Abstract
Small noncoding RNAs (sRNA) can function as posttranscriptional activators of gene expression to regulate stress responses and metabolism. We here describe the mechanisms by which two sRNAs, GlmY and GlmZ, activate the Escherichia coli glmS mRNA, coding for an essential enzyme in amino-sugar metabolism. The two sRNAs, although being highly similar in sequence and structure, act in a hierarchical manner. GlmZ, together with the RNA chaperone, Hfq, directly activates glmS mRNA translation by an anti-antisense mechanism. In contrast, GlmY acts upstream of GlmZ and positively regulates glmS by antagonizing GlmZ RNA inactivation. We also report the first example, to our knowledge, of mRNA expression being controlled by the poly(A) status of a chromosomally encoded sRNA. We show that in wild-type cells, GlmY RNA is unstable due to 3′ end polyadenylation; whereas in an E. coli pcnB mutant defective in RNA polyadenylation, GlmY is stabilized and accumulates, which in turn stabilizes GlmZ and causes GlmS overproduction. Our study reveals hierarchical action of two well-conserved sRNAs in a complex regulatory cascade that controls the glmS mRNA. Similar cascades of noncoding RNA regulators may operate in other organisms.
Author Summary
Hierarchical action of regulators is a fundamental principle in gene expression control, and is well understood in protein-based signaling pathways. We have discovered that small noncoding RNAs (sRNAs), a new class of gene expression regulators, can also act hierarchically and form a regulatory cascade. Two highly similar sRNAs function after transcription to activate the Escherichia coli glmS mRNA, which codes for an essential function in amino-sugar metabolism. It is somewhat unusual for two sRNAs to act upon the same target mRNA, and despite their seeming homology, these two sRNAs (GlmY and GlmZ) employ different molecular mechanisms and function hierarchically to activate glmS expression: GlmZ directly activates glmS translation via disruption of an mRNA structure that inhibits translation, whereas GlmY controls the processing of GlmZ to prevent the inactivation of this direct activator. We also found that GlmY is itself controlled by an RNA processing event (3′ end polyadenylation), which typically destabilizes bacterial RNA. Our data unequivocally demonstrate that E. coli glmS is exceptionally dependent on RNA-based mechanisms for its genetic control. Given the large number of noncoding RNAs of unknown function, we believe that similar regulatory RNA cascades may operate in other organisms.
A regulatory RNA cascade that posttranscriptionally activates the glmS mRNA is identified, with two highly similar small noncoding RNAs acting hierarchically in a manner thus far known only in protein-based regulatory circuits.
Introduction
The posttranscriptional regulation of glmS expression, a gene encoding an essential enzyme (glucosamine-6-phosphate [GlcN-6-P] synthase) in amino-sugar metabolism, has recently attracted much attention. In Bacillus subtilis, the 5′ untranslated region (5′ UTR) of glmS mRNA contains a ribozyme that undergoes self-cleavage in the presence of GlcN-6-P, thereby destabilizing the glmS mRNA [1]. This cis-encoded mechanism of glmS mRNA control and metabolite sensing seems to be highly conserved among Gram-positive species [2]. Whereas Gram-negative bacteria such as E. coli lack the glmS ribozyme [2], recent studies have shown that E. coli glmS is posttranscriptionally controlled by multiple trans-encoded factors, including small noncoding RNAs (sRNAs) and several proteins [3–5].
Functional characterization of two E. coli sRNAs, GlmY and GlmZ (a.k.a. SroF/tke1 [6,7] and RyiA/SraJ [8,9], respectively; Figure 1A) identified these as positive regulators of glmS mRNA; constitutive expression of either sRNA causes overproduction of GlmS protein [3,4]. In addition, the bacterial Sm-like protein, Hfq, a facilitator of sRNA–mRNA interactions [10,11], has been implicated in GlmZ RNA function. Hfq associates with GlmZ RNA in vivo [8], and is required for the GlmZ-mediated up-regulation of GlmS protein synthesis [4].
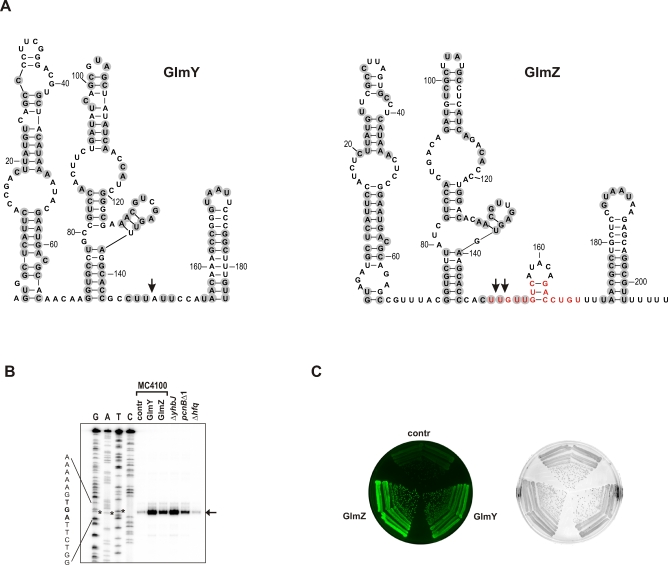
Figure 1. GlmY and GlmZ Are Related sRNAs and Promote GlmS Synthesis.
(A) Consensus structures of the E. coli GlmY (184 nt) and GlmZ (207 nt) RNAs (see [3] and Figure S1, respectively, for sequence alignments). Vertical arrows indicate previously mapped 3′ processing sites [6,9]. Grey circles denote nucleotides conserved between the E. coli GlmY and GlmZ RNAs. The GlmZ residues involved in glmS mRNA binding (see Figure 3A below) are shown in red.
(B) Primer extension analysis detects the 5′ end of monocistronic glmS mRNA (vertical arrow) that results from processing of dicistronic glmUS mRNA at the glmU stop codon (UGA; denoted by asterisks). Total RNA was prepared from early stationary phase cultures of E. coli strain MC4100 carrying control vector pJV300 (lane “contr”), or the sRNA expression plasmids, pPL-GlmY (lane “GlmY”) or pPL-GlmZ (lane “GlmZ”), and from strains JVS-8018 (ΔyhbJ), JVS-2058 (pcnBΔ1), or Δhfq::cat, as indicated. A 5′ end-labeled primer complementary to the glmS coding region was annealed to RNA, followed by reverse transcription and separation of the obtained cDNAs on a sequencing gel along with a corresponding sequencing ladder (lanes G, A, T, and C). Shown is an autoradiogram of the relevant section of the gel.
(C) Both GlmY and GlmZ activate a translational glmS::gfp fusion. Colony fluorescence of E. coli carrying a glmS::gfp fusion plasmid (pJU-172) in combination with plasmid pJV300, pPL-GlmY, or pPL-GlmZ. Labeling as in (B). Shown are images of a LB agar plate obtained in the fluorescence mode (left) or the visible light mode (right).
The PAP I and YhbJ proteins negatively regulate glmS mRNA expression; E. coli strains carrying functional disruptions of the respective genes overproduce glmS mRNA and GlmS protein [4,5]. PAP I (poly(A) polymerase I encoded by pcnB) is the main enzyme responsible for 3′ end RNA polyadenylation [12], which typically destabilizes bacterial transcripts by facilitating the 3′ → 5′ exonucleolytic degradation of RNA with a structured 3′ end [13,14]. Interestingly, glmS mRNA is markedly stabilized in a PAP I–deficient strain, i.e., E. coli pcnBΔ1 [5]. However, it has been unclear whether glmS mRNA is a substrate of PAP I, and whether polyadenylation directly destabilizes this mRNA in wild-type cells. YhbJ is a protein of unknown function, and appears to act upstream of GlmZ to negatively regulate glmS mRNA [4]. That is, the GlmS overproduction observed in an E. coli ΔyhbJ mutant strain is lost upon glmZ deletion [4].
Collectively, these observations suggested that multiple factors encoded by unlinked loci of the E. coli chromosome control glmS epistasis at the posttranscriptional level. However, whether these factors acted in concert or independently, and how GlmS protein synthesis was ultimately activated, remained to be determined. Here, we show that the regulatory GlmY and GlmZ sRNAs display significant homology of sequence and structure. Despite this apparent homology, we have found that the two RNAs act hierarchically such that GlmY acts upstream of GlmZ, and only GlmZ directly activates glmS mRNA translation. We propose that the main role of GlmY in this novel regulatory RNA cascade is to antagonize YhbJ-dependent inactivation of GlmZ RNA, i.e., the bona fide glmS activator. We also show that PAP I controls the levels of GlmY, and that loss of GlmY RNA polyadenylation causes the previously observed GlmS overproduction in PAP I–deficient E. coli. The hierarchical action of noncoding RNAs described here reveals a novel principle in RNA-based regulatory circuits previously only known in protein-based systems.
