Abstract
Glomerulopathy with fibronectin (FN) deposits (GFND) is an autosomal dominant disease with age-related penetrance, characterized by proteinuria, microscopic hematuria, hypertension, and massive glomerular deposits of FN that lead to end-stage renal failure. The genetic abnormality underlying GFND was still unknown. We hypothesized that mutations in FN1, which encodes FN, were the cause of GFND. In a large Italian pedigree with eight affected subjects, we found linkage with GFND at the FN1 locus at 2q32. We sequenced the FN1 in 15 unrelated pedigrees and found three heterozygous missense mutations, the W1925R, L1974R, and Y973C, that cosegregated with the disease in six pedigrees. The mutations affected two domains of FN (Hep-II domain for the W1925R and the L1974R, and Hep-III domain for the Y973C) that play key roles in FN–cell interaction and in FN fibrillogenesis. Mutant recombinant Hep-II fragments were expressed, and functional studies revealed a lower binding to heparin and to endothelial cells and podocytes compared with wild-type Hep-II and an impaired capability to induce endothelial cell spreading and cytoskeletal reorganization. Overall dominant mutations in FN1 accounted for 40% of cases of GFND in our study group. These findings may help understanding the pathogenesis of proteinuria and glomerular FN deposits in GFND and possibly in more common renal diseases such as diabetic nephropathy, IgA nephropathy, and lupus nephritis. To our knowledge no FN1 mutation causing a human disease was previously reported.
Keywords: genetics, proteinuria, extracellular matrix, kidney, podocytes
Glomerulopathy with fibronectin (FN) deposits (GFND) is a hereditary kidney disease (MIM 601894) with proteinuria, microscopic hematuria, and hypertension that lead to end-stage renal failure (ESRF) in the second to sixth decade of life. The condition was recognized as a distinct disease entity by Strom et al. (1). Light microscopy demonstrated enlarged glomeruli with deposits in the mesangium and subendothelial space, with scant immunoreactivity for immunoglobulins or complement factors (1). The most striking finding in this disease is strong immune reactivity of the glomerular deposits to FN (1), an adhesive high-molecular-weight dimeric glycoprotein that is part of extracellular matrix (2).
Clustering of the disease within families (1, 3) indicates a genetic origin for GFND, and segregation is consistent with an autosomal dominant pattern of inheritance with age-related penetrance. However, the genetic abnormality underlying GFND was still unknown (3–5). By whole-genome linkage analysis in a large pedigree, a gene locus for GFND was mapped on 1q32, within a 4.1-cM interval that contains a cluster of genes involved in the regulation of complement activation (RCA) (6). However, mutational analysis and functional studies failed to find any abnormality (7).
Here, we investigated the genetic basis of GFND. Results of linkage analysis excluded the 1q32 locus and revealed a region on 2q34 containing the FN1 gene, encoding FN, as a previously undescribed locus for the disease. By sequence analysis, we found heterozygous FN1 mutations that cosegregate with the disease in six of 15 unrelated pedigrees. We studied the molecular implications of the genetic defects to the pathogenesis of proteinuria and FN glomerular deposits in GFND.
Results and Discussion
Studies in Pedigree F233.
Clinical description.
This is an Italian family that has been partially described (1) and was updated in the present paper. Overall, eight subjects in this pedigree [three previously described (1) and five newly reported in this paper] were affected by the disease in accordance with the criteria described in Methods. Five of them underwent renal biopsy showing enlarged glomeruli with extensive deposits in the mesangium and subendothelial space that stained very strongly for FN. FN was mainly stained by an antibody detecting both plasma and cell-derived FN and to much lesser degree by an antibody specific for only cell-FN (8), suggesting that the FN that accumulated in the glomeruli was mainly derived from the plasma (9). By electron microscopy, the deposits were mainly granular (1, 8).
The clinical course of the affected subjects is shown in Table 1. The index case, subject 717, is a 14-year-old boy who was referred in 2005 to the Department of Nephrology of Ospedali Riuniti di Bergamo with nephrotic-range proteinuria (7.46 g per day), low serum albumin, and severe hypertension. He received a multidrug treatment titrated against urinary protein excretion [see supporting information (SI) Text] and after 2 years, urinary protein excretion is about one-third the baseline values, blood pressure is completely normalized, and the renal function is stably normal (Table 1).
Table 1.
