Abstract
Starch and pullulan-modifying enzymes of the α-amylase family (glycoside hydrolase family 13) have several industrial applications. To date, most of these enzymes have been derived from isolated organisms. To increase the number of members of this enzyme family, in particular of the thermophilic representatives, we have applied a consensus primer-based approach using DNA from enrichments from geothermal habitats. With this approach, we succeeded in isolating three new enzymes: a neopullulanase and two cyclodextrinases. Both cyclodextrinases displayed significant maltogenic amylase side activity, while one showed significant neopullulanase side activity. Specific motifs and domains that correlated with enzymatic activities were identified; e.g., the presence of the N domain was correlated with cyclodextrinase activity. The enzymes exhibited stability under thermophilic conditions and showed features appropriate for biotechnological applications.
Microorganisms, including bacteria, fungi, and yeasts, have been reported to grow on starch as carbon and energy sources and thereby produce starch-modifying and degrading enzymes, which are of importance for biotechnological applications. These enzymes are glycoside hydrolases (GHs) capable of hydrolyzing glycosidic bonds between two or more carbohydrates or between a carbohydrate and an aglycon moiety. GHs are to date classified by amino acid similarities into 108 families (8, 9). Hydrolysis of starch demands the coordinated activity of a number of different enzymes. Most of the known starch-modifying enzymes can be found in GH family 13 (GH13) (18), including α-amylases, which are industrially used enzymes (7, 31), as well as enzymes with a broad spectrum of other starch-modifying and degrading activities, e.g., pullulanases, α-1,6-glucosidases, branching enzymes, maltogenic amylases, neopullulanases, and cyclodextrinases (CDases) (13, 18). Despite the diversity of activities in GH13, the amino acid sequences of members in this family show four highly conserved regions around the active center (18).
Recently, a variety of hyperthermophilic bacterial and archaeal species producing thermophilic enzymes of the GH13 family and showing pronounced thermostability and optima, appropriate for different industrial applications, have been described (17).
To extend our knowledge on thermophilic GH13 members, we aimed to establish a method based on CODEHOP (consensus-degenerate hybrid oligonucleotide primer) that allows the isolation of GH13 members directly from environmental DNA. With this method, we were able to isolate and characterize three enzymes that were characterized with respect to their molecular and catalytic properties. Furthermore, a detailed analysis of structure-function relationships was performed.
MATERIALS AND METHODS
Sample collection, enrichments, and DNA isolation.
The DNA samples were obtained from in situ enrichments in hot springs from the southern part of Iceland (see Table 3). In situ enrichments were performed using sterile permeable 1-liter polyethylene flasks, which were inoculated with untreated hot spring water from the relevant hot spring and supplemented with 0.1% starch. The flasks were closed to prevent medium and biomass loss, and the permeability allowed gases and ions from the hot spring to flow into and out of the bottle. The flasks were maintained with 500 g of weight in the hot springs. Enrichments were centrifuged at 10,000 × g for 30 min at 4°C; the pellets were homogenized with a sterile glass mortar, and DNA was extracted as described by Marteinsson et al. (20).
TABLE 3.
Designations, sources, similarities, and functions of the new enzymes from the GH13 family
| Enzyme | Source | Location (conditions) | Closest match for:
|
Closest structure-determined homolog by similarity
|
Main activity (exptl, this work) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conserved fragment, regions I and II
|
Complete sequence
|
||||||||||||
| Enzyme | Organism | Enzyme | Organism | Swiss-Prot accession no. | Identity (%) | Enzyme | Organism | Swiss-Prot accession no. | Identity (%) | ||||
| Amy1 | Environmental DNA (oligotrophic enrichment) | Hveragerdi (60-65°C, pH 7-8) | CDase | Alicyclobacillus acidocaldarius | CDase | Alicyclobacillus acidocaldarius | Q9WX32 | 80 | Maltogenic amylase | Thermus sp. | O69007 | 42 | CDase |
| Neopullulanase 2/ TVAII | T. vulgaris | Q08751 | 41 | ||||||||||
| Amy29 | Environmental DNA (oligotrophic enrichment) | Hveragerdi (60-65°C, pH 7-8) | Neopullulanase | Synechocystis sp. | Neopullulanasea | T. thermophilus | Q5SI17 | 64 | Maltogenic amylaseb | Thermus sp. | O69007 | 44 | Neopullulanase |
| Amy132 | Environmental DNA (oligotrophic enrichment) | Grensdalur (65°C, pH 6-7) | Neopullulanase | G. stearothermophilus | CDase | Anoxybacillus flavithermus | Q5BLZ6 | 76 | Neopullulanase | G. stearothermophilus | P38940 | 67 | CDase |
Domain composition similar to that for query ABA′C.
Domain composition: NABA′C.
Conserved-region PCR with CODEHOP primers.
