Abstract
The [PIN+] prion, a self-propagating amyloid form of Rnq1p, increases the frequency with which the [PSI+] or [URE3] prions arise de novo. Like the prion domains of Sup35p and Ure2p, Rnq1p is rich in N and Q residues, but rnq1Δ strains have no known phenotype except for inability to propagate the [PIN+] prion. We used solid-state NMR methods to examine amyloid formed in vitro from recombinant Rnq1 prion domain (residues 153–405) labeled with Tyr-1–13C (14 residues), Leu-1–13C (7 residues), or Ala-3–13C (13 residues). The carbonyl chemical shifts indicate that most Tyr and Leu residues are in β-sheet conformation. Experiments designed to measure the distance from each labeled residue to the next nearest labeled carbonyl showed that almost all Tyr and Leu carbonyl carbon atoms were ≈0.5 nm from the next nearest Tyr and Leu residues, respectively. This result indicates that the Rnq1 prion domain forms amyloid consisting of parallel β-strands that are either in register or are at most one amino acid out of register. Similar experiments with Ala-3–13C indicate that the β-strands are indeed in-register. The parallel in-register structure, now demonstrated for each of the yeast prions, explains the faithful templating of prion strains, and suggests as well a mechanism for the rare hetero-priming that is [PIN+]'s defining characteristic.
Three genetic criteria designed to identify infectious proteins (prions), resulted in the finding that two yeast nonchromosomal genes, [URE3] and [PSI+], are prions of Ure2p and Sup35p, respectively (1). One of these outre genetic properties was that overproduction of the prion-forming protein should increase the frequency of the prion arising de novo (1). In fact, both [URE3] (1) and [PSI+] (2) show this property. However, Derkatch et al. observed that, although overproduction of Sup35p induced the appearance of [PSI+] in some strains, it was ineffective in doing so in others (3). This factor needed for [PSI+]-inducibility behaved as a nonchromosomal gene, and so was dubbed [PIN+] (3).
[PIN+] has all of the genetic properties of a prion (4, 5), and was shown to be a self-propagating amyloid form of Rnq1p (4, 6), a protein rich in asparagine (N) and glutamine (Q), which had been shown to form self-propagating aggregates in vivo (7). Deletion of the RNQ1 gene produces no evident phenotype (7), and the only known consequence of [PIN+] is its priming effect on the other prions. Like its prion seeding effect, [PIN+] seeds aggregate formation by polyQ (Huntingtin) (8, 9) and is thus required for Huntingtin-induced toxicity in yeast (9). Although Rnq1p and Sup35p form largely separate amyloid aggregates in vivo (10), it is likely that the mechanism of priming of [PSI+] generation by [PIN+] is a direct, but rare, addition of Sup35p to the fibrils of Rnq1p (11). Several variants of [PIN+] are recognized (12, 13), distinguished by the efficiency with which they induce the appearance of [PSI+] and [URE3]. As has been shown for [PSI+] (14), it is likely that variants of [PIN+] are different amyloid structures, although the precise nature of such differences remains unknown for any system at present.
Aggregates formed in vitro from Rnq1p are filamentous and stain with thioflavin T (7). Circular dichroism indicates that these aggregates are high in β-sheet content and, on exposure to proteinase K, show partial resistance (6). These studies indicate that the Rnq1p aggregates are amyloid fibers. Moreover, these fibers formed in vitro from recombinant Rnq1p are infectious for yeast spheroplasts, efficiently transmitting the [PIN+] gene (6).
Because amyloid is neither soluble nor crystalline, neither solution NMR nor x-ray crystallography is suitable for elucidating its detailed structure, although the study of amyloid-like microcrystals of short oligopeptides has revealed details of packing and sidechain interactions that are relevant to amyloid structures (15, 16). Electron spin resonance studies have been used to study amylin (IAPP) and α-synuclein (17, 18) as well as amyloid of PrP (19). Solid-state NMR has been particularly useful for the elucidation of the structure of amyloids (reviewed in ref. 20). Selective labeling of particular amino acid residues with magnetic nuclear isotopes 13C and 15N, combined with pulse sequences designed to determine distances between labeled nuclei have provided structural information which has allowed construction of detailed evidence-based models of several amyloids. Various fragments of the Alzheimer's disease-linked Aβ have shown parallel and antiparallel structures (21–24). The full-length Aβ1–40 has a parallel in-register structure (25–27), as does amylin (17, 28), α-synuclein (18), and an amyloid of PrP (19). Solid-state NMR has also been used to obtain detailed information on the HETs protein prion domain (residues 218–289) (29, 30), and residues 105–115 of transthyretin (31).
