Abstract
The asexual blood stage of Plasmodium falciparum is comprised of morphologically distinct ring, trophozoite and schizont stages. Each of these developmental stages possesses a distinct pattern of gene expression. Regulation of P. falciparum gene expression is thought to occur, at least in part, at the promoter level. Previously, we have found that although the RNA of the P. falciparum hrp3 gene is only seen in ring-stage parasites, deletion of a specific sequensce in the 5’ end of the promoter region decreased ring-stage expression of hrp3 and enabled detection of its transcripts in trophozoite-stage parasites. In order to investigate this stage specific regulation of gene expression, we employed a series of nested deletions of the 1.7-kb hrp3 promoter. Firefly luciferase gene was used as a reporter to evaluate the role of promoter sequences in gene regulation. Using this approach, we identified a ring-stage specific regulatory region on the hrp3 promoter located between -1.7-kb and -1.1-kb from the ATG initiation codon. Small 100–150 bp truncations on this enhancer-like region failed to uncover discrete regulatory sequences, suggesting the multipartite nature of this element. The data presented in this study demonstrates that stage specific promoter activity of the hrp3 gene in P. falciparum blood stage parasites is supported, at least in-part, by a small promoter region that can function in the absence of a larger chromosomal context.
Keywords: Plasmodium, transcription, regulation, histidine-rich protein 3, stage specificity, expression
1. Introduction
Plasmodium falciparum infection remains a major disease burden in the developing world [1, 2]. Therefore, it is critically important to understand the details of regulation of gene expression during the parasite development. Infection in the human host results in a series of morphologically distinct stages that include ring, trophozoite, schizont and merozoite, during the parasite development within the mature erythrocyte. Transit through developmental stages is linked to strict regulation of distinct stage specific patterns of gene expression. Even housekeeping genes thought to be expressed throughout the cell cycle, exhibit complex patterns of developmental expression [3–7].
In the parasite, regulation of gene expression seems to occur at the transcriptional and post-transcriptional level. While transcriptome and proteome analyses indicate that some genes are regulated post-transcriptionally [8, 9], other genes are regulated at the level of transcription. For example, analyses of 5’ end upstream regions of the asexual and sexual stages have been shown by transfection studies that they function as promoters by supporting reporter gene expression [10–16]. Moreover, functional and bioinformatic studies of sexual and asexual Plasmodium promoters have uncovered putative cis-acting regulatory elements possibly involved in gene regulation [17–22]. In addition, it has been recently shown that the expression of parasite variant protein PfEMP1, is regulated at the level of transcription initiation [23]. Taken together, these results indicate the presence of transcriptional gene regulation in the parasite. Although, basal and stage specific cis-acting elements in the parasite promoters remain poorly described owing to the high A+T content of its genome.
During our studies on promoter recombination using hrp3 nested promoter deletions, we identified a region between -1.7-kb and -1.1-kb, from the ATG initiation codon, involved in ring stage specificity [24]. Unexpectedly, further promoter deletion switched the steady state accumulation of the reporter gene mRNA from ring to trophozoite stage of the parasite development. These results suggest the presence of stage specific cis-acting regulatory elements on the hrp3 promoter.
P. falciparum histidine-rich protein-2 and 3 (hrp2, 3) as well as the Knob associated histidine-rich protein (kahrp or hrp1) comprise the histidine-rich protein family. The members of this family are expressed at the early stage of the parasite development [25, 26]. We took advantage of the previously reported hrp3 promoter nested deletions [24] to further characterize the putative ring specific element. The hrp3 promoter region between -1.7-kb and -1.1-kb, from the ATG initiation codon, is involved in ring stage specific gene expression [24]. This region was placed upstream of the calmodulin (cam) promoter; it specifically increased the reporter activity in ring-stage parasites but had little effect on trophozoite parasites, suggesting that this enhancer-like region contains stage specific promoter elements. This presumptive element did not show homology to other Plasmodium promoter regions, suggesting that its function may be regulated through other mechanism rather than being sequence specific, such as transcription initiation or nuclear localization. Although, sequence-dependent regulation cannot be rule out. Here we identified in P. falciparum an enhancer-like region, carrying stage-specific hrp3 promoter elements. Our findings contribute to the understanding of the regulation of gene expression in the malaria parasite.
2. Experimental procedures
2.1. Plasmid constructs
The hrp3 promoter deletions were made using Erase-a-base® (Promega) and pHRPCAT [10] containing the hrp3 promoter driving the chloramphenicol acetyl transferase (CAT) gene. Kpn I-Dra III-digested pHRPCAT was incubated with exonuclease III (exo III) at predetermined times to obtain specific-sized promoters of 1.1-kb and 0.6-kb. The firefly luciferase (FFL) gene was amplified by PCR from the pVLH plasmid [15] using oligonucleotides 5’-GACATGCATGAAGACGCCAAAAACATAAAG-3’ and 5’-GACAAGCTTGCTTACAATTTGGACTTTCCG-3’, (boldfaced nucleotides represent Nsi I and Hind III restriction sites, respectively). The FFL was then used to replace the CAT gene in exoIII-digested pHRPCAT to generate pH1.7FL, pH1.1FL and pH0.6FL. Short hrp3 promoter deletions were generated by PCR using the specific oligonucleotides (Table S1, supplementary information).
All pcamSluc-derived constructs were engineered by first creating a Sal I site at the 5’ end of the cam promoter in the pcamGFP plasmid [27]. Secondly, the firefly luciferase gene substituted the GFP gene to generate pcamSluc. Subsequently, the hrp3 promoter region from -1.7-kb to -1.4-kb (fragment A) was amplified by PCR using the oligonucleotides HAF (5’-ACGCGTCGACCGCCCAATCATTATTTTATG-3’) and HAR (5’-ACGCGTCGACCCATAAAATATAAAAATAATTTG-3’). In addition, the region A was divided using PCR into fragment A1 from -1.7-kb to -1.1-kb with the oligonucleotides HAF and HA1R (5’-ACGCGTCGACCATTTATATTTATATTAAGAG-3’), and fragment A2 from -1.4-kb to -1.1-kb using the HA2F (5’-ACGCGTCGACGAATATATTCATAATTATAATATTG-3’) and HAR oligonucleotides, boldfaced nucleotides in all primers represent the Sal I restriction site. These fragments were placed into pcamSluc to generate pAhrpcamSluc, pA1hrpcamSluc and pA2hrpcamSluc. We then amplified the hrp3 promoter region from -1.1-kb to -0.6-kb (fragment B) by PCR using the oligonucleotides HBF (5’-ATCGTCGACTATGTATATGTATGTATTTTAAAATATAATAAAATG -3’) and HBR (5’-ATCGTCGACGTATGGATAGATTTTATTTTTAAAAAATAATAAATTTTATTATATTC-3’). Subsequently, fragment B was divided by PCR into fragment B1 -1.1-kb to -0.8-kb using HBF and HB1R (5’-ATCGTCGACATTATGAATATAAGAATATTCCATCTATCTTATG -3’) oligonucleotides; and fragment B2 from -0.8-kb to -0.6-kb using HB2F (5’-ATCGTCGACAATATTCAAAAAATAACAGATTTAAACCCTCAAAAATATAG -3’) and HBR. A Sal I restriction site (boldfaced nucleotides) was engineered at the 5’ and 3’ end on each PCR product. Fragments B, B1 and B2 were then inserted into Sal I-digested pcamSluc to generate pBhrpcamSluc, pB1hrpcamSluc and pB2hrpcamSluc respectively. A Renilla luciferase plasmid was constructed as reported by Militello and Wirth [28]. The Renilla reniformis luciferase gene was amplified by PCR using the plasmid pRL-null (Promega) as template and forward oligonucleotide 5’-TTAATGCATATGCTTCGAAAGTTTATGATC-3’ and reverse oligonucleotide 5’-TTCAAGCTTATTGTTCATTTTTGAGAACTCGC-3’ containing engineered restriction enzyme sites (Nsi I and Hind III, boldfaced nucleotides). Nsi I/Hind III-digested PCR fragment was used to replace the GFP gene in pcamGFP to generate pcamRLuc plasmid.
