Abstract
Cancer stem cells (CSCs) are critical for the initiation, propagation, and treatment resistance of multiple cancers. Yet functional interactions between specific signaling pathways in solid organ “cancer stem cells,” such as those of the liver, remain elusive. We report that in regenerating human liver, two to four cells per 30,000–50,000 cells express stem cell proteins Stat3, Oct4, and Nanog, along with the prodifferentiation proteins TGF-β-receptor type II (TBRII) and embryonic liver fodrin (ELF). Examination of human hepatocellular cancer (HCC) reveals cells that label with stem cell markers that have unexpectedly lost TBRII and ELF. elf+/− mice spontaneously develop HCC; expression analysis of these tumors highlighted the marked activation of the genes involved in the IL-6 signaling pathway, including IL-6 and Stat3, suggesting that HCC could arise from an IL-6-driven transformed stem cell with inactivated TGF-β signaling. Similarly, suppression of IL-6 signaling, through the generation of mouse knockouts involving a positive regulator of IL-6, Inter-alpha-trypsin inhibitor-heavy chain-4 (ITIH4), resulted in reduction in HCC in elf+/− mice. This study reveals an unexpected functional link between IL-6, a major stem cell signaling pathway, and the TGF-β signaling pathway in the modulation of mammalian HCC, a lethal cancer of the foregut. These experiments suggest an important therapeutic role for targeting IL-6 in HCCs lacking a functional TGF-β pathway.
Keywords: hepatocellular cancer, spectrin, embryonic liver fodrin, Smads, Stat3
Although the existence of cancer stem cells (CSCs) was first proposed >40 years ago (1, 2), only in the past decade have these cells been identified in hematological malignancies and, more recently, in solid tumors that include breast, prostate, brain, and colon (3). Exploration of the difference between CSCs and normal stem cells is crucial not only for the understanding of tumor biology but also for the development of specific therapies that effectively target these cells in patients (4). Yet, the origin of CSCs and the mechanisms by which they arise remain elusive. For tumors containing a subpopulation of CSCs, there are at least two proposed mechanisms for how the CSCs could have arisen: oncogenic mutations that inactivate the constraints on normal stem cell expansion or, alternatively, in a more differentiated cell, oncogenic mutations could generate continual proliferation of cells in cell cycle that no longer enter a postmitotic differentiated state, thereby creating a pool of self-renewing cells in which further mutations can accumulate. The plasticity of such cells is reflected by recent studies where pluripotent stem cells could be induced from embryonic or adult fibroblasts by introducing four factors, Oct3/4, Sox2, c-Myc, and Klf4, under embryonic stem cell culture conditions (5). Potentially biologically significant pathways that modulate these stem/progenitor cells in cancer tissues could be identified through dual roles in embryonic stem cell development and tumor activation or suppression (4).
Multiple signaling networks orchestrate the development and differentiation of embryonic stem (ES) and somatic stem cells into functional neuronal, hematopoietic, mesenchymal, and epithelial lineages. Among these, the signaling mechanisms activated by TGF-β family proteins have emerged as key players in the self-renewal and maintenance of stem cells in their undifferentiated state, the selection of a differentiation lineage, and the progression of differentiation along individual lineage (4). Through gene knockout experiments and observation of ES cells, TGF-β-family proteins have emerged as bifunctional regulators of the maturation of cells in each of the lineages mentioned above and as suppressors of carcinogenesis (6). When TGF-β signaling is disrupted, the imbalance can result in an undifferentiated phenotype, and cancer may ensue (7). TGF-β-family signals are conveyed through two types (types I and II) of transmembrane receptor serine-threonine kinases, which form a complex at the cell surface. Ligand binding to this complex induces a conformational change that results in phosphorylation and activation of type I receptors by type II receptors. Activation of Smad transcription factors ensues and results in their nuclear translocation and activation or repression of gene expression.
Smad activation is modulated by various receptors or Smad-interacting proteins that include ubiquitin and small ubiquitin-related modifier (SUMO) ligases and multiple adaptor proteins that include Smad anchor for receptor activation (SARA), filamin, and ELF. ELF, a β-spectrin, first isolated from foregut endodermal stem cell libraries, is crucial for the propagation of TGF-β signaling (8). Specifically, ELF associates with Smad3 presenting it to the cytoplasmic domain of the TGF-β Type I receptor complex, followed by heteromeric complex association with Smad4, nuclear translocation, and target gene activation (9). Depending on the differentiated state of the target cell, the local environment, and the identity and dosage of the ligand, TGF-β proteins promote or inhibit cell proliferation, apoptosis, and differentiation. TGF-β-family signaling is most prominent at the interface between development and cancer in gut epithelial cells. Inactivation of at least one of the TGF-β signaling components (such as the TGF-β receptors, Smad2 or Smad4) occurs in almost all gastrointestinal tumors (7, 10). Smad2+/−/smad3+/− double heterozygous and elf−/− homozygous mice all showed defective liver development, and elf+/− mice are now observed to develop dramatic spontaneous HCCs. Genetic studies thus identify TBRII and ELF as functional suppressors of HCC formation (11).