Results
Similarities of GlmY and GlmZ RNAs
E. coli glmS is transcribed as part of a dicistronic glmUS operon mRNA [15]. Ribonuclease E cleaves the glmUS transcript within the glmU stop codon; this event generates a monocistronic glmS mRNA which strongly accumulates in E. coli pcnBΔ1 or ΔyhbJ, and causes GlmS overproduction [4,5]. We used primer extension assays to test whether constitutive GlmY or GlmZ expression also induces the accumulation of monocistronic glmS mRNA. To this end, E. coli strain MC4100 (wild type) was transformed with plasmids pPL-GlmY or pPL-GlmZ carrying the glmY or glmZ genes, respectively, under control of a constitutive PLlacO1 promoter. Figure 1B shows that the expression of either sRNA promotes the accumulation of a monocistronic glmS mRNA species identical to that observed in ΔyhbJ or pcnBΔ1 mutant strains.
We previously established several glmS::gfp fusion plasmids as reporters of GlmY activity [3]. Of these, plasmid pJU-172 constitutively expresses a glmS::gfp fusion mRNA spanning the C-terminal glmU sequence (last 17 codons), the glmS 5′ UTR (161 nucleotides [nt]), and the first seven glmS codons translationally fused to gfp. This fusion is highly activated in the presence of the glmY expression plasmid, pPL-GlmY, suggesting that GlmY activates the glmS gene at the posttranscriptional level in a process that involves the glmS 5′ UTR. To test whether this fusion was also activated by GlmZ, the colony fluorescence of E. coli carrying pJU-172 in combination with a control plasmid (pJV300), or pPL-GlmY or pPL-GlmZ, was compared (Figure 1C). GlmZ activated the glmS::gfp fusion at least as strongly as GlmY, indicating that the cloned fragment of glmS mRNA contained the determinants for regulation by either sRNA.
The primary GlmY and GlmZ RNAs are of similar length (184 and 207 nt, respectively), and both sRNAs are subject to 3′ cleavage which yields an approximately 150-nt species [6,9]. Alignment of γ-proteobacterial glmZ sequences combined with RNA structure prediction (Figure S1) suggests that GlmZ adopts a secondary structure very similar to that of GlmY RNA, and identified a high degree of sequence identity between the E. coli GlmY and GlmZ RNAs (Figure 1A). Therefore, the two sRNAs seem homologous in sequence and structure, and both promote the accumulation of monocistronic glmS mRNA that underlies GlmS overproduction.
Hierarchy, and Not Redundancy, in the Action of GlmY and GlmZ sRNAs
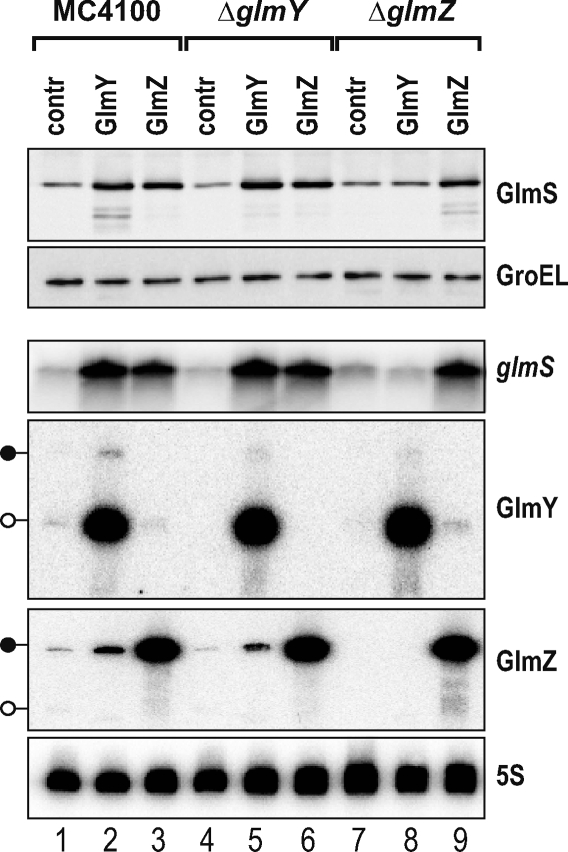
Given their apparent homology, we tested whether the two sRNAs could complement each other's function. Wild-type E. coli and isogenic ΔglmY or ΔglmZ mutant strains were transformed with pJV300, pPL-GlmY, or pPL-GlmZ, and cellular GlmS protein and glmS mRNA levels were determined upon growth to early stationary phase (Figure 2). GlmZ, which accumulated as the primary 207-nt RNA, increased glmS mRNA and GlmS protein levels in all three genetic backgrounds (lanes 3, 6, and 9). In contrast, GlmY, which accumulated as the processed 148-nt RNA, activated glmS only in the wild-type (lane 2) and ΔglmY (lane 5) backgrounds, and totally failed to up-regulate glmS in the ΔglmZ strain (lane 8). This specific failure was also observed when the glmS::gfp fusion was used as reporter of glmS activation in the tested backgrounds (unpublished data). Plasmid pPL-GlmY conferred the same GlmY RNA levels to the ΔglmZ as to the other two tested strains (Figure 2), which ruled out the possibility that absence of glmZ prevented GlmY accumulation and, consequently, glmS activation. Therefore, we concluded that despite their apparent homology, the two sRNAs are not functionally redundant; instead, they act in a hierarchical manner such that GlmY requires GlmZ to bring about glmS activation.
Figure 2. GlmY-Mediated glmS Activation Strictly Requires glmZ .
Western and northern blot analysis of strains MC4100, JVS-8030 (ΔglmY), or JVS-8024 (ΔglmZ), each of these strains carrying plasmids pJV300 (“contr” lanes), pPL-GlmY (“GlmY” lanes), or pPL-GlmZ (“GlmZ” lanes). Whole-cell protein and total RNA samples were prepared from early stationary phase cultures. Western blots were probed with sera specific for GlmS (top panel) or GroEL (loading control; second panel from top) proteins. For northern blot detection of glmS mRNA (third panel from top), 12 μg of total RNA per lane was separated on a 1.5% agarose-formaldehyde gel. To detect GlmY and GlmZ, 5 μg of total RNA was separated on a denaturing 5% PAA gel (fourth and fifth panels from top) as shown. The 5S rRNA (bottom panel) probing confirmed equal amounts of RNA in each sample. Black lollipops indicate the primary GlmY or GlmZ transcripts, whereas open lollipops indicate the processed, approximately 150-nt species of these sRNAs.
Previous work had shown that the proteins Hfq and YhbJ are involved in glmS mRNA regulation [4]. We thus expressed GlmY and GlmZ in a larger array of E. coli mutants, including single or combined deletions of glmY, glmZ, hfq, and yhbJ (Figure S2). The results, in brief, showed that hfq is absolutely required for glmS activation by either sRNA, even in the absence of the negatively acting yhbJ. Expression of the two sRNAs in ΔyhbJ did not further elevate the already high glmS mRNA in this background. A ΔyhbJ ΔglmY ΔglmZ triple mutant displayed wild-type glmS mRNA levels; in this YhbJ-deficient mutant, GlmZ activated the glmS mRNA, whereas GlmY did not. Taken together, these experiments argued against the existence of a glmZ-independent pathway of GlmY action on glmS. Moreover, they argued that GlmZ acted independently of yhbJ, and likely directly upon glmS mRNA.
Mechanism of glmS Activation by GlmZ RNA
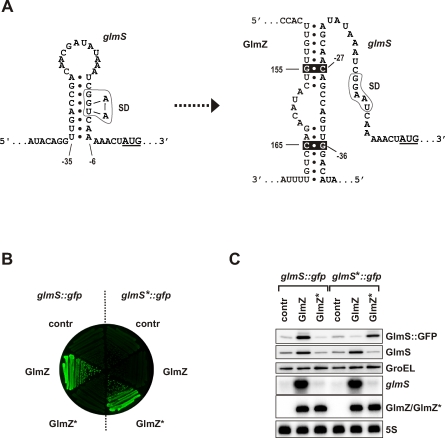
To understand the obvious hierarchy of the two sRNAs, we next addressed the molecular mechanism by which GlmZ activates the glmS mRNA. Work on rpoS mRNA activation by DsrA sRNA [16,17] had established an “anti-antisense” mechanism by which an sRNA competes with the formation of a stem-loop structure that normally sequesters the Shine-Dalgarno (SD) sequence of the target mRNA and inhibits its translation. Preliminary RNA structure probing experiments (unpublished data) and in silico prediction by others [4] prompted us to consider an anti-antisense mechanism of glmS mRNA activation. That is, the glmS SD is predicted to be occluded in a hairpin formed by the 5′ UTR of glmS mRNA (Figure 3A, left). We predicted residues 150–157 and 163–169 of GlmZ RNA, which are located in an accessible, single-stranded region (Figure 1A), to base-pair with the 5′ UTR of glmS mRNA (Figure 3A, right). This alternative pairing would disrupt the glmS hairpin, free the SD, allow ribosome access, and consequently promote glmS mRNA translation. Notably, this mechanism would be specific for GlmZ since the residues to interact with glmS are not conserved in GlmY species (Figure 1A and [3]).