For the primer construction, amino acid sequences of various amylolytic enzymes were retrieved from protein sequence databases and aligned using CLUSTAL_X version 1.8 (29). Sixteen different forward primers and seven different reverse primers, aimed to target sequences encoding conserved amino acid sequence regions A (or I) and B (or II) (18), were constructed. The primers were designed according to the CODEHOP strategy (26): they were degenerate at a 3′ core region of 11 or 12 nucleotides, across four codons encoding highly conserved amino acids in regions A and B. In contrast, they were nondegenerate at a 5′ region (consensus clamp region) of 18 to 25 nucleotides, with the most probable nucleotide predicted for each position. The degeneracy of the primer pools ranged from 16-fold to 32-fold, and the primers were 29 to 32 bp in length. The primers were used in a matrix of PCRs where every forward primer was used with every reverse primer. Primers resulting in isolation of the gene fragments described in this paper are listed in Table 1 and Fig. 1A and B. C-F corresponds to region A and C-R to region B. Other screening primers will be described elsewhere.
TABLE 1.
PCR primers used during the amplification of gene fragments and for the amplification of full-length genesa
| Primer group and designation | Nucleotide sequence |
|---|---|
| CODEHOP primers for PCR amplification | |
| C1-f | 5′-GGTGATGCTGGACGCGrtnttyaayca |
| C2-f | 5′-GCATGTTATGCTGGATGCAgtnttyaayca |
| C3-f | 5′-CATTAAAGTTTATGTGGATGCGgtnathaayca |
| C4-f | 5′-TCATGCCCGTCACCgarcaycaycc |
| C5-r | 5′-GTTGGCCACGTCCATCckccanccrtc |
| C6-r | 5′-GATCAACTTAATTAGCAACATCCATTckccanccrtc |
| C7-r | 5′-AATATGTTTCACCGCATCAAATckraanccrtc |
| C8-r | 5′-GCCCCGCTGGGTGtcrtgrttntc |
| Gene-specific primers for cloning | |
| Amy1-F | 5′-CGCCAATTGATGGAGCTTGTGTGGCAG |
| Amy1-R | 5′-CGGGAAGCTTTCACTGGTTGTGAAATCCGTC |
| Amy29-F | 5′-CGCGAATTCAAAATGACACCCGATTGGGTCAAAG |
| Amy29-R | 5′-GCCGGATCCCTACCCTTGAACCCGCCAAAC |
| Amy132-F | 5′-GGGAATTCATTAATGTTAAAAGAAGCGATTTACCA |
| Amy132-R | 5′-CGCAGATCTTATTCAGCGGACGTAGCTTGTA |
Nucleotide positions with introduced variations are shown in lowercase. Restriction sites in the gene-specific primers for cloning are underlined.
FIG. 1.
(A) Schematic drawing of the domain composition of the GH13 enzymes. The domain length (number of residues) is indicated below the respective domain, and the domains are color coded, with the N domain typical of CDases, neopullulanases, and maltogenic amylases in pink, the A domain in gray, the B domain in yellow, and the C domain in green. The positions of the four conserved regions in the α-amylase family are shown in brown, and the regions used in the initial amplifications are marked with arrows. res, residues. (B) Multiple sequence alignment of amino acid sequences of the cyclodextrin-degrading enzymes (Amy1 and Amy132), the neopullulanase (Amy29), and three structure-determined enzymes from the “neopullulanase-like” group, which are identified by their Swiss-Prot accession numbers in the alignment (P38940, neopullulanase from Geobacillus stearothermophilus; O69007, maltogenic amylase from a Thermus sp.; Q08751, neopullulanase [TVAII] from Thermoactinomyces vulgaris). The alignment was generated with ClustalW, using default parameters (28). The secondary structure elements of the respective enzymes are shown. The domains are indicated and coded (N domain [pink], A domain [gray], B domain [yellow], C domain [green]). The four conserved regions of the GH13 family (I to IV) and the additional conserved regions of cyclodextrin-degrading enzymes as well as the “specificity-determining” region are marked with boxes. Amino acids forming a cyclodextrin binding pocket in TVAII are shaded light gray. Amino acids proposed for Ca2+ binding are indicated in pink in the TVAII and Geobacillus nucleoprotein sequences, and the calcium-binding loop is underlined. Triangles mark the positions of catalytic residues.
The PCR was carried out with DyNAzyme DNA polymerase (Finnzymes, Espoo, Finland) with a PTC-0225 MJ Research thermal cycler. The reaction mixture was first denatured at 95°C for 5 min, followed by 30 cycles of denaturing at 95°C for 50 s, annealing at 52°C for 50 s, and extension at 72°C for 3 min and finally an extension for 7 min at 72°C to enhance formation of A overhangs. PCR products were separated on gels and purified using GFX spin columns (Amersham Biosciences, Piscataway, NJ). The gene fragments with sizes of 300 to 600 bp were selected and were cloned into conventional pUC-based sequencing vectors by the TA-cloning method (1). Eight to 12 clones from each band were sequenced with M13 forward and reverse primers on ABI 3700 DNA sequencers, using a BigDye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, CA). Each gene fragment was designated with the abbreviation Amy (denoting its origin in the α-amylase family) and a sequential number. This nomenclature is for clarity kept throughout the paper.
Amplification and analysis of selected full-length genes.