Development of HCC occurs through progression of liver injury initiated by chronic hepatitis, extensive alcohol intake, or toxins, sequentially resulting in liver cirrhosis, dysplastic lesions, and finally, invasive liver carcinoma (12). Recent studies suggest these agents can target liver progenitor cells [oval cells in rodents and hepatic progenitor cells (HPC) in humans], leading to their expansion and transformation (13, 14). A considerable proportion of HCCs express one or more HPC marker not present in normal, mature hepatocytes (15, 16). Similarly, HPCs occur in HCC precursors such as small cell dysplastic foci and hepatocellular adenoma (17). These findings suggest that human liver tumors can be derived from hepatic stem cells rather than from mature cell types.
In this report, evidence suggests that human HCC could arise as a direct consequence of dysregulated proliferation of hepatic progenitor cells in a setting where TGF-β has been disrupted. Using human HCC and mouse genetic models, we show that lesions in the TGF-β pathway normally result in a “homeostatic” activation of the IL-6 pathway, that appears to be critical to the development of hepatic cancer.
Results
Hepatic Stem Cells Are Found in Normal Liver and HCCs.
To search for hepatic stem cells, we studied five patients with monthly posttransplantation liver biopsies. Living donor liver transplantation offers a unique opportunity to examine the regeneration of the human liver, a process presumed to involve the recruitment of hepatocytes and later, hepatic progenitor cells (18). The surgical procedure involves resection and transplant of a lobe representing 55–60% liver mass from a donor to a recipient, which, by 3 months, grows to 85% of original mass (19). We hypothesized that, at the end of liver regeneration, there would be an expanded population of liver progenitor/stem cells that were long-term label-retaining (20). A broad microarray and protein analysis approach led us to focus on 40 proteins to be further characterized by immunohistochemical and confocal immunofluorescence labeling of living donor liver-transplanted and human HCC tissues. We ultimately directed our search for cells expressing five of these proteins: Oct4, Nanog, Stat3, TBRII, and ELF. Both Oct4 (21) and Nanog (22) have been shown to be expressed in embryonic and pluripotent stem cells; Stat3 appears to be essential for embryonic visceral endoderm development and for self-renewal of pluripotent embryonic stem cells (23, 24); both TBRII and ELF have been implicated in early embryonic development of the foregut and in endodermal malignancies (9). Serial sections were examined by immunohistochemistry to help determine the local microscopic anatomy of the visualized cells (i.e., relationship to portal structures, etc.) and the number of cells comprising the cluster. We identified a cluster of two to four cells out of the entire 30,000- to 50,000-cell population of living donor liver-transplanted specimens that expressed Stat3, Oct4, and Nanog and TGF-β signaling proteins, TBRII and ELF. These cells, in supplemental models, also stained positively for both a hepatocytic cell lineage marker, albumin, and a cholangiocytic cell lineage marker, cytokeratin-19 (CK19), along with phosphorylated histone H3, a marker for active proliferation [Figs. 1 and 2 A–D; supporting information (SI) Figs. 5 and 6] (20). These putative progenitor/stem cells were generally found localized in the portal tract region surrounded by a “shell” of six to seven cells expressing TBRII, ELF, and albumin, but not Nanog or Oct4, reflecting a more differentiated phenotype (Fig. 2 A–D and SI Fig. 6). These findings, together with the known role of the TGF-β signaling pathway in liver development, led us to hypothesize that: (i) these Stat3+, Oct4+, Nanog+, TBRII+, and ELF+ cells represent the progenitor/stem cell pool that becomes activated during the regenerative process; and (ii) TBRII and ELF may be involved in the initiation of hepatocyte differentiation of Stat3+/Oct4+ progenitor/stem cells. To our knowledge, the demonstration of a hepatic progenitor/stem cell in postembryonic human liver is previously undescribed.
Fig. 1.