The Ure2 and Sup35 prion proteins of yeast each consist of an N-terminal Q/N-rich amyloid-forming prion domain, a functional C-terminal domain and a connecting domain (refs. 32 and 33, reviewed in ref. 34). Solid-state NMR studies of amyloids of the prion domains of Sup35 (35) and Ure2p (36) as well as Ure2p10–39 (37) have shown that each is a parallel, in-register β-sheet structure, meaning that, for example, residue 35 of one molecule is aligned opposite residue 35 of the preceding molecule and the subsequent molecule in the fibril.
Residues 153–405 of Rnq1p are quite Q/N rich (7) and are sufficient to serve as a prion domain (38), although some shorter segments contained in this region are also able to propagate the [PIN+] prion (38). We report here that Rnq1153–405 has a parallel in-register β-sheet structure, suggesting a mechanism for the cross-seeding of one prion by another.
Results
Fibril Formation by Rnq1153–405.
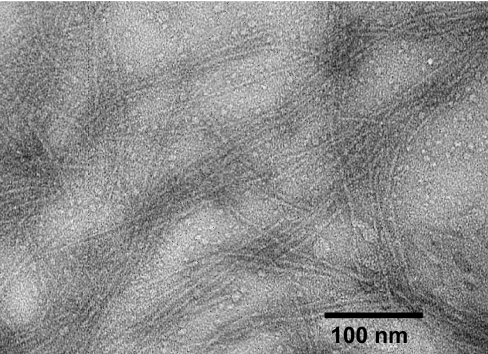
Fibers are reported to comprise only a small fraction of structures observed among aggregates of Rnq1132–405 formed in the absence of urea (11). Our fibril preparations, prepared in 4M urea, essentially as described by Sondheimer and Lindquist (7) and by Patel and Liebman (6) to make infectious aggregates, produced mostly fibrils (Fig. 1). Fibrils were regular, nonbranching with a diameter of 6–8 nm.
Fig. 1.
Transmission electron micrographs of Rnq1153–405 fibrils negatively stained with uranyl acetate.
X-Ray Fiber Diffraction Shows β-Sheet Structure.
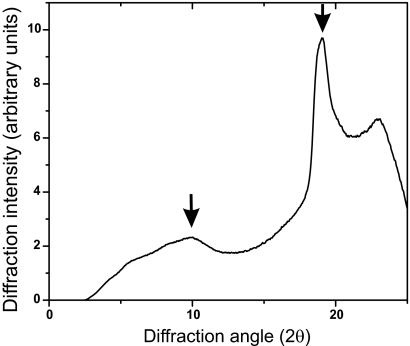
Amyloid fibers of Rnq1153–405 in water were exposed to x-rays and the diffraction pattern was observed (Fig. 2). A sharp line at 2θ = 19° (19.038) indicates a primary spacing of 0.47 nm, the spacing between strands of a β-sheet. A broader peak at 10° (10.023) reflects a spacing of 0.87 nm, usually interpreted as reflecting the distance between layered β-sheets. The filamentous structure and the presence of the characteristic 0.47 nm reflection by x-ray fiber diffraction confirm that Rnq1153–405 forms an amyloid structure.
Fig. 2.
x-ray fiber diffraction of fibrils of Rnq1153–405. The major sharp peak at 2θ = 19° indicates a distance of 4.7 Å, the distance between β strands.
Solid-State NMR Chemical Shifts Indicate β-Sheet Conformation.