Figure 3. GlmZ Directly Acts on the glmS mRNA to Enhance GlmS Synthesis.
(A) Predicted anti-antisense mechanism by which GlmZ activates the E. coli glmS mRNA. The left panel shows the 5′ UTR of the glmS mRNA in an unbound, “inactive” state in which an intrinsic hairpin sequesters the glmS SD (boxed) and thus inhibits translation. Residues −42 to +3 relative to the AUG start codon, which is underlined, are shown. The right panel shows glmS mRNA in its “activated” form, i.e., upon base-pairing of residues 150–157 and 163–169 of GlmZ RNA with the glmS 5′ UTR, which disrupts the inhibitory hairpin and liberates the glmS SD. The positions of point mutations introduced in glmS (C−27 → G, G−36 → C; glmS* allele) and GlmZ (G+155 → C, C+165 → G; GlmZ* allele), which are expected to maintain base pairing in the glmS*/GlmZ* duplex, are indicated by inverse (white on black) print.
(B) Colony fluorescence of strain JVS-8113 (ΔglmY ΔglmZ) carrying either the glmS::gfp or glmS*::gfp fusion plasmids, each in combination with plasmid pJV300 (“contr”), pPL-GlmZ (“GlmZ”), or pPL-GlmZ* (“GlmZ*”). Shown is an image of a LB agar plate obtained in the fluorescence mode.
(C) The strains shown in (B) were grown to early stationary phase and subjected to western blot (top three panels) and northern blot (bottom three panels) analysis. The plasmid-encoded GlmS::GFP fusion protein was detected using α-GFP antibodies (top panel); the chromosomally encoded glmS mRNA and GlmS protein, and the plasmid-encoded GlmZ RNA (as indicated) were detected as in Figure 2.
To test the validity of the above predictions, we mutated the 5' UTR of a glmS::gfp fusion. In the resulting mutant, glmS*::gfp, two point mutations (C−27 → G and G−36 → C; Figure 3A) disrupt the putative GlmZ binding site in the glmS 5' UTR while leaving the nucleotides required for the formation of the putative inhibitory hairpin intact. We subsequently measured the effect of these mutations on fusion activation by pPL-GlmZ, and observed that the glmS*::gfp fusion was no longer activated (Figure 3B and 3C), suggesting that GlmZ no longer binds to glmS mRNA and promotes its translation. In the reciprocal experiment, we constructed mutant plasmid pPL-GlmZ* by altering sRNA residues G+155 → C and C+165 → G (Figure 3A). While pPL-GlmZ* no longer up-regulated the parental glmS::gfp fusion or the chromosomally encoded glmS mRNA, the compensatory mutations facilitated activation of the glmS*::gfp fusion (Figure 3B and 3C). These results demonstrate that an anti-antisense mechanism underlies glmS mRNA activation by GlmZ sRNA.
Full-Length GlmZ Is the Active RNA Species and Enhances glmS Translation
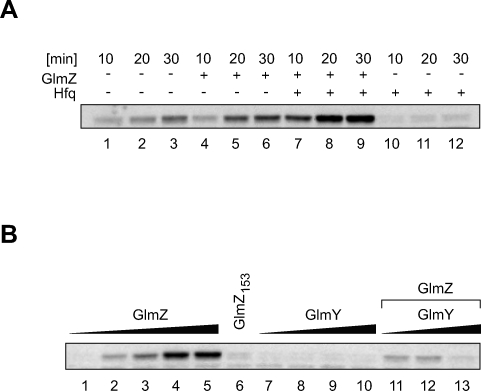
To obtain direct evidence for translational glmS activation, in vitro synthesized glmS::gfp fusion mRNA was incubated with reconstituted 70S ribosomes in the presence or absence of GlmZ RNA, and GlmS::GFP fusion protein synthesis was monitored over the course of 30 min (Figure 4A). Since glmS activation in vivo strictly requires Hfq (Figure S2 and [4]), the in vitro reactions were also performed in the presence of purified Hfq protein. We found that in the presence of 10-fold excess of GlmZ RNA over target mRNA, GlmS::GFP protein synthesis was activated 1.8-fold (comparing 20-min time points; lane 2 versus 5). However, when Hfq was added to the reaction (lanes 7–9), GlmZ promoted 14.8-fold activation of glmS::gfp translation (compare lanes 8 and 11; note that Hfq by itself rather repressed mRNA translation). To the best of our knowledge, this is the first time that translational mRNA activation by a regulatory RNA has been demonstrated in vitro.
Figure 4. GlmZ, Not GlmY, Enhances glmS Translation In Vitro.
(A) In vitro synthesized glmS::gfp fusion mRNA (0.1 μM) was translated with reconstituted 70S ribosomes in the absence (lanes 1–6) or presence (lanes 7–12) of Hfq (0.5 μM) and absence (lanes 1–3 and 10–12) or presence (lanes 4–9) of GlmZ RNA (1 μM). Protein samples were prepared upon incubation for 10, 20, or 30 min, and subjected to western blot analysis using monoclonal α-GFP antibodies to detect the synthesized GlmS::GFP fusion protein.
(B) In vitro translation of glmS::gfp mRNA (0.1 μM) in the presence of Hfq (0.5 μM) for 15 min. Reactions contained increasing amounts of GlmZ RNA (0, 0.1, 0.2, 0.5, and 1 μM; lanes 1–5), GlmY RNA (0.1, 0.2, 0.5, and 1 μM; lanes 7–10), the 3′ truncated GlmZ153 RNA (1 μM; lane 6), or 0.1 μM GlmZ RNA with increasing amouts of GlmY RNA (0.1, 0.2, and 0.5 μM; lanes 11–13). Note that with fixed GlmZ and increasing GlmY concentrations, GlmS synthesis actually decreases (lanes 11–13). This may be explained by competition of GlmY with GlmZ for Hfq-binding, which would inhibit GlmZ-glmS mRNA pairing and, consequently, GlmS synthesis.
The GlmY and Processed GlmZ RNAs Are Not Activators In Vitro
We had previously established that in vivo, the primary GlmZ transcript is subject to 3′ end RNA processing, which generates a 153-nt species, here named GlmZ153 [9]. Kalamorz et al. [4] observed that GlmZ RNA processing is abrogated in a ΔyhbJ strain, and concomitantly, full-length GlmZ and glmS mRNA accumulate; this led the authors to hypothesize that only full-length GlmZ, and not GlmZ153, regulates glmS mRNA. Indeed, sequence inspection shows that GlmZ153 lacks the above proposed glmS target binding site (Figures 1A and 3A). Similarly, GlmY does not possess the glmS target site (Figure 1A). Consequently, both GlmZ153 and GlmY should be silent in our in vitro system of glmS activation. To validate this prediction, glmS::gfp mRNA was translated for 15 min along with the full-length GlmZ, GlmZ153, or GlmY RNAs (Figure 4B). Quantification of GlmS::GFP confirmed that glmS::gfp mRNA was activated by GlmZ in a dose-dependent manner (lanes 1 to 5), and that 10-fold excess of GlmZ over mRNA strongly promoted glmS::gfp translation (lane 5). In contrast, 10-fold excess of the GlmZ153 (lane 6) or GlmY (lane 10) RNAs failed to promote glmS::gfp translation. Moreover, addition of GlmY to reactions spiked with full-length GlmZ RNA did not further elevate protein synthesis (lanes 11–13). Thus, only full-length GlmZ RNA is a direct glmS activator, and GlmY most likely achieves glmS activation by a translation-independent mechanism.