Following sequencing of the obtained target gene fragments, upstream and downstream flanking regions the selected fragments were amplified from the corresponding genomic DNA in a series of nested PCRs, using one gene-specific, 5′-biotin-labeled primer and one arbitrary primer (Arb1 or Arb2 [see Table 1]), targeting the unknown flanking sequence. The PCR product was purified with streptavidin-coded Dynabeads (Invitrogen) and further with QIAquick PCR purification spin columns (Qiagen, Hilden, Germany) prior to a second PCR with a nested gene-specific primer upstream of the previous one and a primer (Arb3 [see Table 3]) targeting the 5′ consensus sequence of the previously used arbitrary primer. The PCR product of the latter amplification was cloned and sequenced as described above, and the sequence information was used to make new gene-specific primers for the next nested PCR amplification until the complete genes were obtained. Similarity searches by BLAST were performed on the NCBI server (http://www.ncbi.nlm.nih.gov). The ClustalW tool on the EBI server (http://www.ebi.ac.uk/clustalw) was used to create multiple sequence alignments which were displayed using Gene doc 2.6.02. Pairwise alignments were made using LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html).
Cloning and expression in Escherichia coli.
The complete genes were PCR amplified (Expand high-fidelity PCR system; Roche Diagnostics, Mannheim, Germany), digested (as were the vectors) (Table 2) with appropriate restriction enzymes (New England Biolabs, Beverly, MA), treated with bacterial alkaline phosphatase, ligated to the vectors by using T4 DNA ligase (Invitrogen Life Technologies, Frederick, MD), and transformed into E. coli Nova Blue cells (Novagen) by using standard procedures (27). Inserts from positive clones were fully sequenced using the T7 forward and T7 reverse primers and a BigDye Terminator v2.0 DNA sequencing kit (PE Applied Biosystems) on an ABI 3100 sequencer (PE Applied Biosystems). Those containing correct gene inserts were transformed into the E. coli expression strains (Table 2). The genes were expressed at 37°C from shake flask cultivations in LB medium and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) or 1 mM IPTG at optical densities at 580 nm of 0.6 to 0.7 or produced in a 2.5-liter bioreactor by substrate-limited fed-batch cultivation according to Ramchuran et al. (23). After 2 to 3 h of induction, the cells were harvested by centrifugation at 8,000 × g at 4°C for 20 min.
TABLE 2.
Designations, expression vectors, and expression strains for the selected genes encoding GH13 enzymes
| Clone | Vector (REa sites for cloning) | Tag | E. coli expression strain |
|---|---|---|---|
| amy1 | pET32a (MfeX, HinDIII) | N-terminal His tag | Origami (DE3)pLys |
| amy29 | pBTac1 (EcoRI, BamHI) | None | BL21(DE3)-RIL codon plus |
| amy132 | pJOE3075 (Asex, BglA1) | None | BL21(DE3)-RIL codon plus |
RE, restriction endonuclease.
Purification of recombinant enzymes.
Cell extract was prepared from frozen cells suspended in an extraction buffer. Upon passing through a French pressure cell at 1.3 × 108 Pa, cells were disrupted and cell debris and unbroken cells were removed by centrifugation for 30 min at 48,000 × g at 4°C. Amy1 and Amy132 were heat treated for 30 min at 50°C. Amy29 was heat treated at 45°C and 55°C for 60 min each. Heat precipitation was followed by an additional centrifugation step (48,000 × g, 4°C, 30 min).
Amy1.
The supernatant obtained after heat treatment was applied to a Ni-nitrilotriacetic acid column, and protein was eluted with increasing imidazole concentrations. After ultrafiltration, the protein solution was applied to a Superdex 200 HiLoad 16/60 column equilibrated with 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM dithioerythritol (DTE). The His tag was removed by enterokinase digestion (2.5 mg protein, 50 μl 50 mM CaCl2, 172.5 μl 10 mM Tris-HCl, pH 8.0, and 2.5 μl enterokinase [1 mg/ml in 10 mM Tris-HCl, pH 8.0] for 24 h at 37°C).
Amy29.
The heat-treated supernatant diluted in 50 mM Tris-HCl, pH 9.0, was applied to a DEAE column and eluted by an increasing NaCl gradient, and a separation on an SP-HiTrap column followed. After ultrafiltration, the protein solution was applied to a Superdex 200 HiLoad 16/60 column. It was found that incubation of the enzyme at 30°C led to loss of the His tag, making enterokinase treatment unnecessary.
Amy132.
The supernatant obtained after heat treatment was applied to a Q-Sepharose HiLoad column and eluted by an increasing NaCl gradient. After ultrafiltration, the protein solution was applied to a Superdex 200 HiLoad 16/60 column, followed by separation on a UNO Q1 column (Bio-Rad) by use of an increasing NaCl gradient.
Protein methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 12% gels was performed according to Laemmli (15). Protein concentrations were determined by the method of Bradford (3), with bovine serum albumin fraction V as the standard. Native molecular mass was determined by gel filtration chromatography on a Superdex 200 column. A calibration of the column was performed with the high-molecular-weight and low-molecular-weight kits from GE Healthcare.
Determination of enzyme activities and kinetic parameters.