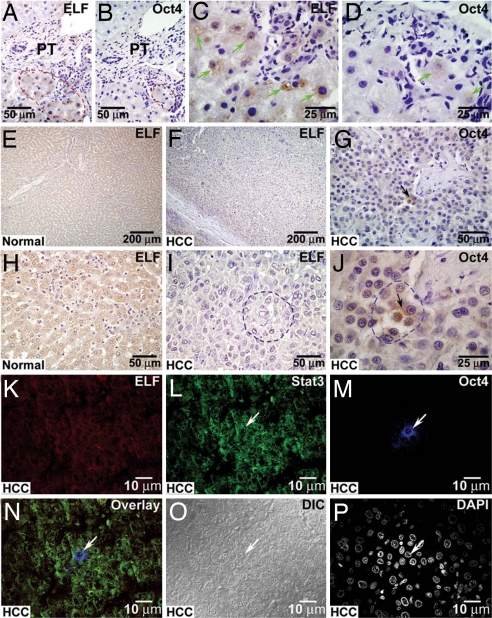
Identification of liver progenitor/stem cells in posttransplant human liver tissues. Immunohistochemical labeling of posttransplant human liver tissue taken from living donor liver transplant 4 weeks after transplantation. The tissue is labeled for the presence of ELF (A, arrows) and Oct4 (B, arrows). Sections are taken consecutively to enable identical localization. (C–N) Confocal images of human liver at 3 months after living-related liver transplantation. (C–E) The tissue is labeled with stem cell proteins Stat3 and Oct4 and prodifferentiation TGF-β signaling component ELF. (F) These proteins coexpress in a small cluster of two to four cells. (G) DAPI represents nuclear labeling. (H) Differential interference chromatography (DIC) represents a transmission image of this cluster of cells. (I–K) Regenerative liver tissue from another liver transplant is labeled with p-histone H3 (Ser10), Oct4, and ELF. (L) These proteins coexpress in this cluster of progenitor-like cells. (M and N) DAPI represents nuclear labeling (M), and DIC represents transmission images (N). Arrows point to the nuclei of the progenitor-like cells. (Scale bars for all figures are in micrometers.)
Fig. 2.
Identification of liver progenitor/stem cells in posttransplant human liver and HCC tissues. Immunohistochemical labeling of posttransplant human liver tissues taken from living donor liver transplant recipient 4 weeks after transplantation. (A–D) The tissue is labeled for the presence of ELF (A and C) and Oct4 (B and D). Sections are taken consecutively to enable identical localization. (E–J) Equivalent areas are marked by red dotted lines, and green arrows point to the positive labeling. Immunohistochemical labeling of normal human liver (E and H) and HCC tissues (F, G, I, and J). The loss of ELF is evident when comparing the immunohistochemical labeling of normal (E and H) and HCC samples (F and I). Strikingly, there are small pockets of three to four Oct4 positively stained cells present in the midst of transformed hepatocellular cells (G and J, arrows). These Oct4-positive cells are stained negatively for ELF (I, area marked by blue dotted line). (K–M) Confocal images of human HCC labeled to highlight prodifferentiation TGF-β signaling component ELF (K) and progenitor cell proteins Stat3 and Oct4 (L and M). (N, white arrow) The overlay image demonstrates a cell that labeled positively for Stat3 and Oct4 but lacks nuclear expression of ELF. (O) DIC represents transmission image of this cluster of cells. (P) DAPI represents nuclear labeling. The white arrow points to the nucleus of the HCC progenitor/stem cell lacking ELF. PT represents portal tract.
Ablating TGF-β Signaling Results in Spontaneous HCCs.
Our observations demonstrating the presence of TGF-β signaling components TBRII and ELF in human hepatic stem cells led us to explore the impact of these components of the TGF-β pathway on liver development and tumorigenesis. As we have reported, mice homozygous for elf (elf−/−) undergo midgestational death with hypoplastic livers (9). Heterozygous elf+/− mice, however, spontaneously developed tumors of the liver with an incidence of 40% (SI Fig. 7). Liver lesions included early centrilobular steatosis and dysplasia in most sections, with nuclear disarray and stratification, mitosis and apoptosis, proceeding to poorly differentiated carcinoma (SI Fig. 7 A and E–H). We hypothesized that the interruption of the TGF-β pathway resulted in hepatocellular carcinoma through disruption of a normal pattern of cellular differentiation by hepatic progenitor/stem cells.
We examined human HCC tissue specimens from 10 individuals. In 9 of the 10 HCC tissues, we observed a small strongly positive cluster of three to four Oct4+ cells that was negative for TBRII and ELF (Fig. 2 E–J and SI Fig. 8). Cells with this phenotype were never observed during our surveys of either normal liver or of biopsies from regenerating organs. We speculate that these Stat3+/Oct4+-positive human HCC cells that have lost TGF-β signaling proteins have the potential to give rise to HCCs (Fig. 2 K–P).