2.2. Parasite transfection
P. falciparum transient transfections were performed as described in Deitsch et al 2001. Experimental plasmids (100 µg per transfection) were combined with control plasmids, either 20 µg of CAT-containing plasmid (pcamCAT, [27]) or 10 µg of Renilla luciferase-containing plasmid (pcamRluc), and electroporated into the red blood cell as described previously [29]. Percoll-purified schizonts were added to DNA-loaded erythrocytes. After 12–14 hours post-invasion (hpi), half of the culture was taken (ring-parasites sample). The remaining half was kept in culture and processed after 32–36 hpi for the trophozoite sample.
2.4. Firefly, Renilla luciferase and CAT assays
Transfected parasites were first released from the red cells by 0.01% saponin lysis and released-parasites were lysed (Promega’s lysis buffer) 12–14 hpi (ring parasites) or 32–34 hpi (trophozoite parasites). The parasite extract was incubated at 60° C for 10 min., cleared by centrifugation and combined with [14C]chloramphenicol (at 0.05mCi/ml) and n-Butyryl-CoA for 3 hours to determine the CAT activity. After incubation, samples were mixed with xylene and the aqueous phase containing acetylated [14C]chloramphenicol was quantified in a liquid scintillation counter. CAT activity was used to normalize the firefly luciferase activity (Fig. 1 and 2B). The rest of the experiments were normalized using Renilla luciferase as a control. Parasites transfected with luciferase genes were quantified in a single tube Sirius luminometer (Berthold Detection Systems) as recommended by the manufacturer (Promega dual luciferase assay). Firefly and Renilla luciferase activities were reported as arbitrary light units.
Figure 1.
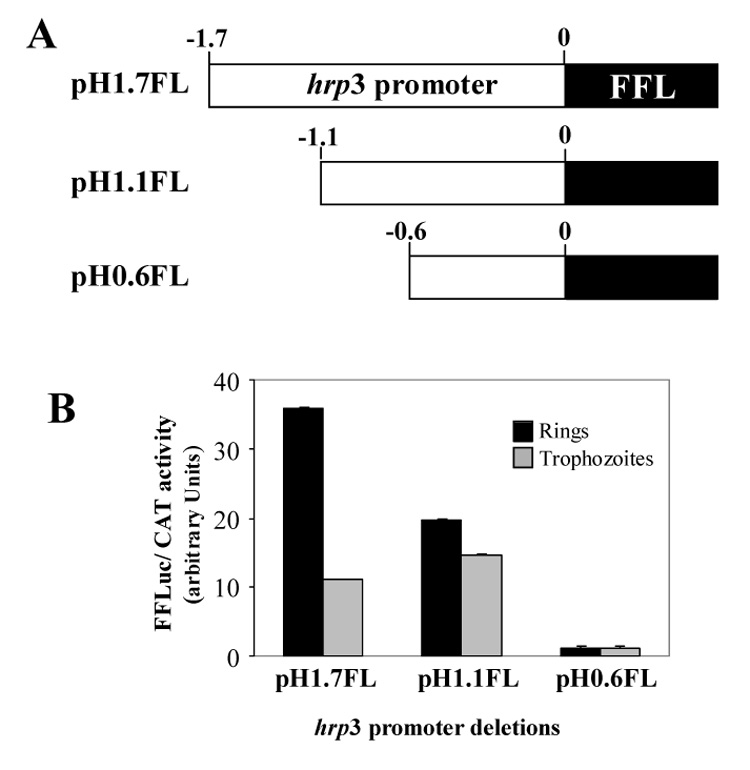
Deletion of hrp3 promoter reduced the ring parasite firefly luciferase activity. A) Schematic representation of hrp3 promoter truncations driving firefly luciferase (FFL). B) Transient expression of luciferase gene in ring and trophozoite parasites. FFL activity was normalized by cotransfection using Chloramphenicol acetyltransferase gene (CAT) as described in Material and Methods. Bars are the mean of four independent experiments ± SD.
Figure 2.