Clinical data from GFND affected subjects of pedigree F233
| Patient sex/age* | Data at first observation |
Followup |
Data at last observation |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age,yr | Proteinuria, g/24 hr | S. creat., mg/dl | Hypert./microh. | Age at N-range proteinuria | Age at hypert./microh. | Age at ESRF | Age at TR | Age, yr | Proteinuria g/24 h | S. creat., mg/dl | Hypert./microh. | |
| 725‡ | 59 | Nephrotic range | Normal | +/? | 59 | 59/? | 74 | — | 73† | 2.5† | 4.0† | +/?† |
| F/77 | ||||||||||||
| 719 | 44 | 300 mg/dl | 1.4 | −/? | 46 | 46/46 | — | — | 54 | 1.06 | 1.55 | −/+ |
| M/55 | ||||||||||||
| Died‡ | 40 | ? | 2.2 | +/? | — | 40/? | — | — | — | — | — | — |
| M | ||||||||||||
| 720 | 35 | Nephrotic range | 1.5 | +/? | 35 | 35/— | — | — | 46 | 1.42 | 2.52 | +/− |
| M/48 | ||||||||||||
| 716‡ | 18 | 4.16 | 1.4 | −/− | 18 | 19/— | 32 | 37 | 41 | 0.2¶ | 1.22¶ | −/− |
| M/42 | ||||||||||||
| 721 | 27 | 0.74 | 0.8 | −/+ | — | —/27 | — | — | 40 | 4.77 | 1.0 | −/+ |
| M/40 | (Normal) | |||||||||||
| 723‡ | 16 | ? | Normal | −/? | 24 | 30/? | 34 | 35 | 35 | ? | 1.5¶ | ? |
| M/39 | ||||||||||||
| 717‡ | 12 | 7.46 | 0.7 | +/+ | 12 | 12/12 | — | — | 14§ | 2.31§ | 0.7§ | −/+§ |
| M/14 | (Normal) | |||||||||||
Nephrotic range (N-range) proteinuria: >3.5 g per 24 h, S. creat, serum creatinine; normal values, 0.6–1.3 mg/dl; died, deceased at the age of 47 years because of rupture of subarachnoid aneurysm; —, not applicable.
*Age at present.
†Data before ESRF.
‡Biopsy diagnosis.
§Values recorded after 2 years of multidrug renoprotective treatment.
¶Measured after kidney transplantation.
Three of the other affected subjects (716, 723, and 725) developed ESRF, and two of them (716 and 723) received a renal transplant. Three years after transplantation, an allograft biopsy in subject 716 showed recurrence of the disease in the transplant (Table 1). In subject 723, the graft was functioning well at 1 year follow-up.
Identification of a locus for GFND.
We performed haplotype analysis in pedigree F233 by using 10 polymorphic microsatellite DNA markers spanning 22 Mb along the RCA cluster at the 1q32 locus (6) (see SI Text and SI Fig. 4).
Seventeen subjects were haplotyped. Segregation of GFND in this family was consistent with autosomal dominant inheritance and age-related penetrance. Because the disease has progressive manifestations, the absence of the disease could not be determined with certainty in the four healthy subjects of the third generation (all <35 years of age). Data were first evaluated on the basis of “affecteds-only” strategy. None of the haplotypes cosegregated with GFND and linkage analysis by GENEHUNTER software gave a multipoint logarithm of odds (lod) score less than −2 throughout the chromosomal area. In further analyses, liability classes were assigned according to age at examination, as described in Methods. Results of two-point and multipoint linkage analyses confirmed the exclusion of 1q32 as disease locus in this pedigree (SI Fig. 4).
We further performed haplotype analysis on loci including genes encoding other proteins of the complement system, namely complement factor D (CFD) and complement C3 (19p13) and CD59 (11p13), with negative results in both regions (data not shown).
We then looked at the locus of the candidate gene FN1 at 2q34, because FN1 encodes FN, the main component of glomerular deposits in GFND. Eighteen subjects (including the deceased subject for whom DNA was obtained from autopsy material) were haplotyped in a 37-Mb interval between markers D2S2167 and D2S2297 (Fig. 1a). All subjects affected with GFND shared the same haplotype, and none of the unaffected subjects inherited this allele except for subject 733 in the third generation that was unaffected at the time of analysis but was still at risk because of the young age (24 years). Of note, subject 733 had a mild progressively worsening proteinuria (from 0.18 g per 24 h in 2006 to 0.30 g per 24 h in 2007) but normal renal function. Recombination events were found between markers D2S2387 and D2S2289 in the second generation and between markers D2S2359 and D2S2297 in the third generation (Fig. 1a). With the affecteds-only model, linkage analysis gave a maximum multipoint lod score of 1.8. Two-point linkage analysis with liability classes (see Methods) resulted in Zmax = 3.084 (θ = 0) for markers D2S128 and D2S2361, which is very close to maximum values obtained with SLink simulation (SI Table 2). Consistently, multipoint linkage analysis with the eight microsatellite markers gave a Zmax plateau of 3.084 on the same markers (Fig. 1a).