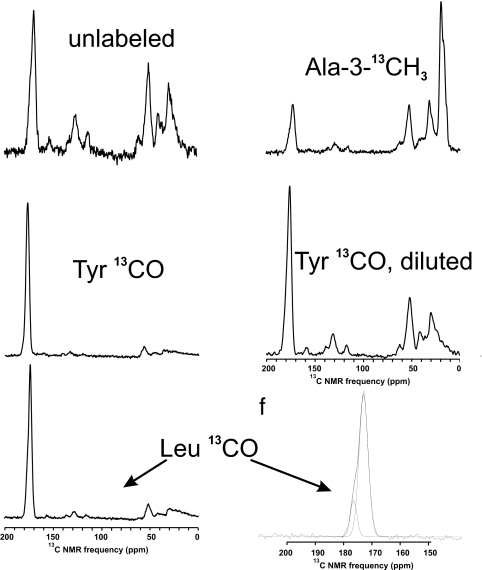
One-dimensional solid-state NMR spectra of wet fibrils of Rnq1153–405 labeled with Tyr-1–13C, Leu-1–13C or Ala-3–13C are shown in Fig. 3. Tyr-1–13C labeled wet amyloid showed a single carbonyl peak at 172.2 ppm, whereas the shift expected for random coil structure is 174.2 ppm (39), suggesting that the Tyr residues detected are in β-sheet structure. The same fibrils, when dry, showed a second minor peak at 175.5 ppm whose intensity suggests that 2.9 (of 15) residues are not in β-sheet structure, but are mobile when the sample is wet. Wet or dry amyloid formed of Rnq1153–405 labeled with Leu-1–13C gave two carbonyl peaks, a minor peak centered at 176.7 ppm, which is possibly due to a sum of the average of four natural abundance carbonyl carbons per molecule, and a major peak at 173.3 ppm (Fig. 3). The expected random coil chemical shift for Leu-1–13C is 175.9 ppm, indicating that most Leu residues are in β-sheet structure.
Fig. 3.
Solid-state NMR spectra of samples used in this work. Spectra of Rnq1153–405 labeled with Tyr-1–13C, Leu-1–13C, or Ala-3–13C were recorded at 9.39 T at 20 kHz MAS. The carbonyl peak of the Leu-1–13C-labeled sample was asymmetric, and could be deconvoluted into two Gaussian peaks.
As expected, only 67% labeling Rnq1153–405 with Ala-3–13C was achieved (see Methods), but it was critical for our purpose that substantial labeling of other residues be avoided. Indeed, in the 1D spectrum (Fig. 3) the methyl region (13 alanine atoms labeled 67% + 22 methyls in MIVLT residues with 1.1% natural abundance) gave an intensity of 100 (arbitrary units), from which the intensity per fully labeled residue is calculated to be 11.2 units. Based only on natural abundance, we expect the 365 carbonyl carbons to have a total intensity of 50 units and observed 27 units. The alpha carbon region (253 atoms × 0.011 × 11.2) should produce 31 units and we observed 31 units. These results suggest that there was little leakage of label into residues other than alanine.
PITHIRDS-CT Data Indicate 0.5-nm Nearest Neighbor 13C–13C Distances for Most Labeled Sites.
In solution NMR, differences in local magnetic fields due to chemical shift anisotropies and dipole–dipole couplings are averaged out by rapid molecular tumbling and very sharp lines result. In solid-state NMR, there is little molecular motion and so magic-angle spinning (MAS) of the sample as a whole is necessary to prevent these effects from producing excessive line broadening and dephasing of spins. However, because of their sharp distance dependence (as 1/r3), these same dipole–dipole couplings are useful for measuring distances and bond angles. Thus, “dipolar recoupling” methods have been developed to restore some of the dipole–dipole couplings that would otherwise be averaged out by MAS. The dipolar recoupling method used here is called PITHIRDS-CT and has the virtue of being relatively insensitive to errors in pulse calibration (40). A π pulse each rotation lasting 1/3 of the rotation period (hence PITHIRDS) recouples the 13C–13C interactions that would otherwise have been averaged out by MAS.