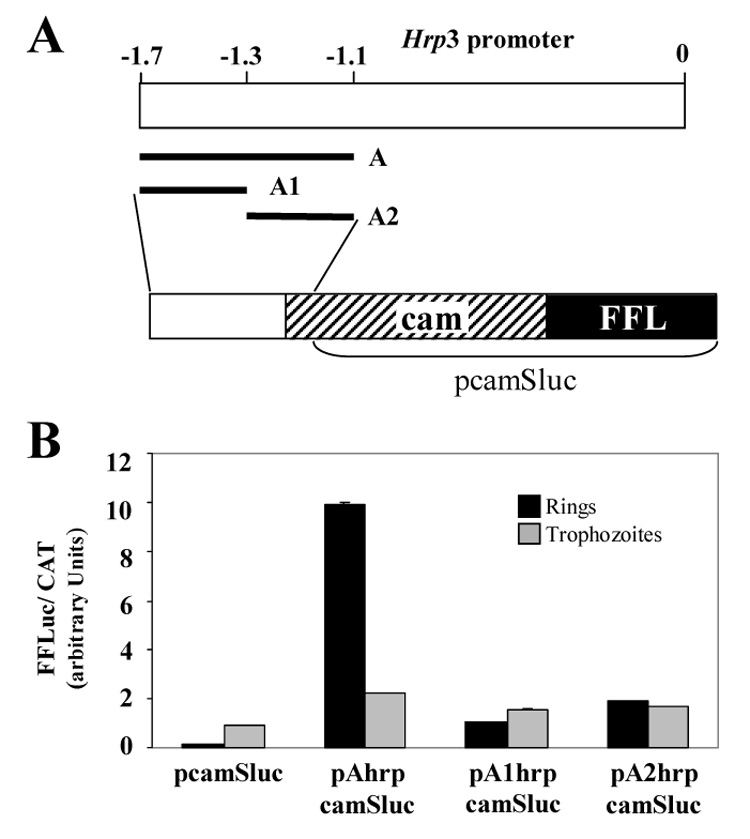
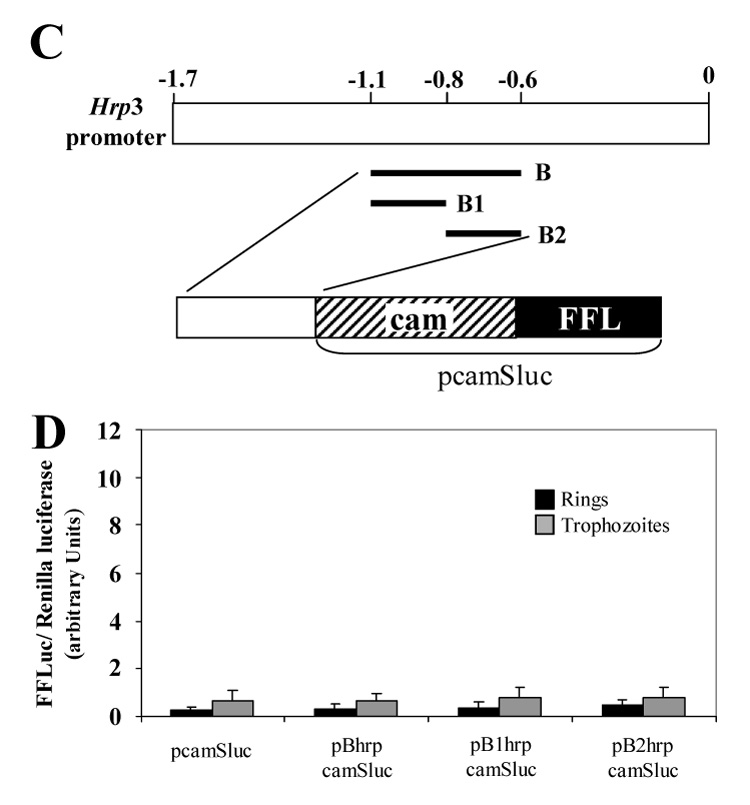
Ring specific cam-dependent luciferase activity is stimulated by hrp3 region between -1.7 to -1.1. A) Schematic representation of the 1.7 kb hrp3 promoter region depicting the different hrp3 fragments placed upstream of the cam 5’ end region. In addition, the organization of the hybrid promoter is shown. B) Transient luciferase activity was measured in ring and trophozoite parasites and normalized to CAT activity. The control plasmid (pcamSluc) contains cam promoter and firefly luciferase gene (FFL). pAhrpcamSluc, pA1hrpcamSluc, pA2hrpcamSluc represent plasmids with hrp3 A, A1 and A2 fragments placed upstream of the cam promoter respectively. Bars are the mean of three independent experiments ± SD. C) Schematic representation of the 1.7 kb hrp3 promoter region depicting the different hrp3 fragments placed upstream the cam 5’ end region. In addition, the general organization of the hybrid promoter is shown. D) Transient luciferase activity was measured in ring and trophozoite parasites and normalized by cotransfection with a plasmid carrying the Renilla luciferase gene driven by the cam promoter. The control plasmid (pcamSluc) contains the cam promoter driving firefly luciferase gene (FFL). pBhrpcamSluc, pB1hrpcamSluc, pB2hrpcamSluc represent plasmids with hrp3 B, B1 and B2 fragments placed upstream the cam promoter respectively. Bars are the mean of four independent experiments ± SD.
2.5. Mapping of the hrp3 transcription start site
The hrp3 transcription start site (TSS) was determined by 5’ rapid amplification of cDNA ends (5’ RACE, Invitrogen) using total RNA from non-transfected and pH1.7HG-transfected parasites [24]. Total RNA from transfected and non-transfected parasites was isolated using TRIzol® LS reagent (Invitrogen). Five micrograms of total RNA from non-transfected parasites was reverse transcribed using the oligonucleotide 5’-GTAATGCATTGAATTTTATAAAAAAGCAAAATTATTTA-3’ (boldface nucleotides represents a Nsi I site) complementary to the region from -350 nt to -320 nt of the hrp3 promoter. For RNA from pH1.7HG-transfected parasites, 5 µg of total RNA was reverse transcribed using a specific oligonucleotide to the human DHFR gene, 5’-CCAAATCAATTTCTGGAAAAAACGTGTCAC-3’ complementary to the region 434 nt to 464 nt from the ATG initiation codon. cDNA generated from non-transfected and pH1.7HG-transfected parasites was then amplified by PCR using nested reverse oligonucleotide 5’-ATCATGCATTTTATTATTTTATTTTATTAATATAAGAC-3’ (boldface nucleotides represents a Nsi I site) complementary to the hrp3 promoter region from -543 nt to -514 nt and forward oligonucleotides provided by the manufacturer (Invitrogen). PCR products were cloned into pCR2.1® vector (Invitrogen), DNA was extracted from 15 different recombinant colonies for each group and sequenced to determine the transcription start site.
3. Results
3.1. The hrp3 region between -1.7-kb and -1.1-kb regulates ring-stage specific gene expression.
We generated nested deletions on the histidine-rich protein 3 promoter (hrp3) to evaluate the role of promoter sequences on gene expression. For this purpose, the CAT gene from pHRPCAT [10] was replaced by the firefly luciferase (FFL) gene in hrp3 promoter deletions (pH1.7FL, pH1.1 FL and pH0.6FL, Fig. 1A). These constructs containing FFL and the hrp3 promoter deletions were subsequently used to transiently transfect malaria parasites. FFL activity from the hrp3 promoter deletion, was normalized to CAT activity by co-transfecting the pcamCAT plasmid [27]. The FFL activity in ring parasites transfected with the 1.1-kb hrp3 promoter (pH1.1FL) lacking the enhancer-like region between -1.7-kb and -1.1-kb, was approximately 55 % of that of the full length promoter (Fig. 1B, compare pH1.7FL and pH1.1FL, rings). In contrast, FFL activity remained constant in trophozoite parasites transfected with the 1.7-kb and the 1.1-kb hrp3 promoters respectively (Fig. 1B, compare pH1.7FL and pH1.1FL, trophozoites). Further deletion to 0.6-kb greatly decreased FFL activity in both ring and trophozoite parasites (Fig. 1B, pH0.6FL). Taken together, these data indicate the presence of a ring stage specific enhancer-like element between -1.7-kb and -1.1-kb of the hrp3 promoter.