Fig. 1.
FN1 mutations in GFND. (a) (Upper) Haplotype analysis at FN1 locus in pedigree F233. Microsatellite loci are on the left. (Lower) Multipoint linkage analysis by GENEHUNTER (model: autosomal dominant transmission with age-related penetrance, analysis with liability classes). For markers D1S128 and D1S2361, the maximum lod score was Zmax = 3.084, as indicated. *, subjects previously published. ‡, biopsy-proven GFND. Solid symbols, affected individuals; crossed symbols, deceased; violet arrow, proband: the whole FN1 was sequenced; red dots, FN1 mutation carriers. ¶, subjects screened for the FN1 mutation and for SNP segregation; un, unavailable. (b) Schematic diagram of fibronectin. Fibronectin monomer consists of type I (blue), II (green), and III (orange) repeats and the alternatively spliced sites EDI, EDII, and IIICS. The three main heparin-binding domains and the binding sites for integrins are shown. Positions of the GFND-associated mutations are indicated by arrows. (c Upper) Pedigrees of the other five families with FN1 mutations. (Lower) The number of affected subjects, the mutation, and the origin of all of the six mutated families, are reported.
Identification of an FN1 mutation.
We sequenced the exons and flanking intronic regions of FN1 (NC_000002, gi:51511462). A list of primers used for sequencing is given in SI Table 3.
A heterozygous 5773T>A missense mutation in exon 36, which causes a tryptophan-to-arginine substitution (W1925R) in the III13 repeat (Fig. 1 b and c), was identified in all affecteds and in subject 733 (Fig. 1a) and was not found in the other nine subjects of the pedigree or in any of 100 healthy subjects.
We then screened FN1 in 14 additional GFND pedigrees and FN1 mutations were found in five of them, as reported below.
Studies in Pedigree F656.
The family is from New Zealand with six affected subjects, the father and five of seven siblings. FN deposition was confirmed by biopsy in four cases. The affected children presented with proteinuria at a wide age range of 14, 16, 26, 43, and 47 years, respectively. Three patients developed ESRF at ages 33, 34, and 35 years and received either successful renal transplantation (two cases) or hemodialysis. Sequencing of FN1 revealed a heterozygous missense mutation (5921T>G) in exon 37 in the affecteds, leading to an L1974R change in repeat III13 (Fig. 1 b and c). The mutation segregated with the affected status and was not found in the mother, the two healthy siblings and 180 healthy control individuals.
Studies in Pedigree F468.
This is an Italian family previously described (1, 8) with three affected subjects. The proband is a 53-year-old woman who developed proteinuria in the nephrotic range at 25 years of age. Presently, proteinuria is still in the nephrotic range, and renal function is moderately reduced. Her daughter developed nephrotic range proteinuria at 14 years of age (1, 8). Presently, at the age of 25 years, renal function is normal. In both affected subjects, biopsy confirmed diagnosis of GFND, and electron microscopy examination revealed mainly granular deposits in the subendothelial space and in the mesangium with few fibrils. A paternal aunt of the proband died at 37 years of age of nephropathy. The proband's father died of cancer at 30 years of age.
A heterozygous 2918A>G mutation causing a Y973C change in repeat III4 was found in the index case and in her affected daughter but not in the unaffected father (Fig. 1 b and c and SI Fig. 5) or in any of 230 healthy Caucasian controls.
Studies in Pedigree F546.
The family is from The Netherlands and includes two affected subjects (10). The index case is a male who developed low-grade proteinuria at 29 years of age. At 38 years, proteinuria was in the nephrotic range, and a renal biopsy revealed GFND. In his first child, proteinuria (2.8 g per 24 h) was detected at the age of 21 years. Renal biopsy showed glomerular lesions similar to those seen in his father with massive deposits in the mesangial area and to a lesser extent in the subendothelium (10) that showed a fine granular pattern alternating with areas containing more regularly arranged fibrils. Immunofluorescence showed strong staining for plasma FN (10). At the last update, the other son, 21 years old, was healthy.
The heterozygous 2918A>G mutation was found in the index case and in the two sons (Fig. 1 b and c, and SI Fig. 5). Because of the age-related penetrance of GFND, the unaffected son can be considered still at risk.
Studies in Pedigree F663.
In a German pedigree with four affected siblings, biopsies in two cases confirmed the diagnosis of GFND. Proteinuria was observed in all four cases. One patient (proteinuria at 18 years of age) developed ESRF at 33 years. In the oldest patient (42 years), serum creatinine levels are in the range 2–3 mg/dl.
