Abstract
Epidemiological studies indicate that consumption of green-yellow vegetables rich in chlorophyll, vitamin C, vitamin E, and carotenoids reduce the risk of cancer. We sought to examine the antigenotoxic and antioxidant properties of chlorophyll-rich methanol extracts of Angelica keiskei, Oenanthe javanica, and Brassica oleracea (kale). In the Salmonella mutagenicity assay, A. keiskei caused dose-dependent inhibition against three heterocyclic amine mutagens in the presence of S9, O. javanica was antimutagenic only at the highest concentration in the assay (2 mg/plate), and B. oleracea showed no consistent inhibitory activity at non-toxic levels. None of the extracts were effective against three direct-acting mutagens in the absence of S9. Extracts of A. keiskei and, to a lesser extent O. javanica, inhibited two of the major enzymes that play a role in the metabolic activation of heterocyclic amines, based on ethoxyresorufin-O-deethylase and methoxyresorufin-O-demethylase assays in vitro. All three plant extracts were highly effective in assays which measured ferric reducing/antioxidant power, oxygen radical absorbance capacity, and Fe2+/H2O2-mediated DNA nicking. Finally, using the ‘comet’ assay, all three plant extracts protected against H2O2-induced genotoxic damage in human HCT116 colon cancer cells. These findings provide support for the antigenotoxic and antioxidant properties of chlorophyll-rich extracts of A. keiskei, O. javanica, and B. oleracea, through mechanisms that include inhibition of carcinogen activation and scavenging of reactive oxygen species.
Keywords: Antimutagen, antioxidant, heterocyclic amines, phytochemical, comet assay, D NA breaks
1. Introduction
The human diet contains a great variety of carcinogens, as well as many natural anticarcinogens. Numerous epidemiological studies have shown that intake of green-yellow vegetables rich in chlorophyll, vitamin C, vitamin E, and carotenoids reduce the risk of cancer and other chronic diseases [1–4]. An inverse association between the consumption of vegetables and the formation of epithelial cancers may be stronger for raw vegetables than cooked vegetables [5]. Previous work showed the beneficial effects of green juice made of raw Angelica keiskei and Brassica oleracea (kale) on nitrite scavenging in vitro, growth of cancer cells in culture, and plasma lipid and antioxidant status in smokers [6–8]. A growing interest exists in establishing the various health benefits of raw green-yellow vegetable extracts, including their potential antioxidant and antigenotoxic properties.
Based on these initial studies, we sought to examine the antimutagenic and antioxidant properties of methanol extracts of A. keiskei, O. javanica, and B. oleracea, including studies with cooked meat heterocyclic amines, which are potent mutagens and multi-organ carcinogens [9]. Heterocyclic amines have been shown to respond in vitro and in vivo to a broad array of antimutagens and cancer chemopreventive agents, as well as various tumor promoters [10].
2. Materials and methods
2.1. Mutagens and other chemicals
The heterocyclic amines 2-amino-3-methylimidazo[4,5-f]quinoline (IQ), 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx), and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) were from Toronto Research Chemicals Inc. (North York, Ontario, Canada), as were the directing-acting mutagens 2-hydroxyamino-3-methylimidazo[4,5-f]quinoline (N-OH-IQ) and 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-OH-PhIP). All other chemicals, including the direct-acting mutagen 4-nitro-1,2-phenylenediamine (NPD), were from sources reported previously [11–13]. Gallic acid and ascorbic acid were purchased from Sigma-aldrich Co. (St. Louis, Missouri, USA).
2.2. Analysis of total phenolics
The phenolics were extracted twice with 5 mL of extraction solvent (methanol:acetate acid: H2O at 50:3.7:46.3) from ~100 mg of each dry vegetable powder. The first extraction was carried out with rocking for 5 h and the second was run overnight at 4°C. The total phenolic content of the extracts were assessed with the Folin-Ciocalteu reagent according to the modification for plant materials developed by Singleton et al. [14]. Briefly, the extracts were mixed with the Folin-Ciocalteu phenol reagent, Na2CO3 and H2O and absorbance at 760 nm measured 2 h after incubation at room temperature using a Bekman DU 530 spectrophotometer (USA). The total phenolic content was determined from lyophilized material with the results presented on a dry weight basis and are expressed as μmol Gallic Acid Equivalents (GAE/g).
2.3. Determination of Vitamin C
Ascorbic acid and total vitamin C (ascorbic acid plus dehydroascorbic acid) were determined by high-performance liquid chromagraphy (HPLC) using a modification in the procedure of Sanchez-Mata et al. [15]. Two grams of freeze dried vegetables were homogenized with 10 mL of an extraction solution (3% metaphosphoric acid plus 8% acetic acid). The resulting mixture was centrifuged, filtered, and adjusted to 25 mL with distilled water. Filtered sample through a 0.45-μm membrane filter was analyzed by HPLC. Waters 2690 alliance separation module and a symmetry C18 stainless steel column (250×4.6 i.d. mm) was used for analysis. The solvent system used was an isocratic gradient of a 0.01% solution of H2SO4, adjusted to pH 2.5–2.6. The flow rate was fixed at 1.0 mL/min. A Waters 996 UV-visible photodiode array detector was set at 245 nm. Chromatographic data and UV-visible spectra were collected, stored, and integrated using Waters Empower. Identification of the ascorbic acid was carried out by HPLC by comparing the retention time and UV-visible absorption spectrum with those of the standard. The ascorbic acid content was calculated on the basis of a calibration curve and expressed as mg/gram of sample on dry weight basis.
2.4. Preparation of methanol extracts
Methanol extracts of A. keiskei, B. oleracea, and O. javanica (Pulmuone Co., Korea), were prepared as reported before [6–8]. Fresh vegetables were cleaned and freeze dried. The powdered sample (2 g) was extracted twice with 100 mL methanol at room temperature(~23°C) for 12 h. Extracts were recovered by centrifugation at 1,000 ×g for 10 minutes and filtered through whatman No. 2 filter paper. Filtered extracts were concentrated to dryness under nitrogen gas and stored at −20 °C until they were utilized in the following assays.
2.5. Salmonella mutagenicity assays
Methanol extracts were compared for their inhibitory activities in the Salmonella mutagenicity assay [16]. The basic methodology, using heterocyclic amine mutagens in the presence and absence of S9, was as reported elsewhere [11–13]. In brief, 50 μL of mutagen (or DMSO vehicle alone), 0.1 mL of an overnight culture of Salmonella typhimurium strain TA98, 50 μL of plant extract (0.1–2 mg/plate equivalent starting material), and 0.2 mL phosphate-buffered saline or 0.2 mL of 10% Aroclor-induced rat liver S9 mix were preincubated for 30 minutes at 37°C. Following the addition of 2 mL top agar, the mixture was poured onto minimal glucose plates, and His+ revertant colonies were counted 2 days later using a Sorcerer Colony Counter (Perceptive Instruments, Haverhill, England). To rule out possible toxic and/or other confounding effects, we routinely confirmed the presence of a normal background lawn of growth, spontaneous counts in the usual range, and high cell viability. No toxicity was detected for data reported here. For purposes of comparison among the different plant extracts and mutagens, data on His+ revertants/plate were expressed as ‘percent of control’.