3.2. The hrp3 region between -1.7-kb and -1.1-kb specifically stimulates ring-stage activity on heterologous cam promoter
To investigate whether the hrp3 enhancer-like region between -1.7-kb and -1.1-kb contains bona fide ring specific elements we performed gain-of-function experiments. In order to test this possibility we selected a 600-nucleotide long calmodulin (cam) promoter. This promoter is active at all stages of the parasite growth with a peak of expression at 40 hours post-invasion [30, 31]. Our rationale is that if this region contains ring specific enhancer elements it would specifically increase the cam promoter-mediated luciferase activity in ring parasites. Conversely, if this region does not contain an enhancer element, it would not have an effect on cam-mediated luciferase activity. It is also possible that gene stage specificity in the hrp3 promoter requires the combined action of promoter elements located both in the -1.7-kb and -1.1-kb region and downstream in the hrp3 promoter. Therefore, the use of cam will test whether or not the -1.7-kb and -1.1-kb region contains a bona fide enhancer-like element. For this test, we placed the hrp3 region from -1.7-kb to -1.1-kb, fragment A (Fig. 2A and S1) upstream of the cam promoter driving the FFL activity. Transient transfection of ring-stage parasites (12–14 hpi) showed that FFL activity driven by the hybrid cam promoter containing hrp3 fragment A was approximately 55 fold higher (Fig. 2B) than the FFL activity driven by the cam promoter alone (Fig. 2B, compare pcamSluc and pAhrpcamSluc, rings). To further refine the location of stage specific elements in the hrp3 promoter, we divided fragment A into 5’end fragment, A1 and a 3’ end fragment A2 (Fig. 2A and S2, orange and blue areas respectively, supplementary information). These fragments were placed upstream the cam promoter and the hybrid promoter used to transfect P. falciparum. Neither subfragment stimulated the ring FFL activity to levels similar to that seen when fragment A was used (Fig. 2B, compare pAhrpcamSluc, pA1hrpcamSluc and pA2hrpcamSluc, rings). FFL activity was also measured in trophozoite parasites collected 32–34 hpi to examine the effect of hrp3 fragments on cam-dependent FFL activity. Figure 2B shows that the presence of the hrp3 fragment in pAhrpcamSluc and its subfragments in pA1hrpcamSluc and pA2hrpcamSluc had no significant effect on trophozoite stage cam-dependent FFL activity when compared with the control (cam promoter alone in pcamSluc).
To further confirm that fragment A specifically increased cam-dependent promoter activity in ring-stage parasites, we placed a 500 bp hrp3 fragment normally located between -1.1-kb and -0.6-kb (or fragment B, Fig. 2C and S1, supplementary information), upstream of the cam promoter in the pcamSluc plasmid to generate pBhrpcamSluc. In this and subsequent experiments, FFL activity was normalized to Renilla luciferase (RL) activity [28]. Using RL instead of CAT allows a single sample to be used to determine experimental and control luciferase activities. In contrast to region A, region B did not increase luciferase activity driven by the hrp3-cam hybrid promoter in either ring- or trophozoite-stage parasites (Fig. 2D, compare pcamSLuc and pBhrpcamSLuc). Similarly, the subfragments B1 and B2 (pB1hrpcamSLuc and pB2hrpcamSLuc respectively) that span the hrp3 promoter regions from -1.1-kb to -0.8-kb and -0.8-kb to -0.6-kb respectively (Fig. 2C and S2, green and yellow areas respectively, supplementary information) did not stimulate luciferase activity mediated by the hrp3-cam hybrid promoter (Fig 2D). Together, these results indicate that the hrp3 region between -1.7-kb and -1.1-kb contains a bona fide ring specific enhancer-like element.
In order to map the putative regulatory elements in the hrp3 promoter enhancer region, we generated a series of 100–150 bp nested deletions on the hrp3 promoter from -1.7-kb to -0.7-kb. We generated hrp3 5’ end truncations by using the polymerase chain reaction (PCR) and specific oligonucleotides (Table S1. Supplementary information). Nested truncations were placed upstream of the firefly luciferase gene (Fig. 3) and used to transfect malaria parasites. Luciferase activity was then measured at 12–14 hpi (ring-stage) and normalized by cotransfection with a plasmid carrying Renilla luciferase. Figure 3 shows a step-wise decrease of FFL activity in ring-stage parasites as the promoter was shortened. In contrast, promoter truncations had no effect (and remained relatively constant) on trophozoite-stage transient FFL activity (32–34 hpi, Fig. 3). These results strongly suggest that regulation of gene expression in ring-stage parasites in the hrp3 promoter is mediated by a regulatory element that may have a multipartite structure.
Figure 3.
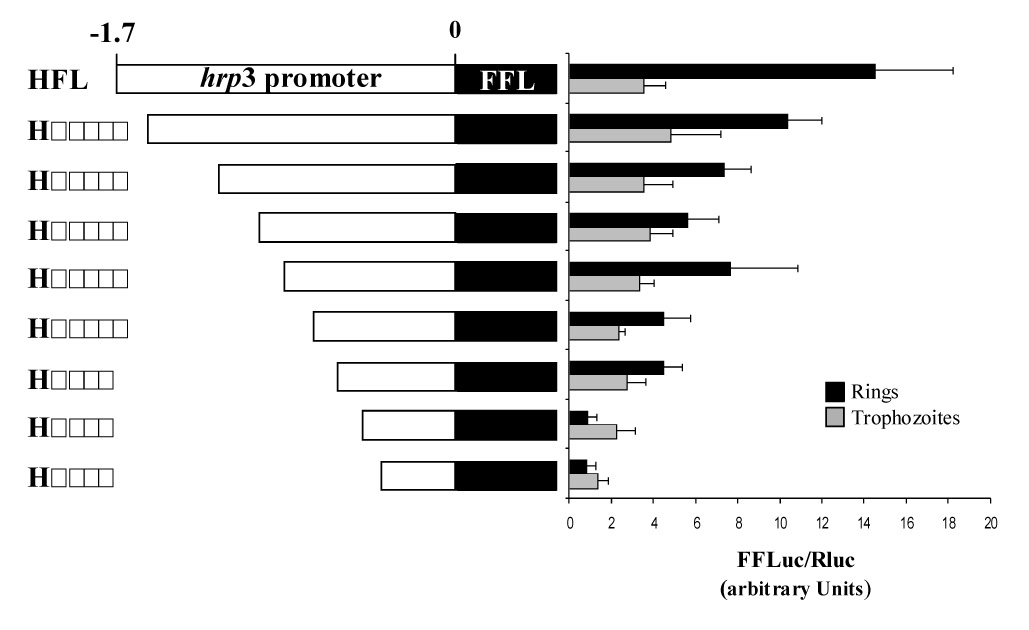
A putative multipartite element on hrp3 promoter controls ring specific expression of a reporter gene. Schematic representation of hrp3 deletions (left side of panel). The constructs’ name indicates the promoter size after deletion. hrp3 promoter was deleted by PCR and placed upstream the firefly luciferase gene. Constructs were transfected into P. falciparum. Firefly and Renilla luciferase activities were determined 12–14 hours post-invasion (hpi) (ring) and 32–34 hpi (trophozoite). Bars are the mean of four independent experiments ± SD.
