Abstract
Bacterial artificial chromosome (BAC) vectors are important tools for microbial genome research. We constructed a novel BAC vector, pUvBBAC, for replication in both gram-negative and gram-positive bacterial hosts. The pUvBBAC vector was used to generate a BAC library for the facultative intracellular pathogen Listeria monocytogenes EGD-e. The library had insert sizes ranging from 68 to 178 kb. We identified two recombinant BACs from the L. monocytogenes pUvBBAC library that each contained the entire virulence gene cluster (vgc) of L. monocytogenes and transferred them to a nonpathogenic Listeria innocua strain. Recombinant L. innocua strains harboring pUvBBAC+vgc1 and pUvBBAC+vgc2 produced the vgc-specific listeriolysin (LLO) and actin assembly protein ActA and represent the first reported cloning of the vgc locus in its entirety. The use of the novel broad-host-range BAC vector pUvBBAC extends the versatility of this technology and provides a powerful platform for detailed functional genomics of gram-positive bacteria as well as its use in explorative functional metagenomics.
With the rapid increase in the number of completed microbial genome sequences, interest in the manipulation and functional characterization of whole genomes is being revolutionized. Methods and technologies that permit the cloning and manipulation of large chromosomal regions, such as the genomic islands which impart either virulence or novel metabolic properties to pathogenic and other microorganisms, are rapidly becoming valuable tools to study in detail properties encoded within these regions (41).
The development of bacterial artificial chromosomes (BACs) has provided an important genetic tool for the cloning and mapping of complex genomes. Different cloning vectors based on low-copy-number replicons either from the bacterial F plasmid (45) or bacteriophage P1 (23, 49) have been developed. These provide a powerful resource in molecular biology, because they allow the cloning of several tens to hundreds of kilobases of contiguous DNA sequences (55) and are stable and easy to handle (31). Their particular use is in the study of functional genome segments that are otherwise too large to be cloned into other more conventional vectors. BAC-based libraries of genomic DNA from numerous viruses (1, 4, 8, 29-31, 42, 43) and plant (9, 15, 27, 32, 47, 53), animal (10, 34, 50, 55), and fungal (20, 39, 54) species have been generated and are now established technologies in large-scale sequencing projects. Classical and molecular genetic techniques are being used in conjunction with BAC recombinants for the introduction of reporter systems into mammalian organisms, the in vivo complementation of mutations, and in vivo and in vitro reverse genetic technologies that introduce point mutations (25), targeted deletions, or new sequence elements into BAC vectors (21). More recently, in vivo recombination (“recombineering”) (37) technology employing either the bacterial recA mutant or the bacteriophage red gam mutant recombination has been used for the directed manipulation of large BAC vectors.
The use of BAC technology in the study of the function of prokaryote genomes is limited (41, 46, 52). Major applications involve the generation of large fragment libraries directly from microbes in natural environments providing access to local metagenomic DNA (40, 51). A BAC vector has also been used to express Bacillus cereus genomic DNA (41). The expression of heterologous B. cereus DNA in Escherichia coli occurred at a low frequency but was sufficient to allow the detection of antibiotic, pigment, and enzymatic activities. BAC vectors that have recently been described allow the shuttling of large DNA segments between E. coli and genetically amenable Streptomyces hosts, where they integrate into the host chromosome (48). This genetic system overcomes the potential limitations of heterologous gene expression and permits the manipulation and identification of novel antibiotic-producing gene clusters.
We are interested in the study of virulence and survival factors as well as the comparative genome analysis of the gram-positive facultative intracellular pathogen Listeria monocytogenes EGD-e and related species. L. monocytogenes is an opportunistic facultative intracellular gram-positive microorganism and a deadly cause of food-borne infections. A distinguishing feature of pathogenic Listeria strains is their ability to grow in host cells and propagate by cell-to-cell spread in the infected host. The responsible genes are clustered on a chromosomal locus designated a virulence gene cluster (vgc) or Listeria pathogenicity island 1 (LIPI-1). The vgc codes for listeriolysin (LLO), required to breach the phagolysosomal membrane of the infected host cell; PlcA, a phosphatidylinositol phospholipase C, acts together with LLO to enable bacterial escape from the phagolysosome, while the phosphatidylcholine phospholipase C lecithinase activity encoded by plcB is required for the dissolution of host membranes during cell-to-cell spread. Mpl, a metalloprotease is involved in the conversion of pro-phosphatidylcholine phospholipase C into its active form, and ActA, the actin assembly-inducing protein precursor, is involved in actin cytoskeleton rearrangements and polymerization-inducing bacterial movement within the infected host cytosol, whereas the regulatory factor A (PrfA) controls the expression of the vgc members (19).
The generation of a BAC vector capable of replication in a broad range of gram-negative and gram-positive host bacteria would be desirable, because it would couple the cloning, sequencing, and ordered mapping of BAC-based libraries with functional genomic analysis in the appropriate host. Here we describe the construction of a novel BAC vector capable of replication in E. coli and Listeria spp. and its use in the generation of chromosomal libraries and functional genomics in these species.
MATERIALS AND METHODS
Bacterial strains.
L. monocytogenes EGD-e serotype 1/2a, Listeria innocua 6a (NCTC11288), and E. coli DH10β (Invitrogen) were used in this study (Table 1).
TABLE 1.
Strains, plasmids, BACs, and primers used in this study
| Strain, plasmid, BAC, or primer | Genotype, characteristics, or sequence | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10β | F′ mcrAΔ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX174 deoR recA1 φaraD139Δ(ara leu)7697 galU galK λ−rpsL endAl nupG | Invitrogen |
| L. monocytogenes | Serotype 1/2a, EGD-e | 17 |
| L. innocua | Serotype 6a, NCTC11288 | Laboratory collection |
| Plasmids and BACs | ||
| pSOG7 | Broad-host-range shuttle vector for gram-negative and gram-positive bacteria | 35 |
| pCOP4 | Derivate of pIP501 from Streptococcus agalactiae | 5 |
| pCR-XL-TOPO | Cloning vector for E. coli | Invitrogen |
| pBeloBAC11 | BAC cloning vector for E. coli | 24 |
| pUvBBAC | Derivative of pBeloBAC11 cloning vector for replication in E. coli and Listeria spp. | This work |
| pUvBBAC+vgc1 | pUvBBAC carrying chromosomal fragment of L. monocytogenes EGD-e with vgc | This work |
| pUvBBAC+vgc2 | pUvBBAC carrying chromosomal fragment of L. monocytogenes EGD-e with vgc | This work |
| Primers | ||
| P1 | AATTCCTCGAGAATTTCACACAGGAAACAGC | This study |
| P2 | AATTCCTCGAGCGATCACTCATCATGTTC | This study |
| P3 | AATTCCTCGAGTGTAGAAGGAGGGTGAAACC | This study |
| P4 | CGGGATCCCGTGTGGGAACTAAATTATACG | This study |
| P5 | TCCCATGGCCTAATAATGCCAAATACCG | This study |
| P6 | CGAAGATCTCGTACGCGTTCATGAAAATGCTTCTG | This study |
| P7 | CGGGATCCCGCCTCCTTTGATTAGTATATTC | This study |
| P8 | ATCCTGCAGCGTGATACGCTAATACAACC | This study |
| T7 | TAATACGACTCACTATAGGG | New England Biolabs |
| SP6 | ATTTAGGTGACACTATAG | New England Biolabs |
| 16S rRNA gene (probe) | FAM-CGTATTACCGCGGCTGCTGGCAC-TAMRA | 28 |
| 16S rRNA gene (forward) | TCCTACGGGAGGCAGCAGT | 28 |
| 16S rRNA gene (reverse) | GGACTACCAGGGTATCTAATCCTGTT | 28 |
| hly (probe) | FAM-CGAGTTCATCCGCGTGTTTCTTTTCG-TAMRA | This study |
| hly (forward) | TGCAAGTCCTAAGACGCCA | This study |
| hly (reverse) | CACTGCATCTCCGTGGTATACTAA | This study |
Construction of pUvBBAC.