GlmY Promotes the Accumulation of Full-Length GlmZ RNA
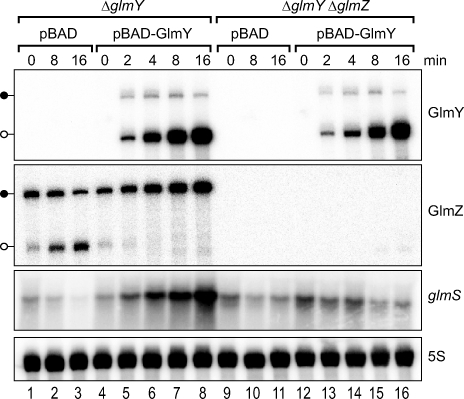
Given the above results, we speculated that GlmY activated glmS indirectly by means of increasing the amount of the full-length GlmZ RNA. If so, GlmZ levels should be elevated upon GlmY expression, which is indeed observed in bacteria carrying plasmid pPL-GlmY (compare lanes 1 and 2 in Figure 2). To determine whether GlmY counteracts GlmZ RNA processing, we studied the effects of induced glmY expression. To this end, glmY was cloned under control of an arabinose-inducible PBAD promoter, and the resulting plasmid, pBAD-GlmY, or an “empty” pBAD control vector was introduced into E. coli ΔglmY. The resulting strains were cultured to early stationary phase, and glmY expression was induced with arabinose (Figure 5). Northern blot analysis of samples from the pBAD control strain extracted within 16 min of induction (lanes 1–3) showed that the inducer arabinose itself had a considerable effect on GlmZ and glmS expression. That is, it induced GlmZ RNA processing (see position of open lollipop for GlmZ in Figure 5), and reduced the levels of both full-length GlmZ RNA and glmS mRNA. (Note that this effect is not observed using an IPTG-inducible system; see Figure S3. Since the araD mutation of MC4100 impairs arabinose metabolism [18], this indicates that accumulation of phosphorylated sugar may serve as input signal for the GlmYZ/glmS regulatory circuit.) In contrast, induction of GlmY expression from plasmid pBAD-GlmY fully suppressed GlmZ RNA processing (lanes 4–8); moreover, concomitant to the appearance of GlmY RNA, the levels of both the GlmZ full-length species and the glmS mRNA increased. Interestingly, induction of pBAD-GlmY (lanes 4–8) primarily yields the processed GlmY species (148 nt), thus, cleavage of the primary GlmY RNA (184 nt) seems to occur rapidly and completely. The same experiment, but performed in a ΔglmY ΔglmZ double mutant, confirmed that GlmY-dependent glmS mRNA accumulation strictly requires the presence of GlmZ (lanes 9–16).
Figure 5. Induction of GlmY Expression Antagonizes GlmZ RNA Processing.
E. coli MC4100 ΔglmY (JVS-8030) or ΔglmY ΔglmZ (JVS-8113) strains were transformed with control vector, pBAD, or an arabinose-inducible glmY expression plasmid, pBAD-GlmY (as indicated above the panels). Transformants were grown to early stationary phase and treated with 0.2% l-arabinose to induce glmY expression. RNA was prepared prior to or at the indicated time intervals (in minutes) upon induction, and subjected to northern analysis to detect changes in GlmY, GlmZ, and glmS mRNA expression. Black lollipops indicate the primary GlmY or GlmZ transcripts, whereas open lollipops represent the approximately 150-nt processed species of these sRNAs.
GlmY Is Essential for GlmZ Stabilization and GlmS Overproduction in a pcnB Mutant
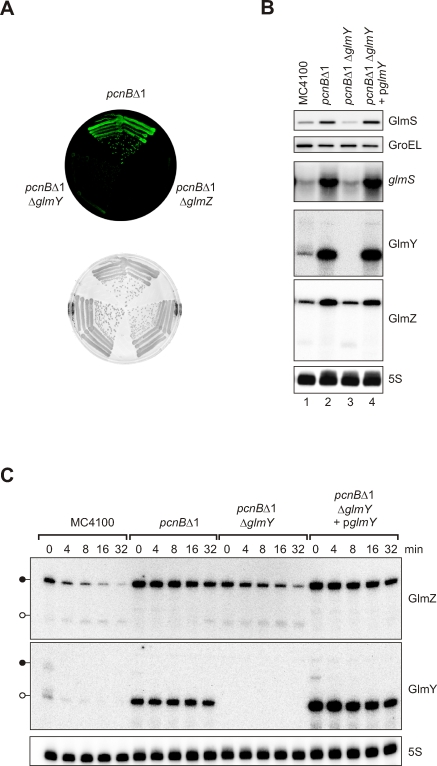
Given that GlmY antagonized the processing/degradation of GlmZ, it was likely that GlmZ accumulation resulted from increased GlmZ RNA stability. To prove GlmY-dependent RNA stability changes, we sought to identify a genetic background in which the GlmZ RNA and glmS mRNA levels change in response to presence or absence of the chromosomal glmY gene. Generally, two mutations were known to alter glmS mRNA levels, i.e., ΔyhbJ and pcnBΔ1 [4,5]. Deletion of glmY in these mutant backgrounds (shown for pncBΔ1 in Figure 6A) revealed that glmY is strictly required for glmS mRNA activation in pcnBΔ1, as judged by a loss of glmS::gfp fusion activity in the pcnBΔ1 ΔglmY double mutant. Northern blot probing (Figure 6B) confirmed that glmS mRNA highly accumulated in E. coli pcnBΔ1, and also revealed a strong increase in GlmY and GlmZ RNA levels in the absence of polyadenylation (lane 2). However, the pcnB effect is totally lost upon glmY deletion (lane 3), suggesting that PAP I acts upstream of GlmY. Ectopic glmY expression restored the pcnB effect to the pcnBΔ1 ΔglmY double mutant strain (lane 4).
Figure 6. GlmY and GlmZ Accumulate in a pcnB Mutant and Promote GlmS Synthesis.
(A) Activity of the glmS::gfp fusion in strains JVS-2058 (pcnBΔ1), JVS-8217 (pcnBΔ1 ΔglmY), or JVS-8242 (pcnBΔ1 ΔglmZ). Shown are images of an LB agar plate obtained in the fluorescence mode (top) or the visible light mode (bottom).
(B) GlmY is essential for GlmS overproduction upon loss of PAP I–mediated polyadenylation. Western and northern blot analysis to detect GlmS protein, glmS mRNA, and GlmY and GlmZ sRNAs (as indicated) in the same strains as in (A), but without the glmS::gfp fusion plasmid, and additionally, the pcnBΔ1 ΔglmY double-mutant strain complemented with plasmid pglmY (carrying the glmY gene with its own promoter). Samples were prepared from early stationary phase cultures.
(C) Northern blot analysis of GlmY and GlmZ RNA decay upon transcription block in the same strains as in (B). Bacteria were grown to early stationary phase, and RNA samples were withdrawn prior to or at the indicated time-points (in min) upon treatment with the inhibitor, rifampicin.
Rifampicin-treatment experiments showed that the accumulation of processed GlmY and of full-length GlmZ in pcnBΔ1 cells was accompanied by increased stability of these RNA species (Figure 6C). Specifically, the half-life of GlmY was increased from approximately 4 min (MC4100) to greater than 30 min (pcnBΔ1). Likewise, GlmZ was stabilized approximately 5-fold (∼4 min vs. ∼20 min). In addition, GlmZ RNA processing was suppressed. If GlmY acts as a stabilizer of GlmZ RNA, deletion of glmY in the pcnBΔ1 strain should reverse the aforementioned GlmZ stabilization. In line with this prediction, GlmZ half-life was reduced to approximately 8 min in the pcnBΔ1 ΔglmY double mutant. Upon complementation of the pcnBΔ1 ΔglmY strain with a glmY expression plasmid, GlmZ again decayed with the same half-life (∼20 min) as in the pcnBΔ1 single mutant. Together with the observation that GlmY expression from plasmid pPL-GlmY cannot bypass a ΔglmZ mutation in pcnBΔ1 (unpublished data), these experiments corroborated that GlmY RNA activates glmS mRNA by antagonizing GlmZ RNA decay.
Loss of GlmY RNA Polyadenylation Causes GlmS Overproduction in E. coli pcnBΔ1
GlmS overproduction in E. coli pcnBΔ1 had been postulated to result from stabilization of no longer 3′ polyadenylated glmS mRNA [5]. However, the fact that a glmS::gfp fusion having a different 3′ end was equally activated in pcnBΔ1 (Figure 6A) was not in favor of this postulate. Moreover, the ΔglmY and ΔglmZ mutations abolished glmS mRNA accumulation in pcnBΔ1 (Figure 6B and unpublished data), yet it was hard to conceive how the GlmYZ sRNAs could affect polyadenylation of glmS mRNA.
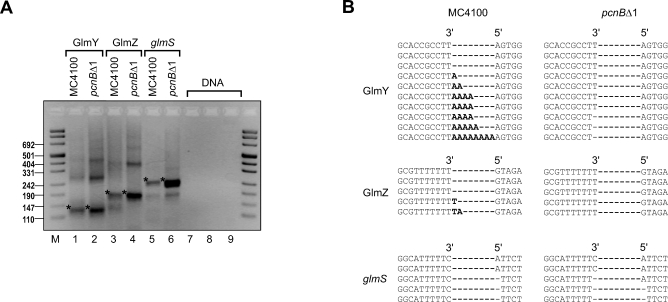
To determine whether GlmY, GlmZ or glmS transcripts were polyadenylated, we used an RNA circularization assay previously developed to assess the poly(A) status of chloroplast RNAs [19,20]. Briefly, total RNA of the MC4100 and pcnBΔ1 strains was treated with tobacco acid pyrophosphatase (to convert primary 5′ triphosphate to monophosphate groups), and subjected to 3′/5′ end circularization by T4 RNA ligase, and cDNA synthesis. Subsequently, 3′/5′ junction fragments were amplified by PCR (Figure 7A), cloned, and sequenced. This analysis revealed homopolymeric tails composed of up to eight adenosines at the 3′ end of the processed GlmY RNA (148-nt species) in cDNA of MC4100; no such poly(A) tails were found in cDNA from pcnBΔ1 cells (Figure 7B). In contrast to GlmY, no poly(A) tails were detected at the 3′ ends of GlmZ or glmS RNA, irrespective of whether MC4100 or pcnBΔ1 cDNA was analyzed (Figure 7B). Thus, GlmY seems to be the only RNA for which differential stabilities and expression levels in wild type versus pcnBΔ1 (Figure 6B and 6C) correlate with the presence or absence of 3′ end poly(A) tails. Therefore, GlmZ stabilization and, consequently, GlmS overproduction in pcnBΔ1 are best explained by a loss of polyadenylation of the processed GlmY RNA.