The heterozygous 2918A>G mutation was found in all four affected siblings but not in the unaffected mother (Fig. 1 b and c and SI Fig. 5). DNA from the father was unavailable (deceased, cause unknown), thus we could not establish the origin of the mutation.
Studies in Pedigree F1098.
The family is from Japan (11). The index case is a boy who developed high-grade proteinuria and microscopic hematuria at the age of 3 years. Renal biopsy revealed enlarged glomeruli with deposition of granular material in glomeruli that stained brightly with anti-FN antibodies (11). Family history was remarkable for renal disease in 12 members that had mild hematuria and/or proteinuria but neither nephrotic syndrome nor renal insufficiency. No relative had undergone renal biopsy (11). It was hypothesized that the index case has a different disease than anybody in this pedigree that segregates a mild nephropathy (11).
The heterozygous 2918A>G mutation was found in the index case but not in any of nine subjects in this family. Specifically, the mutation was not found in the mother or in any of the other three siblings in either of the father's two haplotypes (DNA from the father was not available), indicating that it occurred de novo in the proband (Fig. 1 b and c and SI Fig. 5).
Of note, the Y973C change was found in four pedigrees from different ethnic backgrounds (Italy, The Netherlands, Germany, and Japan; Fig. 1 b and c). Haplotype analysis in the four pedigrees showed no haplotype sharing, making a common founder origin very unlikely (SI Fig. 5).
Overall, we have studied 15 different families, including most of the published (1, 8, 10, 11) and additional unpublished pedigrees, six of which (40%) carried FN1 mutations. All affected subjects carried the mutation, and segregation was compatible with autosomal dominant inheritance (Fig. 1c). In the six probands carrying FN1 mutations, we also found several SNPs of which the nine coding SNPs and the two previously undescribed intronic SNPs are reported in SI Table 2. Sequencing of family members for nonsynonomous SNPs revealed that rs17449032 does not segregate with GFND, whereas rs1250259 segregated in families F468 and F546 (SI Table 4) but not in family F233 (SI Fig. 6).
Partial protein sequence alignment among multiple species showed that residues W1925, L1974, and Y973 are highly conserved (SI Fig. 7). These findings provide evidence that mutations in FN1 are responsible for GFND.
Functional Studies on GFND-Associated FN1 Mutations.
Fibronectin is secreted in a dimeric form and plays a role in cell-matrix contact processes such as cell attachment and spreading, cell migration, control of cell cytoskeleton and morphology and differentiation (9, 12, 13). It also participates in extracellular matrix formation, hemostasis, and thrombosis (12). All of these biological activities imply interaction of FN with cells and with extracellular material via binding sites for integrins, heparin, and heparan-sulfate proteoglycans.
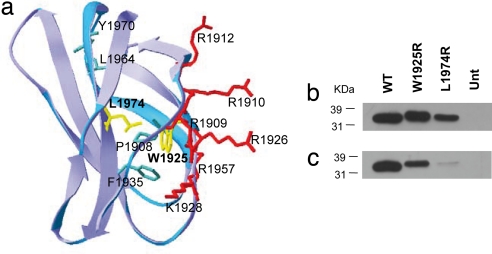
Each FN monomer consists of homologous modules classified as type I, II, or III repeats (Fig. 1b). The C-terminal III12–14 repeats (Hep-II) contain the main binding site for heparin (14, 15) and III13 accounts for ≈98% of Hep-II activity. It comprises a “cationic cradle” with six positively charged residues and a hydrophobic core that consists of P1908, W1925, F1936, L1964, and Y1970 (15, 16). The W1925R mutation introduces a basic amino acid in the Hep-II hydrophobic core. Similarly, the L1974R mutation introduces a basic amino acid very close to the Hep-II hydrophobic core (Fig. 2a). The two mutations could theoretically increase the Hep-II affinity for heparin, by providing additional cationic charge to the domain; however, they could also alter the folding of the domain and impair its function.
Fig. 2.
Expression of recombinant wild-type and mutant Hep-II domains of fibronectin. (a) Structure of FN III13. Amino acid residues are color marked for positively charged (red), hydrophobic core (green), and residues W1925 and L1974 (yellow). (b and c) WT and mutant purified recombinant proteins were analyzed by SDS/PAGE on 12% gels and visualized by Western blotting with either an antibody anti-His (C-term) (b) or an antifibronectin mAb against the Hep-II domain. (c) Position of standards (kDa) are shown. Equal amounts (5 μg each) of WT and mutant proteins were loaded. Separate lanes were labeled with Coomassie blue as control for loading. Unt, untransfected.