We combined different genetic features from plasmids pSOG7 and from BAC vector pBeloBAC11 to construct the broad-host-range vector pUvBBAC (Fig. 1). At first, a 4.3-kb PCR fragment carrying the determinant for erythromycin resistance, ermC, and replication elements copR, repR, and oriR from pIP501 from Streptococcus agalactiae (group B) (22) was amplified from pSOG7 (35) (Table 1; also see Table S1 in the supplemental material), a pCOP4 (5) (Table 1) derivative with oligonucleotide primers (P1 and P2) (Table 1) generating XhoI restriction sites at both the 5′ and 3′ ends of the fragment. After digestion with XhoI, the fragment was ligated into pBeloBAC11. Ligation was performed by adding 15 μl XhoI-digested pBeloBAC11 DNA, 7.5 μl XhoI-digested 4.3-kb PCR fragment, 1.5 μl T4 ligase (1 U/μl; Invitrogen), and 6 μl 5× T4 ligase buffer (Invitrogen) to the reaction mixture. After overnight incubation at 14°C, the ligation mixture was electroporated into E. coli DH10β cells (Invitrogen). Recombinant clones were examined for the relative orientation of the PCR-amplified insert within pBeloBAC11. The relative orientation was determined by selective restriction digests using the restriction endonucleases XhoI, BamHI, HindIII, and KpnI and confirmed by the sequencing of the cloned fragment.
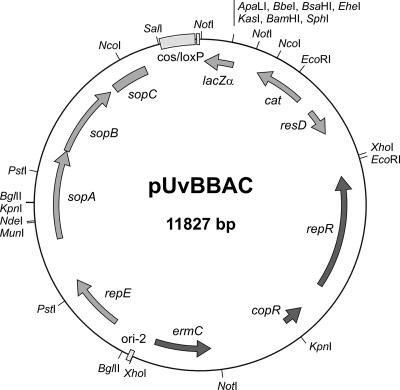
FIG. 1.
Map of the novel hybrid BAC vector pUvBBAC. This derivative of the second-generation BAC cloning vector pBeloBAC11 replicates in both E. coli and a broad range of gram-positive hosts. Recombinant clones are selectable with both chloramphenicol and erythromycin. Regions deriving from pBeloBAC11 are displayed in light gray. Dark gray regions are from pSOG7. The origin, replication, and partition functions of the mini-F derivate are indicated as ori-2; repE; and sopA, sopB and sopC, respectively. ResD is a putative resolvase of the F plasmid also known as protein D. The vector harbors a 5′-end truncated version of ResD. Additional features include the cos and loxP sites, which are required for packaging lambda particles if desired; the loxP site includes the cleavage site for the Cre recombinase. Selection markers in the E. coli host include the chloramphenicol acetylase gene (cat) and the lacZα gene for insert screening by alpha-complementation. The replication and copy number functions derived from pIP501 are indicated as repR and copR, respectively; the selection marker ermC encoding a methylase gene is also indicated.
Generation of BAC library.
All strains and derivatives of E. coli were grown in Luria-Bertani broth (LB), while Listeria strains were cultivated in brain heart infusion broth (BHI). Unless otherwise indicated, bacteria were cultivated at 37°C. Antibiotics were used at the following concentrations: erythromycin at 300 μg/ml for E. coli or 5 μg/ml for Listeria spp. and chloramphenicol at 20 μg/ml for E. coli or 5 μg/ml for Listeria spp. BAC-based vectors were generally isolated from 2-liter cultures following overnight growth with antibiotic selection. The BAC vector was purified using the large-scale QIAfilter plasmid mega kit, according to the protocol supplied by the manufacturer (Qiagen). The DNA pellet of the purified BAC vector was resuspended in 200 μl double-distilled H2O and stored at −20°C. For restriction digestion, pUvBBAC was completely cleaved at 37°C with BamHI (Roche Diagnostics) and dephosphorylated using calf intestine alkaline phosphatase (Roche Diagnostics). The dephosphorylation reaction was blocked by adding 2 μl EGTA (200 mmol/liter) at 65°C for 10 min, and hot phenol extraction was applied using a prewarmed mixture (65°C) of Roti-phenol-chloroform/isoamyl alcohol (25:24:1) mixture (Roth) for 1.5 min. DNA precipitation of the reaction mixture was performed to purify the dephosphorylated vector again. Genomic DNA of L. monocytogenes EGD-e was embedded in agarose plugs (44) using InCert low-melting-point agarose (FMC). Agarose-embedded DNA (2- to 5-μg) plugs were equilibrated twice with 100 μl BamHI restriction enzyme buffer (Roche Diagnostics) under gentle shaking for 30 min at room temperature. Plugs with genomic DNA were partially cleaved with 2.5 units BamHI (Roche Diagnostics) for 30 min, loaded onto a 1% pulsed-field (PF) certified agarose gel (Bio-Rad) and subjected to PF gel electrophoresis (PFGE) in 0.5× Tris-borate-EDTA (TBE) buffer using the Chef-DR II system (Bio-Rad) at 12°C, 6 V/cm for 18 h with 90-s pulses. Following electrophoretic separation, agarose slices harboring chromosomal fragments with sizes of 90 to 150 kb were excised from the gel. These agarose slices were loaded onto a SeaPlaque (GTG) agarose gel for additional PFGE at 12°C, 4 V/cm for 16 h with 5-s pulses in 0.5× TBE buffer. Following the second separation, gel slices with sized DNA fragments of 97 to 145.5 kb were cut out of the gel. A molecular-mass size standard, low-range PFG marker (New England Biolabs) was used. The agarose gel slices were then equilibrated twice with TNE (10 mM Tris, 40 mM NaCl, 1 mM EDTA) buffer for 30 min at 4°C. Agarose digestion was achieved by treating the gel slices for 1 h at 45°C with GELase (Epicenter), according to the instructions of the vendor. Two microliters (∼50 ng) of dephosphorylated pUvBBAC vector was added to a final volume of 250-μl ligation mixture. This includes the 218 μl cooled GELase-digested DNA plugs, 5 μl of T4 ligase (1 U/μl; GE Healthcare), and 25 μl 10× T4 ligase buffer (GE Healthcare). After incubation at 14°C for two days, the ligation mixture was electroporated or chemically transformed into the appropriate bacteria.