2.6. CYP1A1 and CYP1A2 activity
Cytochrome P4501A1 (CYP1A1) and CYP1A2 activities were determined in ethoxyresorufin-O-deethylase (EROD) and methoxyresorufin-O-demethylase (MROD) assays, respectively [11]. Conversion of substrate (7-ethoxyresorufin or methoxyresorufin) to product (resorufin) was measured at 37 °C in a Hitachi F2500 fluorescence spectrophotometer, with excitation at 530 nm and emission at 585 nm. Assays were performed in duplicate, with various concentrations of plant extract. Controls also were included to verify that changes produced by plant extracts were not from simple quenching of fluorescence.
2.7. ORAC and FRAP assays
The oxygen radical absorbance capacity (ORAC) assay measures the relative efficiency of antioxidants to protect the fluorescent protein β-phycoerythrin (PE) against peroxidation [17]. Peroxyl radicals generated at a constant rate using the thermolabile azo-initiator ABAP (2,2′-azobis 2-methyl-propionamidine dihydrochloride) were related to the water-soluble vitamin E analog Trolox (6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid). The ORAC assay was carried out in a microplate fluorometer Cytofluor 400 (PerSeptive Biosystems, Framingham, MA), with Trolox diluted to a final concentration of 0, 10, 20 and 40μM, in triplicate. The kinetic change in PE fluorescence was recorded at 2 minute intervals for 2 hours (excitation 485 nm, emission 585 nm).
The ferric reducing/antioxidant power (FRAP) assay determines the ability of extracts to reduce Fe3+/tripyridyltriazine complex (Fe3+–TPTZ) to its colored form, Fe2+/TPTZ [17,18]. In brief, test sample or standard (Trolox, final concentrations of 0, 62.5, 125, 250 and 500 μM were added to the wells of a 96-well plate in duplicate, followed by addition of pre-warmed Fe3+–TPTZ reagent. After 15 minutes incubation at 37°C, absorbance was read at 550 nm and the FRAP value was calculated from the standard calibration curve.
2.8. Plasmid DNA nicking
Conversion of double-stranded supercoiled circular DNA to relaxed or linear ‘open’ DNA was determined based on the methodology previously described [20]. Thus, plasmid pUC19 was incubated with 0.1 mM H2O2 and 0.1 mM Fe2+ in 150 mM NaCl, pH 7.9, for 30 minutes at 37°C, in the presence and absence of plant extract. Following electrophoresis in 0.8% agarose gel at 100 V for 1 hour and ethidium bromide staining, closed circular, linear, and relaxed forms of pUC19 were quantified on an AlphaInnotech photodocumentation system.
2.9. Comet assay
Antigenotoxic activity of the three plant extracts was studied using the single cell gel electrophoresis (‘comet’) assay, a sensitive technique that has been applied to studies of DNA damage and its prevention by antioxidants in people, as well as in human colon cancer cells in vitro [21–28]. In the present investigation, HCT116 human colon cancer cells from the American Type Culture Collection (Manassas, VA) were grown in McCoy’s 5A medium supplemented with 10% fetal calf serum and 1% penicillin/streptomycin, at 37°C in a 95% humidified incubator containing 5% CO2. Confluent cells were trypsinized, re-seeded at a density of 1 × 105 cells in 24-well plates, and cultured for another 48 hours at 37°C. Cells were pre-incubated with or without each plant extract and 0.1 mM H2O2 on ice for 30 minutes, they were washed in phosphate-buffered saline (PBS), trypsinized, and suspended in 1% low melting point agarose. Following slide preparation and exposure to ice-cold lysis solution (2.5 M NaCl, 100 mM disodium EDTA, 10 mM Tris, 10% DMSO, and 1% Triton X-100) for 1 hour, slides were placed in a submarine gel electrophoresis unit containing 300 mM NaOH and 1 mM EDTA, pH 13. Forty 40 minutes later, electrophoresis was performed at 20 V (300 mA) for 20 minutes, and slides were then immersed in neutralizing buffer (0.4 M Tris–HCl, pH 7.5). After staining with ethidium bromide, 50 cells/slide (3 slides/treatment) were scored using an inverted fluorescence microscope (Olympus IX-70A). Data were expressed relative to positive controls treated with H2O2, which were assigned an arbitrary value of 1.0.
2.10. Statistical analysis
Values were expressed as mean ± standard deviation and analyzed statistically using SPSS statistical package for WINDOWS (version 10.0; SPSS Inc, Chicago). All data were obtained from three separated experiments. P values of <0.05 were considered statistically significant.
3. Results
3.1. Antimutagenic activity
Initially, extracts of A. keiskei, O. javanica and B. oleracea were tested for antimutagenic activity against the heterocyclic amines IQ, MeIQx, and PhIP in the presence of an exogenous metabolic activation system (Table 1). No signs of toxicity were observed under the conditions reported here. A. keiskei produced dose-dependent inhibition against all three mutagens, whereas O. javanica showed a more complex pattern, with slight, non-statistically significant enhancement against two of the mutagens (MeIQx and PhIP) at low concentrations in the assay but antimutagenic activity against all three mutagens at the highest concentration of 2 mg/plate. B. oleracea was the least effective of the three plant extracts, and indeed at one or more concentrations in the assay tended to enhance mutagenicity. When ranked in terms of their potencies against three heterocyclic amines in the presence of S9, the relative order of antimutagenic activity was as follows: A. keiskei > O. javanica ≫ B. oleracea.
Table 1.