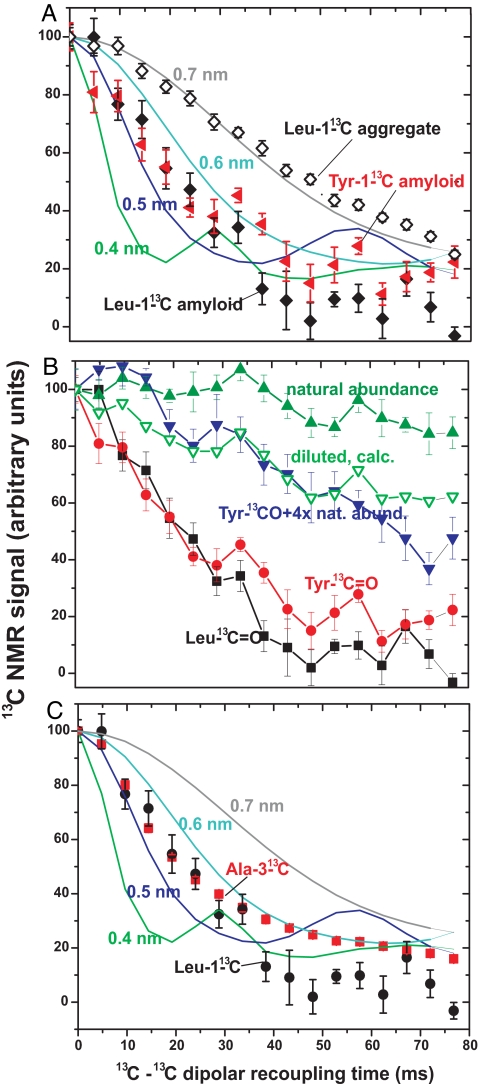
The prion domain of Rnq1 (residues 153–405) has 15 tyrosine and 7 leucine residues, each evenly distributed so that no two tyrosines or two leucines have fewer than two intervening residues (>0.9 nm). The rate of decay of the 13C NMR signal is proportional to 1/r3 where r is the distance to the next nearest 13C atom. In fibers made from molecules labeled with Leu-1-13C or with Tyr-1–13C, the signal decay is rapid indicating, based on the standard curves, a distance of ≈0.5 nm from the next nearest labeled carbonyl carbon (Fig. 4A). Rnq1153–405 fibrils labeled with Leu-1–13C were dissolved in 8 M guanidine HCl and then precipitated with methanol. The resulting nonspecific aggregate showed a substantially decreased rate of signal decay, indicating that the specific structure of the fibers is responsible for the rapid decay of signal, not simple aggregation (Fig. 4A).
Fig. 4.
Dipolar recoupling experiments indicate parallel structure of Rnq1p153–405. (A) Dipolar recoupling experiments with Rnq1153–405 labeled with Leu-1–13C or Tyr-1–13C. Rate of signal decay is inversely proportional to the cube of the distance to the next nearest 13C. Standard curves for 0.4, 0.5, 0.6, and 0.7 nm are simulations carried out as described (28). (B) Dilution of Tyr-1–13C-labeled Rnq1153–405 with unlabeled Rnq1153–405 reduces the signal decay indicating that the next nearest Tyr-1–13C is on another Rnq1153–405 molecule. Natural abundance shows the decay from an unlabeled sample with the natural 1.1% 13C signal. (C) The rate of signal decay of Ala-3–13C indicates an in-register β-sheet structure (see text).
The natural abundance (1.1%) 13C in an unlabeled sample shows the expected slow signal decay because the rare 13C atoms are far from each other (Fig. 4B). Forming amyloid from fully Tyr-1–13C-labeled Rnq1153–405 diluted with four parts of unlabeled Rnq1153–405 decreased the rate of decay dramatically, indicating that the decay is due to intermolecular interactions and not intramolecular interactions (Fig. 4B). Assuming that the amyloid structure is a parallel in-register β-sheet, each labeled molecule is adjacent to two others. The “diluted, calc.” curve was calculated assuming that only if both adjacent molecules are unlabeled will the labeled molecule show the decay rate of the natural abundance (unlabeled) molecule. The decay rate of the Tyr-1–13C diluted sample showed a rate of signal decay essentially the same as this expected rate (Fig. 4B).
