K into the nucleus. In the other group of cells which we define as originally possessing a ‘low’ nuclear signal, its recovery after bleaching was more rapid. The signal returned from 50% to 95% of the initial level within 12 min (Figure 6A; labelled IP5K fast). This is faster than fluorescence recovery in control cells overexpressing linear fused multimers of EGFP, containing three (3GFP; [42]) and four (4GFP; [43,44]) EGFP subunits which have previously been shown to enter and leave the nucleus only by passive diffusion [42]. For example, nuclear fluorescence of 3GFP recovered from 57% to approx. 80% within 16 min (Figure 6B; 3GFP), and fluorescence of 4GFP recovered from 50% to 67% within 16 min (Figure 6B; 4GFP). Since IP5K has nearly the same molecular mass as 3GFP and enters the nucleus faster than 3GFP, we propose that IP5K must shuttle actively between nucleus and cytosol. Since active transport would presumably employ nuclear import and export motifs, we conducted additional experiments to identify such sequences.
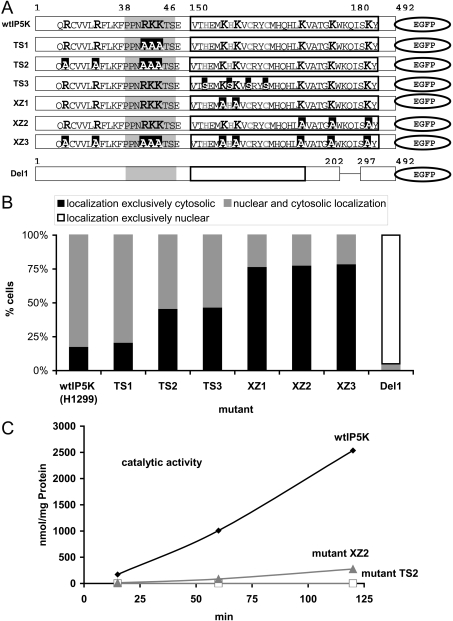
We first searched for putative NLS (nuclear localization sequences) by employing the prediction algorithm PSORTII. One cluster of basic amino acids in the N-terminal part of IP5K (amino acids 38–44: PPNRKKT) fits the consensus sequence of the canonical NLS of SV40 large T antigen. A set of point mutants was generated in the N-terminal part of the IP5K–EGFP construct by QuikChange™ mutagenesis in order to characterize essential amino acids for NLS activity (see Figure 7A for details). Transfected H1299 cells (160–200 cells; obtained from three to four independent experiments) were inspected for each mutant. Unexpectedly, mutation to an alanine residue of the three basic amino acids in the putative NLS had almost no effect on the nuclear localization. In 79% of cells this mutant (termed ‘TS1’; Figures 7A and 7B and 8A1–A4) was distributed in a similar manner to EGFP–IP5K (Figures 7A–7C and 8G1–G4) which showed strong nuclear localization in 82% of transfected H1299 cells. Replacement of the N-terminally neighbouring basic amino acids R28A, R33A and K36A led to a mutant (termed ‘TS2’) showing exclusively cytosolic localization in approx. 46% of transfected cells (Figures 7A–7C and Figures 8B1–B4).
Figure 7. Mutagenesis and fluorescence imaging of IP5K revealing nuclear localization and nuclear export domains.
(A) Graphical scheme of IP5K mutants. The putative canonical NLS is marked in grey; the putative zinc cluster is highlighted by a black box. Basic amino acids are shown in bigger-font bold letters, putative zinc-binding amino acids are in grey and mutated amino acids are shown as white letters on a black background. Deletion is marked by a black line. Mutants are TS1 (R41A/K42A/K43A); TS2 (R28A/R33A/K36A/R41A/K42A/K43A); TS3 (H152S/H156S/C159S/C162S); XZ1 (K155A/K157A); XZ2 K173A/K175A/K179A); XZ3 (R28A/R33A/K36A/R41A/K42A/K43A/K155A/K157A/K173A/K175A/K179A); Del1 (deletion of amino acids 202–297. (B) H1299 cells were transfected with the IP5K mutants. At 24 h after transfection 160–200 cells (from three to four independent experiments) were photographed and the intracellular distribution of IP5K was defined as being exclusively cytosolic (black bars), nuclear plus cytosolic (grey bars) or exclusively nuclear (white bar). (C) Mutants TS2 (grey and white squares) and XZ2 (grey triangles) as well as the wild-type IP5K (black) were bacterially expressed and their catalytic activity was tested in vitro using approx. 0.1 μg of enzyme.