Activated IL-6 Signaling Is Found in HCC with Impaired TGF-β Signaling.
To obtain a molecular signature of hepatic cancer that arises when TGF-β signaling is inactivated and to define the intracellular pathways that are engaged, we performed a series of microarray and proteomic analyses in elf+/−, elf+/−/itih4−/−, and itih4−/− tissues. Significantly increased expression of the IL-6/Stat3, WNT, and CDK4 signaling pathways was observed (Fig. 3 A and B; SI Figs. 9 A–F and 10 B and E and SI Table 1 and data not shown). The previously described association of increased IL-6 signaling activity in hepatic tumorigenesis (25, 26) led us to focus our attention on the IL-6 pathway.
Fig. 3.
Decreased incidence of hepatocellular cancer is observed by genetic modulation of IL-6-stat-3 signaling. (A and B) Heatmap microarray assay illustrating gene expression in mouse liver or HCC tissues. Targeted disruption of the ITIH-4 gene and generation of itih4−/− mice. Exp1: elf+/− liver tissue vs. wild-type liver tissue; Exp2: itih4−/− liver tissue vs. wild-type liver tissue; Exp3: elf+/−/itih4−/− liver tissue vs. wild-type liver tissue. The signal gradients are located below each image. (C) The targeting vector for itih4 gene; the targeting strategy deletes a 1.8-kb SmaI-ClaI fragment that contains second and third exons of the itih4 gene. (D) Southern blot analysis shows ES cells heterozygous (i151, i155, and i160) with correct homologous recombination events within the itih4 locus. Genomic DNA from these clones was digested with EcoRV, followed by Southern blot using a 1.8-kb fragment 3′ to the targeting vector. (E) Immunoblot analysis using the antibody specific for ITIH4 shows loss of ITIH4 expression in itih4−/− and elf+/−/itih4−/− liver tissue lysates compared with the wild-type and elf+/− samples. (F) Kaplan–Meier tumor-free survival curves of wild-type (control), elf+/−, itih4−/−, and elf+/−/itih4−/− (experimental) animals.
Down-Regulation of the IL-6 Pathway by itih4−/− Ablation Inhibits HCC Formation.
How might the increased activity of the IL-6 pathway in HCC associated with impaired TGF-β signaling be linked to the cancer phenotype? We hypothesized that the increased activity of the IL-6 pathway, occurring in hepatic progenitor/stem cells lacking competent TGF-β circuitry, directly resulted in disturbed growth and differentiation of these liver precursors. To test the hypothesis that the increased activity of the IL-6 pathway was a critical step in tumorigenesis and not a consequence, we attempted construction of a mouse defective in IL-6 signaling on a heterozygous elf background. However, stat3-null mice are embryonic lethal, and IL-6 null mice were similarly too fragile to intercross to obtain a homozygous IL-6-null on a heterozygous elf background (27, 28).
We recently engineered a mouse in which the gene for IL-6 regulated protein, itih4, has been deleted (29). ITIH4 is a member of a liver-restricted serine protease inhibitor family, expressed in hepatoblasts, and is a biomarker of foregut cancers of uncertain function (30–32). Mice homozygous for the itih4 mutation (itih4−/−) are normal and fertile, suggesting that the itih4 mutation does not show dominant effects (Fig. 3 C–E). Surprisingly, however, IL-6/Stat3 signaling is one of the most significantly suppressed pathways we detected in the itih4−/− liver tissues (Fig. 3 A, B, and E; SI Figs. 5 and 6 and SI Table 1), far more than the WNT or CKD4 pathways. Interestingly, hepatocytes remained well differentiated in the itih4−/− mice. We suspect that ITIH4, an acute-phase protein, might be associated in a positive-feedback loop with IL-6 (33–35), a regulatory property that characterizes several acute-phase gene products.
To explore the role of IL-6 activation in HCC associated with ELF deficiency, we generated mouse intercrosses between elf+/− mice and itih4−/− mice (Fig. 3 C and D). An examination of elf+/−/itih4−/− mice for tumor development revealed that only 1 of 25 (4%) elf+/−/itih4−/− mice developed HCC, compared with 10 of 25 (40%) elf+/− mice that developed HCC (Fig. 3F). The tumor that developed in the elf+/−/itih4−/− mouse was 0.4 cm3 in size, small compared with the larger 3- to 4-cm3 tumors seen in the elf+/− mice (Fig. 3F).