To investigate the functional effects of the W1925R and L1974R mutations on Hep-II binding to heparin and cells, we generated III12–14wt, III12–14W1925R, and III12–14L1974R poly-His-tagged recombinant fragments and expressed them in Sf9 cells. Recombinant wild-type and mutant proteins were expressed and secreted by insect cells and were detected as a band of ≈34 kDa on Western blot (Fig. 2 b and c). Wild-type and mutant proteins gave a similar staining with the anti-His antibody, indicating they were expressed at the same level. By contrast, a fainter staining of the mutant proteins was observed with the anti-FN antibody, which suggests that the mutations cause changes in epitope recognition.
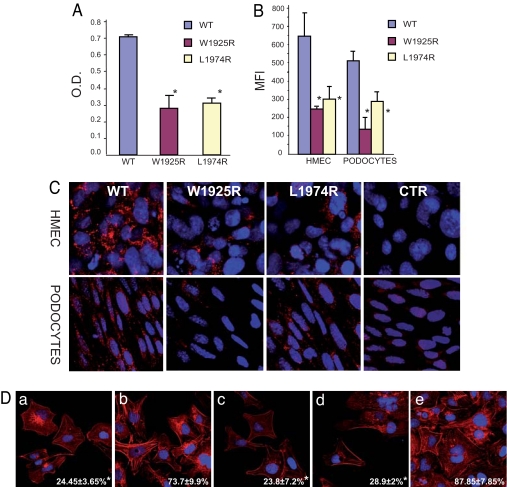
Binding of the III12–14W1925R and the III12–14L1974R purified recombinants to immobilized heparin (by ELISA) showed a significative (P < 0.01) reduction in respect to the wild-type (Fig. 3A). As a consequence, binding of the mutants to endothelial cells (human dermal microvascular endothelial cells line, HMEC-1) and immortalized mouse podocytes was strongly reduced (P < 0.01 vs. wild type), as documented by FACS and confocal microscopy analysis (Fig. 3 B and C). Similar results were obtained with human umbilical vein endothelial cells (HUVEC, SI Fig. 8).
Fig. 3.
Mutations in FN Hep-II domain cause reduced binding to heparin, endothelial cells, and podocytes and impair stress fiber formation. (A) Binding of III12–14wt, III12–14W1925R, and III12–14L1974R to heparin by ELISA. O.D., optical density. (B and C) Binding of WT and mutant poli-His-tagged III12–14W1925R and III12–14L1974R recombinants added to human endothelial cells (HMEC) and mouse podocytes (b) FACS analysis. (C) Confocal microscopy; original magnification, ×600). Staining was done with an anti-His antibody plus FITC-conjugated (FACS) or Cy3-conjugated (confocal, red) secondary antibodies. MFI, median fluorescence intensity. (D) HMEC were plated on a 120-kDa N-terminal FN fragment in the absence (a) or presence of III12–14wt (b), III12–14W1925R (c), III12–141974R (d), or full length FN (e), in serum-free medium for 3 h and then labeled with rhodamine-phalloidin to visualize stress fibers. The percent of stress-positive cells (mean ± SD) is shown in the bottom (white numbers). Data are mean ± SD of three independent experiments. *, P < 0.01 vs. WT. O.D. and MFI values were calculated after subtracting values recorded with addition of buffer alone (blanks).
The initial attachment and spreading of cells to FN is mediated by the interaction of the RGD-containing domain in the FN III9–10 repeats with α5β1 cell integrin (17). But further progression to the formation of actin stress-fibers and focal contacts requires binding of Hep-II to cell-surface proteoglycans and integrins (18–20). We therefore hypothesized that lower heparin affinity and cell binding of the III12–14W1925R and III12–14L1974R resulted in impaired capability to trigger stress-fiber formation and cell spreading. This possibility was tested in vitro in HMEC plated on a layer of 120-kDa fragments of FN containing the RGD domain but lacking the Hep-II domain, in the presence of the III12–14 recombinants (19) (Fig. 3D). HMEC plated on the 120-kDa fragment were able to attach, however, they showed few actin stress fibers, indicating they were not completely spread. If the III12–14wt was added to the media, the percent of stress fiber-positive cells was enhanced 3-fold (19), whereas this increase did not occur when either mutants were added at the same concentration (P < 0.01 vs. wild type; Fig. 3D).
The third GFND-associated mutation, Y973C, affects another heparin-binding domain of FN, Hep-III, located in the III4–5 repeats. It shares with Hep-II the capability of promoting stress-fiber and focal-adhesion formation (21). The Y973C mutation introduces an additional cysteine in III4, which could affect protein folding and function through the formation of abnormal Cys–Cys bonds.