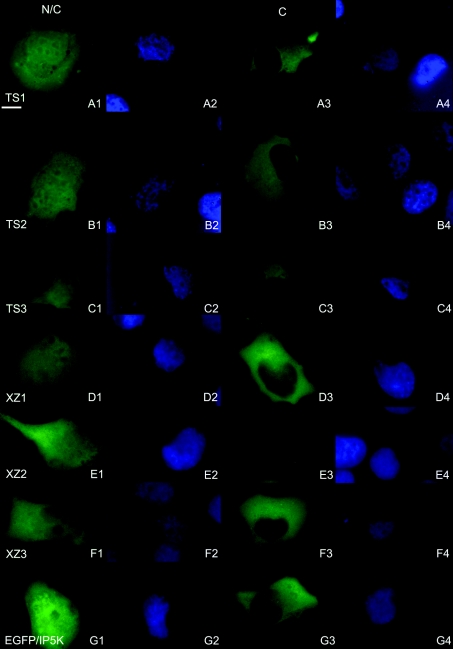
Figure 8. Intracellular localization of IP5K-NLS-mutants.
Mutants TS1–3 and XZ1–3 were overexpressed in H1299 cells for 24 h. After fixation, nuclei were stained with DAPI (A2–G2 and A4–G4). Representative cells for each mutant with nuclear as well as cytosolic localization (N/C) and with exclusively cytosolic localization (C) are shown: (A1–4) TS1; (B1–4) TS2; (C1–4) TS3; (D1–4) XZ1; (E1–4) XZ2; (F1–4) XZ3; (G1–4) IP5K–EGFP. In epi-fluorescence images, scale bar represents 10 μm.
We noticed a predicted H2C2H2 zinc-cluster motif within the IP5K sequence. We investigated the functional significance of this motif by mutating four putative zinc-binding amino acids (His152, His156, Cys159 and Cys162) to serine residues. This mutant, designated ‘TS3’, showed exclusively cytosolic localization in 47% of cells (Figures 7A, 7B and 8C1–8C4). Mutant TS3 therefore exhibited weakened nuclear uptake to the same extent as mutant TS2. An even higher effect was observed with mutants in which basic amino acids adjacent to the H2C2H2 peptide segment were changed to an alanine residue. Mutation to an alanine residue of either Lys155 plus Lys157 (mutant ‘XZ1’; Figures 7A, 7B and 8D1–D4) or Lys173 plus Lys175 plus Lys179 (mutant ‘XZ2’; Figures 7A–7C and 8E1–E4) led to an exclusively cytosolic fusion protein in 80% of transfected cells. A combination of all mutations (mutant ‘XZ3’, R28A/R33A/K36A/R41A/K42A/K43A/K153A/K55A/K173A/K175A/K179A; Figures 7A, 7B and 8F1–F4) did not further increase the percentage of cells showing exclusive cytosolic localization of the fusion protein. Our conclusion is that the investigated amino acids in the segment 150–180 (black box in Figure 7A) are involved in nuclear uptake of IP5K, possibly by actively mediating nuclear import or by binding to a nuclear protein that sequesters IP5K in the nucleus. Furthermore, the same region of IP5K seems to be important for nucleolar uptake, since mutants TS3 and XZ1–3 did not enter nucleoli. In contrast, TS1 and TS2 only showed a slight reduction in nucleolar uptake.
To investigate whether catalytic activity is necessary for its nuclear uptake, wild-type IP5K as well as the mutants TS2 (R28A/R33A/K36A/R41A/K42A/K43A) and XZ2 (K173A/K175A/K179A) were recombinantly expressed in bacteria and conversion of Ins(1,3,4,5,6)P5 into InsP6 was measured. Although mutant XZ2 retained 20% of the wild-type activity, mutant TS2 was completely inactive (Figure 7C). Since mutant TS2 is still imported into the nucleus in 54% of H1299 cells this experiment shows that catalytic activity is not necessary for nuclear uptake of IP5K.