Microarray profiles of itih4−/− and elf+/−/itih4−/− liver tissues indicated a significant suppression of IL-6 signaling (Fig. 3 A and B and SI Table 1). Similarly, immunoblot and immunohistochemical analyses showed that expression of IL-6/Stat3 is decreased in the itih4−/− and elf+/−/itih4−/− liver tissues (SI Figs. 9 and 10), whereas, in contrast, IL-6 is activated in elf+/− mice (SI Table 1). Stat3 phosphorylation is also dramatically decreased in the itih4−/− and elf+/−/itih4−/− liver tissues (Fig. 4 A–D and SI Fig. 9). The disruption of IL-6/Stat3 signaling in the liver tissues of elf+/−/itih4−/− mutant liver tissue was similar to that seen in the itih4−/− liver tissues.
Fig. 4.
Analysis of gene expression in mouse liver tissues and human HCC tissues and cell lines. (A, C, and D) Immunohistochemical labeling demonstrates low/absent expression of phosphorylated Stat3 in normal (wild-type) mouse liver (A), ITIH4−/− liver (C), and Elf+/−/ITIH4−/− liver (D). (B) In contrast, Elf+/− HCC liver tissue shows increased expression of P-Stat3. (E and F) Immunohistochemical detection shows increased expression of Stat3 in human HCC tissues (F, arrows) compared with normal liver tissues (E). (H and G) Phosphorylated-Stat3 is also increased in human HCC tissues (H, arrows) compared with normal liver tissues (G). (Scale bar is in micrometers.)
Consistent with the results obtained in mice, we demonstrated marked elevation of expression of Stat3, p-Stat3, and ITIH4 by immunohistochemistry in human HCC tissues (Fig. 4 E–H; SI Fig. 11 and SI Table 1). In addition, markedly increased Stat3 and p-Stat3 expression was observed in SNU-398 cells derived from a human HCC cell line that does not express TBRII and ELF (SI Fig. 12). These data support our hypothesis that increased IL-6 signaling characterizes human HCC.
Increased Liver Cell Proliferation and Decreased Apoptosis Are Observed with Inactivation of TGF-β Signaling in elf+/− Mice.
How does increased IL-6 signaling result in HCC in the setting where the TGF-β pathway is disrupted? Immunohistochemical analysis of epithelial proliferation by labeling the mouse liver tissues with antibody specific to p-histone H3 (Ser10) showed a significant decrease in the mitotic labeling in itih4−/− (SI Fig. 13B) and elf+/−/itih4−/− (SI Fig. 13C) mutant liver tissues compared with normal wild-type and elf+/− epithelium (SI Fig. 13 A and D). This suggests that hepatocyte proliferation is inhibited in the TGF-β-inactivated state by the disruption of itih4 and, therefore, that IL-6 by some mechanism increases the proportion of proliferating hepatocytes in the absence of TGF-β.
Suppression of the IL-6/Stat3 pathway and TGF-β signaling in elf+/−/itih4−/− cells might be expected to impact on hepatocyte apoptosis (25, 36). Epithelial apoptosis in the mouse liver tissues was examined by using the apoptotic marker, anti-active Caspase-3. In wild-type control mice, apoptosis was noted in hepatocytes (SI Fig. 13E), but few apoptotic cells were seen in itih4−/− and elf+/− mice compared with elf+/−/itih4−/− mutant liver tissue (SI Fig. 13 F–H). We conclude that impairment of TGF-β signaling in liver results in suppression of apoptosis, which can partially be restored by an increase in the IL-6 pathway. In a setting where both the IL-6 and TGF-β circuits are defective, normal apoptosis is physiologically depressed.
Discussion
Clonal studies indicate that hepatocarcinogenesis arises from dysfunctional liver stem cells, and this is further supported by transformation of p53-null hepatic progenitor cells that give rise to HCC (37). A considerable proportion of HCCs express one or more HPC markers, and both hepatocyte and biliary cell markers such as albumin, CK7, and CK19 that are not present in normal mature hepatocytes (15, 16). HCCs that express these HPC markers carry a significantly poorer prognosis and higher recurrence after surgical resection and liver transplantation (38). Fifty-five percent of small dysplastic foci (<1 mm in size), early premalignant lesions, are comprised of progenitor cells and intermediate hepatocytes (39). Indeed, progenitor-like side populations of Huh7 and PLC/PRF/5 cells (human HCC cell lines), with hepatocytic and cholangiocytic lineages, give rise to persistent aggressive tumors upon serial transplantation in immunodeficient NOD/SCID mice (40).