3.3. Location of the hrp3 promoter on an episome did not affect transcription start site (TSS) selection
Previous studies have shown that the location of a promoter on a plasmid rather than in the chromosome can affect its expression. For example, early studies of the regulation of glycophorin binding protein 130 (gbp130) showed that the loss of trophozoite stage specificity was correlated with rearrangement of the chromatin structure on the plasmid copy [12]. In contrast, P. falciparum stage specific promoters such as histidine-rich protein-3 (hrp3) [25], PfEMP1 (var) [15] and P. berghei apical membrane antigen-1 promoter (Pbama1) [32] used to drive reporter gene expression on plasmids, maintained a strict developmental regulation despite the change in location. Together, these results indicate that regulatory mechanisms in the parasite might not be entirely dependent on chromosomal location. To ensure that the hrp3 promoter behaves similarly whether on a chromosome or a plasmid, the hrp3 TSS was mapped by 5’ rapid amplification of cDNA end (RACE). Our rationale was that if the TSS is preserved, it means episomal location does not affect selection of the TSS. The TSS selection determines the length of the 5’ untranslated region (UTR) in the nascent transcripts. Therefore, identification of the TSS will reveal the promoter structure. Because transient transfection is likely to yield low level of mRNA, we decided to map the TSS on a P. falciparum stable cell line transfected with pH1.7HG [24]. In this plasmid the hrp3 promoter drives the expression of the GFP-hDHFR (HDGFP) fusion gene. Both hrp3 and HDGFP mRNAs were characterized by a heterogeneous 5’ end, which is consistent with other 5’ end termini mapped in Plasmodium genes [33]. The TSS (on the basis of sequencing 15 clones) from the endogenous hrp3 mapped to a 130 bp long region between -750 bp and -626 bp from the ATG initiation codon (Fig. S3, closed box. Supplementary information). Similarly, all 5’ end mRNA termini diagnostic of TSS on the episomal hrp3 promoter were mapped within this 130 bp region. Our data demonstrate that TSS selection was not significantly modified by episomal context.
As in other Plasmodium genes [33], endogenous and episomal transcription start sites start with adenosine (Fig. 4, position 11 in the logo). This is consistent with the preference of purine nucleotides at the 5’ end terminus of eukaryotic mRNAs [33]. The analysis of the DNA sequences surrounding each TSS (Fig. 4, 10 nucleotides upstream and downstream the TSS) showed no major difference between the endogenous and plasmid TSS. However, thymine nucleotides were more frequent in the upstream region of the endogenous TSS (Fig. 4). No nucleotide preference is seen at the downstream region, except for thymine at positions 12 and 18 (Fig. 4). This pattern seems to be slightly different in the plasmid TSS, containing predominately adenine at the upstream region and thymine at the downstream region of the TSS (Fig. 4). Further studies will be needed in order to determine the relevance, if any, of this difference on the regulation of gene expression. However, these minor differences on the TSS region are likely not biologically significant.
Figure 4.
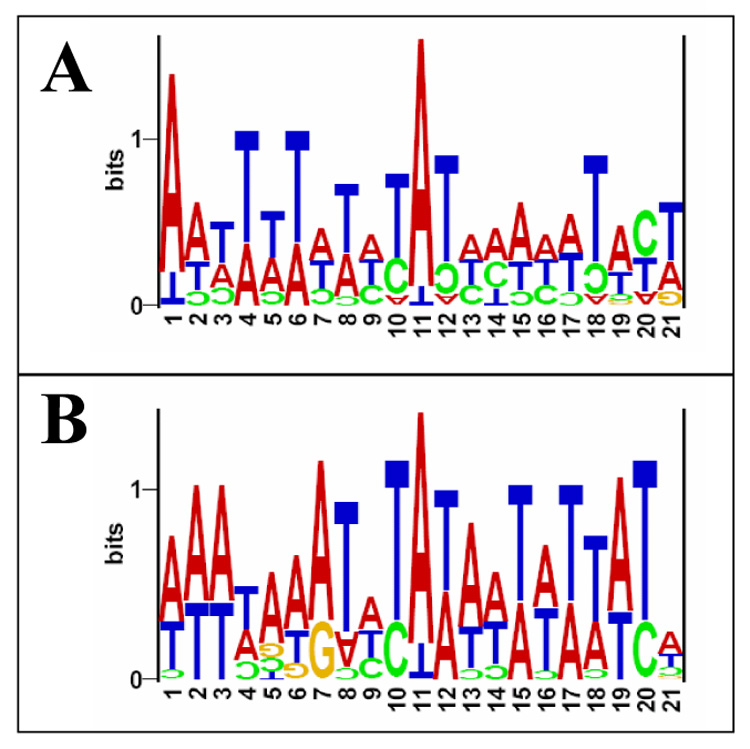
Logo visualization of hrp3 promoter transcription start site. cDNA, from endogenous and episomal located promoter, was cloned in E. coli and recombinant plasmids were purified from 15 independent colonies for each group. Recombinant plasmids were then sequenced and used to determine the 5’ end of cloned cDNA that represents the TSS. The logo sequence shows alignment of 15 different TSS for endogenous and episomal located hrp3 promoter. The first transcribed nucleotide (position 11) was adenosine in most cases. Ten nucleotide upstream and downstream of the TSS in endogenous (A) and episomal (B) were also included. The height of each letter is proportional to the frequency of nucleotides in each position. The total letter height indicates the amount of information (bits) contained in the nucleotides at that position.
4. Discussion
In this report we characterized a promoter region responsible for ring stage specific expression in a P. falciparum gene. This region between - 1.7-kb and -1.1-kb had a marked effect on stage specific expression independent of the chromosomal context. The structure of this enhancer-like element appears to be multipartite. Consistent with our hypothesis, bioinformatic analysis of Plasmodium genome showed that most Plasmodium genes seem to have a cluster of cis-acting regulatory elements (four or five) associated with or as part of their promoters [21]. van Noort et al. 2006 [21] identified a number of motifs that significantly correlated with mRNA expression. None of these motifs were found in the hrp3 promoter, suggesting that they might not be relevant for hrp3 promoter regulation. The hrp3 enhancer region also contains a CCAAT at position -1487 bp to -1483 bp. This was not surprising since the parasite genome encodes the complete CCAAT-box binding complex [34]. However, CCAAT-box containing fragment A1 (Fig 2A and Fig. S2) failed to stimulate ring luciferase activity in the hybrid cam promoter, suggesting that CCAAT-box is not involved in ring stage specificity. Alternatively, CCAAT-box might require other regulatory elements (multipartite structure) that are absent in fragment A1. A TGTATATG motif similar to a schizont/gametocyte motif reported previously [9] is also present in the fragment B1 at position -1109 bp to -1102 bp of the hrp3 promoter (Fig. S2. Supplementary information). However, fragment B or B1 that contain the TGTATATG motif failed to stimulate ring luciferase activity in the hybrid cam promoter. Conversely, fragment A which lacks this motif still conferred specific ring-stage stimulation on the cam-mediated FFL (Fig. 2B and S2). Altogether, these results suggest that, the TGTATATATG motif is not involved in regulation of promoter stage specificity in the parasite.