Many proteins exhibiting nuclear accumulation are not only actively imported into nuclei, but in addition are actively exported. This shuttling of proteins across the nuclear membrane allows the cell to regulate the cytosolic and nuclear levels of the protein. We therefore next looked for evidence that IP5K might have a domain regulating its nuclear export.
An alignment of ten vertebrate IP5K and 18 fungal Ipk1p sequences revealed two large peptide inserts (amino acids 202–297 and amino acids 335–389) that are specific to the vertebrate isoforms. Deletion of the first insert led to a mutant ‘Del1’ that was almost exclusively nuclear in approx. 95% of transfected cells (Figures 7A and 7B). Amino acids 202–297 thus appear to comprise a domain necessary for nuclear export of human IP5K. A representative H1299 cell overexpressing mutant ‘Del1’ of the IP5K–EGFP fusion protein is shown in Figures 9(A)–9(C).
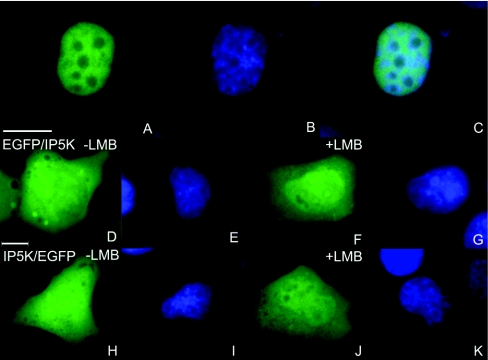
Figure 9. Intracellular localization of the IP5K mutant Del1 and nuclear export inhibition of IP5K by LMB treatment.
Intracellular localization of mutant Del1 (deletion of amino acids 202–297) in H1299 cells, 24 h after transfection, is shown in (A). Nuclei were stained with DAPI (B). Either EGFP–IP5K (D–G) or IP5K–EGFP (H–K) was overexpressed in H1299 cells for 24 h. Then either 20 nM LMB (F, G, J and K) or vehicle (D, E, H and I) were added for an additional 5 h. Nuclei were stained with DAPI (E, G, I and K). In epi-fluorescence images, scale bars represent 10 μm.
To investigate whether nuclear export of IP5K is mediated by an exportin-1-dependent export mechanism we incubated H1299 cells, in which either IP5K–EGFP or EGFP–IP5K were overexpressed, with LMB (leptomycin B), a specific exportin-1 inhibitor [45]. Images of 25 cells each were recorded and the mean fluorescence in nuclei and cytosol was determined (representative cells are shown in Figures 9D–9K). Without LMB the mean ratio of nuclear compared with cytosolic IP5K fluorescence was 1.1 for the N-terminal and 1.0 for the C-terminal fusion protein. After LMB treatment the mean ratio was increased to 1.4 (P=0.0036, n=25) and 1.3 (P=0.0036, n=25) respectively. Since LMB has a smaller effect upon nuclear export than the deletion of the nuclear export domain, it is possible that there are exportin-1-regulated and exportin-1-independent mechanisms involved in the export of IP5K out of the nucleus.
DISCUSSION
In the present study we describe some new aspects of the intracellular localization of IP5K in mammalian cells. We show that IP5K is concentrated in both the nucleolus as well as the euchromatin fraction. The targeting of IP5K to these locations is shown by site-directed mutagenesis to be controlled by amino acid sequences within IP5K; we have identified a nuclear import signal and a peptide sequence which regulates the nuclear export of IP5K. Furthermore, IP5K is also shown to co-localize with mRNA, both within the nucleus and in cytoplasmic SGs. Overall, our results point to IP5K making multifunctional contributions to the overall control of gene expression.
Confocal immunofluorescence analysis of overexpressed as well as endogenous IP5K shows its distribution in the nucleus to be inversely correlated with the intensity of DAPI staining. DAPI is a DNA-binding AT-specific fluorochrome that associates only weakly with euchromatin [37], which represents the fraction of chromosomes that are transcriptionally active. To enable the transcription process, heterochromatin needs to be decondensed by histone deacetylases and other DNA-binding proteins that form chromatin remodelling complexes which, interestingly, are regulated by the IP5K product InsP6 [23–25]. Figure 5 shows that the nuclei of cells treated with RNAi against IP5K exhibit an altered chromatin organization, which provides evidence that a tightly controlled and localized production of InsP6 may be essential for normal nuclear functions.