Electroporation was performed using 30 μl E. coli DH10β (Invitrogen) and 2.25 μl of ligation reaction mixture, which were mixed in a disposable electroporation cuvette (Invitrogen) and stored on ice for 5 min. For electroporation, a Gene Pulser (Bio-Rad) was prepared with the following parameter settings: 1.3 kV was selected for voltage, 200 Ω for resistance, and 25 μF for capacitance. Transformed cells were subsequently merged with 1 ml SOC (0.5% yeast extract, 2.0% tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) medium and incubated under gentle shaking at 37°C for 3 h. After the transformation mixture was placed onto LB plates containing 20 μg/ml chloramphenicol, 50 μg/ml X-Gal, and 50 μg/ml IPTG (isopropyl-β-d-thiogalactopyranoside), plates were incubated overnight at 37°C. Colorless colonies harboring the pUvBBAC recombinants were picked in single wells of 96-well microtiter plates containing storage medium. A LB storage medium with 20 μg/ml chloramphenicol and 5% (vol/vol) glycerol was used. Microtiter plates were incubated without shaking at 37°C for 20 to 24 h under high humidity conditions and subsequently stored at −80°C. Millipore Montage BAC96 miniprep kits were utilized to isolate pUvBBAC from randomly selected E. coli DH10β recombinants, according to the recommendation of the vendor. pUvBBAC insert sizes were determined by the control restriction digestion of 5 μl BAC DNA with 5 units NotI. The digested recombinant BAC DNA was loaded onto a 1% PF certified agarose gel (Bio-Rad) and separated using PFGE at 12°C, 5 V/cm for 18 h with an initial pulse of 2 s and a final pulse of 12 s in 0.5× TBE buffer. Gels were stained with ethidium bromide (0.5 μg/ml) for 1 h and then were destained with double-distilled H2O for 10 min before visualization and documentation. A molecular-mass size standard, low-range PFG marker (New England Biolabs) was used.
Screening of BAC library.
The E. coli DH10β library carrying pUvBBAC with chromosomal DNA fragments of L. monocytogenes EGD-e was screened according to the method of Barloy-Hubler et al. (2). Briefly, in a first screen, four 96-well microtiter plates of recombinants were pooled together in one single microtiter plate and were checked by PCR using specific oligonucleotides for a gene of interest. After agarose gel electrophoresis and the detection of potential positive clone mixtures, e.g., within a specific well of the combined plate, the corresponding wells of all four microtiter plates harboring the positive clone were streaked to obtain single colonies and rechecked in a second round of amplification with the same marker gene primer pair again. For PCR screening (95°C for 30 s, 55°C for 30 s, 72°C for 1 min, 30 cycles) of pooled clones, oligonucelotide pair P3/P4 for the hly gene encoding LLO was used (Table 1). For identification of the internal regions of vgc, the primer pairs P5/P6 (Table 1) and P7/P8 (Table 1) were applied. BAC vectors harboring the vgc pUvBBAC+vgc1 and pUvBBAC+vgc2 were purified as described above, and BAC end sequences were determined using T7 and SP6 sequencing primers (Table 1).
Standard sequencing reaction mixtures consisted of 2 μl of ABI Big Dye sequencing reaction mixture, 2 μl 5× sequencing buffer, 100 to 300 ng of BAC DNA, and 10 pmol of either primer Sp6 or T7 for pUvBBAC end sequencing.
Reactions were performed in an MJ Research thermocycler for 75 cycles of 95°C for 30 s, 50°C for 10 s, and 60°C for 4 min. The reaction products were cleaned up with the Montage SEQ96 sequencing reaction cleanup kit (Millipore), according to the recommendation of the vendor, and loaded onto the ABI Prism 3100 genetic analyzer (Applied Biosystems).
Transformation of Listeria innocua with pUvBBAC harboring the vgc.
Electrocompetent Listeria innocua cells were prepared as described previously (36). Transformation was performed using 50 μl electrocompetent L. innocua cells and 2.5 μg purified BAC vectors pUvBBAC+vgc1 and pUvBBAC+vgc2, which were mixed in a disposable cuvette (Invitrogen) and stored on ice for 5 min. For electroporation, a Gene Pulser (Bio-Rad) was prepared with the following parameter settings: 1.0 kV was selected for voltage, 400 Ω for resistance, and 25 μF for capacitance. Transformed cells were subsequently mixed with 1 ml BHI plus 0.5 M sucrose and incubated at 37°C for 3 h. After the transformation mixture was placed onto BHI plates containing 5 μg/ml erythromycin, plates were incubated overnight at 37°C for L. innocua containing pUvBBAC and for 3 days for L. innocua recombinants harboring pUvBBAC+vgc1 and pUvBBAC+vgc2.
Determination of PCN.
The plasmid copy numbers (PCN) of pUvBBAC+vgc1 and pUvBBAC+vgc2 per L. innocua genome were determined as described previously (26). Briefly, total genomic DNA were isolated as described by Pitcher et al. (38), and plasmid DNA of L. innocua strains harboring vgc1 and vgc2 were purified using the Bacmax DNA purification kit as recommended by Epicenter. The isolated DNA samples were serially diluted 1:10 up to 1:100,000. The DNA concentration of each dilution step was measured using the ND-1000 spectrophotometer (NanoDrop Technologies). The threshold cycle (CT) values of the corresponding template dilutions of the 16S rRNA gene and LLO (hly) were determined by real-time PCR using the ABI Prism 7700 instrument (Applied Biosystems). Reverse, forward, and probe primers for the detection of the 16S rRNA gene and hly are listed in Table 1. Standard curves, CT value versus concentration in pg, were generated using SigmaPlot version 6.0. The mean PCN per genome were determined from the same CT value for all dilution steps (n = 5) as described in Lee et al. (26). Altogether, three independent experiments were performed.
Determination of hemolytic activity.
For the detection of the hemolytic activity of Listeria strains, enterohemolysin agar with blood (Oxoid) was utilized. Strains were grown on these agar plates for about 24 h at 37°C before being kept overnight at 6°C to allow for a more intensive development of hemolysis.
Preparation of culture supernatants for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
For the preparation of both culture supernatants and bacterial cell wall proteins, Listeria strains were grown overnight at 37°C, diluted appropriately, and cultured to an optical density at 600 nm of 1.0. After centrifugation, culture supernatants (45 ml) were precipitated with 10% trichloroacetic acid, washed twice with acetone, and dissolved in 200 μl of 1 M Tris-HCl (pH 8.8). Bacterial pellets were suspended in 1.8 ml of phosphate-buffered saline (PBS), containing 1% SDS. They were subsequently incubated for 45 min at 37°C with gentle shaking. Following centrifugation, supernatants were precipitated with 10% trichloroacetic acid as well, washed twice with acetone, and dissolved in 200 μl of 1 M Tris-HCl (pH 8.8).