Pathogenetic Hypothesis.
The above reported FN1 mutations, and their functional consequences could explain the severe proteinuria and microscopic hematuria that precede progressive renal disease in GFND. Indeed, disturbance in cell spreading and cytoskeleton in glomerular endothelial cells and podocytes due to impaired interaction with mutant FN, is expected to alter the glomerular size-selectivity properties and induce abnormal protein trafficking (22, 23).
FN is present in plasma as a soluble form (pFN) or deposited in extracellular matrix as insoluble organized fibrils (cellular FN) (9). Renal biopsy specimens of GFND patients showed extensive FN deposits in the mesangium and subendothelial space that are mainly granular with only some admixture of irregularly arranged fibrils (1, 8, 10). By using specific antibodies for pFN and cellular FN, it was documented that glomerular FN deposits are mainly derived from plasma and to a lesser extent from resident glomerular cells (1, 8, 10).
The Hep-II and -III domains play a main role in regulating FN assembly into organized fibrils in extracellular matrix, through complex FN–FN and FN–cell surface proteoglycan interactions (18, 24–26). In addition, interaction between the Hep-II domain and the III2–3 repeats keeps pFN in a compact soluble form preventing its deposition in extracellular matrix (27). Based on this evidence and on the results of functional studies presented here, we suggest that GFND-associated mutations in FN1 impair the control of the assembly of FN into fibrils and the balance between soluble and insoluble FN, which could explain the abnormal incorporation of nonfibrillary pFN in the glomerular matrix that has been documented in renal biopsy specimens of patients participating to the present study.
GFND is a very rare disease (1, 8, 10, 11, 28) however, the present data may have implications for the understanding of the pathogenesis of more common renal diseases characterized by FN glomerular deposits, such as diabetic nephropathy (29), IgA nephropathy (30), and lupus nephritis (31).
Methods
Patients and Diagnosis.
We collected blood samples from all available affected and nonaffected members of 15 pedigrees for isolation of genomic DNA after informed consent. The definition of affected status was made on the basis of either a renal biopsy or of a clinical history compatible with a GFND status (with proteinuria, microhematuria, hypertension, and slowly decreasing renal function), with at least one individual with biopsy proven GFND in each pedigree (1). None of the affected subjects presented clinical or laboratory evidence of systemic lupus, cryoglobulinemia, diabetes mellitus, amyloidosis, or autoimmune disease. For the definition of absence of disease, the following criteria were met: absence of significant proteinuria, of hematuria, normal blood pressure, and normal renal function (6). All protocols included in these studies have been approved by Institutional Review Boards. Informed written consent was obtained from all participating subjects, according to the declaration of Helsinki.
DNA Analysis.
We performed linkage analysis in pedigree F233 on the candidate chromosomal region 1q32, 19p13, 11p13, and 2q34 (SI Text).
Haplotypes were reconstructed by using the GENEHUNTER package (Version 1.2). Autosomal dominant transmission with age-related penetrance was assumed on the basis of clinical and pedigree data (6). Liability classes were assigned as follows: affected subjects and unaffected subjects of the first and second generations were assigned liability class 1, with penetrance vector {0.0, 0.99, and 0.99}. Unaffected individuals in the third generation (all <35 years of age) were assigned liability class 2, with penetrance vector {0.0, 0.50, and 0.75}. The disease gene frequency in the general population was set at 0.0001 (6). Two-point and multipoint linkage analyses were performed by GENEHUNTER by using both the affecteds-only and liability classes models. SLink simulations were done by the FASTLINK package.
We sequenced the exons and the flanking intronic regions of FN1 (NC_000002, gi:51511462). The gene has three regions subjected to alternative splicing (EDI, EDII, and IIICS), with the potential to produce 20 different transcript variants. We chose variant 1 that represents the longest transcript and encodes the longest isoform (NM_212482) (see SI Text and SI Table 3). The amino acid numbering is referred to the translation start site (Met + 1), and the nucleotide number is referred to the A of the ATG start codon (www.ncbi.nlm.nhi.gov, NM_212482).
Expression and Functional Studies.
The two mutations affecting the Hep-II domain were introduced into human complementary DNA (cDNA) encoding repeats III12–14 of FN and expressed as His-tagged fusion proteins in Sf9 cells by the Baculovirus system (see SI Text).
Binding of wild-type and mutant III12–14 recombinants to heparin was assessed by ELISA (32). The capability of wild-type and mutant III12–14 recombinants to bind endothelial cells (HMEC and HUVEC) and podocytes (immortalized mouse podocytes from Peter Mundel, Mount Sinai School of Medicine, New York) (33) was evaluated by FACS and confocal microscopy analysis (see SI Text) (34).