Characterization of enzymes included the determination of kinetic parameters, such as pH and temperature optima as well as thermal stability. Furthermore, the influences of cations (Ca2+, Co2+, Mn2+, Ni2+, and Mg2+, each at 0.1 to 10 mM) and product formation were analyzed. Kinetic parameters were calculated from Lineweaver-Burk plots. General carbohydrate hydrolysis was tested with maltooligosaccharides and polysaccharides at concentrations of 0.05 to 1% at the optimal pH and temperature. Product formation from certain substrates was studied via thin-layer chromatography (TLC). The assay contained 0.5% substrate, optimum buffer (containing DTE and 1 mM CaCl2 if necessary), and protein and was incubated for up to 60 min at the optimal temperature. Two to five microliters from the respective vial was applied to the matrix. Transglycosylation activity was determined, with combinations of 100 mM glucose, maltose, and maltotriose as substrates (either alone or in combinations of two or three sugars) and protein. The assays were incubated for 20 min at 55°C, and 5-μl aliquots were applied to a thin-layer plate.
CDase activity (cyclodextrin → maltooligodextrin) was determined as liberation of reducing sugars from 0.5% of each α-, β-, or γ-cyclodextrin (γ-CD) in 50 mM sodium acetate at the optimal pH (with 1 mM DTE and 10 mM CaCl2) and 1 to 10 μg protein with the dinitrosalicylic acid assay (see below). Neopullulanase activity (pullulan → panose) was determined as the liberation of reducing sugars from 0.5% pullulan in 50 mM sodium acetate at the optimal pH (with 1 mM DTE and 10 mM CaCl2) and 1 to 10 μg protein via the dinitrosalicylic assay. To discriminate panose and isopanose, a hydrolysis by glucoamylase was performed and aliquots were applied to a TLC plate (11).
The pH dependence levels of all enzymes were measured between pH 2.0 and 8.0 by using 100 mM citric acid (pH 2.0 to pH 4.5), sodium acetate (pH 4.5 to pH 5.5), piperazine (pH 5.5 to pH 7.5), and triethanolamine (pH 7.5 to pH 8.0). The temperature dependence levels of the enzyme activities were measured between 20°C and 90°C in 100 mM sodium acetate at the optimal pH. The stability of the purified enzymes against thermal inactivation was tested in sealed vials which were incubated at between 55°C and 100°C for up to 120 min at their optimal pHs. Potential stabilizing additives were tested [1 M NaCl, 1 M (NH4)2SO4, 1 to 5 mM CaCl2, or 1 M MgCl2]. After the enzymes were cooled on ice, the remaining enzyme activity was tested at the apparent temperature optimum. The cation specificity (0.1 to 10 mM each) was examined after dialysis against 20 to 50 mM EDTA at the optimal pH for 24 h, followed by dialysis against buffer without EDTA.
Analytical procedures.
Reducing sugars were determined using 3,5-dinitrosalicylic acid according to Miller (21). As a standard, 7 mM glucose was used. Starch was determined using a 1-ml iodine solution (0.01% iodine, 0.1% potassium iodide in 3.8 mM HCl) mixed with a 20-μl sample (up to 0.5% starch). Extinction was measured at 660 nm. α-CD was measured as a methylorange-α-CD inclusion complex (19). β-CD was measured as a phenolphthalein-β-CD inclusion complex (6). Mono- and oligosaccharides were separated on silica 60 plates (Merck), with butanol-ethanol-water (5:3:2) as the mobile phase. For visualization, the plates were dipped in 5% H2SO4 and baked for 15 min at 120°C. A mix of saccharides (1 to 13 glucose units; 0.1%) was used as the standard. To discriminate panose and isopanose, a hydrolysis by glucoamylase was performed and aliquots were applied to a TLC plate (11).
Nucleotide sequence accession numbers.
The sequences of the genes encoding the selected enzymes were deposited in GenBank at the NCBI under accession numbers EU427449 to EU427451.
RESULTS
Isolation of gene fragments from environmental DNA after in situ enrichment.
Two of the four highly conserved regions of the GH13 family (Fig. 1B) were used to design and construct primers for PCR amplification of gene fragments from DNA. Ten environmental samples were screened. After PCR amplification, positive samples were selected using agarose electrophoresis, and those revealing bands with sizes of 300 to 600 bp were selected, as they showed the expected fragment lengths between the conserved A (or I) and B (or II) regions. All the positive fragments were cloned, and 8 to 12 clones from each band were sequenced. To make a selection for full-length gene retrieval, the annotated activity of the closest match was recorded (Tables 3 and 4). For this work, fragments similar to “neopullulanases (EC 3.2.1.135) and CDases (EC 3.2.1.54),” a group of enzymes that are indistinguishable at the sequence level, were chosen.
TABLE 4.
Identities as revealed after pairwise alignments of the amino acid sequences by use of LALIGN
| Enzyme | Identity (%) for indicated enzyme
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall
|
Between catalytic ABA domains
|
Between C domains
|
Between N domains
|
|||||||||
| Amy132 | Amy29 | Amy1 | Amy132 | Amy29 | Amy1 | Amy132 | Amy29 | Amy1 | Amy132 | Amy29 | Amy1 | |
| Amy132 | 100 | 31.2 | 34.9 | 100 | 44.0 | 43.6 | 100 | 17.4 | 11.0 | 100 | 26.4 | |
| Amy29 | 100 | 31.2 | 100 | 40.9 | 100 | 22.5 | ||||||
| Amy1 | 100 | 100 | 100 | 100 | ||||||||
Full-length gene isolation and analysis.