In this study, we demonstrate the cluster of two to four putative progenitor/stem cells per 30,000–50,000 cells in regenerating liver; expressing stem cell markers Stat3, Oct4, and Nanog; and TGF-β signaling proteins TBRII and Smad adaptor protein, ELF, and progenitor cell markers by labeling for both hepatocytic cell lineage markers (albumin) and cholangiocytic cell lineage markers (CK19), along with phosphorylated histone H3, a marker for active proliferation. These putative progenitor/stem cells are generally found localized in the portal tract region surrounded by a “shell” of six to seven cells expressing TBRII, ELF, and albumin, but not Nanog or Oct4, the latter reflecting a more differentiated phenotype, further supporting the role of the progenitor cell in self-renewal and differentiation. We observed small strongly positive clusters of Oct4+ cells that were negative for TBRII and ELF in HCCs (Fig. 2 E–J and SI Fig. 8). Given the important role of TGF-β signaling in liver development and in suppression of hepatocarcinogenesis supported by genetic studies, the Stat3+/Oct4+-positive human HCC cells that have lost TGF-β signaling proteins are likely the HCC progenitor/stem cells that give rise to HCCs. We suggest that these “tumorigenic” progenitor cells might potentially be attractive targets for therapeutic intervention in HCC.
Several TGF-β signaling components are bona fide tumor suppressors with the ability to constrain cell growth and inhibit cancer development at its early stages. Inactivation of at least one of these components (such as the TGF-β receptors, Smad2, or the common mediator Smad4) occurs in almost all gastrointestinal tumors (6, 10, 41). For example, the early embryonic lethality in smad4−/− mice is consistent with the role of Smad4 in normal gut endoderm development. However, the specific roles of the TGF-β pathway in vivo human progenitor systems are unknown. As illustrated in this study, changes in TGF-β signaling drive the selection of defined differentiation pathways and their progression of differentiation in liver tissue.
Deregulation of TGF-β signaling may contribute to impaired differentiation and allow for the development of cancers, linking the differentiation of stem cells with suppression of carcinogenesis. In addition to the loss of TGF-β signaling that occurs in HCC, development of this cancer appears to require IL-6. In turn, increased ITIH4, an IL-6 target, appears to be a critical mediator of hepatocarcinogenesis (see schematic in SI Fig. 14). Therefore, this study reveals a surprising and important functional role of the serine protease inhibitor ITIH4 in hepatocellular transformation, previously identified as an IL-6 regulated biomarker for cancers of the foregut with no known function. Further support comes from current therapeutics in cancer that involve successful strategies at blocking IL-6 signaling (42). Modulation of IL-6 signaling in cancer progenitor cells may provide an important approach for new therapeutics in cancers with poor prognosis such as HCC.
Experimental Procedures
Construction of the Targeting Vector and Generation of Mice Carrying Mutations.
Targeting vector.
Recombinant phage-containing genomic DNA of the itih4 locus was isolated from a 129/SvEv mouse library by using PK7R, a piece of itih4 cDNA, as a probe. The finished construct, p-itih4Neo, is shown in Fig. 3C. This targeting strategy deletes a 1.8-kb SmaI-ClaI fragment that contains the second and third exons of the itih4 gene.
Homologous recombination in ES cells and generation of germ-line chimeras.
TC1 ES cells were transfected with NotI digested p-itih4Neo and selected with G418 and FIAU. ES cell clones that were resistant to both G418 and FIAU were picked and analyzed by Southern blotting for homologous recombination events within the itih4 locus (Fig. 3D). Details are in SI Text.
Confocal Laser-Scanning Immunofluorescence Microscopy.
Colocalization studies were performed with anti-ELF, -Stat3, and -Oct4 by using human regenerating liver and HCC tissues. Normal wild-type, elf+/−, itih4−/−, and elf+/−/itih4−/− mutant livers and HCC tissues were also used for the confocal microscopy. Peptide-specific monoclonal mouse and rabbit polyclonal primary antibodies were visualized with tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG or FITC-conjugated goat anti-mouse IgG. Samples were analyzed with a Bio-Rad MRC-600 confocal microscope (Bio-Rad), with an ILT model 5470K laser (Ion Laser Technology) as the source for the crypton-argon ion laser beam.
Generation of Mouse Embryo-Derived Fibroblasts (MEFs).
MEFs harboring the null-allele elf and itih4 and wild-type were derived as described (9).
Immunoblot Assay.