For spreading assays, HMEC cells were coated on glass coverslips previously coated overnight with human 120-kDa FN alpha-chymotryptic fragment lacking the Hep-II domain with or without purified wild-type III12–14 and mutant III12–14W1925R and III12–14L1974R recombinant proteins. As positive control, cells were seeded on glass coverslips coated with intact FN (19). At the end of the incubation period, the F-actin filaments were stained with rhodamine-phalloidin and cells were examined by using inverted confocal laser microscopy (see SI Text) (33).
Statistical Analysis.
Data are reported as mean ± SD. Results of functional assays on wild-type and mutant III12–14 fragments of FN1 were compared by Student's t test for unpaired data.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Gaia Pianetti for excellent technical assistance in sequencing; Drs. Francesca Mari, Mario De Marchi, Gianna Mazzucco, and Mario Carmellini for help in genetic counseling of the families, sample collection, and clinical data discussion; and Fabio Sangalli and Dr. Simona Buelli for assistance with confocal microscopy experiments. We thank Andrea Hartmann for excellent assistance with the heparin- and HUVEC-binding experiments. This work was supported by a grant from Fondazione ART Associazione Per La Ricerca Sui Trapianti (Milan, Italy). F.C. is a recipient of a fellowship from ART, and M.N. and G.R. are supported by National Institutes of Health (NIH) Grant DK71221. F.H. is supported by NIH Grant DK039255-16 and is the Frederick G. L. Huetwell Professor and a Doris Duke Distinguished Clinical Scientist. P.F.Z. acknowledges support from the Deutsche Forschungsgemeinschaft and from KIDNEEDS.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707730105/DC1.
References
- 1.Strom EH, Banfi G, Krapf R, Abt AB, Mazzucco G, Monga G, Gloor F, Neuweiler J, Riess R, Stosiek P, et al. Kidney Int. 1995;48:163–170. doi: 10.1038/ki.1995.280. [DOI] [PubMed] [Google Scholar]
- 2.Kornblihtt AR, Pesce CG, Alonso CR, Cramer P, Srebrow A, Werbajh S, Muro AF. FASEB J. 1996;10:248–257. doi: 10.1096/fasebj.10.2.8641558. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrandt F, Strahm B, Prochoroff A, Cybulla M, Gemperle O, Krapf R, Brandis M. Am J Med Genet. 1996;63:323–327. doi: 10.1002/(SICI)1096-8628(19960503)63:1<323::AID-AJMG54>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Kundu GC, Yuan CJ, Ward JM, Lee EJ, DeMayo F, Westphal H, Mukherjee AB. Science. 1997;276:1408–1412. doi: 10.1126/science.276.5317.1408. [DOI] [PubMed] [Google Scholar]
- 5.Vollmer M, Krapf R, Hildebrandt F. Nephrol Dial Transplant. 1998;13:2417–2418. doi: 10.1093/ndt/13.9.2417. [DOI] [PubMed] [Google Scholar]
- 6.Vollmer M, Jung M, Ruschendorf F, Ruf R, Wienker T, Reis A, Krapf R, Hildebrandt F. Am J Hum Genet. 1998;63:1724–1731. doi: 10.1086/302162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vollmer M, Kremer M, Ruf R, Miot S, Nothwang HG, Wirth J, Otto E, Krapf R, Hildebrandt F. Genomics. 2000;68:127–135. doi: 10.1006/geno.2000.6292. [DOI] [PubMed] [Google Scholar]
- 8.Mazzucco G, Maran E, Rollino C, Monga G. Hum Pathol. 1992;23:63–68. doi: 10.1016/0046-8177(92)90013-s. [DOI] [PubMed] [Google Scholar]
- 9.Yamada KM, Kennedy DW. J Cell Biol. 1979;80:492–498. doi: 10.1083/jcb.80.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assmann KJ, Koene RA, Wetzels JF. Am J Kidney Dis. 1995;25:781–791. doi: 10.1016/0272-6386(95)90555-3. [DOI] [PubMed] [Google Scholar]
- 11.Niimi K, Tsuru N, Uesugi N, Takebayashi S. Pediatr Nephrol. 2002;17:363–366. doi: 10.1007/s00467-002-0833-2. [DOI] [PubMed] [Google Scholar]