Antimutagenic activities of Angelica keiskei, Oenanthe javanica and Brassica oleracea extracts against three heterocyclic amines
| His+ revertants/plate
|
|||
|---|---|---|---|
| IQa | MeIQxb | PhIPc | |
| A. keiskei | |||
| Control (−)d | 50.33 ± 5.03 | 38.00 ± 7.94 | 50.33 ± 5.03 |
| Control (+)e | 1962.00 ± 139.15 | 966.67 ± 57.00 | 1663.00 ± 20.78 |
| 0.1 mg/plate | 1858.67 ± 105.55 | 960.67 ± 52.55 | 1447.00 ± 10.20** |
| 1.0 mg/plate | 1558.00 ± 54.37** | 692.67 ± 48.00** | 1321.00 ± 11.30** |
| 2.0 mg/plate | 1090.33 ± 164.27** | 391.67 ± 58.94** | 747.67 ± 61.52** |
| O. javanica | |||
| Control (−) | 34.33 ± 5.51 | 45.67 ± 3.21 | 30.00 ± 14.73 |
| control (+) | 1078.00 ± 171.76 | 766.00 ± 75.45 | 1376.67 ± 123.14 |
| 0.1 mg/plate | 1067.00 ± 77.49 | 737.33 ± 17.62 | 1422.33 ± 166.17 |
| 0.5 mg/plate | 725.00 ± 101.82* | 828.00 ± 59.09 | 1587.67 ± 55.23 |
| 1.0 mg/plate | 1006.67 ± 87.67 | 695.00 ± 48.66 | 1303.00 ± 138.39 |
| 2.0 mg/plate | 604.67 ± 183.47** | 541.67 ± 45.49** | 1111.00 ± 137.02* |
| B. oleracea | |||
| control (−) | 34.33 ± 5.51 | 45.67 ± 3.21 | 30.00 ± 14.73 |
| control (+) | 1078.00 ± 171.76 | 766.00 ± 75.45 | 1376.67 ± 123.14 |
| 0.1 mg/plate | 1234.33 ± 240.56 | 638.33 ± 68.24 | 1372.00 ± 57.86 |
| 0.5 mg/plate | 1173.67 ± 231.23 | 880.00 ± 184.47 | 1468.67 ± 166.43 |
| 1.0 mg/plate | 1149.33 ± 96.93 | 695.33 ± 43.25 | 1513.33 ± 82.44 |
| 2.0 mg/plate | 818.33 ± 76.16 | 677.33 ± 92.50 | 1577.00 ± 299.60 |
2-amino-3-methylimidazo[4,5-f]quinoline.
2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline.
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine.
Negative control; DMSO vehicle alone group.
Positive control; mutagens treated group.
P < 0.01 relative to positive control group.
P < 0.05 relative to positive control group.
Three direct-acting mutagens were next examined in the absence of S9, namely, N-OH-IQ, N-OH-PhIP, and NPD (Table 2). A. keiskei at the highest concentration of 2 mg/plate inhibited N-OH-IQ but had no statistically significant effect against N-OH-PhIP or NPD. No antimutagenic activity was seen with O. javanica or B. oleracea against the three direct-acting mutagens, and indeed the former extract produced a slight enhancement of N-OH-PhIP mutagenicity and the latter augmented the mutagenicity of N-OH-IQ. These data suggested no consistent antimutagenic (or co-mutagenic) effects for the plant extracts against direct-acting mutagens under the assay conditions used here.
Table 2.
Antimutagenic activities of Angelica keiskei, Oenanthe javanica and Brassica oleracea extracts against two directing-acting heterocyclic amines and mutagens
| His+ revertants/plate
|
|||
|---|---|---|---|
| NPDa | N-OH-IQb | N-OH-PhiPc | |
| A. keiskei | |||
| Control (−)d | 28.33 ± 7.02 | 28.33 ± 7.02 | 14.00 ± 5.29 |
| Control (+)e | 3086.00 ± 161.11 | 2378.00 ± 219.97 | 1114.67 ± 113.59 |
| 0.1 mg/plate | 2655.33 ± 95.00 | 1791.00 ± 292.74* | 1047.67 ± 68.49 |
| 1.0 mg/plate | 2568.00 ± 174.11 | 1654.00 ± 257.03** | 1014.67 ± 201.53 |
| 2.0 mg/plate | 2192.00 ± 216.52* | 691.67 ± 72.61** | 1050.00 ± 148.92 |
| O. javanica | |||
| Control (−) | 77.00 ± 30.64 | 16.00 ± 7.00 | 22.67 ± 6.35 |
| Control (+) | 2479.67 ± 161.92 | 2553.33 ± 226.62 | 1338.00 ± 53.84 |
| 0.1 mg/plate | 2206.67 ± 260.16** | 2514.33 ± 90.78 | 1106.33 ± 150.98 |
| 0.5 mg/plate | 1973.67 ± 139.33** | 2620.67 ± 131.07 | 1181.33 ± 40.27 |
| 1.0 mg/plate | 2016.67 ± 115.79** | 2514.33 ± 90.78 | 1370.00 ± 150.44 |
| 2.0 mg/plate | 2062.67 ± 44.66** | 2826.67 ± 543.57 | 1676.33 ± 477.63 |
| B. oleracea | |||
| Control (−) | 77.00 ± 30.64 | 16.00 ± 7.00 | 22.67 ± 6.35 |
| Control (+) | 2479.67 ± 161.92 | 2553.33 ± 226.62 | 1338.00 ± 53.84 |
| 0.1 mg/plate | 2062.67 ± 44.66 | 3476.33 ± 158.47** | 1262.33 ± 89.95 |
| 0.5 mg/plate | 2016.67 ± 115.79 | 3383.33 ± 204.59** | 1204.67 ± 26.35* |
| 1.0 mg/plate | 1973.67 ± 139.33 | 4101.67 ± 338.92** | 1280.00 ± 41.76 |
| 2.0 mg/plate | 2206.67 ± 260.16 | 5128.33 ± 125.01** | 1488.67 ± 85.64* |
4-nitro-1,2-phenylenediamine (NPD).
2-hydroxyamino-3-methylimidazo[4,5-f]quinoline.
2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine.
Negative control; DMSO vehicle alone group.
Positive control; mutagens treated group.
P < 0.01 relative to positive control group.
P < 0.05 relative to positive control group.
3.2. Inhibition of cytochrome P450 activity
Heterocyclic amines such as IQ and PhIP are metabolically activated by CYP1A2, generating N-OH-IQ and N-OH-PhIP, whereas CYP1A1 catalyzes both N-hydroxylation and ring hydroxylation reactions. The activities of CYP1A1 and CYP1A2 were followed in EROD and MROD assays, respectively (Figs 1, 2); A. keiskei and O. javanica produced concentration dependent inhibition in both assays, whereas B. oleracea was the least effective of the three plant extracts tested. At the highest concentration in the assay, inhibition of both EROD and MROD activities was in the relative order A. keiskei > O. javanica ≫ B. oleracea, which agreed favorably with the order of antimutagenic activity against IQ, MeIQx, and PhIP in the Salmonella assay (Table 1).
Fig. 1.