The data shown in Fig. 4 were collected with hydrated samples. Similar data with dried samples show slightly less rapid decay [supporting information (SI) Fig. 7], suggesting that a fraction of labeled residues are not in parallel in-register β-sheet structure and become mobile, and thus invisible, on hydration. To estimate this fraction, we assume the non-β-sheet residues decay at the rate of natural abundance 13C (Fig. 4B). With this assumption, the data indicate that 3.7 of 15 tyrosine and one of seven leucine residues are not in β-sheet, consistent with the results of 1D spectra described above.
Ala-3–13C-Labeled Rnq1153–405 Rapid Signal Decay Shows Parallel In-Register Structure.
Although the results with Tyr-1–13C and Leu-1–13C molecules strongly suggest an in-register parallel β-sheet structure, the results are actually compatible with a structure out of register by a single amino acid residue. However, successive “R” groups of amino acids in a β-strand point approximately in opposite directions, so that Rnq1153–405 labeled in the methyl group of the 13 Ala residues would show rapid decay indicative of a 0.5-nm nearest neighbor distance for the in-register case, but a much slower decay indicating a >0.8 nm distance if the structure is out of register by a single residue. In fact, the rapid decay indicating in-register structure was observed (Fig. 4C).
Two-Dimensional 13C–13C, 1H–13C, and 15N–13C Solid-State NMR Spectra of Rnq1153–405.
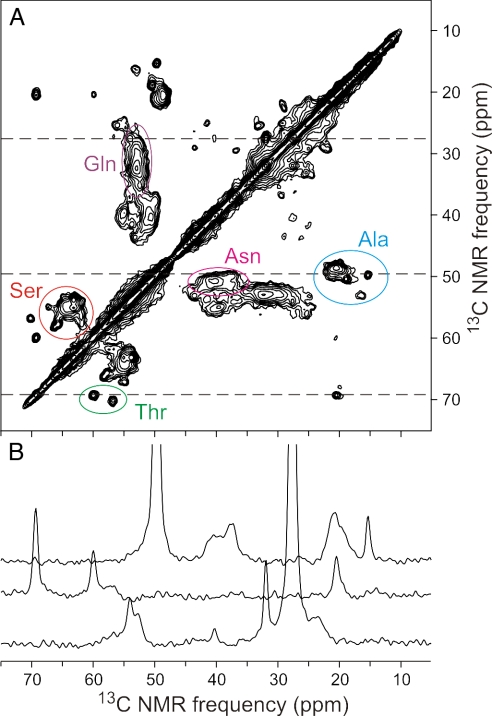
Two dimensional NMR data can indicate the degree of sample heterogeneity, which residues are structured, and, potentially, detailed atomic level structure. Fig. 5 shows the aliphatic part of a 13C–13C 2D experiment (see Methods) using uniformly 13C,15N-labeled Rnq1p153–405. The complete 2D 13C–13C, 15N–13C and 1H-13C NMR spectra are shown in SI Figs. 8–10. Because Rnq1p153–405 has 67 Q, 42 G, 41 N, and 39 S residues, the spectrum is dominated by several areas of overlapping signals (Fig. 5A). However, two alanine αβ resonances and the two threonine αβ peaks are detected with peak widths ≈0.7 ppm, indicating a relatively homogeneous structure at those sites, compared with many amyloids that show peak widths of 1.5–2.5 ppm. Although there are two proline residues, no peaks attributable to proline were identified, suggesting that these residues were mobile or unstructured.
Fig. 5.
Two-dimensional NMR of Rnq1153–405. (A) Aliphatic region of 2D 13C–13C spectrum of U–15N,13C–Rnq1153–405 fibrils, with partial residue-type assignments of Cα/Cβ crosspeaks, obtained at a 17.00-kHz MAS frequency with a total of 55,296 scans and a maximum t1 value of 4.896 ms. Contour levels increase by successive factors of 1.5. (B) One-dimensional slices from the 2D spectrum, at positions indicated by dashed lines.