- 12.Hynes RO, Yamada KM. J Cell Biol. 1982;95:369–377. doi: 10.1083/jcb.95.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hormann H. Klin Wochenschr. 1982;60:1265–1277. doi: 10.1007/BF01727483. [DOI] [PubMed] [Google Scholar]
- 14.Barkalow FJ, Schwarzbauer JE. J Biol Chem. 1991;266:7812–7818. [PubMed] [Google Scholar]
- 15.Sharma A, Askari JA, Humphries MJ, Jones EY, Stuart DI. EMBO J. 1999;18:1468–1479. doi: 10.1093/emboj/18.6.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachchidanand, Lequin O, Staunton D, Mulloy B, Forster MJ, Yoshida K, Campbell ID. J Biol Chem. 2002;277:50629–50635. doi: 10.1074/jbc.M208956200. [DOI] [PubMed] [Google Scholar]
- 17.Sechler JL, Schwarzbauer JE. Cell Adhes Commun. 1997;4:413–424. doi: 10.3109/15419069709004458. [DOI] [PubMed] [Google Scholar]
- 18.Santas AJ, Peterson JA, Halbleib JL, Craig SE, Humphries MJ, Peters DM. J Biol Chem. 2002;277:13650–13658. doi: 10.1074/jbc.M111361200. [DOI] [PubMed] [Google Scholar]
- 19.Peterson JA, Sheibani N, David G, Garcia-Pardo A, Peters DM. J Biol Chem. 2005;280:6915–6922. doi: 10.1074/jbc.M406625200. [DOI] [PubMed] [Google Scholar]
- 20.Woods A, Longley RL, Tumova S, Couchman JR. Arch Biochem Biophys. 2000;374:66–72. doi: 10.1006/abbi.1999.1607. [DOI] [PubMed] [Google Scholar]
- 21.Moyano JV, Maqueda A, Albar JP, Garcia-Pardo A. Biochem J. 2003;371:565–571. doi: 10.1042/BJ20021344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tryggvason K, Patrakka J, Wartiovaara J. N Engl J Med. 2006;354:1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- 23.Ballerman BJ. Nephron Physiol. 2007;106:19–25. [Google Scholar]
- 24.Bultmann H, Santas AJ, Peters DM. J Biol Chem. 1998;273:2601–2609. doi: 10.1074/jbc.273.5.2601. [DOI] [PubMed] [Google Scholar]
- 25.Maqueda A, Moyano JV, Hernandez Del Cerro M, Peters DM, Garcia-Pardo A. Matrix Biol. 2007;126:642–651. doi: 10.1016/j.matbio.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Mao Y, Schwarzbauer B. Matrix Biol. 2005;25:389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Johnson KJ, Sage H, Briscoe G, Erickson HP. J Biol Chem. 1999;274:15473–15479. doi: 10.1074/jbc.274.22.15473. [DOI] [PubMed] [Google Scholar]
- 28.Sato H, Matsubara M, Marumo R, Soma J, Kurosawa K, Taguma Y, Saito T. Am J Kidney Dis. 1998;31:E3. doi: 10.1053/ajkd.1998.v31.pm10074583. [DOI] [PubMed] [Google Scholar]
- 29.Janssen B, Hohenadel D, Brinkkoetter P, Peters V, Rind N, Fischer C, Rychlik I, Cerna M, Romzova M, de Heer E, et al. Diabetes. 2005;54:2320–2327. doi: 10.1681/01.asn.0000926760.87704.9b. [DOI] [PubMed] [Google Scholar]
- 30.Mattii L, Segnani C, Cupisti A, D'Alessandro D, Moscato S, Meola M, Barsotti G, Marino M, Bianchi F, Dolfi A, et al. Nephron Exp Nephrol. 2005;101:e16–e23. doi: 10.1159/000086035. [DOI] [PubMed] [Google Scholar]
- 31.Baelde HJ, Eikmans M, van Vliet AI, Bergijk EC, de Heer E, Bruijn JA. J Pathol. 2004;204:248–257. doi: 10.1002/path.1653. [DOI] [PubMed] [Google Scholar]
- 32.Opperman M, Manuelian T, Jozsi M, Brandt E, Jokiranta TS, Heinen S, Meri S, Skerka C, Gotze O, Zipfel PF. Clin Exp Immunol. 2006;144:342–352. doi: 10.1111/j.1365-2249.2006.03071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morigi M, Buelli S, Angioletti S, Zanchi C, Longaretti L, Zoja C, Galbusera M, Gastoldi S, Mundel P, Remuzzi G, et al. Am J Pathol. 2005;166:1309–1320. doi: 10.1016/S0002-9440(10)62350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manuelian T, Hellwage J, Meri S, Caprioli J, Noris M, Heinen S, Jozsi M, Neumann HPH, Remuzzi G, Zipfel PF. J Clin Invest. 2003;111:1181–1190. doi: 10.1172/JCI16651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.