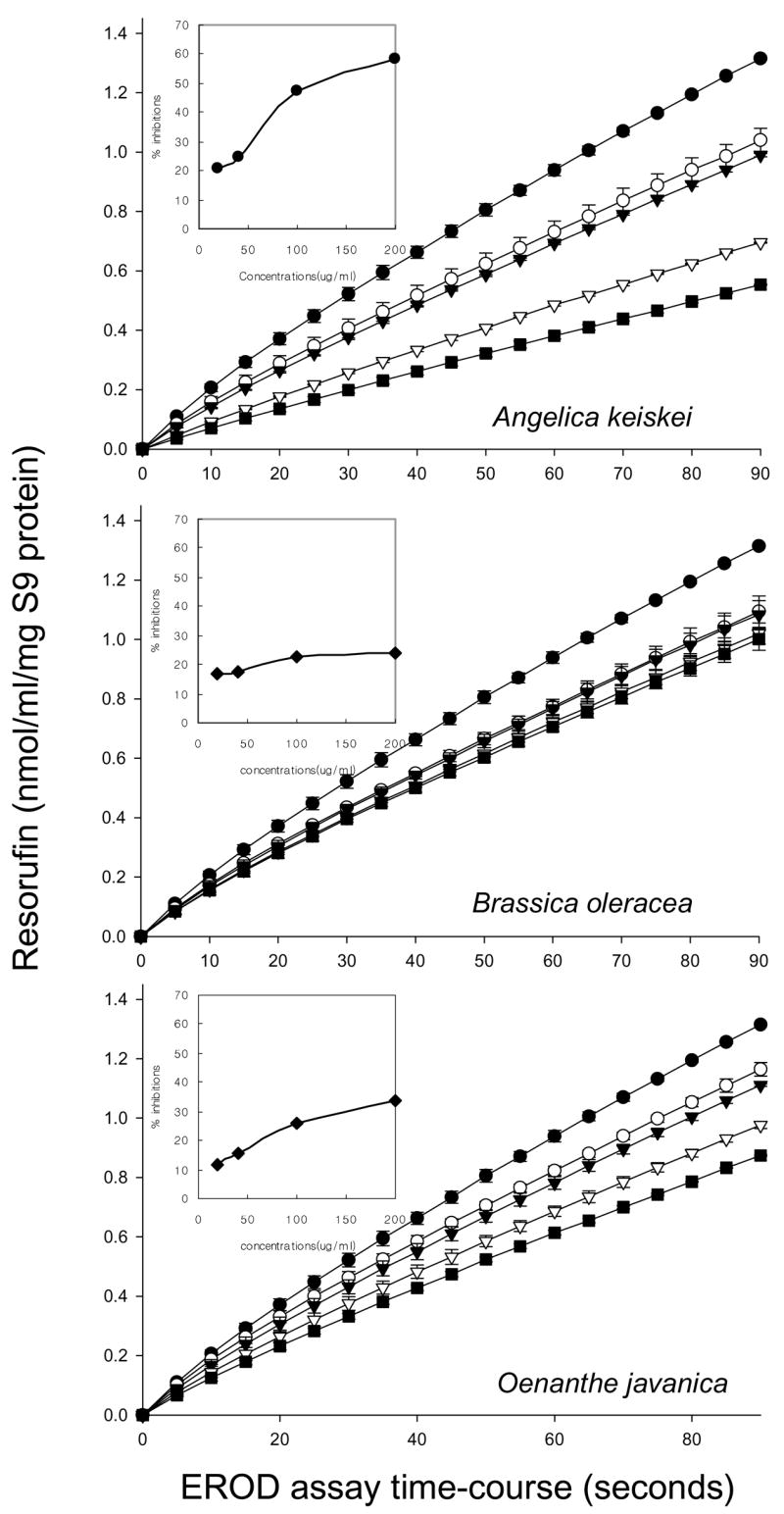
Inhibition of 7-ethoxyresorufin O-deethylase (EROD) activities by Angelica keiskei, Oenanthe javanica and Brassica oleracea extracts. Inset: Percent inhibition vs. concentration of each vegetable extracts. Percent inhibition was calculated from the initial slopes, in which each vegetable extracts addition was compared with the control. Data points and error bars indicate mean ± S.D. (n = 3). Symbols: ● control, ○ 25 μg/mL, ▼ 50 μg/mL. ▽ 100 μg/mL, ■ 200 μg/mL.
Fig. 2.
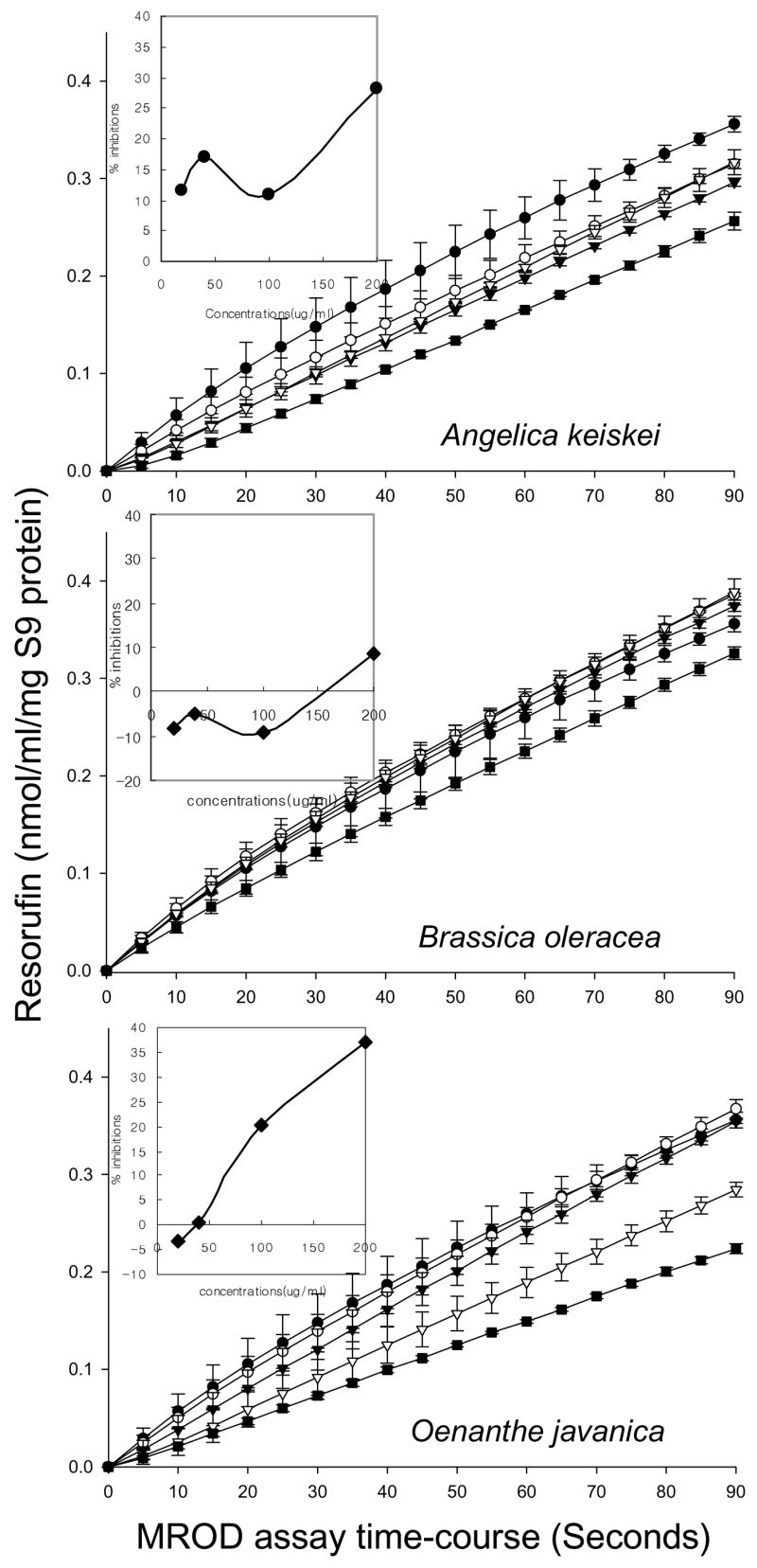
Inhibition of methoxyresorufin O-demethylase (MROD) activities by Angelica keiskei, Oenanthe javanica and Brassica oleracea extracts. Inset: Percent inhibition vs. concentration of each vegetable extracts. Percent inhibition was calculated from the initial slopes, in which each vegetable extracts addition was compared with the control. Data points and error bars indicate mean ± S.D. (n = 3). Symbols: ● control, ○ 25 μg/mL, ▼ 50 μg/mL. ▽ 100 μg/mL, ■ 200 μg/mL.