Supernatant and cell wall proteins (∼10 μg) were further analyzed by immunoblotting. SDS-PAGE was done with 12.5% polyacrylamide gels. A prestained SDS marker (Bio-Rad) served as a molecular weight marker. For immunoblotting reactions, the size-resolved proteins were transferred electrophoretically by a semidry method to Immobilon-P transfer membranes (Millipore). The membrane carrying the supernatant proteins was subsequently probed with a monoclonal anti-LLO antibody, M275 (diluted 1:5 in TBS-Tween), (14) for 2 h at room temperature by gentle agitation, and the second membrane with the fraction containing cell surface proteins was incubated with an equal mixture of two monoclonal anti-ActA antibodies, N81/N111 (33), (diluted 1:5,000 in TBS-Tween) and treated under the conditions reported above. Subsequently, the membranes were washed twice with 25 ml TBS-Tween to remove unbound primary antibody. A horseradish peroxidase-conjugated anti-mouse secondary antibody (Santa Cruz Biotechnology) was utilized for 3 h at room temperature, and then the blots were rinsed thrice with 25 ml TBS-Tween. Finally, the blots were developed with ECL Western blotting detection reagent (GE Healthcare) as recommended by the vendor.
In vitro invasion assay of J774 macrophage cells.
J774 macrophage cells were grown in Dulbecco's minimal essential medium (DMEM) (Gibco) supplemented with 10% fetal calf serum (PAA Laboratories) and 2 mM glutamine (Biochrom AG). For the infection assay, J774 cells were cultured to a semiconfluent layer in 24-well plates. Bacterial cultures were incubated in BHI overnight. Following a 1:10 fresh medium dilution, bacterial cultures were grown for 2 h and diluted to an optical density at 600 nm of 0.1 with DMEM. Bacteria were added at a multiplicity of infection of 10 per well. After incubation for 20 min, supernatant was discarded and infected J774 cells were washed twice with 1× PBS. DMEM was replaced by medium supplemented with 20 μg/ml gentamicin to kill the remaining extracellular bacteria. Cells were further incubated at 37°C for 1, 2, 4, and 8 h. The supernatant fluids were discarded, and the cells were washed three times with 1× PBS and lysed with 0.2% (vol/vol) Triton X-100 in distilled water. Cells were incubated with 0.2% Triton X-100 in distilled water for 20 min at room temperature and then thoroughly mixed to lyse the cells completely. The lysate was diluted 10 times in 1× PBS and plated onto BHI agar plates using the Autoplate 3000 spiral plating system (Spiral Biotech). After 24 h of incubation at 37°C, the number of bacterial colonies was counted and the total number of CFU determined.
The absolute numbers of infecting cells for each bacterial strain were determined by calculating the percentage of intracellular bacteria recovered (in CFU) after lysis of the macrophage cells with reference to the total number of bacteria in the inoculum.
Immunofluorescence.
At 4 h postinfection, J774 cells on glass coverslips were fixed with 3.7% formaldehyde diluted in 1× PBS for 10 min and permeabilized with 0.2% (vol/vol) Triton X-100 in 1× PBS for 1 min. After the coverslips were rinsed with 1× PBS, they were incubated with Oregon Green 488-conjugated phalloidin (Invitrogen) and an anti-ActA-specific monoclonal antibody mixture of N81/N111 antibodies (33). Primary antibody was detected by incubation with a Cy3-conjugated secondary antibody (Dianova). Samples were examined with an immunofluorescence microscope (Axiophot; Zeiss), and images were captured and processed using KS 300 software (Zeiss).
Nucleotide sequence accession number.
The complete sequence of pUvBBAC is available from the EMBL database under accession number AJ509853.
RESULTS
Construction of a hybrid BAC vector capable of replicating in gram-positive and gram-negative hosts.
pBeloBAC11 (24) is a second-generation mini-F-based BAC cloning vector with lacZα for facilitating the detection of recombinants and the cat gene for transformant selection with chloramphenicol. The F factor carries genes that are essential for regulating its own replication and for copy number control in the cell. The ori-2 sequence and RepE protein are required for the unidirectional replication of the F factor, while the SopA and SopB proteins ensure faithful transfer to progeny and low-copy-number maintenance in the transformed cell (45).
The novel shuttle BAC vector pUvBBAC (Fig. 1), which is capable of replication in gram-positive hosts, was generated by the addition of replication functions from the broad-host-range plasmid pIP501 as well as the ermC resistance gene from pE194 to pBeloBAC11 to give pUvBBAC (see Materials and Methods). Subsequent analyses revealed that the pUvBBAC vector replicates in different Listeria species, such as L. monocytogenes EGD-e and L. innocua. Both resistance markers are selectable in gram-positive and -negative hosts. Thus, recombinant E. coli strains were selectable with either erythromycin at 300 μg/ml and/or 20 μg/ml chloramphenicol, and for the selection of correspondent Listeria strains, 5 μg/ml erythromycin and/or 5 μg/ml chloramphenicol were used.
Generation of a BAC library for L. monocytogenes EGD-e.
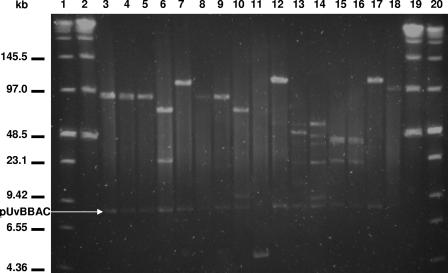
To construct the BAC library for L. monocytogenes EGD-e, the pUvBBAC vector was digested with the restriction endonuclease BamHI and ligated to chromosomal DNA, partially digested with the same enzyme and excised and purified from agarose gels after sizing using PFGE. The excised DNA fragments were sized to obtain inserts ranging from 97 to 145.5 kb. After transformation of the E. coli DH10β host strain with the respective ligation mixtures, a BAC library for L. monocytogenes EGD-e was established with 350 clones. To estimate insert sizes, we selected randomly and examined 16 recombinants from the library. Following BAC purification from these E. coli clones, NotI restriction digestion of purified pUvBBAC vectors carrying large chromosomal DNA fragments of L. monocytogenes EGD-e was carried out. We observed insert sizes in a range of 68 to 178 kb (Fig. 2) after the separation of those fragments by PFGE. Confirmation of the presence of large insert sizes was obtained from the end sequencing of an additional 49 BAC recombinants which had an average of ∼92 kb each. Direct transformation of electrocompetent Listeria cells with ligation mixtures was poor and irreproducible and never led to the detection of more than a few colonies. The transformation of electrocompetent E. coli cells with aliquots of the same ligation mixture generated several tens to hundreds of colonies per ligation and was used in establishing the library. Sequence analyses from 49 of these pUvBBAC clones were mapped to the chromosome of L. monocytogenes EGD-e and covered ∼42% of the chromosome of L. monocytogenes EGD-e (see Table S2 in the supplemental material; Fig. 3A).
FIG. 2.
PFGE of NotI-digested BAC DNA obtained from the L. monocytogenes EGD-e pUvBBAC library. Insert sizes range from 68 to 178 kb. Lanes 1 and 20 contain the low-range PFG marker (New England Biolabs), while the lambda ladder PFG marker (New England Biolabs) is depicted in lanes 2 and 19.
