Abstract
We developed a novel multiplex PCR assay for rapid identification and discrimination of the USA300 and USA400 strains and concomitant detection of Panton-Valentine leukocidin genes, with simultaneous discrimination of methicillin-resistant Staphylococcus aureus strains from methicillin-susceptible S. aureus strains, S. aureus strains from coagulase-negative staphylococci, and staphylococci from other bacteria.
USA300 and USA400 are the predominant community- associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains circulating in North America (8, 12), being implicated in outbreaks of infections associated with significant morbidity and mortality (1, 4, 6). These strains belong to multilocus sequence types (MLST) ST8 and ST1 and staphylococcal protein A (spa) types t008 and t128, respectively, and both strains carry Panton-Valentine leukocidin (PVL) genes and the staphylococcal cassette chromosome mec (SCCmec) type IVa element. However, molecular characterization of these strains can be time consuming and technically laborious. We have designed a multiplex PCR (M-PCR) assay capable of accurately distinguishing USA300 from USA400 strains while simultaneously detecting PVL genes and discriminating MRSA strains from methicillin-susceptible S. aureus (MSSA) strains, S. aureus strains from coagulase-negative staphylococci (CoNS), and staphylococci from other bacteria.
(This work was presented in part at the 107th General Meeting of American Society for Microbiology, 21-25 May, 2007, Toronto, Canada [abstract C-238].)
Sequence alignment and primer design.
Primers for 16S rRNA (Stapy756-F and Staph750-R) (15), thermostable nuclease (nuc) (Nuc-1 and Nuc-2) (15), mecA (mecA147-F and mecA147-R) (14), and PVL genes lukS-PV/lukF-PV (Luk-PV-1 and Luk-PV-2) (7) were as previously described. New sets of primers for strains USA300 and USA400 and for prophage φSa2usa/φSa2mw were designed based on comprehensive analysis and alignment of individual Staphylococcus sp. genomes currently available in the GenBank database. Gene targets for each primer pair are as follows: USA300 strain primers arcA-F (5′-GCAGCAGAATCTATTACTGAGCC-3′) and arcA-R (5′-TGCTAACTTTTCTATTGCTTGAGC-3′) target the arcA gene on the arginine catabolic mobile element (ACME); USA400 strain primers MW756-F (5′-TGGTTAGCTATGAATGTAGTTGC-3′) and MW756-R (5′-GTCCATCCTCTGTAAATTTTGC-3′) target the gene locus MW0756 on νSa3 of strain MW2; and φSa2mw/φSa2usa prophage primers phi-int-F4 (5′-CAAATTTTGAAAACTTTACGC-3′) and phi-int-R4 (5′-TCCAGGATTAAAAGAAGCG-3′) target the MW1409 gene locus of the USA400 MW2 strain.
Development of an M-PCR assay for typing MRSA isolates and distinguishing USA300 and USA400 strains.
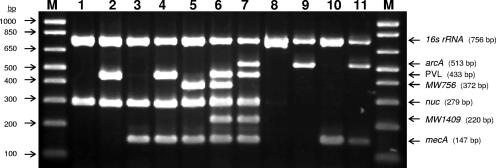
The assay specifically involved targeting the Staphylococcus genus-specific 16S rRNA gene sequence (serving to distinguish Staphylococcus from other bacteria and acting as an internal PCR control), the S. aureus-specific nuc gene, the methicillin resistance determinant mecA, the PVL genes, the phage marker MW1409, the USA400 genomic island gene locus (MW0756), and the USA300 ACME cassette gene (arcA). Amplification in a single M-PCR produced distinct bands corresponding to molecular sizes of 147, 220, 279, 372, 433, 513, and 756 bp for mecA, MW1409, nuc, MW0756, PVL, arcA, and 16S rRNA, respectively (Fig. 1).
FIG. 1.
Novel multiplex PCR assay identifying USA300 and USA400 community-associated MRSA strains, detecting PVL and mecA genes, and simultaneously discriminating S. aureus from CoNS. The optimized M-PCR was performed as follows: 4.65 μl of template DNA was prepared as previously described (15) in a 25-μl final reaction volume containing 0.30, 0.3, 0.4, 0.32, 0.16, 1.17, and 0.16 μM for each of the 16S rRNA, arcA, lukS-PV/lukF-PV, MW0756, nuc, MW1409, and mecA primers, respectively, with the thermocycling conditions set at 94°C for 4 min followed by 10 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 45 s and another 25 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 45 s. The PCR amplicons were visualized using a UV light box after electrophoresis on a 2% agarose gel containing 0.5 μg/ml ethidium bromide. Lane 1, strain ATCC 29213 (PVL− MSSA); lane 2, strain ATCC 49775 (PVL+ MSSA); lane 3, Canadian epidemic MRSA control strain CMRSA2 (PVL− non-USA300 and non-USA400 MRSA); lane 4, strain C1538 (PVL+ non-USA300 and non-USA400 MRSA); lane 5, strain C2901 (PVL− USA400); lane 6, Canadian epidemic CA-MRSA USA400 control strain CMRSA7 (PVL+ USA400); lane 7, Canadian epidemic CA-MRSA USA300 control strain CMRSA10 (PVL+ USA300); lane 8, strain CNS99-PF5 (PVL− and arcA− MS-CoNS); lane 9, strain CNS99-PF7 (PVL− but arcA+ MS-CoNS); lane 10, strain CNS99-PF6 (PVL− and arcA− MR-CoNS); lane 11, strain CNS99-PF8 (PVL− but arcA+ MR-CoNS); lanes M, 1 kb Plus DNA ladder (Invitrogen). Refer to Table 1 for details of each strain.