3.3. Activity of green vegetable extracts in FRAP and ORAC assays
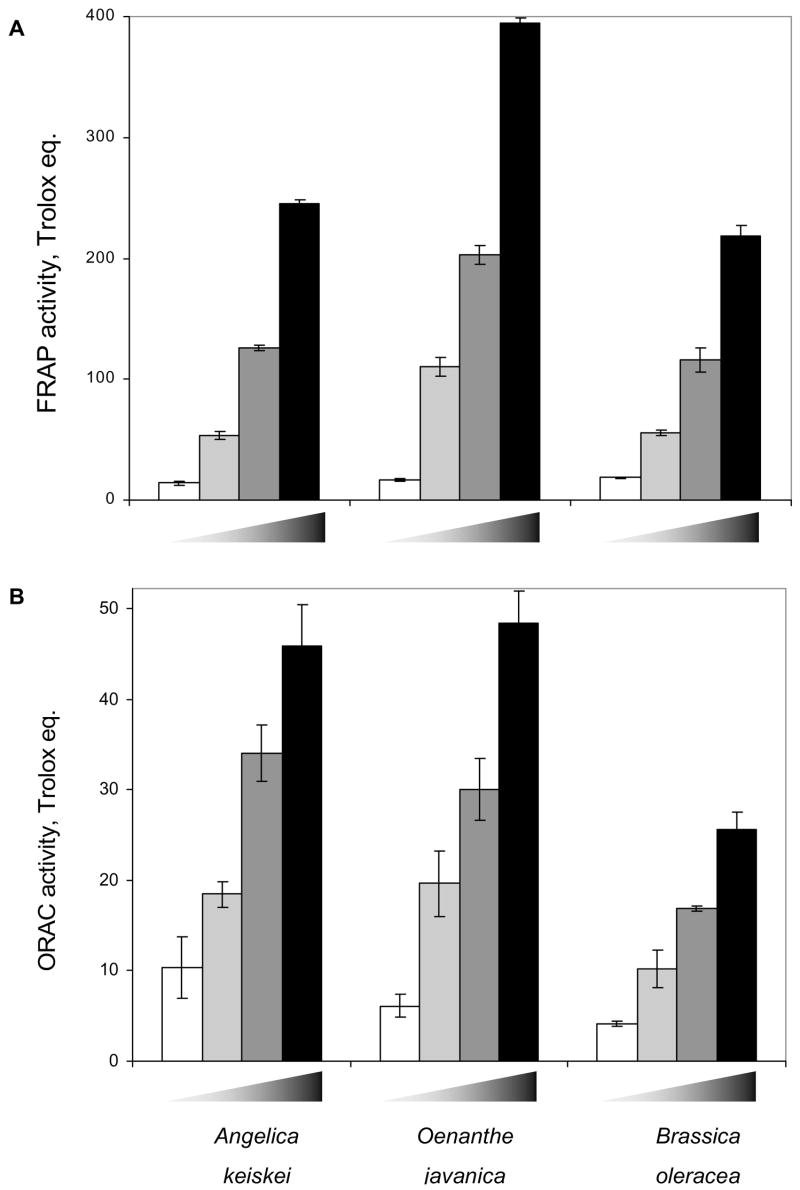
Although the vegetable extracts lacked any consistent electrophile-scavenging activity against the direct-acting mutagens N-OH-IQ, N-OH-PhIP and NPD in the Salmonella assay (Table 2), further tests were undertaken for possible antioxidant properties of three vegetable extracts. Total phenolics and vitamin C concentration were determined. O. javanica had highest phenolic concentrations than A. keiskei and B. oleracea. However, vitamin C contents were not significantly different among three samples (Table 3). Antioxidant properties, as defined by their activities in FRAP and ORAC assays were determined (Fig. 3). All three vegetable extracts exhibited concentration-dependent ferric reducing/antioxidant power (Fig. 3A), and oxygen radical absorbance capacity (Fig. 3B). In conformance with the total phenolics, the relative order of antioxidant activities in these assays was as follows: O. javanica >A. keiskei > B. oleracea.
Table 3.
Total phenolics and total ascorbic acid concentrations of Angelica keiskei, Oenanthe javanica and Brassica oleracea
| Moistures(%)a | Total phenolics (umolGAE/g)b | Ascorbic acid(mg/g)c | |
|---|---|---|---|
| A. keiskei | 92.08 ± 0.43 | 675.00 ± 17.68 | 13.00 ± 0.06 |
| O. javanica | 94.42 ± 0.40 | 1070.00 ± 81.32 | 12.37 ± 2.62 |
| B. oleracea | 92.76 ± 0.46 | 510.00 ± 10.61 | 11.48 ± 0.13 |
Moisture, lipids and protein content were determined using the standard AOAC methods 7(33).