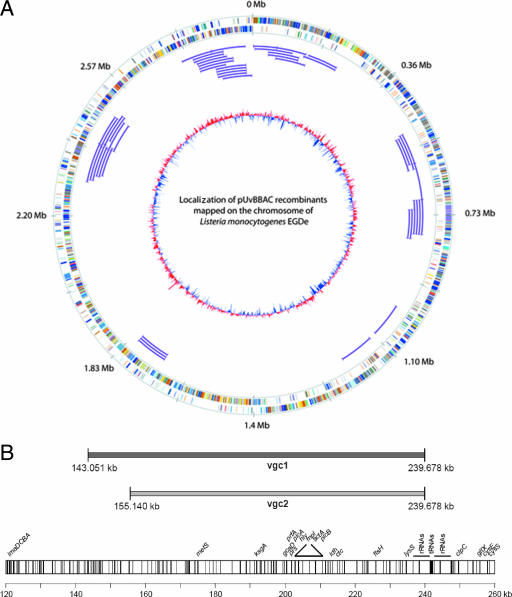
FIG. 3.
(A) Mapping and visualization of pUvBBAC recombinants on the L. monocytogenes EGD-e chromosome using GenomeViz (16). The first (outer) circle represents the scale in megabases, starting with the origin of replication at position 0. The second and third circles show the distribution of coding sequences of the leading and lagging strands and indicate the classification of the clusters of orthologous groups (COG) for each gene and the distribution of COG classes within the genome. The color scheme and categories are according to the convention of the COG database. The chromosomal localization of the mapped pUvBBAC inserts are indicated as thin purple lines. The inner circle indicates the deviations of the GC content averages, with values greater than zero in red and less than zero in blue. (B) Alignment of the chromosomal DNA inserts in recombinants harboring vgc1 and vgc2 to the genome of L. monocytogenes EGD-e. The exact positions of genomic fragments derived from DNA sequencing of either end of the inserted DNA are indicated. All of the known open reading frames within this region are depicted as individual bars.
Identification and isolation of BACs expressing the vgc of Listeria monocytogenes EGD-e.
Following the characterization of the listerial pUvBBAC library, the L. monocytogenes EGD-especific pUvBBAC library was screened for clones harboring the pUvBBAC vector with the vgc locus of L. monocytogenes EGD-e. A PCR-based pooled screening method (2, 11) for pUvBBAC library clones was used to identify those recombinants harboring the vgc locus. In a first round, four 96-well microtiter plates consisting of 350 recombinants were pooled together in one microtiter plate and were checked by PCR using the specific oligonucleotides P3 and P4 (Table 1) for the hly gene located in the vgc. Two pools displayed the expected 1,873-bp-sized PCR product of the hly gene after agarose gel electrophoresis. These pooled clones were streaked to obtain single colonies and rechecked with the same primer pair. After a second round of hly gene amplification, two clones harboring the hly gene were identified. Confirmation that these pUvBBAC vectors indeed harbored the entire vgc locus was achieved by amplification of the internal regions of the vgc locus with primer pairs P5/P6 (2,255 bp) and P7/P8 (3,889 bp) (Table 1) located within the genes for plcA-prfA and for hly-actA. This finding indicates that the entire vgc locus was present in these two BAC clones. Both BACs harboring the vgc locus were isolated, and BAC end sequences were determined by sequencing using T7 and SP6 primers. Comparison of these sequences with the Listeria genome sequence (17) revealed that recombinant pUvBBAC+vgc1 harbors an insert of 96.628 bp (positions 143.051 to 239.678) and pUvBBAC+vgc2 an insert of 84.539 bp (positions 155.140 to 239.678) (Fig. 3B).
For functional genomic analysis, a nonpathogenic L. innocua strain was transformed with purified BAC DNA. Following 3 days of incubation at 37°C, recombinant L. innocua clones appeared on BHI agar plates containing 5 μg/ml erythromycin and 5 μg/ml chloramphenicol. A total of 80 recombinant L. innocua clones were collected from plates with L. innocua(pUvBBAC+vgc1), but only five clones were obtained for L. innocua(pUvBBAC+vgc2). Control transformation with the empty vector pUvBBAC resulted in a relatively high transformation rate of more than 3 × 104 recombinants per plate. Hemolytic activity was detected only in the supernatants of L. innocua recombinants carrying pUvBBAC+vgc1 or pUvBBAC+vgc2.
Characterization of recombinant L. innocua strains.
Following the demonstration of the presence of the vgc from L. monocytogenes EGD-e in both recombinant strains, L. innocua (pUvBBAC+vgc1) and L. innocua(pUvBBAC+vgc2), we examined both for the expression and activity of several virulence factors encoded by the vgc locus.
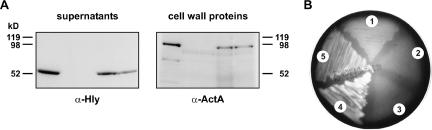
As a first step, we used TaqMan quantitative real-time PCR and determined the PCN of L. innocua harboring vgc to be ∼5 copies per genome equivalent (26). Following this, we looked for the expression of LLO and ActA (Fig. 4A), key determinants for listerial survival and motility within the infected cells. Immunoblot analysis revealed the presence of LLO in the supernatant fractions of both recombinant L. innocua vgc-harboring strains. Similarly, membrane-anchored protein ActA was found to be present in cell wall fractions of these strains, hence matching the respective cellular locations of these proteins in the L. monocytogenes EGD-e strain. As expected, none of these factors was expressed by either the parental L. innocua strain or a derivative transformed with the pUvBBAC vector alone.
FIG. 4.
Demonstration of virulence gene expression and detection of hemolysis for recombinant L. innocua strains. (A) Bacterial culture supernatants and cell wall proteins were separated by SDS-PAGE and further analyzed by immunoblotting. (B) Culture supernatants were probed with an anti-Hly antibody, and the fraction containing cell surface proteins was used for the detection of ActA. Hemolytic activity was determined on blood agar. Lanes: 1, L. monocytogenes EGD-e; 2, L. innocua; 3, L. innocua(pUvBBAC); 4, L. innocua(pUvBBAC+vgc1); 5, L. innocua(pUvBBAC+vgc2). The experiment was repeated three times independently, and one representative immunoblot is shown here.
On blood agar plates, vgc-harboring L. innocua recombinants showed higher hemolytic activity than the L. monocytogenes wild-type strain EGD-e (Fig. 4B). L. innocua and L. innocua(pUvBBAC) were devoid of hemolytic activity. These data correlate well with both the determined copy numbers of these plasmids and the levels of LLO detected by immunoblotting.