For assaying endogenous TBRII, ELF, ITIH4, IL-6, Stat3, pStat3, protein lysates of human HCC cells (SNU-182 (CRL-2235), SNU-398 (CRL-2233), and SNU-449 (CRL-2234) (American Type Culture Collection), MEFs, and normal wild-type, elf+/−, itih4−/−, and elf+/−/itih4−/− mutant liver and HCC tissues were immunoblotted with the indicated antipeptide or antiphospho-specific antibodies (Santa Cruz Biotechnology, Invitrogen, and Abcam).
Histological Analysis and Antibody Staining.
Mice exhibiting overt pathological signs were killed and underwent autopsy. Normal liver and HCC tissues were dissected, fixed with 4% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned at 6 μm. Sections were stained with H&E or subjected to immunohistochemical analysis with antibodies. Immunohistochemical staining was performed with primary antibodies against ELF, Oct4, ITIH4, Stat3, pStat3, pHistone H3 (Ser10), and Caspase-3 (Santa Cruz Biotechnology, Invitrogen, Promega, and Abcam).
Detection of Proliferating Cells.
Proliferating cells were labeled with BrdU-labeling and detection kit (Invitrogen). BrdU (1 ml/100 g of body weight) was injected (i.v.) into 18.5-day postcoitum pregnant mice and 4 h later, fetal stomachs were fixed with 4% paraformaldehyde, embedded in paraffin, and sectioned at 6 μm. The proliferating cells were also identified by anti-pHistone H3 (Ser10) mitotic marker labeling.
Detection of Apoptotic Cells.
Apoptotic cells were detected by the TUNEL method with a MEB STAIN Apoptosis Kit Direct (MBL, 8445) and with anti-Caspase3 antibody (Promega). Tissues were then fixed and analyzed by using immunofluorescence microscopy.
Tumor Cells and Tissues.
Elf+/− mice were intercrossed with itih4−/− mice to obtain elf+/−/itih4−/−mice. Liver and HCC tissues were collected and cultured as described (43). Two different elf+/− HCC cancer cell lines were tested in different experiments, and the results obtained were also independent of passage number. Representative data are shown. The diagnosis of paraffin-mounted tissue biopsies from human HCC and normal liver were microscopically confirmed by pathologists, and an indirect immunoperoxidase procedure was used for immunohistochemical localization of Oct4, TBRII, and ELF protein as described above.
Microarray.
Custom-designed 44K human 60-mer oligo microarrays (Agilent Technologies) were used for the array experiments. Total RNA was extracted from mouse liver and HCC tissues and MEFs by using RNeasy kit (Qiagen).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Drs. John Kim Jessup, Stephen Byers, and Richard Schlegel for critical review of and helpful suggestions with the manuscript; and Tiffany Blake, Rupen Amin, Varalakshmi Katuri, Ed Flores, Eugene Volpe, and Merlyn Deng for excellent technical expertise, manuscript preparation, and help with immunohistochemistry. This work was supported by the National Institutes of Health [Grants RO1 CA106614A (to L.M.), RO1 DK56111 (to L.M.), RO1 CA4285718A (to L.M.), and RO1 DK58637 (to B.M.)], a Veterans Administration Merit Award (to L.M.), an R. Robert and Sally D. Funderburg Research Scholar award (to L.M.), and the Benn Orr Scholar Award (to L.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705395105/DC1.
References
- 1.Bergsagel DE, Valeriote FA. Cancer Res. 1968;28:2187–2196. [PubMed] [Google Scholar]
- 2.Salsbury AJ. Cancer Treat Rev. 1975;2:55–72. doi: 10.1016/s0305-7372(75)80015-6. [DOI] [PubMed] [Google Scholar]
- 3.Morrison SJ, Kimble J. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 4.Mishra L, Derynck R, Mishra B. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Massague J, Blain SW, Lo RS. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 7.Mishra L, Shetty K, Tang Y, Stuart A, Byers SW. Oncogene. 2005;24:5775–5789. doi: 10.1038/sj.onc.1208924. [DOI] [PubMed] [Google Scholar]
- 8.Mishra L, Cai T, Yu P, Monga SP, Mishra B. Oncogene. 1999;18:353–364. doi: 10.1038/sj.onc.1202313. [DOI] [PubMed] [Google Scholar]