Total phenolics are expressed as μmol Gallic Acid Equivalents (GAE/g) based on dry weights.
Ascorbic acid contents are based on dry weights.
Fig. 3.
Antioxidant capacity of A. keiskei, O. javanica and B. oleracea extracts. A ferric reducing/antioxidant power (FRAP) and B oxygen radical absorbance capacity (ORAC) was determined relative to Trolox, a water-soluble vitamin E analog. Results are given as mean ± SD. Wedge symbol: increasing concentration of each extract (1, 5, 10, 20 μg/mL).
3.4. Inhibition of DNA nicking
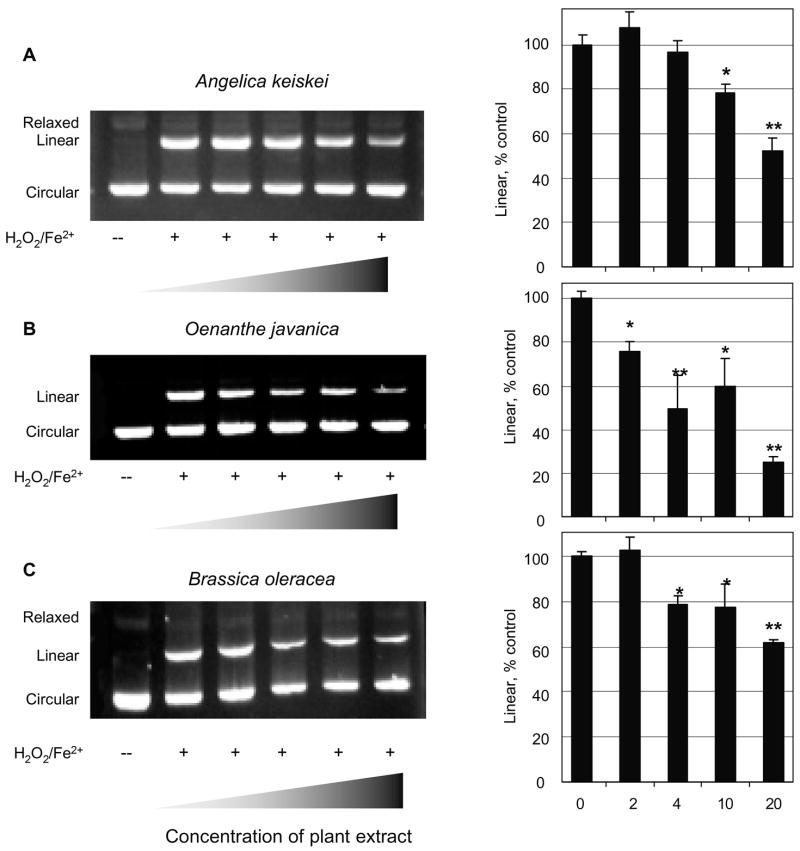
The closed circular form of plasmid pUC19 was converted efficiently to the nicked linear form after treatment with Fe2+/H2O2, and in some experiments the relaxed form also was detected as a minor band (Fig. 4). Each of the plant extracts attenuated the formation of nicked DNA in vitro. For example, when the linear form was normalized to the corresponding control treated with Fe2+/H2O2 but no plant extract, A. keiskei inhibited DNA nicking by ~50% at 20 μg/mL (bar chart, Fig. 4A), O. javanica inhibited by p>70% (Fig. 4B) and B. oleracea inhibited by 40% (Fig. 4C). Inhibition of DNA nicking corresponded favorably with the relative order of activity in the FRAP and ORAC assays (Fig. 3).
Fig. 4.
Inhibition of DNA nicking by A. keiskei, O. javanica and B. oleracea. pUC19 was incubated in the absence (−) or presence (+) of H 2O2/Fe2+ in order to convert closed circular plasmid to nicked linear and/or relaxed forms. The linear form of DNA was quantified relative to controls treated with H 2O2/Fe2+ but no plant extract, and expressed as ‘percent control’ to facilitate comparison among groups. Wedge symbol: increasing concentration of each extract (0, 2, 4, 10, 20 μg/mL). *P < 0.05 and **P < 0.01 by ANOVA.
3.5. Antigenotoxic effects in human colon cancer cells
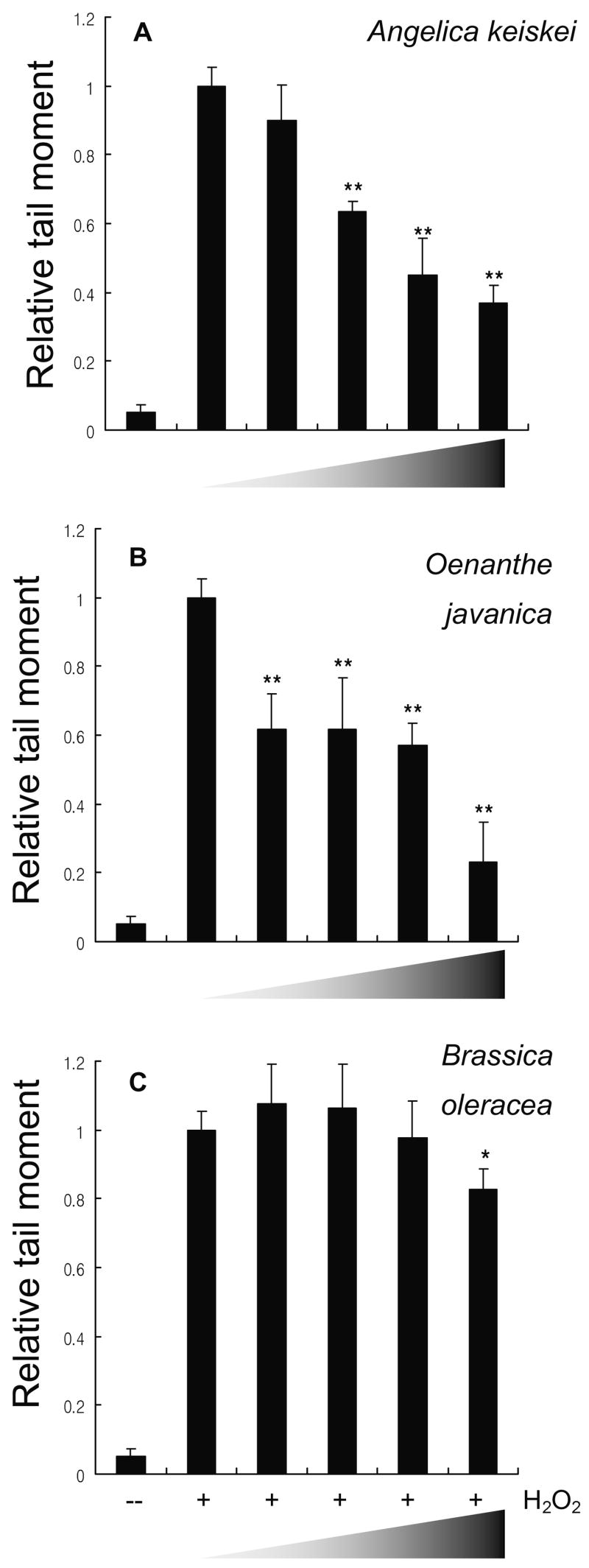
Finally, since plant extracts inhibited DNA nicking induced by Fe2+/H2O2 in vitro, we treated HCT116 human colon cancer cells with H2O2 and examined the genotoxic damage using the ‘comet’ assay (Fig. 5). The median tail moment in controls treated with H2O2 alone (no vegetable extract) was assigned an arbitrary value of 1.0 in order to compare among the three treatment groups; extracts of A. keiskei (Fig. 5A) and O. javanica (Fig. 5B) produced significant, dose-dependent protection, whereas B. oleracea was effective only at the highest concentration tested of 20 μg/mL (Fig. 5C). Little damage was detected in untreated cells.