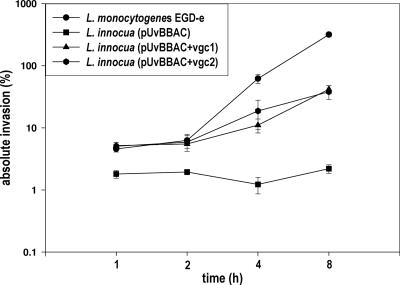
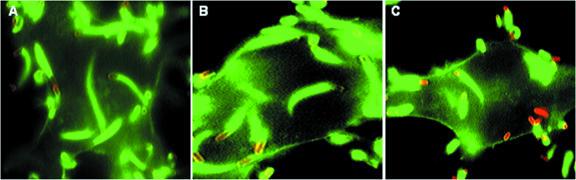
An in vitro infection assay with J774 macrophages was performed to reveal that recombinant L. innocua strains harboring pUvBBAC+vgc1 and pUvBBAC+vgc2 are capable of intracellular growth after escape from the phagosome and of recruiting actin for bacterial movement and multiplication within the cytosol of the eukaryotic host cells. L. innocua strains bearing pUvBBAC, pUvBBAC+vgc1, and pUvBBAC+vgc2 were monitored for growth in J774 macrophages 1, 2, 4, and 8 h following infection as described above. In comparison to the wild-type strain, L. monocytogenes EGD-e, the complemented L. innocua strains pUvBBAC+vgc1 and pUvBBAC+vgc2 grew more slowly, as can clearly be seen from comparisons at the time points 4 h and 8 h postinfection (Fig. 5). Double immunofluorescence staining for the bacterial ActA protein and host actin revealed that the recombinant L. innocua strains pUvBBAC+vgc1 and pUvBBAC+vgc2 are able to accumulate actin filaments around the bacterial cell. Some bacteria harbor long actin tails indicative of vigorous intracytoplasmic motility (Fig. 6A, B, and C).
FIG. 5.
Intracellular growth rates of L. monocytogenes EGD-e and recombinant L. innocua strains in J774 macrophage. Bacteria were incubated with macrophage at a multiplicity of infection of 10 for 20 min, after which gentamicin (20 μg/ml) was added and bacterial growth monitored at 1, 2, 4, and 8 h postinfection.
FIG. 6.
Intracellular actin accumulation and motility of L. monocytogenes EGD-e (A), L. innocua(pUvBBAC+vgc1) (B) or L. innocua(pUvBBAC+vgc2) (C) 4 h after the infection of J774 macrophage cells. Listeriae were detected with monoclonal antibody N81/N111 against ActA and visualized with a Cy3-labeled secondary antibody. Actin filaments of the host cell were stained with Oregon Green 488-conjugated phalloidin.
DISCUSSION
In this study, we have developed a BAC vector capable of replication in both gram-negative and gram-positive bacterial hosts. The BAC library harboring chromosomal DNA inserts between 68 and 178 kb from L. monocytogenes EGD-e was constructed and used in sequencing and mapping strategies for these species. BACs of ∼100 kb were readily transferable using transformation procedures described herein and could be stably propagated in the host. In a library comprising L. monocytogenes EGD-e inserts, we detected two BACs harboring the entire vgc. The transfer of pUvBBAC+vgc1 and pUvBBAC+vgc2 to a nonpathogenic L. innocua strain conferred on the strain the ability to express LLO and the actin-assembling inducing protein actA, two important virulence genes from L. monocytogenes EGD-e. Recombinant L. innocua strains harboring vgc exhibited multiple properties and showed intracellular motility following egress from the phagolysosome. Although L. innocua vgc-harboring recombinants grew better than L. innocua in tissue culture cells, their growth rate was significantly lower than that of L. monocytogenes EGD-e, and they appeared incapable of cell-to-cell spread.
Despite the widespread use of BAC libraries in the construction of large contiguous segments from eukaryotic genomes, their use in studying prokaryotic biology is fairly limited. One of the major limitations of this technology is that it is currently widely applicable only for functional genomic studies in gram-negative bacteria, in particular E. coli. Thus, E. coli BAC vector pBeloBAC11 (24) has been used for the generation of chromosomal libraries of Mycobacterium tuberculosis (7) and Treponema pallidum (46) in sequencing projects, and the expression of heterologous DNA from the Bacillus cereus BAC library has been detected at a low frequency in E. coli (41). In this study, we extend the use of this technology to gram-positive host bacteria by using a low-copy-number pIP501 replicon that is capable of replicating in a wide variety of hosts, including the genera Staphylococcus, Bacillus, Streptococcus, Lactococcus, and Listeria. pUvBBAC retains all the features of the parental pBeloBAC11 and carries in addition the pIP501 repR and copR genes and an erythromycin-resistance gene (ermC) that can be used as a selectable marker in gram-positive and gram-negative hosts.
Replication elements repR and oriR have been shown to be necessary and sufficient for the replication of pIP501. The RepR protein is essential for the initiation of plasmid replication, while oriR provides the putative initiation site of leading strand synthesis. A third region, localized 5′ to pIP501 repR and named copR, is known to negatively regulate PCN. Taken together, these three replication functions confer a broad host range in gram-positive hosts to pIP501 and a low copy number of about 5 to 10 in Bacillus subtilis.
We also found that the chloramphenicol transacetylase gene on pBeloBAC11 imparts selectable resistance in both hosts. Based on studies done previously in Streptococcus and B. subtilis, we estimated a copy number of ∼5 per genome equivalent (6). Following analysis using a highly accurate TaqMan method developed by Lee et al. (26), we were able to confirm that the PCN in L. innocua harboring vgc inserts to be ∼5 copies per genome.
We constructed a BAC library comprising chromosomal DNA from L. monocytogenes EGD-e. The L. monocytogenes EGD-e library had an average size of ∼100 kb; some recombinants had inserts that were almost 200 kb in size. As described previously, BACs appear to maintain heterologous DNA more stably than other cloning systems (31). For the pUvBBAC library, we were not able to detect chimeric recombinants or to identify any integration by single crossover of recombinants into the chromosome of L. monocytogenes. From end-sequencing studies done on 49 recombinant plasmids in our L. monocytogenes EGD-e BAC libraries, we observed that our library covers ∼42% of the total genome (see Table S2 in the supplemental material; Fig. 3A). Therefore, we estimated a total of 150 clones for a minimal overlap library of the L. monocytogenes genome. Currently all our BAC libraries are first generated in commercially available E. coli strains (e.g., DH10β) before transfer to the appropriate Listeria sp. for functional analysis. Transformation rates from primary ligation mixes are poor in Listeria spp., ranging from none to a few colonies, unlike in E. coli where several hundred colonies can be obtained from a single ligation. Nevertheless, once recovered in the gram-negative host, BACs can be transferred singly or in bulk using the transformation procedure described here. Although we have not used recombination-deficient (recA) Listeria strains as recipients, propagation of BACs in L. innocua, in which most of our experiments were performed, was not a problem. No instability, deletions, or loss of BAC inserts was detected despite extensive propagation in these strains.