Utilization of 5′-biotin-labeled, gene-specific primers in combination with an arbitrary primer targeting the unknown flanking sequence was shown to be a successful method for retrieving full-length genes. The gene walking was continued until a probable start codon with potential ribosome binding sites at the 5′ end was detected and the stop codon at the 3′ end of the gene sequence was passed.
The primary structures of all three enzymes were subjected to a BLAST search, upon which the most similar enzyme as well as the most similar three-dimensional (3D)-structure-determined enzyme was identified, and the domain organization and presence of conserved motifs were analyzed (Table 3). An alignment was used to identify domains and conserved regions (Fig. 1A and B).
Amy1 and -132, composed of domains N, A, B, and C, are all considered to be members of the “neopullulanase-like” group (Fig. 1A) (22). Amy29 encoded only domains A, B, and C but otherwise resembled the above-mentioned group at the sequence level, with deviation in the specificity-determining region between regions I and II (Fig. 1B).
The domains were also separately searched, showing that in the case of Amy1 the best match to the full-length enzyme was identical to the best match of the individual domains. For the others, the best matches varied but were all identical to candidates within the same specificity group. This was also the case for the 3D-structure-determined enzymes, where the best-matching enzymes varied between the domains, but the domains were all from “neopullulanase-like” enzymes. In the catalytic (ABA) domain, the four regions typical of GH13 were conserved in the enzymes, as were the strictly conserved catalytic residues (shown with TVAII numbering): D325 in β strand 4 in region II, E354 in β strand 5 in region III, D421 in β strand 7 in region IV, and the strictly conserved residue R323 (Fig. 1B). The highly conserved His residues at positions 244 and 420 were also found in the enzymes. An additional conserved region (located between regions I and II at the end of the B domain) is sometimes proposed as a fingerprint region for specificity. In this case, Amy1 and -132 show the signature MPKL, typical of the “neopullulanase-like” group, while Amy29 displayed the sequence LPKF at this position. Three additional conserved regions in the catalytic domain have been defined for cyclodextrin-degrading enzymes and were also found in all sequences, despite the different domain composition of Amy29 (Fig. 1B).
Biochemical characterization.
The obtained genes were expressed in E. coli, and the respective proteins were purified to apparent homogeneity and examined with respect to their substrate spectra, cation dependence, and thermophilic properties. Rate dependence on substrate concentrations of all proteins followed Michaelis-Menten kinetics. The protein encoded by amy29 was predominantly pullulan degrading, and the other enzymes were preliminarily annotated as CDase or neopullulanase, within the “neopullulanase-like” group, and should, based on their kinetic data, all be classified as CDases. However, significant side activities show the presence of also the preliminary annotated activity.
amy29 encodes a neopullulanase.
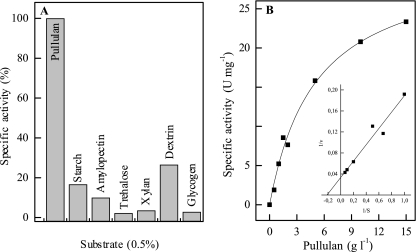
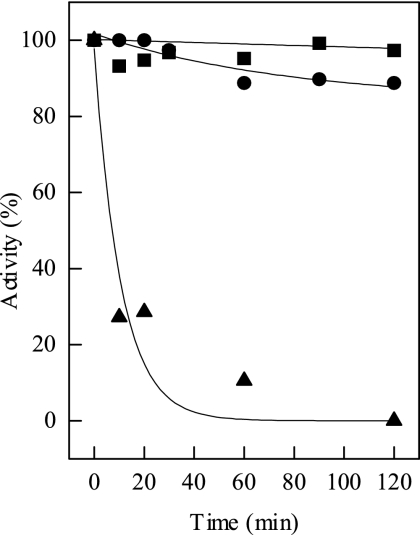
The enzyme encoded by amy29 catalyzed the conversion of polysaccharides into maltooligosaccharides (Fig. 2). The preferred substrate was pullulan. Sugars with fewer than six glucose units and cyclic sugars were not used. Amy29 hydrolyzed only α-1,4-glycosidic bonds. TLC experiments revealed that pullulan was degraded only to panose, defining the enzyme as a neopullulanase. Amy29 differs from the “neopullulanase-like” group of enzymes due to its lack of cyclodextrin degradation as well as the absence of the typical N domain. These data indicate that Amy29 should be classified as a neopullulanase (EC 3.2.1.135). Amy29 turned out to be highly thermostable (Fig. 3). The molecular, catalytic, and thermophilic properties of Amy29 are summarized in Table 5.
FIG. 2.
Activity of Amy29 at 65°C with different substrates (A) and with various concentrations of pullulan (B). A specific activity of 100% corresponds to 15 U mg−1. No activity was obtained for amylase or cyclodextrin. The insert shows the double-reciprocal Lineweaver-Burk plot.
FIG. 3.
Thermostability of Amy29. The enzyme was incubated at 60°C (▪), 80°C (•), and 90°C (▴). One hundred percent activity corresponds to 25 U mg−1 at 55°C.
TABLE 5.