Fig. 5.
Antigenotoxic effects of A. keiskei, O. javanica and B. oleracea in HCT116 human colon cancer cells. Cells were incubated in the absence (−) or presence (+) of 0.1 mM H2O2 and DNA damage was quantified via the comet assay. Median tail moment in positive controls treated with H2O2 but no plant extract was assigned an arbitrary value of ‘1.0’ in order to facilitate comparison among treatment groups. Wedge symbol: increasing concentration of each extract (0, 1, 5, 10, 20 μg/mL) *P < 0.05 and **P < 0.01 by ANOVA.
4. Discussion
The present investigation has demonstrated that chlorophyll-rich methanol extracts of A. keiskei, O. javanica and B. oleracea exhibited antimutagenic and antioxidant activities in vitro. These three vegetable species were chosen for study because of their widespread use in popular health drinks, such as Korean green juice, and because they are known to contain high levels of antioxidants and phytochemicals, such as chlorophyll, β-carotene, vitamin E, vitamin C, Se and Zn [6–8,29]. Under the present conditions, A. keiskei was most effective against the three indirect-acting heterocyclic amines IQ, MeIQx and PhIP in the Salmonella assay, as well as being most inhibitory in the EROD and MROD assays, but had no effect against direct-acting mutagens in the absence of S9. These findings imply that one or more constituents in A. keiskei interfered with the enzymes which metabolically active heterocyclic amines, without affecting the direct-acting mutagens, such as by electrophile-scavenging [10]. Although O. javanica and B. oleracea were less effective than A. keiskei under the present conditions, we cannot exclude the possibility that other cultivars of these three plants might exhibit greater inhibitory activity.
Indeed, in prior studies [6–8], extract of kale (B. oleracea) was more effective than A. keiskei in preventing the oxidation of linoleic acid during storage, which is indicative of antioxidant activity, as well as exhibiting higher nitrite scavenging in vitro. We confirmed the high antioxidant activity of all three plant extracts in ORAC and FRAP assays, and demonstrated inhibition of H2O2-induced DNA damage, assessed directly using pUC19 in vitro and indirectly using the comet assay in HCT116 colon cancer cells. In contrast to the results obtained in the Salmonella assays, O. javanica was generally the most potent of the three plant extracts in the various assays used here in order to assess antioxidant properties.
Although, as mentioned above, different cultivars are likely to exhibit different inhibitory potencies in the various assays, in the present investigation A. keiskei was most effective against heterocyclic amine mutagens, whereas O. javanica had the highest antioxidant potential. These findings suggest that, indeed, a ‘cocktail’ of plant extracts is likely to prove more beneficial than an extract containing a single plant species, since this would likely provide for complementary protective mechanisms. This is one rationale used for health drinks containing a cocktail of plant species, such Korean green juice, which has high levels of phytochemicals with demonstrated cancer inhibitory activity, such as chlorophylls, indoles and isothiocyanates [1–4,20–32]. Future studies should examine the possible synergistic protective effects of A. keiskei, O. javanica and B. oleracea in combination, and assess the optimal ‘blend’ of these three plant extracts with respect to antimutagenic versus antioxidant effects.
Acknowledgments
We thank Dr. Tian-Wei Yu of the Cancer Chemoprotection Laboratory, Linus Pauling Institute, for invaluable assistance with the various short-term assays. Our thanks also go to Pulmuone Co.Ltd for generous donation of A. keiskei, O. javanica and B. oleracea and supporting the part of personnel expenses involved in the present study. These studies were supported in part by NIH grants CA65525, CA90890, and CA80176.