Having established that pUvBBAC can be used to clone large chromosomal fragments, we looked for recombinants in the L. monocytogenes EGD-e(pUvBBAC) library that harbored the vgc locus. All our previous attempts to clone the entire gene cluster have been unsuccessful, and the successful cloning of only individual virulence genes from this cluster has been reported. In addition, we reasoned that the cloning and transfer of this region to a nonpathogenic L. innocua strain would replicate many of the properties for intracellular survival and growth of the pathogenic parental L. monocytogenes EGD-e strain. Screening of the 350 recombinants from the library using a PCR-based strategy led to the detection of two recombinants, designated vgc1 and vgc2. The BACs were recovered, isolated, and transferred by electro-transformation to a plasmidless L. innocua serotype 6a strain. L. innocua recombinant strains harboring vgc1 and vgc2 were readily identified on the basis of their hemolytic properties and PCR amplification of various regions of the vgc locus. Subsequent immunoblotting studies using monoclonal antibodies confirmed that LLO and ActA are indeed expressed and secreted to the supernatant and anchored in the membrane of the bacteria, respectively. End sequencing of pUvBBAC+vgc1 and pUvBBAC+vgc2 revealed that both BACs had one identical end point and harbored insert sizes of 84.5 kb and 96.6 kb, respectively. Although we determined a similar PCN for both L. innocua vgc recombinants, we observed different levels of LLO produced by pUvBBAC+vgc1 and pUvBBAC+vgc2. Therefore, we suspect that the difference in the expression of LLO for L. innocua harboring pUvBBAC+vgc1 is a result of the different gene content localized on the additional 12.1-kb fragment compared to the pUvBBAC+vgc2 recombinant. Detailed genome analysis of this gene region showed a high number of unknown genes (lmo0142 to lmo0151 and lmo0156), whereas lmo0141 and lmo0157 are truncated genes. lmo0152 encodes an oligopeptide ABC transporter-binding protein, and lmo0153 to lmo0155 code for a high-affinity zinc ABC transporter system. It is known that another OppA homolog (lmo2196) affects the rate of intracellular survival of L. monocytogenes in bone marrow-derived macrophages (3), and other zinc transporters, such as zurA (lmo1447) and lmo1671 (high-affinity zinc uptake system protein), were upregulated during growth in the cytosol of macrophage (12). The role of these ABC transporters in the direct or indirect control of LLO and intracellular survival is not clear and warrants further study.
Both the vgc1 and vgc2 pUvBBAC vectors lack the internalin operon inlAB that is located approximately 242.2 kb away from the vgc. In order to examine if L. innocua strains harboring vgc pUvBBAC vectors indeed possess multiple properties, such as the ability to egress from the phagolysosome, intracellular growth and motility, and intercellular spread, we studied these properties using the J774 macrophage-like cell line. Following phagocytosis, L. innocua strains harboring vgc recombinants displayed the ability to grow and accumulate actin for intracellular mobility within the infected macrophage. When assayed quantitatively for their ability to grow within macrophages, L. innocua vgc strains showed low but significant growth compared to the wild-type L. monocytogenes EGD-e strain. None of these properties was observed with L. innocua strains harboring the pUvBBAC vector alone. Surprisingly, although all of the genes known to be required for cell-to-cell spread are present on the vgc-containing plasmids, neither of these L. innocua recombinants were capable of plaque formation. Cumulatively, these data suggest that although vgc-harboring bacteria qualitatively conferred many of the properties required for virulence, additional factors required for intracellular multiplication are lacking in these strains.
Evidence has been presented to show that the ability of intracellular multiplication is not an intrinsic property of every bacterium. Thus, following microinjection into the host cytoplasm, L. monocytogenes, Shigella flexneri, and enteroinvasive Escherichia coli, but not L. innocua, Yersinia enterocolitica, or Staphylococcus aureus, are capable of intracellular growth (18). Nevertheless, our studies indicate that equipping L. innocua strains with plasmids harboring the vgc facilitates intracellular growth for a bacterium that has not previously been adapted for intracytoplasmic growth.
It has recently been demonstrated that a hexose phosphate uptake protein designated hpt is required for the optimal growth of pathogenic Listeria in the host cytoplasm (13). Tantalizingly, hpt gene expression is regulated by the PrfA protein, a master regulator of virulence gene expression in L. monocytogenes EGD-e. Neither of the vgc pUvBBAC vectors described in this study harbors the hpt gene, which could account for the low intracellular growth rates observed. Thus, the complementation of the vgc-containing recombinants could lead to the identification of further genes required for intracellular growth.
The construction and use of the first BAC vector capable of stable propagation in both gram-positive and gram-negative hosts increase the genetic tools available for the study of a broad range of bacteria. Functional genomic analysis of large chromosomal fragments can now be performed for a large group of bacteria that are important as pathogens (Staphylococcus, Streptococcus, and Enterococcus), in industrial use (Bacillus spp. and Lactobacillus), or even for unculturable bacterial species in the environment which may be accessed by BAC cloning. Studies of this vector reported here have led to the first successful cloning of the listerial vgc pathogenicity island and its functional expression in a nonpathogenic L. innocua strain.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Alexandra Amend-Förster, Kornelia Kirchner, Sylvia Krämer, and Nelli Schklarenko. Thanks to S. Brantl for the kind gift of numerous vectors, including pCOP4 utilized for the construction of pSOG7.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) through the SFB 535 and Bundesministerium für Bildung und Forschung (BMBF) through the ERA-NET PathoGenoMics to T.H. and T.C.
Footnotes
Published ahead of print on 25 January 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2001. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J. Virol. 75:5692-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barloy-Hubler, F., D. Capela, J. Batut, and F. Galibert. 2000. High-resolution physical map of the pSymb megaplasmid and comparison of the three replicons of Sinorhizobium meliloti strain 1021. Curr. Microbiol. 41:109-113. [DOI] [PubMed] [Google Scholar]
- 3.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantl, S., and D. Behnke. 1992. Copy number control of the streptococcal plasmid pIP501 occurs at three levels. Nucleic Acids Res. 20:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brantl, S., and E. G. Wagner. 1996. An unusually long-lived antisense RNA in plasmid copy number control: in vivo RNAs encoded by the streptococcal plasmid pIP501. J. Mol. Biol. 255:275-288. [DOI] [PubMed] [Google Scholar]
- 7.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune, W., M. Messerle, and U. H. Koszinowski. 2000. Forward with BACs: new tools for herpesvirus genomics. Trends Genet. 16:254-259. [DOI] [PubMed] [Google Scholar]
- 9.Budiman, M. A., L. Mao, T. C. Wood, and R. A. Wing. 2000. A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res. 10:129-136. [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, L., J. F. Taylor, R. A. Wing, D. S. Gallagher, S. S. Woo, and S. K. Davis. 1995. Construction and characterization of a bovine bacterial artificial chromosome library. Genomics 29:413-425. [DOI] [PubMed] [Google Scholar]