Figure 7. GlmY, and Not GlmZ or glmS RNAs, Is Polyadenylated In Vivo.
(A) Electrophoretic separation (3% agarose gel) of PCR products obtained on cDNA of 5′–3′ circularized RNA. Total RNA was prepared from E. coli MC4100 or its isogenic pcnBΔ1 mutant (JVS-2058). PCR products corresponding to the expected sizes of processed GlmY RNA (lanes 1 and 2; 130 bp), full-length GlmZ RNA (lanes 3 and 4; 184 bp), and monocistronic glmS mRNA (lanes 5 and 6; 215 bp) are denoted by an asterisk (*). Control reactions were preformed using the same primer pairs as for GlmY (lane 7), GlmZ (lane 8), or glmS (lane 9) but with E. coli DNA as a template. Band sizes of a comigrating DNA marker (M) are given to the left.
(B) Sequence analysis upon cloning of the above PCR products. Alignment of ten GlmY-specific insert sequences reveals the existence of non-templated adenosine residues (nucleotides in bold-face) in cDNA from wild-type E. coli. The absence of these poly(A) tails in pcnBΔ1 cDNA suggests that GlmY undergoes PAP I–mediated polyadenylation. In contrast, no such poly(A) tails were found in GlmZ-specific or glmS-specific cDNA insert sequences (five inserts were sequenced for each primer pair and cDNA).
Discussion
RNA-based regulatory mechanisms are commonly used to control the expression of metabolic genes. In addition to transcriptional and translational attenuators [21], trans-encoded sRNAs and cis-encoded metabolite-sensing aptamers have increasingly been recognized as mRNA regulators in metabolic pathways. Whereas cis-encoded metabolite sensors are prevalent in Gram-positive species, few such elements are known in Gram-negative bacteria [2,22–24]. Conversely, the number of E. coli sRNAs has come close to one hundred [25,26], and fewer sRNAs have been reported from Gram-positive species. Conforming to this obvious bias, the posttranscriptional control of glmS relies on a cis-encoded RNA sensor in B. subtilis, and on trans-encoded riboregulators in E. coli. Interestingly, this is reminiscent of the distinctly different RNA-based mechanisms controlling expression of the tryptophan biosynthetic operon in these two model bacteria [21]. In both cases, i.e., glmS and tryptophanase genes, the riboregulatory mechanisms facilitate feedback control of the encoded metabolic pathways [1,4,21]. The network of regulatory factors including the conserved GlmY and GlmZ sRNAs (ref. [3] and Figure S1) that controls the E. coli glmS mRNA appears to be well suited for this task, for it provides multiple entry points for the environmental signals that determine glmS epistasis.
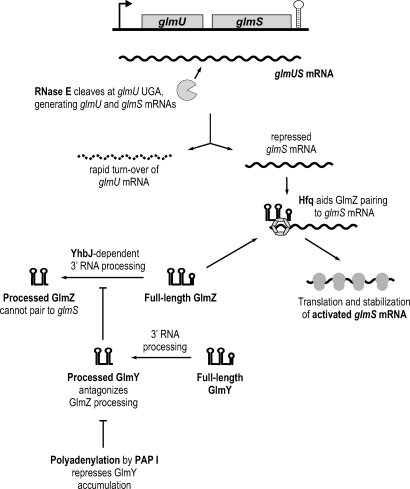
Figure 8 summarizes the results of this and previous studies [3–5] to suggest a model of how this complex regulatory network may operate in E. coli. NagC-controlled transcription yields a dicistronic glmUS mRNA [15] which undergoes RNase E cleavage in the glmU stop codon sequence to generate monocistronic glmS mRNA [4,5]. This cleavage is independent of the activities of GlmY, GlmZ, Hfq, PAP I, or YhbJ, since the monocistronic glmS mRNA is also detected in strains devoid of any of these factors (Figures 1B and S2). Because the remaining glmU mRNA lacking an intact stop codon is rapidly degraded, most of the cellular GlmU protein seems to be synthesized from the primary glmUS operon mRNA [5]. The glmS mRNA is also quickly turned over unless it becomes stabilized by GlmZ. Whether glmS mRNA is stabilized due to enhanced translation, the GlmZ/glmS RNA interaction, or both, is currently unknown. However, given that the half-life of bacterial mRNA is strongly affected by the association with ribosomes [27,28], increased translation may account for most of the observed glmS mRNA stabilization. The RNA chaperone, Hfq, is essential for glmS translational activation by GlmZ, and is also known to associate in vivo with both GlmZ [8,29] and glmS mRNA (A. Sittka, S. Lucchini, K. Papenfort, C. M. Sharma, J. C. Hinton, and J. Vogel, unpublished data). Regarding glmS, the Hfq function seems to be limited to facilitating the GlmZ/glmS interaction since an hfq mutation does not affect basal glmS expression (Figure 1B and [4]) or glmS mRNA stability (unpublished data).
Figure 8. Pathways of glmS Activation by GlmY and GlmZ RNAs in E. coli .
Model summarizing the findings of this and previous studies [3–5]. See Discussion for details.
GlmZ is the only factor to directly activate glmS translation, and we have provided evidence that this process relies upon an anti-antisense mechanism that induces a translation-competent status of the mRNA. Similar to the DsrA-regulated rpoS mRNA [16,17], and other sRNA-activated mRNAs [30,31], the anti-antisense mechanism proposed here disrupts an inhibitory hairpin that sequesters the glmS RBS. Using purified components, we have been able to reconstitute glmS mRNA activation in vitro, proving that the ultimate step in this system requires only the translational machinery and GlmZ RNA.
GlmZ exists in two forms, i.e., the primary 207-nt RNA which can activate glmS, and the processed, inactivated GlmZ153 species. Transcription of glmZ appears to be growth-rate controlled, and to be higher in fast-growing cells [9]. Although the upstream factor(s) that regulate glmZ transcription remain unknown, YhbJ acts downstream in the glmS activator cascade by regulating the availability of full-length GlmZ at the posttranscriptional level. YhbJ acts as a glmS repressor inasmuch as it promotes GlmZ processing [4], and we have demonstrated here that this event generates an inactive GlmZ species. Whether YhbJ, which is conserved in many bacteria [4], is an RNA-binding protein that interacts with GlmZ (or GlmY) is currently unknown, and our attempts to purify soluble YhbJ protein have so far been unsuccessful.
GlmY is also upstream of GlmZ in the herein-described cascade, but in contrast to YhbJ, this RNA acts as a GlmZ/glmS activator. In our current working model (Figure 8), GlmY only acts to overcome YhbJ-mediated GlmZ inactivation, which is supported by the observations that GlmY expression has no glmS effect in ΔyhbJ (Figure S2, lanes 7 and 8) and that glmS overexpression is not consistently affected by secondary mutation of glmY in the yhbJ deletion strain (Figure S4).
At the transcriptional level, the alternative sigma factor, σ54, may control glmY expression [3]. σ54 along with a yet to be identified activator protein could up-regulate glmY to bolster up GlmZ action upon nitrogen starvation. Similar to GlmZ, GlmY RNA is also repressed at the posttranscriptional level, albeit by polyadenylation. Loss of this repression in pncBΔ1 results in GlmY RNA stabilization and GlmS overproduction. However, under normal growth, the YhbJ-dependent GlmZ processing is likely the rate-limiting step since the GlmZ levels in ΔyhbJ are sufficiently high so that GlmY cannot further activate glmS expression (Figure S2).
Nonetheless, little is known as to how RNA polyadenylation fluctuates in response to environmental conditions, and certain transcripts have been shown to become differentially polyadenylated in a growth rate–dependent manner [32]. Moreover, greater than 90% of all E. coli mRNAs expressed during exponential growth are controlled by polyadenlyation [33], but the number of mRNAs that are direct PAP I substrates is unknown. Notably, Joanny et al. [5] discussed the possibility that differential expression of regulatory sRNAs may account for some of these PAP I effects, similar to the stabilization of plasmid-borne cis-antisense RNAs that underlie alterations of plasmid copy number upon loss of polyadenylation [34,35]. It is worthwhile mentioning that similar to GlmY, additional chromosomal E. coli sRNAs are known to be polyadenylated [9]; at least one of these, SraL, is stabilized in a pcnB mutant of Salmonella [36]. Thus, it is reasonable to predict that additional mRNAs will be identified as differentially regulated in response to altered sRNA polyadenylation.