Most Ala αβ cross-peaks (Fig. 5A) have α resonances below and β resonances above the random coil values of 50.8 and 17.4 (relative to TMS), respectively, indicating β-sheet structure. The two isolated peaks are apparently not in β-sheets. For Thr (random coil values α = 60.1, β = 68.1), one residue has shifts suggesting β-sheet and the other does not (Fig. 5A). Likewise for Ser (random coil α = 56.6, β = 62.1), most intensity appears to be in β-sheet, but a few residues are not. Most Asn (random coil α = 51.4, β = 37.2) and Gln (random coil α = 54.0, β = 27.7) intensity is consistent with β-sheet, but the many residues and broad distribution make it impossible to rule out some non-β-sheet structure (Fig. 5A).
Discussion
A self-propagating amyloid of Rnq1p is the basis of the [PIN+] prion of Saccharomyces cerevisiae. These asparagine/glutamine-rich fibers apparently prime the polymerization of the similarly N/Q-rich prion domains of the Sup35 and Ure2 proteins, initiating formation of the [PSI+] and [URE3] prions, respectively. Here we examine the structure of amyloid of Rnq1p, formed under conditions (6) that produce infectious material. Solid-state NMR measurements of nearest-neighbor distances between tyrosine or leucine residues show distances of ≈0.5 nm, indicative of a parallel in-register β-sheet structure, but consistent with a structure out of register by a single amino acid. Similar experiments with Rnq1153–405 labeled with Ala-3–13C show that the structure is, in fact, in-register. Infectious amyloid of Sup35NM (35) and the prion domain of Ure2p (36) likewise have a parallel in-register β-sheet structure, suggesting that [PIN+] priming of [PSI+] or [URE3] is a simple direct (rare) substitution of a Sup35 or Ure2 molecule for a Rnq1 molecule at a growing end of a fibril.
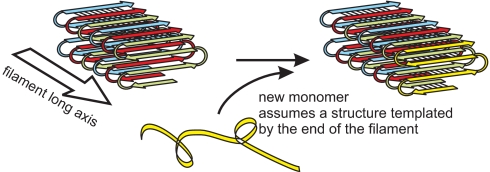
In the parallel in-register β-sheet structure, each residue of the prion domain contacts the same residue in the molecule that was laid down before it and the same residue in the next molecule to join the fibril (Fig. 6). This provides the possibility of passing on structural variations, such as locations of turns of the sheet, lengths of loops, or irregularities in the structure, from parent molecules (already in the fiber) to daughter molecules newly being laid down. This is the essential “templating” mechanism that must be present for these proteins to act as genes. This can explain the prion “variants,” cases where the identical amino acid sequence produces different phenotypes or prion stabilities, doubtless due to differing details of the amyloid structure.
Fig. 6.
Schematic model of fibrils of Rnq1p153–405. There is no information on the lengths or locations of loop regions or on the nature of the contacts between sheets.
This structure also indicates that the sequence of events is not, as often suggested, a primary conformational change of the prion protein followed by aggregation. For example, the monomeric protein could not adopt the structure it has in the fibril because the backbone bonds that drive the structure in the fibril are all intermolecular. It seems evident that the conformational change is coincident with and a consequence of the new molecule joining the fibril, and the details of the conformational change are directed by the structure of the end of the fibril (Fig. 6).
Although amyloid of Rnq1153–405 clearly has a parallel in-register β-sheet structure, two-dimensional 13C–13C NMR experiments show line widths suggestive of significant structural heterogeneity among fibrils, consistent with the known existence of multiple [PIN+] prion variants (12). Most or all of the variants must share the parallel in-register β-sheet structure as a common feature, but the locations of turns or other details may differ among fibrils.
Although amyloids of a number of peptide fragments have been shown to be antiparallel structures, most natural amyloids and amyloids of the prion domains of each of the yeast prions has a parallel in-register structure. The (so far exceptional) HETs prion domain amyloid is proposed to be an intramolecular “pseudo” parallel in-register β-sheet, but in some ways also resembles a β-helix (29, 30).
Further work will be required to more completely define the Rnq1p amyloid structure, the structural differences among prion strains, the structural determinants of interactions with chaperones, and other cellular components.
Methods
Protein Preparation.
Rnq1p153–405 with C-terminal His6 tag was labeled and expressed in E. coli and purified as described in SI Text.
Electron Microscopy.