Molecular, catalytic, and thermophilic properties of Amy29
| Parameter | Result for Amy29 |
|---|---|
| Apparent molecular mass (kDa) | |
| Native | 90 |
| Subunit | 45 |
| Oligomeric structure | α2 |
| Preferred substrate | Pullulan |
| Vmax (65°C) (U mg−1) | 25 |
| Km (65°C) (g liter−1) | 4.5 |
| Product | Panose |
| pH optimum | 5.3 |
| Temp optimum (°C) | 65 |
| Thermostability | |
| Half-life at 65°C (min) | No loss of activity over 120 min |
| Half-life at 80°C (min) | >480 |
| Cation dependence (0.01 to 50 mM) | |
| Mn2+ | Threefold |
| Ca2+ | No effect |
| Cu2+ (below 0.1 mM) | Complete inhibition |
| Effect of EDTA treatment | None |
amy1 and -132 encode CDases.
The enzymes encoded by amy1 and -132 were all found to hydrolyze cyclodextrins, defining the enzymes as CDases. The molecular, catalytic, and thermophilic properties of both enzymes are summarized in Table 6. Besides cyclodextrins, the enzymes hydrolyzed various polymeric sugars (Table 6). The efficiency of cyclodextrin hydrolysis was different for each enzyme. Whereas Amy1 converted starch and cyclic substrates at similar efficiencies, Amy1 showed a clear preference for cyclodextrins over starch.
TABLE 6.
Molecular, catalytic, and thermophilic properties of the proteins with CDase activity
| Parameter | Result for Amy1 | Result for Amy132 |
|---|---|---|
| Apparent molecular mass (kDa) | ||
| Native | 195a | 147 |
| Subunit | 85a | 68 |
| Calculated | 65.8 | 66 |
| Oligomeric structure | α2 | α2 |
| pH optimum | 6 | 5.5-6 |
| Temp optimum (°C) | 55 | 55 |
| Thermostability | ||
| Half-life at 55°C (min) | 30 | No loss of activity over 120 min |
| Half-life at 70°C (min) | 5 | 10 |
| Half-life at 55°C (min) at 1 mM DTE or 1M NaCl | 120 | No stabilization by DTE or NaCl |
| Substrate specificity (%)b | ||
| Starch | 69 | 14 |
| Amylose | 8 | 33 |
| Amylopectin | 82 | 0 |
| α-Cyclodextrin | 100 | 100 |
| β-Cyclodextrin | 69 | 80 |
| Pullulan | 0 | 15 |
| Glycogen | 1 | 0 |
| Maltotetraose | 3 | 1 |
| Maltoheptaose | 10 | 6 |
| Maltodextrin | 15 | 8 |
| Trehalose | 0 | 0 |
| Xylan (birch wood) | 0 | 0 |
| Kinetic constants (starch) | ||
| Vmax (U mg−1) | 47.5 | 150 |
| Km (g liter−1) | 6 | 63 |
| Vmax/Km | 7.9 | 2.3 |
| Kinetic constants (α-cyclodextrin) | ||
| Vmax (U mg−1) | 43 | 332 |
| Km (g liter−1) | 4 | 3.3 |
| Vmax/Km | 10.8 | 100 |
| Kinetic constants (β-cyclodextrin) | ||
| Vmax (U mg−1) | 34 | 200 |
| Km (g liter−1) | 8 | 2.4 |
| Vmax/Km | 4.3 | 83.3 |
With the N-terminal tag.
Determined with 0.5% (Amy132) and 1% (Amy1) substrate.
The data indicate that Amy1 should be designated a CDase with a side activity for starch as a substrate; TLC experiments revealed glucose and maltose as products. For Amy132, formation of maltose was shown as the product from starch and cyclodextrins. Glucose and maltotriose were formed additionally from substrates with odd-numbered glucose units. Additionally, pullulan was degraded. The only product was panose, defining the enzyme as a (maltogenic) CDase with neopullulanase activity. Both Amy29 and Amy132 showed transglycosylation activity; both, Amy1 and Amy132 formed maltose from glucose and maltotriose. In addition, Amy132 formed maltotetraose from maltose and maltotriose.
Both enzymes showed cation-dependent activity. Addition of 1 mM Ca2+ increased the activity of Amy1 by 30%. Treatment with 50 mM EDTA resulted in a total inhibition, which was restored by addition of divalent cations. Ca2+ was replaced effectively by Co2+, Zn2+, Mg2+, Ni2+, and Mn2+. Cu2+, however, inhibited activity at concentrations below 0.1 mM. Treatment of Amy132 with 20 mM EDTA led to an activity loss that could not be reverted by addition of divalent cations. Addition of Cu2+, Zn2+, and Fe2+ inhibited the enzyme at concentrations below 0.1 mM.
The CDases had apparent temperature optima at 55°C but showed moderate thermostabilities (Table 6).
DISCUSSION
The aim of this work was to isolate new genes encoding starch-modifying enzymes from environmental DNA by using samples collected from geothermal habitats in Iceland. The CODEHOP consensus primer strategy was applied (25), and this procedure resulted in the isolation of a number of putatively interesting gene fragments. Subsequently, new genes from GH13 were fully retrieved with extension primers, i.e., using an arbitrary primer coupled to a biotin-labeled, gene-specific primer based on the fragment-derived sequence. This combination of genetic methods led to the successful amplification of a set of genes. Three isolated candidates were characterized after heterologous expression in E. coli. Thus, we successfully isolated new members of the GH13 family exhibiting neopullulanase and CDase activities with interesting properties for industrial applications.