The key finding of this paper is that two strikingly similar sRNAs, GlmY and GlmZ, form a regulatory cascade to regulate one and the same target mRNA. Following the original description of the E. coli CsrB and CsrC sRNAs, two homologous antagonists of CsrA protein [37,38], numerous families of homologous sRNAs have been reported (e.g., [8,39]). These include CsrB-like RNAs of other organisms [40], the E. coli OmrA/B sRNAs controlling outer membrane protein biogenesis [41], and a family of four to five Vibrio sRNAs regulating quorum sensing [42,43]. The common denominator in these systems is that the sRNAs act redundantly, or at least additively, and can to some degree substitute for each other's function. In contrast, the hierarchy of the sRNAs studied here is novel since GlmY strictly requires GlmZ for glmS activation.
Our results have shown that GlmY can antagonize GlmZ 3′ end RNA processing, an event that one may refer to as controlled GlmZ inactivation. Given the related structures of the two sRNAs, this antagonism may involve some kind of molecular mimicry such that GlmY intercepts the factor (YhbJ?) that promotes GlmZ inactivation. If so, this mechanism would bear strong similarity to the activity of small anti-adaptor proteins, e.g., ComS and IraP, which interfere with the controlled proteolysis of transcriptional regulators, ComK or RpoS, respectively [44–46]. In analogy to the studies of ComS or IraP, the development of an in vitro system to study GlmZ degradation will help answer whether GlmY acts by molecular mimicry to save GlmZ from inactivation.
In summary, the GlmYZ system represents a fine example of a regulatory circuit composed of RNA that is no less sophisticated than those composed of proteins. Given the many noncoding RNAs yet to be functionally characterized, we find it reasonable to speculate that hierarchically acting regulatory RNAs will also be discovered in other organisms.
Materials and Methods
Media and growth conditions.
Growth in Luria–Bertani (LB) broth (220 rpm, 37 °C) or on LB agar plates at 37 °C was used throughout this study. A culture grown to an optical density at 600 nm (OD600) of 2 is referred to as an early stationary phase culture. Antibiotics (where appropriate) were used at the following concentrations: 100 mg/ml ampicillin, 50 mg/ml kanamycin, and 20 mg/ml chloramphenicol. For GlmY expression from pBAD-GlmY, cultures were induced with l-arabinose (0.2% final concentration). Rifampicin (Sigma) was added to the cultures at a final concentration of 500 μg/ml to block transcription.
Bacterial strains.
The E. coli strains used in this study are listed in Table 1. MC4100 was used as parental wild-type strain. To construct JVS-2058, the pcnBΔ1::kan allele of SK7988 [47] was moved into MC4100 by P1 transduction. JVS-8018 (ΔyhbJ::kan), JVS-8024 (ΔglmZ::kan), and JVS-8030 (ΔglmY::kan) were constructed by using the one-step gene inactivation protocol with PCR products obtained on pKD4 with primer pairs JVO-1951/-1952, JVO-1947/-1948, and JVO-1661/-1662, respectively (deoxyribonucleotides are listed in Table S1), followed by P1 transduction into MC4100. The kan marker of JVS-8018 was eliminated by using the FLP expression plasmid pCP20 [48], and the ΔglmY::kan (from JVS-8030), ΔglmZ::kan (from JVS-8024), and Δhfq::cat alleles were moved into this strain by P1 transduction to obtain JVS-8076 (Δyhb, ΔglmY::kan), JVS-8078 (Δyhb, ΔglmZ::kan), and JVS-8146 (ΔyhbJ, Δhfq::cat), respectively. Similarly, the kan markers of JVS-8076 and JVS-8030 were eliminated using pCP20, and the ΔglmZ::kan (from JVS-8024) allele moved into the respective strains to construct JVS-8111 (ΔyhbJ ΔglmY ΔglmZ::kan) and JVS-8113 (ΔglmY ΔglmZ::kan).
Table 1.
Bacterial Strains
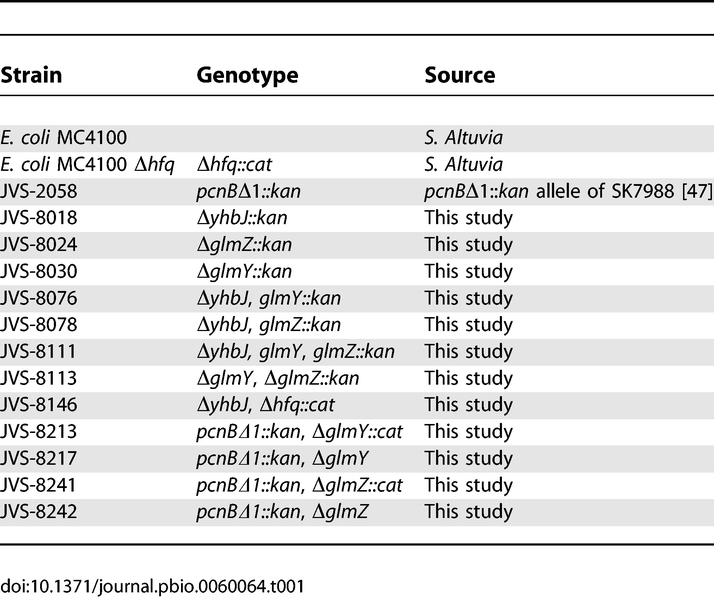
JVS-8213 (pcnBΔ1::kan ΔglmY::cat) and JVS-8241(pcnBΔ1::kan ΔglmZ::cat) were derived from strain JVS-2058 (pcnBΔ1::kan) by using the one-step gene inactivation protocol with PCR products obtained on pKD3 [48] with primer pairs JVO-1661/-1662 and JVO-1947/-1948, respectively. Elimination of the cat markers of JVS-8213 and JVS-8241 resulted in strains JVS-8217 (pcnBΔ1::kan ΔglmY) and JVS-8242 (pcnBΔ1::kan ΔglmZ), respectively.
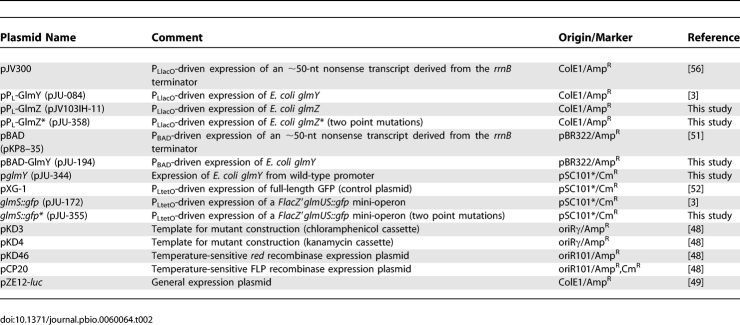
Plasmids.
Plasmids used in this study are listed in Table 2. To construct plasmid pPL-GlmZ, an E. coli glmZ fragment was amplified with oligos jb-103-H and jb-103-I, and inserted by blunt end and EcoRI cloning into vector pZE12-luc [49], following a previously published protocol [50]. Plasmid pPL-GlmZ* was constructed by introduction of two point mutations into pPL-GlmZ by PCR amplification of this plasmid using primers JVO-2609/-2610, followed by self-ligation of the PCR product.
Table 2.
Plasmids
Construction of plasmid pBAD-GlmY (l-arabinose-inducible glmY expression) followed the previously published protocol by [51]. Briefly, vector pBAD-His-myc was amplified by PCR using sense primer JVO-0901 (adds an XbaI restriction site upstream of the rrnB terminator) and antisense primer JVO-0900 (anneals to the −1 position of the PBAD promoter). A glmY-derived PCR product amplified with primers JVO-1399/-0670 was digested with XbaI, and inserted into the above-described plasmid backbone (treated with XbaI) to yield pBAD-GlmY.
For the construction of a low-copy complementation plasmid of glmY expression, the glmY gene including an approximately 360-nt upstream region was PCR amplified with primers JVO-0670/-2019, the resulting product digested with AatII and XbaI, and ligated to the AatII/XbaI digested plasmid pXG-1 [52] to yield pglmY.
The glmS::gfp fusion plasmid, pJU-172, which expresses a FlacZ::glmU-glmS::gfp mini-operon, was constructed by inserting a BfrBI/NheI-digested PCR product obtained with primers JVO-1270/-1294 into the operon GFP fusion vector pXG-30 [52] digested with the same restriction enzymes. In the resulting plasmid, the C-terminal sequence (17 amino acids [aa]) of glmU is fused to the carboxy terminus of a FLAG epitope-tagged, truncated lacZ gene (FlacZ′). The glmS gene (first 7 aa) is fused to the N-terminus of gfp. The corresponding glmS*::gfp fusion plasmid, pJU-355, carrying compensatory base-pair changes was constructed by self-ligation of a PCR product obtained on plasmid pJU-172 and using primers JVO-2605/-2606.