References
- 1.Donaldson MS. Nutrition and cancer: A review of the evidence for an anti-cancer diet. Nutr J. 2004;20:390–396. doi: 10.1186/1475-2891-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Vecchia C. Mediterranean diet and cancer. Public Health Nutr. 2004;7:965–968. doi: 10.1079/phn2004562. [DOI] [PubMed] [Google Scholar]
- 3.He K, Hu FB, Colditz GA, Manson JE, Willett WC, Liu S. Changes in intake of fruits and vegetables in relation to risk of obesity and weight gain among middle-aged women. Int J Obes Relat Metab Disord. 2004;28:1569–1574. doi: 10.1038/sj.ijo.0802795. [DOI] [PubMed] [Google Scholar]
- 4.Smith-Warner SA, Elmer PJ, Fosdick L, Randall B, Bostick RM, Grandits G, Grambsch P, Louis TA, Wood JR, Potter JD. Fruits, vegetables, and adenomatous polyps: the Minnesota Cancer Prevention Research Unit case-control study. Am J Epidemiol. 2002;155:1104–1113. doi: 10.1093/aje/155.12.1104. [DOI] [PubMed] [Google Scholar]
- 5.Link LB, Potter JD. Raw versus cooked vegetables and cancer risk. Cancer Epidemiol Biomarkers Prev. 2004;13:1422–1435. [PubMed] [Google Scholar]
- 6.Chung SY, Cho HS, Ryu JW, Yoon S. Antioxidant nutrients and related biological activities of green yellow vegetable juices. J Korean Assoc Cancer Prev. 1999;4:136–142. [Google Scholar]
- 7.Kim JS, Kim HY, Park YK. The effects of green vegetable juice (Angelica keiskei) supplementation on plasma lipids and antioxidant status in smokers. Korean J Nutr. 2003;6:933–941. [Google Scholar]
- 8.Chung SY, Kim NK, Yoon S. Nitrite scavenging effect of methanol fraction obtained from green yellow vegetable juices. J Korean Soc Food Sci Nutr. 1999;8:342–347. [Google Scholar]
- 9.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dashwood RH. Modulation of heterocyclic amine-induced mutagenicity and carcinogenicity: an A-to-Z guide to chemopreventive agents, promoters, and transgenic models. Mutat Res. 2002;511:89–112. doi: 10.1016/s1383-5742(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 11.Yu TW, Xu M, Dashwood RH. Antimutagenic activity of spearmint. Environ Mol Mutagen. 2004;44:387–393. doi: 10.1002/em.20063. [DOI] [PubMed] [Google Scholar]
- 12.Yu Z, Xu M, Santana-Rios G, Shen R, Izquierdo-Pulido M, Williams DE, Dashwood RH. A comparison of whole wheat, refined wheat and wheat bran as inhibitors of heterocyclic amines in the Salmonella mutagenicity assay and in the rat colonic aberrant crypt focus assay. Food Chem Toxicol. 2001;39:655–665. doi: 10.1016/s0278-6915(01)00012-6. [DOI] [PubMed] [Google Scholar]
- 13.Santana-Rios G, Orner GA, Amantana A, Provost C, Wu SY, Dashwood RH. Potent antimutagenic activity of white tea in comparison with green tea in the Salmonella assay. Mutat Res. 2001;495:61–74. doi: 10.1016/s1383-5718(01)00200-5. [DOI] [PubMed] [Google Scholar]
- 14.Singleton VL, Orthofer R, Lamuela-Raventós RM. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Meth Enzymol. 1999;299:152–178. [Google Scholar]
- 15.Sanchez-Mata MC, Camara-Hurtado M, Diez-Marques C, Torija-Isasa ME. Comparison of high-performance liquid chromatography and spectrofluorimetry for vitamin C analysis of green beans (Phaseolus vulgaris L.) Eur Food Res Technol. 2000;210:220–225. [Google Scholar]
- 16.Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 17.Cao GH, Prior RL. Measurement of oxygen radical absorbance capacity in biological samples. Methods Enzymol. 1999;299:50–62. doi: 10.1016/s0076-6879(99)99008-0. [DOI] [PubMed] [Google Scholar]
- 18.Benzie IF, Strain JJ. Ferric reducing antioxidant power assay: direct measure of total antioxidant activity of biological fluids and modified version for simultaneous measurement of total antioxidant power and ascorbic acid concentration. Methods Enzymol. 1999;299:15–27. doi: 10.1016/s0076-6879(99)99005-5. [DOI] [PubMed] [Google Scholar]
- 19.Benzie IF, Szeto YT. Total antioxidant capacity of teas by the ferric reducing/antioxidant power assay. J Agric Food Chem. 1999;47:633–636. doi: 10.1021/jf9807768. [DOI] [PubMed] [Google Scholar]
- 20.Liang R, Senturker S, Shi X, Bal W, Dizdaroglu M, Kasprzak KS. Effects of Ni(II) and Cu(II) on DNA interaction with the N-terminal sequence of human protamine P2: enhancement of binding and mediation of oxidative DNA strand scission and base damage. Carcinogenesis. 1999;20:893–898. doi: 10.1093/carcin/20.5.893. [DOI] [PubMed] [Google Scholar]
- 21.Mastaloudis A, Yu TW, O’Donnell RP, Frei B, Dashwood RH, Traber MG. Endurance exercise results in DNA damage as detected by the comet assay. Free Radic Biol Med. 2004;36:966–975. doi: 10.1016/j.freeradbiomed.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Osswald K, Becker TW, Grimm M, Jahreis G, Pool-Zobel BL. Inter- and intra-individual variation of faecal water: genotoxicity in human colon cells. Mutat Res. 2000;472:59–70. doi: 10.1016/s1383-5718(00)00113-3. [DOI] [PubMed] [Google Scholar]
- 23.Pool-Zobel BL, Adlercreutz H, Glei M, Liegibel UM, Roser S. Isoflavanoids and lignans have different potentials to modulate oxidative genetic damage in human colon cells. Carcinogenesis. 2000;21:1247–1252. [PubMed] [Google Scholar]
- 24.Pool-Zobel BL, Bub A, Muller H, Wollowski I, Rechkemmer G. Consumption of vegetables reduces genetic damage in humans: first results of an intervention trial with carotenoid-rich foods. Carcinogenesis. 1997;18:1847–1850. doi: 10.1093/carcin/18.9.1847. [DOI] [PubMed] [Google Scholar]
- 25.Pool-Zobel BL, Bub A, Schroder N, Rechkemmer G. Anthocyanins are potent antioxidants in vitro but do not reduce oxidative DNA damage within human colon cells. Eur J Nutr. 1999;38:227–234. doi: 10.1007/s003940050065. [DOI] [PubMed] [Google Scholar]
- 26.Pool-Zobel BL, Leucht U. Induction of DNA damage in human colon cells derived from biopsies by suggested risk factors of colon cancer. Mutat Res. 1997;375:105–116. doi: 10.1016/s0027-5107(97)00006-7. [DOI] [PubMed] [Google Scholar]
- 27.Sherman MP, Aeberhard EE, Wong VZ, Simmons MS, Roth MD, Tashkin DP. Effects of smoking marijuana, tobacco or cocaine alone or in combination on DNA damage in human alveolar macrophages. Life Sci. 1995;56:2201–2207. doi: 10.1016/0024-3205(95)00208-n. [DOI] [PubMed] [Google Scholar]
- 28.Tachon P, Deflandre A, Giocomone PU. Modulation by L-histidine of H2O2-mediated damage of cellular and isolated DNA. Carcinogenesis. 1994;15:1621–1626. doi: 10.1093/carcin/15.8.1621. [DOI] [PubMed] [Google Scholar]
- 29.Chung SY, Kim HW, Yoon S. Analysis of antioxidant nutrients in green yellow vegetable juice. Korean J Food Sci Technol. 1999;31:880–886. [Google Scholar]
- 30.Dashwood RH. Chlorophylls as anticarcinogens. Int J Oncol. 1997;10:721–727. doi: 10.3892/ijo.10.4.721. [DOI] [PubMed] [Google Scholar]
- 31.Dashwood RH, Xu M. The disposition and metabolism of 2-amino-3-methylimidazo[4,5-f ]quinoline in the F344 rat at high versus low doses of indole-3-carbinol. Food Chem Toxicol. 2003;41:1185–1192. doi: 10.1016/s0278-6915(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 32.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 33.AOAC. Official Methods of Analysis. 15. Association of Official Analytical Chemists; Arlington, VA: 1990. [Google Scholar]