Neopullulanase.
amy29 encodes a novel neopullulanase based on both sequence similarity and activity profile. However, this enzyme is rather unusual since it is composed of domains A, B, and C and is lacking domain N, which is present in most other neopullulanases (10, 16). This moderately thermophilic protein had an amino acid sequence with the highest similarity to neopullulanases from Thermus thermophilus and a Synechocystis sp. There are, to our knowledge, no activity data from these organisms available for comparison. Amy29 showed high activity with pullulan, and the only product formed was panose, clearly defining the enzyme as a neopullulanase (2). The amino acid sequence from Amy29 showed in regions I to IV the typical motifs found for pullulan-hydrolyzing enzymes of the neopullulanase group. Most neopullulanases exhibit additional activity with cyclodextrins (2, 4). This was not the case for Amy29, which is in accordance with the absence of the N-terminal domain, typical for cyclodextrin-degrading enzymes (Fig. 1A).
The activity of the enzyme was dependent on addition of Mn2+. Ca2+ ions, which typically are the activating cations for the GH13 family, did not increase the activity of Amy29. Interestingly, Amy29 has a sequence insertion in the region of the catalytic domain, in the position corresponding to the calcium-binding loop of known neopullulanases (10, 11, 12). This may have led to a loss of Ca2+ binding, as only two of the seven residues (corresponding to G50 and N52) identified as ligands are conserved.
While exhibiting only a moderate temperature optimum (65°C), the enzyme showed a high thermostability. A half-life of 480 min was established at 85°C, i.e., 20°C above the optimum. This thermostability is surprising since even neopullulanases from hyperthermophilic archaea, such as Desulfurococcus mucosus, with a temperature optimum of 85°C, showed a clearly lower stability against thermal inactivation (half-life of 50 min) (5).
CDases.
Cyclodextrin-hydrolyzing enzymes belong to a subfamily of the GH13 family based on amino acid sequence. This subfamily comprises CDases, maltogenic amylases, and neopullulanases (22), which have unique but overlapping substrate and product spectra and are sometimes termed “the neopullulanase-like group.” According to their amino acid sequence conservation, the enzymes encoded by amy1 and amy132 might exhibit any of the three catalytic activities. However, all three activities were detected only in Amy132, while Amy1 showed maltogenic amylase and CDase activity (Table 6). To date, only a few cyclodextrin-hydrolyzing enzymes from bacteria and archaea have been described (16). Recently, we identified two novel CDases from thermophilic bacteria (30). Sequence comparison of the CDases revealed sequence-function relationships, especially for substrate binding. It has been proposed that the presence of a specific N domain and the occurrence of the cyclodextrin binding pocket are determinants of cyclodextrin hydrolysis (24, 30). This N domain, as well as all conserved amino acids that form the cyclodextrin binding pocket, was detected in the sequences of Amy1 and Amy132 (Fig. 1B). Additionally, the conserved regions of cyclodextrin-degrading enzymes of the GH13 family were shown (22). Thus, we classified both enzymes as CDases, based on these features and, most importantly, the activity on cyclodextrins. In contrast, GH13 enzymes which do not contain the N domain and the respective conserved regions do not show CDase activity. This is in accordance with the lack of CDase activity in Amy29, where the N domain is absent (see above).
Amy1 showed calcium-dependent activity and thermostability, while for Amy132, metal ion dependence was only indirectly determined as loss of activity upon EDTA treatment. Residues corresponding to the calcium-binding ligands identified in TVAII from Thermoactinomyces vulgaris (12) and neopullulanase from Geobacillus stearothermophilus (11) were conserved in Amy132, and it is likely that calcium is bound at this position (11). Despite the apparent calcium dependence of Amy1, it was not possible to predict the calcium-binding residues, due to low sequence conservation in the region corresponding to the calcium-binding loop.
Conclusion.
In summary, we were able to obtain enzymes of the GH13 family from environmental DNA of geothermal habitats that showed interesting properties with respect to biotechnological applications. The product spectrum of the neopullulanase (Amy29) may be especially valuable. Panose, the product of neopullulanases, is used as a sweetener (14). Finally, CDases and maltogenic amylases can be used for antistaling purposes in baked products and for the production of maltose and maltodextrin syrups, which is used in many food products. All enzymes were moderately thermophilic and exhibited distinct thermostability at temperatures from 50 to 80°C, which is an advantage in biotechnological process control. Amy29 is of particular interest, showing an extreme thermostability in combination with a broad substrate spectrum. Moreover, all enzymes were active without the addition of cations, which is likely a result of tight calcium binding. This is advantageous since the addition of calcium, which is usually necessary for starch conversion processes, can be avoided or minimized.
Thus, by using the CODEHOP strategy we were able to retrieve enzymes of the GH13 family directly from environmental DNA. This creates the possibility of high-throughput screening to broaden the spectrum of thermophilic enzymes suitable for industrial applications.
Acknowledgments
This work was supported by the European Union (contract number QLK3-CT-2000-01068) and the Swedish Research Council (VR).