- 11.Capela, D., F. Barloy-Hubler, M. T. Gatius, J. Gouzy, and F. Galibert. 1999. A high-density physical map of Sinorhizobium meliloti 1021 chromosome derived from bacterial artificial chromosome library. Proc. Natl. Acad. Sci. USA 96:9357-9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chico-Calero, I., M. Suarez, B. Gonzalez-Zorn, M. Scortti, J. Slaghuis, W. Goebel, and J. A. Vazquez-Boland. 2002. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. USA 99:431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darji, A., K. Niebuhr, M. Hense, J. Wehland, T. Chakraborty, and S. Weiss. 1996. Neutralizing monoclonal antibodies against listeriolysin: mapping of epitopes involved in pore formation. Infect. Immun. 64:2356-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentzbittel, L., A. Abbott, J. P. Galaud, L. Georgi, F. Fabre, T. Liboz, and G. Alibert. 2002. A bacterial artificial chromosome (BAC) library for sunflower, and identification of clones containing genes for putative transmembrane receptors. Mol. Genet. Genomics 266:979-987. [DOI] [PubMed] [Google Scholar]
- 16.Ghai, R., T. Hain, and T. Chakraborty. 2004. GenomeViz: visualizing microbial genomes. BMC Bioinformatics 5:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 18.Goetz, M., A. Bubert, G. Wang, I. Chico-Calero, J. A. Vazquez-Boland, M. Beck, J. Slaghuis, A. A. Szalay, and W. Goebel. 2001. Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc. Natl. Acad. Sci. USA 98:12221-12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hain, T., C. Steinweg, and T. Chakraborty. 2006. Comparative and functional genomics of Listeria spp. J. Biotechnol. 126:37-51. [DOI] [PubMed] [Google Scholar]
- 20.Hamer, L., H. Pan, K. Adachi, M. J. Orbach, A. Page, L. Ramamurthy, and J. P. Woessner. 2001. Regions of microsynteny in Magnaporthe grisea and Neurospora crassa. Fungal Genet. Biol. 33:137-143. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton, C. M. 1997. A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene 200:107-116. [DOI] [PubMed] [Google Scholar]
- 22.Horodniceanu, T., D. H. Bouanchaud, G. Bieth, and Y. A. Chabbert. 1976. R plasmids in Streptococcus agalactiae (group B). Antimicrob. Agents Chemother. 10:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ioannou, P. A., C. T. Amemiya, J. Garnes, P. M. Kroisel, H. Shizuya, C. Chen, M. A. Batzer, and P. J. de Jong. 1994. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat. Genet. 6:84-89. [DOI] [PubMed] [Google Scholar]
- 24.Kim, U. J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H. L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 25.Lalioti, M., and J. Heath. 2001. A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 29:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, C. L., D. S. Ow, and S. K. Oh. 2006. Quantitative real-time polymerase chain reaction for determination of plasmid copy number in bacteria. J. Microbiol. Methods 65:258-267. [DOI] [PubMed] [Google Scholar]
- 27.Lijavetzky, D., G. Muzzi, T. Wicker, B. Keller, R. Wing, and J. Dubcovsky. 1999. Construction and characterization of a bacterial artificial chromosome (BAC) library for the A genome of wheat. Genome 42:1176-1182. [PubMed] [Google Scholar]
- 28.Martin, F. E., M. A. Nadkarni, N. A. Jacques, and N. Hunter. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40:1698-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGregor, A., and M. R. Schleiss. 2001. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Mol. Genet. Metab. 72:15-26. [DOI] [PubMed] [Google Scholar]
- 30.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monaco, A. P., and Z. Larin. 1994. YACs, BACs, PACs and MACs: artificial chromosomes as research tools. Trends Biotechnol. 12:280-286. [DOI] [PubMed] [Google Scholar]
- 32.Mozo, T., K. Dewar, P. Dunn, J. R. Ecker, S. Fischer, S. Kloska, H. Lehrach, M. Marra, R. Martienssen, S. Meier-Ewert, and T. Altmann. 1999. A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat. Genet. 22:271-275. [DOI] [PubMed] [Google Scholar]
- 33.Niebuhr, K., T. Chakraborty, M. Rohde, T. Gazlig, B. Jansen, P. Köllner, and J. Wehland. 1993. Localization of the ActA polypeptide of Listeria monocytogenes in infected tissue culture cell lines: ActA is not associated with actin “comets.” Infect. Immun. 61:2793-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osoegawa, K., M. Tateno, P. Y. Woon, E. Frengen, A. G. Mammoser, J. J. Catanese, Y. Hayashizaki, and P. J. de Jong. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10:116-128. [PMC free article] [PubMed] [Google Scholar]
- 35.Otten, S. 2001. Charakterisierung und identifizierung präferentiell intrazellulär exprimierter gene aus Listeria monocytogenes. Logos Verlag, Berlin, Germany.
- 36.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 37.Payne, C. M., L. J. Mullins, and J. J. Mullins. 1999. Manipulating large genomic clones via in vivo recombination in bacteria. J. Hum. Hypertens. 13:845-848. [DOI] [PubMed] [Google Scholar]
- 38.Pitcher, A., A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guandinium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 39.Randall, T. A., and H. S. Judelson. 1999. Construction of a bacterial artificial chromosome library of Phytophthora infestans and transformation of clones into P. infestans. Fungal Genet. Biol. 28:160-170. [DOI] [PubMed] [Google Scholar]
- 40.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rondon, M. R., S. J. Raffel, R. M. Goodman, and J. Handelsman. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. USA 96:6451-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudolph, J., D. J. O'Callaghan, and N. Osterrieder. 2002. Cloning of the genomes of equine herpesvirus type 1 (EHV-1) strains KyA and racL11 as bacterial artificial chromosomes (BAC). J. Vet. Med. B 49:31-36. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 76:2973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarzkopf, A., C. Cuny, and W. Witte. 1995. Bestimmung der Fragmentmuster der genomischen DNA mittels Pulsfeld-Gelelektrophorese bei Staphylococcus aureus. Bundesgesundheitsblatt 38:215-219. [Google Scholar]
- 45.Shizuya, H., B. Birren, U. J. Kim, V. Mancino, T. Slepak, Y. Tachiiri, and M. Simon. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89:8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smajs, D., M. McKevitt, L. Wang, J. K. Howell, S. J. Norris, T. Palzkill, and G. M. Weinstock. 2002. BAC library of T. pallidum DNA in E. coli. Genome Res. 12:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song, J., F. Dong, and J. Jiang. 2000. Construction of a bacterial artificial chromosome (BAC) library for potato molecular cytogenetics research. Genome 43:199-204. [PubMed] [Google Scholar]
- 48.Sosio, M., E. Bossi, and S. Donadio. 2001. Assembly of large genomic segments in artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 29:E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sternberg, N. 1990. Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc. Natl. Acad. Sci. USA 87:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki, K., S. Asakawa, M. Iida, S. Shimanuki, N. Fujishima, H. Hiraiwa, Y. Murakami, N. Shimizu, and H. Yasue. 2000. Construction and evaluation of a porcine bacterial artificial chromosome library. Anim. Genet. 31:8-12. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki, M. T., O. Beja, L. T. Taylor, and E. F. Delong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 52.Tauch, A., I. Homann, S. Mormann, S. Ruberg, A. Billault, B. Bathe, S. Brand, O. Brockmann-Gretza, C. Ruckert, N. Schischka, C. Wrenger, J. Hoheisel, B. Mockel, K. Huthmacher, W. Pfefferle, A. Puhler, and J. Kalinowski. 2002. Strategy to sequence the genome of Corynebacterium glutamicum ATCC 13032: use of a cosmid and a bacterial artificial chromosome library. J. Biotechnol. 95:25-38. [DOI] [PubMed] [Google Scholar]
- 53.Vanhouten, W., and S. MacKenzie. 1999. Construction and characterization of a common bean bacterial artificial chromosome library. Plant Mol. Biol. 40:977-983. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, H., S. Choi, A. K. Johnston, R. A. Wing, and R. A. Dean. 1997. A large-insert (130 kbp) bacterial artificial chromosome library of the rice blast fungus Magnaporthe grisea: genome analysis, contig assembly, and gene cloning. Fungal Genet. Biol. 21:337-347. [DOI] [PubMed] [Google Scholar]
- 55.Zimmer, R., and G. A. Verrinder. 1997. Construction and characterization of a large-fragment chicken bacterial artificial chromosome library. Genomics 42:217-226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.