Comparative analyses of our data with published studies on gene expression in malaria parasites suggest that the hrp3 cis-acting element between -1.7-kb and -1.1-kb might represent a new mechanism of stage specific regulation. For example, GC-rich promoter elements [35] in pfhsp86, GC-rich elements CAGACAGC in the pgs28 promoter [18] and CGCACAACAC P. falciparum in rif genes [20] have been described. These sequences are not found in blood stage promoters such as hrp3, msp1, var7b and knob-associated histidine rich protein (kahrp), suggesting that they are not involved in regulation of stage specificity in these promoters. Also, it has been reported that intronic regulatory elements regulate allelic expression in centromeric var7b genes [15, 36]. Similarly, hrp3 5’ end sequence contains an intron located 13 bp upstream of the ATG initiation codon [27]. Although, hrp3 intron might play a similar regulatory role as the centromeric var7b genes intron, previous studies has shown that deletion of this intron in the 5’ end hrp3 does not affect reporter expression [10]. However, the role of intronic sequence on hrp3 stage-specific regulation has not yet been assessed.
Sequence comparison between hrp3 and several var promoters using bioinformatics analysis (Hiller and Haldar unpublished) as well as comparison to an extended data set obtained from the Plasmodium database (www.plasmodb.org) of putative ring and/or trophozoite 5’ end regions (Lopez-Estraño, Phan and Garzon unpublished), failed to uncover conserved sequences that may contribute to promoter regulation. The absence of recognizable sequence homology amongst Plasmodium promoters suggests that stage specific regulation may require complex mechanisms that could include transcriptional initiation and/or nuclear position amongst others. A wide variety of eukaryotic genes, including those of Plasmodium, are regulated by DNA structure such as formation of kinks by poly T tracts flanking the core promoter [37–39] and poly(dA:dT) sequences [40], both of which are very abundant in the Plasmodium genome. Other DNA-dependent structures that could play a role in the promoter function such as poly purine-pyrimidine sequences have also been described [41]. A long poly(dA)poly(dT) sequence has been associated with cam promoter basal activity [42]. Much shorter poly(dA)poly(dT) tracts are distributed along the hrp3 promoter, but whether they are involved in stage or basal promoter regulation needs to be determined.
The Plasmodium signals that control gene expression are phylogenetically related and promoters seem to be interchangeable in some degree amongst parasite species [43]. Despite this conservation, P. falciparum promoters are quite diverse in terms of their cis regulatory elements and questions such as what constitutes a minimal promoter, remain to be addressed. Interestingly, deletion of the hrp3 region between -1.7-kb and -1.1-kb turned the truncated promoter constitutive both at the mRNA [24] and at the translation level (luciferase activity, this study). Further deletion to generate the 0.6-kb truncated promoter switched the accumulation of mRNA from ring stage in the 1.7-kb full length promoter to the trophozoite stage in the 0.6-kb truncated promoter [24]. Furthermore, the 0.6-kb promoter did not support luciferase activity in either stage of the parasite development (Fig 1). These results suggest that the region between -1.1 to -0.6-kb might contain transcriptional elements responsible for the basal activity of this promoter. In fact, basal regulatory elements have not been fully characterized in Plasmodium promoters. Although comparisons with other eukaryotic promoter have shown several cis acting elements such as SV40 enhancer elements [44], CAAT boxes [10,35], SP1 or octamer and GC box binding sites (GC boxes) [35] in some parasite promoters, but they were not identified in the -1.1 to -0.6-kb hrp3 region. In addition, sequences that have been recently described as regulatory elements in the parasite genome [9, 18, 20, 21, 34, 35] were not present in the hrp3 promoter. Together, these results indicate that basal regulatory cis acting elements might also be quite diverse in the malaria parasite.
Conservation of the transcriptional start site on the chromosomally and episomally located hrp3 promoter suggests that interaction of transcription factors with their cognate sequences and/or chromatin structure were not altered in the episomal plasmid. This is particularly relevant, since chromatin rearrangement has been associated with the loss of stage specificity in episomally located gbp 130 promoter [12]. In the hrp3 promoter the TSS was heterogeneous, but restricted to a discrete 130 nucleotide region. In contrast, var7b promoter TSS takes place in a specific sequence similar to the Trichomonas vaginalis initiator sequence (TCATA). There are four var-initiator-like sequences at positions -1572, -1560, -1425 and -870 in the hrp3 promoter (Fig. S3. Supplementary information, open box) but none of the HDGFP mRNA 5’ end termini mapped, were located at or close to TCATA positions. Therefore, the presence of an initiator sequence does not necessarily indicate function. Our results suggest the presence of different TSS selection mechanisms for Plasmodium genes. Luciferase activity driven by the 0.6-kb truncated hrp3 promoter was very low compared with the 1.7-kb full length. Analysis of this promoter in stable cell lines by Northern blot [24], showed reporter mRNA in trophozoite but not in ring parasites. Comparison of the 0.6-kb hrp3 truncated promoter in a stable cell line [24] with transient luciferase activity (this study), showed no correlation between the mRNA accumulation of reporter gene and luciferase transient activity in P. falciparum. This result suggests that mRNA translation driven by the 0.6-kb promoter has been impaired. One possibility is the use of a cryptic TSS in the firefly luciferase mRNA; this in turn generates a mRNA with shortened 5’ UTR that might be responsible for the FFL low activity in 0.6-kb hrp3 promoter. In fact, the complete TSS region has been truncated in the 0.6-Kb hrp3 promoter, suggesting that other TSSs have been used (Fig. S3, closed box and position -624, open circle. Supplementary information). For example, it has been shown in different organisms that elements on the 5’ UTR regulate translation of mRNA [53] and therefore changes of the TSS might regulate the translation efficiency on a given gene.
We have identified stage-specific promoter elements that underscore the complex nature of regulation of gene expression in P. falciparum. In addition, our results suggest the use of a multipartite ring specific region in the hrp3 promoter. Future studies will further characterize the mechanism and identify the regulatory elements at play in the malaria parasite and contribute uncover unique pathways that could be exploited for drug development to fight the disease.