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Ausubel, F. M., R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 2.Bertoldo, C., and G. Antranikian. 2002. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 6:151-160. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Doman-Pytka, M., and J. Bardowski. 2004. Pullulan degrading enzymes of bacterial origin. Crit. Rev. Microbiol. 30:107-121. [DOI] [PubMed] [Google Scholar]
- 5.Duffner, F., C. Bertoldo, J. T. Andersen, K. Wagner, and G. Antranikian. 2000. A new thermoactive pullulanase from Desulfurococcus mucosus: cloning, sequencing, purification, and characterization of the recombinant enzyme after expression in Bacillus subtilis. J. Bacteriol. 182:6331-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goel, A., and S. N. Nene. 1995. Modifications in the phenolphthalein method for spectrophotometric estimation of beta cyclodextrin. Starch 47:399-400. [Google Scholar]
- 7.Guzman-Maldonado, H., and O. Paredes-Lopez. 1995. Amylolytic enzymes and products derived from starch: a review. Crit. Rev. Food Sci. Nutr. 35:373-403. [DOI] [PubMed] [Google Scholar]
- 8.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hondoh, H., T. Kuriki, and Y. Matsuura. 2003. Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase. J. Mol. Biol. 326:177-188. [DOI] [PubMed] [Google Scholar]
- 11.Imanaka, T., and T. Kuriki. 1989. Pattern of action of Bacillus stearothermophilus neopullulanase on pullulan. J. Bacteriol. 171:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamitori, S., A. Abe, A. Ohtaki, A. Kaji, T. Tonozuka, and Y. Sakano. 2002. Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 Å resolution and alpha-amylase 2 (TVAII) at 2.3 Å resolution. J. Mol. Biol. 318:443-453. [DOI] [PubMed] [Google Scholar]
- 13.Kuriki, T., and T. Imanaka. 1999. The concept of the α-amylase family: structural similarity and common catalytic mechanism. J. Biosci. Bioeng. 87:557-565. [DOI] [PubMed] [Google Scholar]
- 14.Kuriki, T., M. Tsuda, and T. Imanaka. 1992. Continous production of panose by immobilized neopullulanase. J. Ferment. Bioeng. 73:198-202. [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lee, H. S., M. S. Kim, H. S. Cho, J. I. Kim, T. J. Kim, J. H. Choi, C. Park, H. S. Lee, B. H. Oh, and K. H. Park. 2002. Cyclomaltodextrinase, neopullulanase, and maltogenic amylase are nearly indistinguishable from each other. J. Biol. Chem. 277:21891-21897. [DOI] [PubMed] [Google Scholar]
- 17.Leuschner, C., and G. Antranikian. 1995. Heat-stable enzymes from extremely thermophilic microorganisms. World J. Microbiol. Biotechnol. 11:95-114. [DOI] [PubMed] [Google Scholar]
- 18.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 19.Mäkelä, M. J., and T. K. Korpela. 1988. Determination of the catalytic activity of cyclomaltodextrin glucanotransferase by maltotriose-methylorange assay. J. Biochem. Biophys. Methods 15:307-318. [DOI] [PubMed] [Google Scholar]
- 20.Marteinsson, V. T., S. Hauksdottir, C. F. Hobel, H. Kristmannsdottir, G. O. Hreggvidsson, and J. K. Kristjansson. 2001. Phylogenetic diversity analysis of subterranean hot springs in Iceland. Appl. Environ. Microbiol. 67:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 22.Park, K. H., T. J. Kim, T. K. Cheong, J. W. Kim, B. H. Oh, and B. Svensson. 2000. Structure, specificity and function of cyclomaltodextrinase, a multispecific enzyme of the alpha-amylase family. Biochim. Biophys. Acta 1478:165-185. [DOI] [PubMed] [Google Scholar]
- 23.Ramchuran, S. O., E. Nordberg Karlson, S. Velut, L. De Maré, P. Hagander, and O. Holst. 2002. Production of heterologous thermostable glycoside hydrolases and the presence of host-cell proteases in substrate limited fed-batch cultures of Escherichia coli BL21(DE3). Appl. Microbiol. Biotechnol. 60:408-416. [DOI] [PubMed] [Google Scholar]
- 24.Rashid, N., J. Cornista, S. Ezaki, T. Fukui, H. Atomi, and T. Imanaka. 2002. Characterization of an archaeal cyclodextrin glucanotransferase with a novel C-terminal domain. J. Bacteriol. 184:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose, T. M., J. G. Henikoff, and S. Henikoff. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, S., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a labratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner, P., A. Labes, O. H. Fridjonsson, G. O. Hreggvidson, P. Schonheit, J. K. Kristjansson, O. Holst, and E. Nordberg Karlsson. 2005. Two novel cyclodextrin-degrading enzymes isolated from thermophilic bacteria have similar domain structures but differ in oligomeric state and activity profile. J. Biosci. Bioeng. 100:380-390. [DOI] [PubMed] [Google Scholar]
- 31.van der Maarel, M. J., B. van der Veen, J. C. Uitdehaag, H. Leemhuis, and L. Dijkhuizen. 2002. Properties and applications of starch-converting enzymes of the alpha-amylase family. J. Biotechnol. 94:137-155. [DOI] [PubMed] [Google Scholar]