Whole-cell protein fractions and Western blot.
Protein sample preparation and western blotting was carried out as described [52]. An amount of whole-cell protein fractions corresponding to 0.01 OD culture volume was separated by 12% SDS-PAGE and transferred onto a PVDF membrane (PerkinElmer). A polyclonal serum was used 1:5,000 in 3% BSA, TBST20 to detect GlmS. GFP and GroEL detection was carried out as described previously [52]. Whenever GlmS and GroEL were detected on the same membrane, blots were first hybridized with α-GlmS, stripped using Roti-Free solution (Roth) according to the manufacturer's protocol, and afterwards hybridized with α-GroEL. Blots were developed using Western Lightning reagent (PerkinElmer), and signals detected with a Fuji LAS-3000 charge-coupled device (CCD) camera.
RNA isolation and Northern blot detection.
RNA using TRIZOL reagent (Invitrogen) or the Promega SV total RNA isolation kit were performed as described [51,52]. Separation of 5 μg of total RNA (samples prepared with TRIZOL) was on 5% polyacrylamide gels containing 8.3 M urea, or of 12 μg of total RNA (samples prepared with Promega SV totalRNA) on 1.5% agarose gels containing 2.2 M formaldehyde, and northern detection was carried out as described [52]. RNA stability experiments were performed as described in [6]. GlmY, GlmZ, and 5S rRNA were detected using 5′ end-labeled oligodeoxyribonucleotide JVO-1730, −1697, and −0322, respectively. glmS mRNA was detected using an RNA probe derived from in vitro transcription of a PCR fragment (primers: JVO-1598/-1599; T7 promoter is added by JVO-1599) in the presence of [α-32P] UTP with Ambion's T7 polymerase Maxiscript kit. Signals were visualized on a phosphorimager (Phosphorimager, FLA-3000 Series; Fuji), and band intensities quantified with AIDA software (Raytest).
In vivo whole-cell colony plate fluorescence imaging.
E. coli strains expressing plasmid-borne gfp fusions were streaked on standard LB plates supplemented with the appropriate antibiotics. After overnight growth, colonies were photographed in a FUJI LAS-3000 image analyzer using a CCD camera with a 510-nm emission filter and excitation at 460 nm.
cDNA synthesis and cloning for 3′ end sequencing.
The procedure for RNA 3′ end sequencing has been described previously [20] and was used with some modifications. A total of 8 μg of DNA-free total RNA prepared from early stationary phase cultures was treated with ten units tobacco acid pyrophosphatase (Epicentre) for 30 min at 37 °C to convert 5′ triphosphate groups of primary transcripts to 5′ monophosphates. Following organic extraction, RNA was treated overnight at 17 °C with 40 units of T4 RNA ligase (New England Biolabs) to circularize transcripts. Following organic extraction and ethanol precipitation, 1.5 μg of self-ligated RNA was converted to cDNA using 100 pmol random hexamer primers and the Superscript III (200 units) reverse transcription kit (Invitrogen) in a 20-μl reaction. Herein, 10-min incubation at 25 °C was carried out before addition of the reverse transcriptase, followed by four subsequent 15-min incubation steps at 42 °C, 50 °C, 55 °C, and 60 °C. After heat inactivation of the reverse transcriptase for 5 min at 85 °C, samples were treated with one unit of RNase H (New England Biolabs) at 37 °C for 20 min. A total of 1 μl of the reaction served as template in a subsequent standard 25-μl PCR reaction using 0.5 units Taq polymerase (New England Biolabs) and primer pairs designed to amplify products representing successful self-ligated GlmY (primer JVO-2349/-2355), GlmZ (primer JVO-2350/-2356), and glmS (primer JVO-2022/-2388) transcripts. PCR products were separated by 3% agarose gel electrophoresis, and fragments of the expected size were excised, purified, and cloned into the pCR 2.1-TOPO vector (Invitrogen). Colony PCR products obtained on positive clones with standard primers M13 Forward and M13 Reverse were sequenced with primer M13 Reverse.
Primer extension.
The processing site within the dicistronic glmUS mRNA was determined by primer extension according to the SuperscriptIII protocol (Invitrogen). In brief, 10 μg of total RNA prepared with the Promega SV total RNA isolation kit from cultures grown to an OD600 of 2 were incubated with 2 pmol of 5′ end-labeled primer JVO-2022 (anneals to positions −19 to −2 relative to the glmS start codon) for 5 min at 65 °C, followed by 5 min on ice. The samples were adjusted to a final volume of 20 μl, containing 1× first strand buffer, 0.5 mM each of dATP, dGTP, dCTP, and dTTP, 5 mM DTT, 20 units superasin RNase inhibitor (Ambion), and 200 units SuperscriptIII (Invitrogen). Reverse transcription was carried out for 60 min at 50 °C, followed by inactivation of the enzyme at 70 °C for 15 min. After treatment with 2 units RNaseH (New England Biolabs), 3 μl of the reactions was separated on a denaturing 6% polyacrylamide gel containing 7 M urea. The sequencing ladder was established by using the CycleReader DNA sequencing kit (Fermentas) according to the manufacturer's protocol with 5′ end-labeled primer JVO-2022 on a PCR product obtained on E. coli K12 DNA with primer pair JVO-1177/-2022.
In vitro translation assay.
Translation reactions were carried out using PURESYSTEM (PGM-PURE2048C; Cosmo Bio Co.) according to the manufacturer's instructions. The 10-μl reactions contained, in addition to 70S ribosomes, 1 pmol glmS::gfp mRNA template and Hfq, as well as GlmY and GlmZ sRNA variants (see Figure 4 legend for final concentration). RNA was denatured 1 min at 90 °C and chilled on ice for 5 min before Hfq was added and preincubation at 37 °C was allowed to proceed for 10 min. Puresystem mix was added and incubation continued at 37 °C for the time indicated in the figure legend. Reactions were stopped with four volumes of ice-cold acetone, kept on ice for 15 min, and proteins were collected by centrifugation (10,000 g, 10 min, 4 °C). Proteins were detected by western blot analysis with monoclonal antibodies against GFP as described [52].
RNAs were synthesized in vitro using the AmpliScribe T7-flash kit (Epicentre) according to the manufacturer's instructions. Templates for glmS::gfp, GlmY, GlmZ, and GlmZ153 T7 in vitro transcription were established by PCR on plasmids pJU-172 (for T7-glmS::gfp), pPL-GlmY (for T7-GlmY), and pPL-GlmZ (for T7-GlmZ and T7-GlmZ153) using primer pairs JVO-1866/-pZE-T1, JVO-1560/-1562, JVO-2130/-2427, and JVO-2130/-2528, respectively.
Supporting Information
(A) Alignment of glmZ genes identified in different enterobacteria. The transcriptional start site (+1) and a putative transcriptional terminator (arrowheads) are denoted for the E. coli glmZ. A putative −10 element of σ70-type promoters is underlined. Sequences were collected from the following genomes (accession numbers in parentheses): E. coli K12 (NC_000913), Shigella flexneri 2a strain 301 (NC_004337), Salmonella typhimurium LT2 (NC_003197), Yersinia pestis CO92 (NC_003143), Erwinia carotovora subsp. atroseptica (NC_004547), and Photorhabdus luminescens subsp. laumondii (NC_005126), and the unfinished genome sequences of Klebsiella pneumoniae strain Kp342 (http://www.tigr.org) and of Serratia marcescens strain Db11 (http://www.sanger.ac.uk). The alignment was computed with Multalign (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html; [53]).
(B) Secondary structure of E. coli GlmZ RNA predicted by Mfold [54]. Residues that are conserved in the GlmZ RNAs shown in (A) are shadowed. Vertical arrows indicate previously mapped 3′ processing sites [9]. The residues predicted by the RNA-Hybrid program [55] to interact with the glmS-leader are highlighted in red.
(54 KB PDF)
Western and northern blot analyses comparing the effects of GlmY and GlmZ expression in MC4100, Δhfq::cat, JVS-8018 (ΔyhbJ), JVS-8146 (ΔyhbJ Δhfq), JVS-8113 (ΔglmY ΔglmZ), or JVS-8111 (ΔyhbJ ΔglmY ΔglmZ). Each of these strains carried plasmids pJV300 (“contr” lanes), pPL-GlmY (“GlmY” lanes), or pPL-GlmZ (“GlmZ” lanes). Whole-cell protein and total RNA samples were prepared from early stationary phase cultures. Western blots were probed with sera specific for GlmS (top panel) or GroEL (loading control; second panel from top) proteins. For northern blot detection of glmS mRNA (third panel from top), 12 μg of total RNA per lane was separated on a 1.5% agarose-formaldehyde gel. To detect GlmY and GlmZ, 5 μg of total RNA was separated on a denaturing 5% PAA gel (fourth and fifth panels from top). The 5S rRNA (bottom panel) probing confirmed equal amounts of RNA in each sample. Black lollipops indicate the primary GlmY or GlmZ transcripts; open lollipops the processed, approximately 150-nt RNA species.