Since euchromatin is where newly synthesized mRNA arises, it is possible that IP5K might also be involved in the process of mRNA maturation and processing. This idea is further supported by the observation that IP5K co-localizes with mRNA in the nucleus (Figures 3A–3C). Interestingly, the co-localization of IP5K and mRNA is not limited to nuclear mRNA, it is also evident in cytoplasmic SGs, which are sites of mRNA storage under certain stress conditions [35].
In our experiments we monitored SGs using the marker proteins PABP and TIAR. SGs were formed in response to treatment of cells with puromycin (Figures 3M–3R), or by overexpression of IP5K (Figures 3D–3I). The latter phenomenon is apparently not caused by IP5K itself, but is a general response to protein overexpression [27]. We observed that the puromycin-induced SGs were more abundant and smaller in size than SGs observed after vector-based protein overexpression. This phenomenon was previously described by Kedersha and Anderson [35], who showed that drug-induced SGs increase in size and decrease in number when cells recover from chemical stress. Alternately, the difference in number and size between puromycin and solely protein overexpression-induced SGs might be due to different kinetics of recovery from both perturbations, since cells overexpressing IP5K were investigated 24 h after stress induction by transfection, whereas puromycin-treated cells were directly examined after incubation without any recovery time.
The fact that the catalytically inactive mutant C162Y is no longer a component of SGs does not rule out a critical role of InsP6 in SG formation, since endogenous active IP5K is still present. It is possible that the mutant does not associate with SGs because the tertiary structure of the protein is compromised, although the lack of catalytic activity may still be relevant.
We have searched for amino acid sequences within IP5K that might regulate its intracellular localization. We identified a region involved in nuclear uptake of IP5K composed of a putative zinc-finger motif (amino acids 150–180) overlapping several basic residues (Lys155, Lys157, Lys173, Lys175 and Lys179) In addition, multiple sequence alignments revealed a novel domain is present in vertebrate IP5K but not fungal IP5K (Ipk1). Deletion of the N-terminal one of two vertebrate-specific peptide inserts in human IP5K (insert 1, residues 202–297) effectively inhibited nuclear export of the modified protein. Since LMB has a smaller effect upon nuclear export than the deletion of the nuclear export domain, it is possible that there are LMB-sensitive and LMB-insensitive mechanisms to export IP5K out of the nucleus.
It is reasonable to assume that the functional significance of the highly organized intracellular targeting of IP5K is related to the biological consequences of a localized production of InsP6. The latter is a polyanionic molecule probably present as a soluble polymagnesium hydrate complex [46] which can also associate with negatively charged membranes [47] and polynucleotide structures by Mg2+ salt bridging. Restricted diffusion might necessitate a sophisticated system of nuclear and extranuclear targeting sites for IP5K so as to generate InsP6 precisely where it is required.
For example, InsP6 plays a role in non-homologous end-joining through its effects upon Ku mobility [21]. InsP6 is also required for protein maturation. For example, in two families of mRNA- and tRNA-editing adenosine deaminases (ADARs and ADATs) InsP6 was found to act as a ‘prosthetic’ group essential for correct protein folding and for catalytic activity [22]. These precedents raise the possibility of InsP6 having this role in the assembly of higher-order multiprotein complexes, such as RNPs. We also propose that there is functional significance to the localization of IP5K in euchromatin, where RNA is generated. Localized fluctuations in InsP6 levels can also regulate chromatin remodelling [23–25]. Finally, InsP6 regulates casein kinase activity [48]; nucleolin, a particularly abundant protein in the nucleolus, is apparently a casein kinase substrate [49]. The nucleolus has other functions such as splicosome assembly [50] and so we should not exclude other roles for IP5K in this organelle. Overall our results suggest a regulatory context for the localized production of InsP6 from IP5K.
The higher organized intracellular targeting of IP5K that we have observed in mammalian cells has not previously been seen in experiments with yeast [16,26]. Most yeast mRNA is without introns and its nuclear export is regulated by mechanisms differing in many aspects from the export of predominantly spliced mRNA of vertebrates [51,52]. This, and the greater size and complexity of vertebrate cells and nuclei, could necessitate a more sophisticated targeting of the putative regulator InsP6. However, we do not rule out the alternative possibility that IP5K may have other functions in addition to its ability to synthesize InsP6.