Supplementary Material
Acknowledgments
We thank Ashok Aiyar for technical assistance with luciferase measurements, N. Luisa Hiller for computational analysis of hrp3 and var promoters. We also thank Dr. Kent Gartner and Dr. Charles Lessman, University of Memphis for critical reading of the manuscript. We are very thankful to the Molecular Resource Center of the University of Tennessee. This work was funded by NIH grants 1R03AI054529 (to KH and CLE), R01HL69630, R01AI39071 (to KH) and Startup funds from The University of Memphis (to CLE). JPS was supported by a fellowship from INSERM (France).
Abbreviations
- dhfr-ts
dihydrofolate reductase-thymidylate synthase
- bp
base pair
- nt
nucleotides
- cam
calmodulin
- msp1
merozoite surface protein 1
- hrp3
histidine-rich protein 3
- hrp2
histidine-rich protein 2
- PfEMP1
Plasmodium falciparum erythrocyte membrane protein 1
- hsp86
heat shock protein 86
- pfs16
Plasmodium falciparum sexual-stage antigen 16
- pfs25
Plasmodium falciparum sexual-stage antigen 25
- RNA
ribonucleic acid
- rRNA
ribosomal ribonucleic acid
- CAT
chloramphenicol acetyl transferase
- PCR
polymerase chain reaction
- hpi
hours post-invasion
- mRNA
messenger ribonucleic acid
- rif
repetitively Interspersed Family
- stevor
subtelomeric variable open reading frame
- HDGFP
human dihydrofolate reductase fused to Aequorea victoria Green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Global. WHO declares emergency against AIDS, TB, malaria. AIDS policy & law. 2006;21:5. [PubMed] [Google Scholar]
- 2.Simooya O. The WHO 'Roll Back Malaria Project': planning for adverse event monitoring in Africa. Drug Saf. 2005;28:277–286. doi: 10.2165/00002018-200528040-00001. [DOI] [PubMed] [Google Scholar]
- 3.Wesseling JG, Snijders PJ, van Someren P, Jansen J, Smits MA, Schoenmakers JG. Stage-specific expression and genomic organization of the actin genes of the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1989;35:167–176. doi: 10.1016/0166-6851(89)90119-9. [DOI] [PubMed] [Google Scholar]
- 4.Waters AP, Syin C, McCutchan TF. Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature. 1989;342:438–440. doi: 10.1038/342438a0. [DOI] [PubMed] [Google Scholar]
- 5.Waters AP. The ribosomal RNA genes of Plasmodium. Adv Parasitol. 1994;34:33–79. doi: 10.1016/s0065-308x(08)60136-0. [DOI] [PubMed] [Google Scholar]
- 6.Li J, McConkey GA, Rogers MJ, Waters AP, McCutchan TR. Plasmodium: the developmentally regulated ribosome. Exp Parasitol. 1994;78:437–441. doi: 10.1006/expr.1994.1051. [DOI] [PubMed] [Google Scholar]
- 7.Fang J, Sullivan M, McCutchan TF. The effects of glucose concentration on the reciprocal regulation of rRNA promoters in plasmodium falciparum. J Biol Chem. 2003 doi: 10.1074/jbc.M308284200. [DOI] [PubMed] [Google Scholar]
- 8.Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, James K, Rutherford K, Harris B, Harris D, Churcher C, Quail MA, Ormond D, Doggett J, Trueman HE, Mendoza J, Bidwell SL, Rajandream MA, Carucci DJ, Yates JR, 3rd, Kafatos FC, Janse CJ, Barrell B, Turner CM, Waters AP, Sinden RE. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 9.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE. Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci U S A. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crabb BS, Cowman AF. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc Natl Acad Sci U S A. 1996;93:7289–7294. doi: 10.1073/pnas.93.14.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horrocks P, Lanzer M. Differences in nucleosome organization over episomally located plasmids coincides with aberrant promoter activity in P. falciparum. Parasitol Int. 1999;48:55–61. doi: 10.1016/s1383-5769(99)00002-1. [DOI] [PubMed] [Google Scholar]
- 13.Dechering KJ, Kaan AM, Mbacham W, Wirth DF, Eling W, Konings RN, Stunnenberg HG. Isolation and functional characterization of two distinct sexual-stage-specific promoters of the human malaria parasite Plasmodium falciparum. Mol Cell Biol. 1999;19:967–978. doi: 10.1128/mcb.19.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voss TS, Thompson JK, Waterkeyn J, Felger I, Weiss N, Cowman AF, Beck HP. Genomic distribution and functional characterisation of two distinct and conserved Plasmodium falciparum var gene 5' flanking sequences. Mol Biochem Parasitol. 2000;107:103–115. doi: 10.1016/s0166-6851(00)00176-6. [DOI] [PubMed] [Google Scholar]
- 15.Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- 16.Crabb BS, Triglia T, Waterkeyn JG, Cowman AF. Stable transgene expression in Plasmodium falciparum. Mol Biochem Parasitol. 1997;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- 17.Militello KT, Dodge M, Bethke L, Wirth DF. Identification of regulatory elements in the Plasmodium falciparum genome. Mol Biochem Parasitol. 2004;134:75–88. doi: 10.1016/j.molbiopara.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Chow CS, Wirth DF. Linker scanning mutagenesis of the Plasmodium gallinaceum sexual stage specific gene pgs28 reveals a novel downstream cis-control element. Mol Biochem Parasitol. 2003;129:199–208. doi: 10.1016/s0166-6851(03)00101-4. [DOI] [PubMed] [Google Scholar]
- 19.Horrocks P, Lanzer M. Mutational analysis identifies a five base pair cis-acting sequence essential for GBP130 promoter activity in Plasmodium falciparum. Mol Biochem Parasitol. 1999;99:77–87. doi: 10.1016/s0166-6851(98)00182-0. [DOI] [PubMed] [Google Scholar]
- 20.Tham WH, Payne PD, Brown GV, Rogerson SJ. Identification of basic transcriptional elements required for rif gene transcription. Int J Parasitol. 2006 doi: 10.1016/j.ijpara.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 21.van Noort V, Huynen MA. Combinatorial gene regulation in Plasmodium falciparum. Trends Genet. 2006;22:73–78. doi: 10.1016/j.tig.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Gunasekera AM, Myrick A, Militello KT, Sims JS, Dong CK, Gierahn T, Le Roch K, Winzeler E, Wirth DF. Regulatory motifs uncovered among gene expression clusters in Plasmodium falciparum. Mol Biochem Parasitol. 2007 doi: 10.1016/j.molbiopara.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Kyes S, Christodoulou Z, Pinches R, Kriek N, Horrocks P, Newbold C. Plasmodium falciparum var gene expression is developmentally controlled at the level of RNA polymerase II-mediated transcription initiation. Mol Microbiol. 2007;63:1237–1247. doi: 10.1111/j.1365-2958.2007.05587.x. [DOI] [PubMed] [Google Scholar]