(210 KB PDF)
E. coli Top10F′ (lacI q; Invitrogen) was transformed with control vector, pJV300, or the IPTG-inducible glmY expression plasmid, pPL-GlmY (as indicated above the panels). Transformants were grown to early stationary phase, and treated with 0.5 mM IPTG to induce glmY expression. RNA was prepared prior to or at the indicated time-intervals (in minutes) upon induction, and subjected to northern analysis to detect changes in GlmY, GlmZ, and glmS mRNA expression. Black lollipops indicate the primary GlmY or GlmZ transcripts, open lollipops the processed, approximately 150-nt species of these sRNAs.
(60 KB PDF)
(A) Activation of a translational glmS::gfp fusion in a ΔyhbJ background requires glmZ, but not glmY. Colony fluorescence of E. coli MC4100 (wt) and mutant strains JVS-8018 (ΔyhbJ), JVS-8076 (ΔyhbJ ΔglmY), JVS-8078 (ΔyhbJ ΔglmZ) harboring plasmid glmS::gfp, or pXG-1 (gfp control plasmid). Images were obtained in the fluorescence (left panel) and visual light (right panel) mode.
(B) Western and northern blot analyses (as in Figure S2) comparing the effects of GlmY and GlmZ expression in MC4100, JVS-8030 (ΔglmY), JVS-8024 (ΔglmZ), JVS-8018 (ΔyhbJ), JVS-8076 (ΔyhbJ ΔglmY), and JVS-8078 (ΔyhbJ ΔglmZ).
(169 KB PDF)
Sequences are given in 5′ → 3′ direction; “P∼” denotes a 5′ monophosphate.
(76 KB DOC)
Acknowledgments
We thank M. Badet-Denisot for GlmS antiserum; S. Kushner for strains; S. Gottesman, C. M. Sharma, K. Papenfort, U. Bläsi, and C. Yanofsky for insightful comments on the manuscript. We are indebted to H. Aiba who kindly told us of the PURESYSTEM.
Abbreviations
- 5′ UTR
5′ untranslated region
- bp
base pair
- nt
nucleotide
- SD
Shine-Dalgarno
- sRNA
small noncoding RNA
Footnotes
Author contributions. JHU and JV conceived and designed the experiments. JU performed the experiments. JHU and JV analyzed the data. JV wrote the paper.
Funding. JHU was supported by grants from Deutsche Forschungsgemeinschaft, Germany (DFG; VO 875/1–1,3).
Competing interests. The authors have declared that no competing interests exist.
References
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci U S A. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Papenfort K, Thomsen J, Schmitz RA, Vogel J. A Conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J Mol Biol. 2007;373:521–528. doi: 10.1016/j.jmb.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Kalamorz F, Reichenbach B, Marz W, Rak B, Görke B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol Microbiol. 2007;65:1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- Joanny G, Le Derout J, Brechemier-Baey D, Labas V, Vinh J, et al. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 2007;35:2494–2502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, et al. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- Møller T, Franch T, Hojrup P, Keene DR, Bachinger HP, et al. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Cao GJ, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci U S A. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- Dreyfus M, Regnier P. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 1995;14:3958–3965. doi: 10.1002/j.1460-2075.1995.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci U S A. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Börner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Hess WR. Complete 5' and 3' end maturation of group II intron-containing tRNA precursors. RNA. 2001;7:285–292. doi: 10.1017/s1355838201001960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA. 2007;13:1141–1154. doi: 10.1261/rna.620507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Iost I, Dreyfus M. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995;14:3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, et al. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiecki J, Wegrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Rep. 2003;4:172–177. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Lin-Chao S, Cohen SN. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci U S A. 1993;90:6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderbom F, Binnie U, Masters M, Wagner EG. Regulation of plasmid R1 replication: PcnB and RNase E expedite the decay of the antisense RNA, CopA. Mol Microbiol. 1997;26:493–504. doi: 10.1046/j.1365-2958.1997.5871953.x. [DOI] [PubMed] [Google Scholar]
- Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, et al. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, et al. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Wickner S, Gottesman S. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev. 2006;20:884–897. doi: 10.1101/gad.1400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay K, Hamoen LW, Venema G, Dubnau D. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 1997;11:119–128. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- Ogura M, Liu L, Lacelle M, Nakano MM, Zuber P. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol Microbiol. 1999;32:799–812. doi: 10.1046/j.1365-2958.1999.01399.x. [DOI] [PubMed] [Google Scholar]
- O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, et al. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, et al. sigma(E)-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Alignment of glmZ genes identified in different enterobacteria. The transcriptional start site (+1) and a putative transcriptional terminator (arrowheads) are denoted for the E. coli glmZ. A putative −10 element of σ70-type promoters is underlined. Sequences were collected from the following genomes (accession numbers in parentheses): E. coli K12 (NC_000913), Shigella flexneri 2a strain 301 (NC_004337), Salmonella typhimurium LT2 (NC_003197), Yersinia pestis CO92 (NC_003143), Erwinia carotovora subsp. atroseptica (NC_004547), and Photorhabdus luminescens subsp. laumondii (NC_005126), and the unfinished genome sequences of Klebsiella pneumoniae strain Kp342 (http://www.tigr.org) and of Serratia marcescens strain Db11 (http://www.sanger.ac.uk). The alignment was computed with Multalign (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html; [53]).
(B) Secondary structure of E. coli GlmZ RNA predicted by Mfold [54]. Residues that are conserved in the GlmZ RNAs shown in (A) are shadowed. Vertical arrows indicate previously mapped 3′ processing sites [9]. The residues predicted by the RNA-Hybrid program [55] to interact with the glmS-leader are highlighted in red.
(54 KB PDF)
Western and northern blot analyses comparing the effects of GlmY and GlmZ expression in MC4100, Δhfq::cat, JVS-8018 (ΔyhbJ), JVS-8146 (ΔyhbJ Δhfq), JVS-8113 (ΔglmY ΔglmZ), or JVS-8111 (ΔyhbJ ΔglmY ΔglmZ). Each of these strains carried plasmids pJV300 (“contr” lanes), pPL-GlmY (“GlmY” lanes), or pPL-GlmZ (“GlmZ” lanes). Whole-cell protein and total RNA samples were prepared from early stationary phase cultures. Western blots were probed with sera specific for GlmS (top panel) or GroEL (loading control; second panel from top) proteins. For northern blot detection of glmS mRNA (third panel from top), 12 μg of total RNA per lane was separated on a 1.5% agarose-formaldehyde gel. To detect GlmY and GlmZ, 5 μg of total RNA was separated on a denaturing 5% PAA gel (fourth and fifth panels from top). The 5S rRNA (bottom panel) probing confirmed equal amounts of RNA in each sample. Black lollipops indicate the primary GlmY or GlmZ transcripts; open lollipops the processed, approximately 150-nt RNA species.
(210 KB PDF)
E. coli Top10F′ (lacI q; Invitrogen) was transformed with control vector, pJV300, or the IPTG-inducible glmY expression plasmid, pPL-GlmY (as indicated above the panels). Transformants were grown to early stationary phase, and treated with 0.5 mM IPTG to induce glmY expression. RNA was prepared prior to or at the indicated time-intervals (in minutes) upon induction, and subjected to northern analysis to detect changes in GlmY, GlmZ, and glmS mRNA expression. Black lollipops indicate the primary GlmY or GlmZ transcripts, open lollipops the processed, approximately 150-nt species of these sRNAs.
(60 KB PDF)
(A) Activation of a translational glmS::gfp fusion in a ΔyhbJ background requires glmZ, but not glmY. Colony fluorescence of E. coli MC4100 (wt) and mutant strains JVS-8018 (ΔyhbJ), JVS-8076 (ΔyhbJ ΔglmY), JVS-8078 (ΔyhbJ ΔglmZ) harboring plasmid glmS::gfp, or pXG-1 (gfp control plasmid). Images were obtained in the fluorescence (left panel) and visual light (right panel) mode.
(B) Western and northern blot analyses (as in Figure S2) comparing the effects of GlmY and GlmZ expression in MC4100, JVS-8030 (ΔglmY), JVS-8024 (ΔglmZ), JVS-8018 (ΔyhbJ), JVS-8076 (ΔyhbJ ΔglmY), and JVS-8078 (ΔyhbJ ΔglmZ).
(169 KB PDF)
Sequences are given in 5′ → 3′ direction; “P∼” denotes a 5′ monophosphate.
(76 KB DOC)