It has been noted previously that overall cellular levels of InsP6 are generally unresponsive to extracellular stimuli that can elicit quite substantially changes in the levels of the other inositol phosphates [53]. This has made it difficult to put many of the biological effects of InsP6 into a regulatory context [53]. The present studies raise the strong possibility of there being considerable microheterogeneity in the synthesis of InsP6. Significant localized changes in InsP6 concentration may arise from local fluctuations in the concentration of IP5K, without overall cellular levels of InsP6 being affected. Thus the present study reveals new and important aspects of IP5K function.
Online data
Acknowledgments
We thank Dr Evita Mohr for providing anti-PABP and Jeff M. Reece and Jeff C. Tucker for help with microscopy techniques. This work was supported by the grants from the DFG (Deutsche Forschungsgemeinschaft) given to G. W. M. (MA 989/3-1 and 3-2 and a project within the Graduiertenkolleg 336) and the NIH (National Institutes of Health)/DFG transition award given to M. A. B.
References
- 1.Irvine R. F. Inositide evolution: towards turtle domination? J. Physiol. 2005;566:295–300. doi: 10.1113/jphysiol.2005.087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.York J. D. Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Shears S. B. How versatile are inositol phosphate kinases? Biochem. J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allbritton N. L., Meyer T., Stryer L. Range of messenger action of calcium ion and inositol 1,4,5-trisphosphate. Science. 1992;258:1812–1815. doi: 10.1126/science.1465619. [DOI] [PubMed] [Google Scholar]
- 5.Remus T. P., Zima A. V., Bossuyt J., Bare D. J., Martin J. L., Blatter L. A., Bers D. M., Mignery G. A. Biosensors to measure inositol 1,4,5-trisphosphate concentration in living cells with spatiotemporal resolution. J. Biol. Chem. 2006;281:608–616. doi: 10.1074/jbc.M509645200. [DOI] [PubMed] [Google Scholar]
- 6.Martelli A. M., Fala F., Faenza I., Billi A. M., Cappellini A., Manzoli L., Cocco L. Metabolism and signaling activities of nuclear lipids. Cell. Mol. Life Sci. 2004;61:1143–1156. doi: 10.1007/s00018-004-3414-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehm M. A., Schreiber I., Bertsch U., Wegner A., Mayr G. W. Identification of the actin-binding domain of Ins(1,4,5)P3 3-kinase isoform B (IP3K-B) Biochem. J. 2004;382:353–362. doi: 10.1042/BJ20031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nalaskowski M. M., Bertsch U., Fanick W., Stockebrand M. C., Schmale H., Mayr G. W. Identification of the actin-binding domain of Ins(1,4,5)P3 3-kinase isoform B (IP3K-B) J. Biol. Chem. 2003;278:19765–19776. doi: 10.1074/jbc.M211059200. [DOI] [PubMed] [Google Scholar]
- 9.Nalaskowski M. M., Windhorst S., Stockebrand M. C., Mayr G. W. Subcellular localisation of human inositol 1,4,5-trisphosphate 3-kinase C: species-specific use of alternative export sites for nucleo-cytoplasmic shuttling indicates divergent roles of the catalytic and N-terminal domains. Biol. Chem. 2006;387:583–593. doi: 10.1515/BC.2006.075. [DOI] [PubMed] [Google Scholar]
- 10.Nalaskowski M. M., Deschermeier C., Fanick W., Mayr G. W. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem. J. 2002;366:549–556. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeds A. M., Sandquist J. C., Spana E. P., York J. D. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J. Biol. Chem. 2004;279:47222–47232. doi: 10.1074/jbc.M408295200. [DOI] [PubMed] [Google Scholar]
- 12.Saiardi A., Nagata E., Luo H. R., Sawa A., Luo X., Snowman A. M., Snyder S. H. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nalaskowski M. M., Mayr G. W. The families of kinases removing the Ca2+ releasing second messenger Ins(1,4,5)P3. Curr. Mol. Med. 2004;4:277–290. doi: 10.2174/1566524043360726. [DOI] [PubMed] [Google Scholar]