- 24.López-Estraño C, Semblat JP, Gopalakrishnan AM, Turner L, Mazier D, Haldar K. Plasmodium falciparum: hrp3 promoter region is associated with stage-specificity and episomal recombination. Exp. Parasitology. 2007 doi: 10.1016/j.exppara.2007.01.020. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheresh P, Harrison T, Fujioka H, Haldar K. Targeting the Malarial Plastid via the Parasitophorous Vacuole. J Biol Chem. 2002;277:16265–16277. doi: 10.1074/jbc.M109331200. [DOI] [PubMed] [Google Scholar]
- 26.Lanzer M, de Bruin D, Ravetch JV. A sequence element associated with the Plasmodium falciparum KAHRP gene is the site of developmentally regulated protein-DNA interactions. Nucleic Acids Res. 1992;20:3051–3056. doi: 10.1093/nar/20.12.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadekoppala M, Cheresh P, Catron D, Ji D, Deitsch K, Wellems TE, Seifert HS, Haldar K. Rapid recombination among transfected plasmids, chimeric episome formation and trans gene expression in Plasmodium falciparum. Mol Biochem Parasitol. 2001;112:211–218. doi: 10.1016/s0166-6851(00)00368-6. [DOI] [PubMed] [Google Scholar]
- 28.Militello KT, Wirth DF. A new reporter gene for transient transfection of Plasmodium falciparum. Parasitol Res. 2003;89:154–157. doi: 10.1007/s00436-002-0721-5. [DOI] [PubMed] [Google Scholar]
- 29.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orfa Rojas M, Wasserman M. Stage-specific expression of the calmodulin gene in Plasmodium falciparum. J Biochem (Tokyo) 1995;118:1118–1123. doi: 10.1093/oxfordjournals.jbchem.a124996. [DOI] [PubMed] [Google Scholar]
- 31.Akompong T, Kadekoppala M, Harrison T, Oksman A, Goldberg D, Fujioka H, Samuel BU, Sullivan D, Haldar K. Trans expression of a P. falciparum hisdine rich protein II (HRPII) reveals sorting of soluble proteins in the periphery of the host erythrocyte and disrupts transport to the malarial food vacuole. J Biol Chem. 2002 doi: 10.1074/jbc.M201968200. [DOI] [PubMed] [Google Scholar]
- 32.Ozwara H, van der Wel A, Kocken CH, Thomas AW. Heterologous promoter activity in stable and transient Plasmodium knowlesi transgenes. Mol Biochem Parasitol. 2003;130:61–64. doi: 10.1016/s0166-6851(03)00141-5. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe J, Sasaki M, Suzuki Y, Sugano S. Analysis of transcriptomes of human malaria parasite Plasmodium falciparum using full-length enriched library: identification of novel genes and diverse transcription start sites of messenger RNAs. Gene. 2002;291:105–113. doi: 10.1016/s0378-1119(02)00552-8. [DOI] [PubMed] [Google Scholar]
- 34.Coulson RM, Hall N, Ouzounis CA. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome research. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su XZ, Wellems TE. Sequence, transcript characterization and polymorphisms of a Plasmodium falciparum gene belonging to the heat-shock protein (HSP) 90 family. Gene. 1994;151:225–230. doi: 10.1016/0378-1119(94)90661-0. [DOI] [PubMed] [Google Scholar]
- 36.Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- 37.Angermayr M, Oechsner U, Gregor K, Schroth GP, Bandlow W. Transcription initiation in vivo without classical transactivators: DNA kinks flanking the core promoter of the housekeeping yeast adenylate kinase gene, AKY2, position nucleosomes and constitutively activate transcription. Nucleic Acids Res. 2002;30:4199–4207. doi: 10.1093/nar/gkf551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter ME. Positive and negative effects of deletions and mutations within the 5' flanking sequences of Plasmodium falciparum DNA polymerase delta. Mol Biochem Parasitol. 2002;122:9–19. doi: 10.1016/s0166-6851(02)00064-6. [DOI] [PubMed] [Google Scholar]
- 39.Burghaus PA, Lingelbach K. Luciferase, when fused to an N-terminal signal peptide, is secreted from transfected Plasmodium falciparum and transported to the cytosol of infected erythrocytes. J Biol Chem. 2001;276:26838–26845. doi: 10.1074/jbc.M100111200. [DOI] [PubMed] [Google Scholar]
- 40.Suter B, Schnappauf G, Thoma F. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 2000;28:4083–4089. doi: 10.1093/nar/28.21.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maiti AK, Brahmachari SK. Poly purine.pyrimidine sequences upstream of the beta-galactosidase gene affect gene expression in Saccharomyces cerevisiae. BMC Mol Biol. 2001;2:11. doi: 10.1186/1471-2199-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polson HE, Blackman MJ. A role for poly(dA)poly(dT) tracts in directing activity of the Plasmodium falciparum calmodulin gene promoter. Mol Biochem Parasitol. 2005;141:179–189. doi: 10.1016/j.molbiopara.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 43.van der Wel AM, Tomas AM, Kocken CH, Malhotra P, Janse CJ, Waters AP, Thomas AW. Transfection of the primate malaria parasite Plasmodium knowlesi using entirely heterologous constructs. J Exp Med. 1997;185:1499–1503. doi: 10.1084/jem.185.8.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz i Altaba A, Ozaki LS, Gwadz RW, Godson GN. Organization and expression of the Plasmodium knowlesi circumsporozoite antigen gene. Mol Biochem Parasitol. 1987;23:233–245. doi: 10.1016/0166-6851(87)90030-2. [DOI] [PubMed] [Google Scholar]
- 45.Mbacham WF, Chow CS, Daily J, Golightly LM, Wirth DF. Deletion analysis of the 5' flanking sequence of the Plasmodium gallinaceum sexual stage specific gene pgs28 suggests a bipartite arrangement of cis-control elements. Mol Biochem Parasitol. 2001;113:183–187. doi: 10.1016/s0166-6851(01)00210-9. [DOI] [PubMed] [Google Scholar]
- 46.Dechering KJ, Thompson J, Dodemont HJ, Eling W, Konings RN. Developmentally regulated expression of pfs16, a marker for sexual differentiation of the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1997;89:235–244. doi: 10.1016/s0166-6851(97)00123-0. [DOI] [PubMed] [Google Scholar]
- 47.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 48.Deitsch KW. Gene silencing and antigenic variation in malaria parasites. Scientific World Journal. 2001;1:650–652. doi: 10.1100/tsw.2001.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 50.Thomson JK, Rubio JP, Caruana S, Brockman A, Wickham ME, Cowman AF. The chromosomal organization of the Plasmodium falciparum var gene family is conserved. Mol. Biochem. Parasitol. 1997;87:49–60. doi: 10.1016/s0166-6851(97)00041-8. [DOI] [PubMed] [Google Scholar]
- 51.Rubio JP, Thompson JK, Cowman AF. The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. Embo J. 1996;15:4069–4077. [PMC free article] [PubMed] [Google Scholar]
- 52.Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci U S A. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5'- and 3'- UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