The formation of fibrils by Rnq1p was confirmed by electron microscopy of negatively stained samples. A fibril suspension was applied to a carbon-coated copper grid for ≈2 min, washed briefly with H2O, stained for ≈2 min with 2% uranyl acetate, blotted, and air dried. The stained samples were examined with an FEI Morgagni transmission electron microscope operating at 80 kV.
Mass Spectrometry.
Incorporation of alanine-3–13C was estimated by mass spectrometric measurements of the chymotryptic peptide SSLASMAQSY, which was ≈67% labeled. There was no evidence for incorporation of 13C into other residues, a conclusion supported by NMR data (see Results).
X-Ray Fiber Diffraction.
Amyloid fibers of Rnq1153–405 were washed extensively with water and packed wet into a 0.7-mm-diameter, thin walled boron-rich glass capillary by centrifugation. The packed fibers were placed in a 0.15418 nm wavelength x-ray beam generated by a rotating anode source operated at 50 kV and 100 mA, coupled with a multilayer focusing optics. Diffraction data were collected in 120-min exposures, during which the sample was continuously rotated around an axis perpendicular to the beam. Diffraction data were recorded on an RaxisIV 2-dimensional imaging plate detector placed 15 cm from the capillary. Background data were collected similarly, but exposing a part of the same capillary that was free of fiber but was filled with water. The data were converted to intensity vs. 2θ plots by circular summation around the center of the two-dimensional images using the R-axis Display Program (Rigaku Americas). Background was subtracted and the resulting plots were displayed by Origin 7 (OriginLab)
Solid-State NMR Methods.
Solid-state NMR experiments on selectively labeled Rnq1p153–405 were carried out at 9.39 T (100.4 MHz 13C NMR frequency) using an InfinityPlus spectrometer (Varian) and magic angle spinning (MAS) NMR probes (Varian) with 3.2 mm diameter rotors. All measurements were at room temperature. 13C NMR spectra were recorded at an MAS frequency of 20.00 kHz with 1H–13C cross-polarization (41) and two-pulse phase-modulated 1H decoupling (42). The dipolar recoupling measurements were carried out at an MAS frequency of 20.00 kHz using the PITHIRDS-CT method (40). Pulsed spin-lock detection was used for improved signal-to-noise ratio (43). Rnq1153–405 fibrils were washed extensively with H2O, lyophilized, packed into rotors (4- to 6-mg sample masses) and rehydrated by addition of ≈5 μl of H2O. Each PITHIRDS-CT data point is the result of 512–576 scans with a 4-s recycle delay.
The original PITHIRDS-CT data, Sraw(t) were corrected for the 1.1% natural abundance of 13C as follows: (i) PITHIRDS-CT data were recorded for an unlabeled sample of Rnq1153–405 (Fig. 4B) and, assuming linear decay, the natural abundance signal is Sna = 100 − 0.208 t, where t is the effective dipolar dephasing time in milliseconds; (ii) the number of natural abundance 13C nuclei per Rnq1153–405 molecule, Nna, is 0.011 × (total sites − labeled nuclei), where total sites is 372 carbonyls or 35 methyls. Labeled nuclei, Nlabel, was 7, 15 and 8.7 for Leu, Tyr, and Ala, respectively, taking account of the 67% labeling of alanines. The corrected PITHIRDS-CT data were S(t) = [Sraw(t) − fna Sna(t)]/(1 − fna), where fna = Nna/(Nna + Nlabel) and Sraw(0) = 100. Standard errors are shown.