- 14.Saiardi A., Caffrey J. J., Snyder S. H., Shears S. B. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- 15.Bennett M., Onnebo S. M., Azevedo C., Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cell. Mol. Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Stephens L. R., Hawkins P. T., Stanley A. F., Moore T., Poyner D. R., Morris P. J., Hanley M. R., Kay R. R., Irvine R. F. Myo-inositol pentakisphosphatases. Structure, biological occurrence and phosphorylation to myo-inositol hexakisphosphate. Biochem. J. 1991;275:485–499. doi: 10.1042/bj2750485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15b.Phillippy B. Q., Ullah A. H., Ehrlich K. C. Purification and some properties of inositol 1,3,4,5,6-pentakisphosphate 2-kinase from immature soybean seeds. J. Biol. Chem. 1994;269:28393–28399. [PubMed] [Google Scholar]
- 16.York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 17.Verbsky J. W., Wilson M. P., Kisseleva M. V., Majerus P. W., Wente S. R. The synthesis of inositol hexakisphosphate. Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J. Biol. Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- 18.Fujii M., York J. D. A role for rat inositol polyphosphate kinases rIPK2 and rIPK1 in inositol pentakisphosphate and inositol hexakisphosphate production in rat-1 cells. J. Biol. Chem. 2005;280:1156–1164. doi: 10.1074/jbc.M412006200. [DOI] [PubMed] [Google Scholar]
- 19.Verbsky J. W., Chang S.-C., Wilson M. P., Mochizuki Y., Majerus P. W. The pathway for the production of inositol hexakisphosphate in human cells. J. Biol. Chem. 2005;280:1911–1920. doi: 10.1074/jbc.M411528200. [DOI] [PubMed] [Google Scholar]
- 20.Verbsky J., Majerus P. W. Increased levels of inositol hexakisphosphate (InsP6) protect HEK293 cells from tumor necrosis factor α- and Fas-induced apoptosis. J. Biol. Chem. 2005;280:29263–29268. doi: 10.1074/jbc.M503366200. [DOI] [PubMed] [Google Scholar]
- 21.Byrum J., Jordan S., Safrany S. T., Rodgers W. Visualization of inositol phosphate-dependent mobility of Ku: depletion of the DNA-PK cofactor InsP6 inhibits Ku mobility. Nucleic Acids Res. 2004;32:2776–2784. doi: 10.1093/nar/gkh592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macbeth M. R., Schubert H. L., Vandemark A. P., Lingam A. T., Hill C. P., Bass B. L. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen X., Xiao H., Ranallo R., Wu W. H., Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 24.Steger D. J., Haswell E. S., Miller A. L., Wente S. R., O'Shea E. K. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rando O. J., Chi T. H., Crabtree G. R. Second messenger control of chromatin remodeling. Nat. Struct. Biol. 2003;10:81–83. doi: 10.1038/nsb0203-81. [DOI] [PubMed] [Google Scholar]
- 26.Odom A. R., Stahlberg A., Wente S. R., York J. D. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 27.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W., Malcolm B. A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 29.Genove G., Glick B. S., Barth A. L. Brighter reporter genes from multimerized fluorescent proteins. BioTechniques. 2005;39 doi: 10.2144/000112056. 814, 816, 818 passim. [DOI] [PubMed] [Google Scholar]
- 30.Horn S., Endl E., Fehse B., Weck M. M., Mayr G. W., Jucker M. Restoration of SHIP activity in a human leukemia cell line downregulates constitutively activated phosphatidylinositol 3-kinase/Akt/GSK-3β signaling and leads to an increased transit time through the G1 phase of the cell cycle. Leukemia. 2004;18:1839–1849. doi: 10.1038/sj.leu.2403529. [DOI] [PubMed] [Google Scholar]
- 31.Segel I. H. Enzyme Kinetics. New York: Wiley; 1975. Enzyme kinetics, behavior and analysis of rapid equilibrium and steady-state enzyme systems; pp. 18–99. [Google Scholar]
- 32.Mayr G. W. A novel metal-dye detection system permits picomolar-range h.p.l.c. analysis of inositol polyphosphates from non-radioactively labelled cell or tissue specimens. Biochem. J. 1988;254:585–591. doi: 10.1042/bj2540585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayr G. W. Mass determination of inositol phosphates by high-performance-liquid-chromatography with postcolumn complexometry (metal-dye-detection) In: Irvine R. F., editor. Methods in Inositide Research. New York: Raven Press; 1990. pp. 83–108. [Google Scholar]