- 9.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Science. 2003;299:574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 10.Weinstein M, Yang X, Deng C. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 11.Kanzler S, Meyer E, Lohse AW, Schirmacher P, Henninger J, Galle PR, Blessing M. Oncogene. 2001;20:5015–5024. doi: 10.1038/sj.onc.1204544. [DOI] [PubMed] [Google Scholar]
- 12.Farazi PA, DePinho RA. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 13.Alison MR. Stem Cell Rev. 2005;1:253–260. doi: 10.1385/SCR:1:3:253. [DOI] [PubMed] [Google Scholar]
- 14.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durnez A, Verslype C, Nevens F, Fevery J, Aerts R, Pirenne J, Lesaffre E, Libbrecht L, Desmet V, Roskams T. Histopathology. 2006;49:138–151. doi: 10.1111/j.1365-2559.2006.02468.x. [DOI] [PubMed] [Google Scholar]
- 16.Van Eyken P, Sciot R, Paterson A, Callea F, Kew MC, Desmet VJ. Hum Pathol. 1988;19:562–568. doi: 10.1016/s0046-8177(88)80205-3. [DOI] [PubMed] [Google Scholar]
- 17.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 18.Fausto N, Campbell JS, Riehle KJ. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 19.Akamatsu N, Sugawara Y, Tamura S, Imamura H, Kokudo N, Makuuchi M. Surgery. 2006;139:765–772. doi: 10.1016/j.surg.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 22.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 23.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niwa H, Burdon T, Chambers I, Smith A. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maione D, Di Carlo E, Li W, Musiani P, Modesti A, Peters M, Rose-John S, Della Rocca C, Tripodi M, Lazzaro D, et al. EMBO J. 1998;17:5588–5597. doi: 10.1093/emboj/17.19.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schirmacher P, Peters M, Ciliberto G, Blessing M, Lotz J, Meyer zum Buschenfelde KH, Rose-John S. Am J Pathol. 1998;153:639–648. doi: 10.1016/S0002-9440(10)65605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano T, Ishihara K, Hibi M. Oncogene. 2000;19:2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 28.Levy DE, Lee CK. J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhanumathy CD, Tang Y, Monga SP, Katuri V, Cox JA, Mishra B, Mishra L. Dev Dyn. 2002;223:59–69. doi: 10.1002/dvdy.1235. [DOI] [PubMed] [Google Scholar]
- 30.Koomen JM, Shih LN, Coombes KR, Li D, Xiao LC, Fidler IJ, Abbruzzese JL, Kobayashi R. Clin Cancer Res. 2005;11:1110–1118. [PubMed] [Google Scholar]
- 31.Ogata H, Chinen T, Yoshida T, Kinjyo I, Takaesu G, Shiraishi H, Iida M, Kobayashi T, Yoshimura A. Oncogene. 2006;25:2520–2530. doi: 10.1038/sj.onc.1209281. [DOI] [PubMed] [Google Scholar]
- 32.Song J, Patel M, Rosenzweig CN, Chan-Li Y, Sokoll LJ, Fung ET, Choi-Miura NH, Goggins M, Chan DW, Zhang Z. Clin Chem. 2006;52:1045–1053. doi: 10.1373/clinchem.2005.065722. [DOI] [PubMed] [Google Scholar]
- 33.Brasier AR, Recinos A, 3rd, Eledrisi MS. Arterioscler Thromb Vasc Biol. 2002;22:1257–1266. doi: 10.1161/01.atv.0000021412.56621.a2. [DOI] [PubMed] [Google Scholar]
- 34.Popa C, Netea MG, Radstake TR, van Riel PL, Barrera P, van der Meer JW. Ann Rheum Dis. 2005;64:1195–1198. doi: 10.1136/ard.2004.032243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray S, Boldogh I, Brasier AR. Gastroenterology. 2005;129:1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 36.Massague J, Chen YG. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 37.Braun L, Mikumo R, Fausto N. Cancer Res. 1989;49:1554–1561. [PubMed] [Google Scholar]
- 38.Wu PC, Lai VC, Fang JW, Gerber MA, Lai CL, Lau JY. J Hepatol. 1999;31:965–966. doi: 10.1016/s0168-8278(99)80303-1. [DOI] [PubMed] [Google Scholar]
- 39.Roskams T. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 40.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H, Taniguchi H. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 41.Roberts A, Mishra L. Oncogene. 2005;24:5667. [Google Scholar]
- 42.Wallner L, Dai J, Escara-Wilke J, Zhang J, Yao Z, Lu Y, Trikha M, Nemeth JA, Zaki MH, Keller ET. Cancer Res. 2006;66:3087–3095. doi: 10.1158/0008-5472.CAN-05-3447. [DOI] [PubMed] [Google Scholar]
- 43.Katuri V, Tang Y, Li C, Jogunoori W, Deng CX, Rashid A, Sidawy AN, Evans S, Reddy EP, Mishra B, et al. Oncogene. 2006;25:1871–1886. doi: 10.1038/sj.onc.1209211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.