Measurements on uniformly 15N,13C-labeled Rnq1 (U-15N,13C-Rnq1) fibrils were performed in a 17.6 T field (747.9 MHz 1H NMR frequency, 188.1 MHz 13C NMR frequency, 75.8 MHz 15N NMR frequency), using a Varian Infinity spectrometer console and a three-channel MAS probe with 1.8 mm rotor diameters (produced by the research group of Ago Samoson, National Institute of Chemical Physics and Biophysics, Tallinn, Estonia). Lyophilized U–15N,13C–Rnq1 fibrils were packed into the MAS rotor and then rehydrated by addition of deionized water until the sample appeared translucent when viewed in the rotor with strong illumination. 2D 13C–13C and 15N–13C NMR spectra were recorded at a 17.00 kHz MAS frequency. Two-dimensional 1H-13C spectra were recorded at a 40.00 kHz MAS frequency. A 2.824-ms (48 rotor periods) finite-pulse RFDR sequence was applied in the mixing period of 2D 13C-13C measurements, with 18.0-μs 13C π pulses. The mixing period in 2D 15N–13C measurements consisted of a 6.00-ms 15N–13C cross-polarization period, optimized for polarization transfer to α-carbons, followed by a 20-ms 13C–13C spin diffusion period. A 200-μs 1H–13C cross-polarization period was used for mixing in 2D 1H–13C measurements. Proton decoupling fields in all measurements were 110 kHz.
Supplementary Material
ACKNOWLEDGMENTS.
R.B.W. thanks Kent Thurber, Anant Paravastu, Sorin Luca, and Kan Hu for frequent advice, encouragement, and rescue; Frank Shewmaker for advice on protein purification; and Eric Anderson for mass spectrometry. This work was supported by the Intramural Program of the National Institute of Diabetes Digestive and Kidney Diseases of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712032105/DC1.
References
- 1.Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in S. cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 2.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 3.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derkatch IL, et al. Dependence and independence of [PSI+] and [PIN+]: A two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN]. Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 6.Patel BK, Liebman SW. “Prion proof” for [PIN+]: Infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+]. J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 8.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 9.Merrin AB, et al. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- 11.Vitrenko YA, Gracheva EO, Richmond JE, Leibman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem. 2007;282:1779–1787. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- 12.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradley ME, Liebman SW. Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 15.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 17.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279:48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 18.Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of α-synuclein fibrils studied by site-directed spin labeling. J Biol Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 19.Cobb NJ, Sonnichsen FD, Mchaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register β-structure. Proc Natl Acad Sci USA. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tycko R. Molecular structure of amyloid fibrils: Insights from solid-state NMR. Q Rev Biophys. 2006;1:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 21.Lansbury PT, Jr, et al. Structural model for the beta-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol. 1995;2:990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 22.Benzinger TL, et al. Propagating structure of Alzheimer's beta-amyloid(10–35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory DM, et al. Dipolar recoupling NMR of biomolecular self-assemblies: Determining inter- and intrastrand distances in fibrilized Alzheimer's β-amyloid peptide. Solid State Nucl Magn Reson. 1998;13:149–166. doi: 10.1016/s0926-2040(98)00086-1. [DOI] [PubMed] [Google Scholar]
- 24.Burkoth TS, et al. Structure of the β-amyloid(10–35) fibril. J Am Chem Soc. 2000;122:7883–7889. [Google Scholar]
- 25.Antzutkin ON, et al. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of beta-sheets in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci USA. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petkova AT, et al. A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luca S, Yau W-M, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007 doi: 10.1021/bi701427q. PMID: 17979302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siemer AB, et al. 13C, 15N resonance assignment of parts of the HET-s prion protein in its amyloid form. J Biomol NMR. 2006;34:75–87. doi: 10.1007/s10858-005-5582-7. [DOI] [PubMed] [Google Scholar]
- 31.Jaroniec CP, et al. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.TerAvanesyan A, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 34.Ross ED, Minton AP, Wickner RB. Prion domains: Sequences, structures and interactions. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- 35.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxa U, et al. Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 37.Chan JCC, Oyler NA, Yau W-M, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitrenko YA, Pavon ME, Stone SI, Liebman SW. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr Genet. 2007;51:309–319. doi: 10.1007/s00294-007-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. H-1, C-13 and N-15 random coil NMR chemical-shifts of the common amino-acids. 1. Investigations of Nearest-Neighbor Effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 40.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid-state NMR. J Chem Phys. 2007;126 doi: 10.1063/1.2437194. 064506. [DOI] [PubMed] [Google Scholar]
- 41.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59:569–590. [Google Scholar]
- 42.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 43.Petkova AT, Tycko R. Sensitivity enhancement in structural measurements by solid state NMR through pulsed spin locking. J Magn Reson. 2002;155:293–299. doi: 10.1006/jmre.2002.2519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.