- 34.Guse A. H., Goldwich A., Weber K., Mayr G. W. Non-radioactive, isomer-specific inositol phosphate mass determinations: high-performance liquid chromatography-micro-metal-dye detection strongly improves speed and sensitivity of analyses from cells and micro-enzyme assays. J. Chromatogr. B Biomed. Appl. 1995;672:189–198. doi: 10.1016/0378-4347(95)00219-9. [DOI] [PubMed] [Google Scholar]
- 35.Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 36.Herold A., Klymenko T., Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 37.Schweizer D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 1976;58:307–324. doi: 10.1007/BF00292840. [DOI] [PubMed] [Google Scholar]
- 38.Miller A. L., Suntharalingam M., Johnson S. L., Audhya A., Emr S. D., Wente S. R. Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J. Biol. Chem. 2004;279:51022–51032. doi: 10.1074/jbc.M409394200. [DOI] [PubMed] [Google Scholar]
- 39.Thomas M. G., Martinez Tosar L. J., Loschi M., Pasquini J. M., Correale J., Kindler S., Boccaccio G. L. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell. 2005;16:405–420. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ives E. B., Nichols J., Wente S. R., York J. D. Biochemical and functional characterization of inositol 1,3,4,5,6-pentakisphosphate 2-kinases. J. Biol. Chem. 2000;275:36575–36583. doi: 10.1074/jbc.M007586200. [DOI] [PubMed] [Google Scholar]
- 41.Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng C., Brownlie R., Babiuk L. A., van Drunen Littel-van den Hurk S. Characterization of the nuclear localization and nuclear export signals of bovine herpesvirus 1 VP22. J. Virol. 2005;79:11864–11872. doi: 10.1128/JVI.79.18.11864-11872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee S., Javier M., Stochaj U. In vivo analysis of nuclear protein traffic in mammalian cells. Exp. Cell Res. 1997;236:346–350. doi: 10.1006/excr.1997.3746. [DOI] [PubMed] [Google Scholar]
- 44.Beetz C., Brodhun M., Moutzouris K., Kiehntopf M., Berndt A., Lehnert D., Deufel T., Bastmeyer M., Schickel J. Identification of nuclear localisation sequences in spastin (SPG4) using a novel tetra-GFP reporter system. Biochem. Biophys. Res. Commun. 2004;318:1079–1084. doi: 10.1016/j.bbrc.2004.03.195. [DOI] [PubMed] [Google Scholar]
- 45.Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 46.Torres J., Dominguez S., Cerda M. F., Obal G., Mederos A., Irvine R. F., Diaz A., Kremer C. Solution behaviour of myo-inositol hexakisphosphate in the presence of multivalent cations. Prediction of a neutral pentamagnesium species under cytosolic/nuclear conditions. J. Inorg. Biochem. 2005;99:828–840. doi: 10.1016/j.jinorgbio.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Poyner D. R., Cooke F., Hanley M. R., Reynolds D. J., Hawkins P. T. Characterization of metal ion-induced [3H]inositol hexakisphosphate binding to rat cerebellar membranes. J. Biol. Chem. 1993;268:1032–1038. [PubMed] [Google Scholar]
- 48.Solyakov L., Cain K., Tracey B. M., Jukes R., Riley A. M., Potter B. V., Tobin A. B. Regulation of casein kinase-2 (CK2) activity by inositol phosphates. J. Biol. Chem. 2004;279:43403–43410. doi: 10.1074/jbc.M403239200. [DOI] [PubMed] [Google Scholar]
- 49.Kim K., Dimitrova D. D., Carta K. M., Saxena A., Daras M., Borowiec J. A. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein a complex formation. Mol. Cell Biol. 2005;25:2463–2474. doi: 10.1128/MCB.25.6.2463-2474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox A. H., Lam Y. W., Leung A. K., Lyon C. E., Andersen J., Mann M., Lamond A. I. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 51.Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546, 563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 52.Stutz F., Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 53.Shears S. B. Assessing the omnipotence of inositol hexakisphosphate. Cell. Signaling. 2001;13:151–158. doi: 10.1016/s0898-6568(01)00129-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.