Validation of M-PCR assay.
The new multiplex PCR assay was validated using 42 representative MRSA/MSSA control strains and 6 ACME-positive (ACME+) or ACME-negative (ACME−) CoNS control strains that had undergone detailed phenotypic and molecular characterization, including analysis of carriage of PVL (7) and other genes, pulsed-field gel electrophoresis (PFGE) fingerprinting (9), SCCmec typing (14), spa typing (5, 10), and MLST (2) and eBURST (3, 11) analyses (Table 1). The MRSA/MSSA strains differed in their genotypic characteristics and represented 10 major clonal complex groups found in the worldwide MLST collection. The assay was capable of accurately and reproducibly discriminating USA300 from USA400 and other MRSA and MSSA strains while simultaneously detecting PVL genes and φSa2mw/φSa2usa phages, with a resultant 100% concordance to genotypic features in all these control strains (Table 1). There were seven PVL+ MRSA and nine PVL+ MSSA strains, belonging to non-USA300/non-USA400 strains with well-diversified genomic backgrounds, and all were positive for the PVL genes but negative for the φSa2mw/φSa2usa phage marker, suggesting that PVL genes in these strains may be carried by phages/plasmids other than φSa2mw/φSa2usa. More interestingly, there were two strains (with non-USA300/non-USA400 PFGE profiles), one a PVL− MRSA strain (CMRSA5) and one a PVL+ MSSA strain (SAF516), that were positive for the phage marker. Since this primer pair is a marker for the PVL-bearing phage φSa2mw/φSa2usa in USA400/USA300, this result suggests that variations of these particular phages can also be present in S. aureus strains other than USA300 and USA400 with or without the PVL genes. Further studies are required to better understand this observation.
TABLE 1.
Molecular and genotypic features of Staphylococcus aureus and CoNS control strainsa
| Strain type and name | PFGE type (pattern)b | spa type | Molecular type | Genotype (presence of indicated gene or marker)c
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Staphylococcus sp. specific (16S rRNA) | S. aureus specific (nuc) | Methicillin resistant (mecA) | PVL specific (PVL) | Phage (MW1409) | USA400 (MW0756) | USA300 (arcA) | ||||
| PVL+ USA300 | ||||||||||
| CMRSA10 | USA300-0114 (A) | t008 | ST8-MRSA-IVa | + | + | + | + | − | + | |
| PMRSA-13 | USA300-0114 (A) | t008 | STnew-MRSA-Ivad | + | + | + | + | + | − | + |
| PMRSA-16 | USA300-0114 (A) | t008 | ST8-MRSA-IVa | + | + | + | + | + | − | + |
| PMRSA-46 | USA300-0114 (A) | t008 | ST8-MRSA-IVa | + | + | + | + | + | − | + |
| PMRSA-12 | USA300 (B) | t008 | ST8-MRSA-IVa | + | + | + | + | + | − | + |
| C21744 | USA300 (C) | t008 | ST8-MRSA-IVa | + | + | + | + | + | − | + |
| PVL− USA400 | ||||||||||
| C2901 | USA400 (B) | t128 | ST1-MRSA-IVa | + | + | + | − | − | + | − |
| C2140 | USA400 (A) | t128 | ST1-MRSA-IVa | + | + | + | − | − | + | − |
| PVL+ USA400 | ||||||||||
| CMRSA7 | USA400 (A) | t128 | ST1-MRSA-IVa | + | + | + | + | + | + | − |
| PMRSA-18 | USA400 (A) | t128 | ST1-MRSA-IVa | + | + | + | + | + | + | − |
| PMRSA-50 | USA400 (A) | t128 | ST1-MRSA-IVa | + | + | + | + | + | + | − |
| C10687 | USA400 (A) | t128 | ST1-MRSA-IVa | + | + | + | + | + | + | − |
| C6413 | USA400 (C) | t128 | ST1-MRSA-IVa | + | + | + | + | + | + | − |
| PVL− MRSA | ||||||||||
| CMRSA1 | USA600 | t004 | ST45-MRSA-II | + | + | + | − | − | − | − |
| CMRSA2 | Like USA100/800 | t002 | ST5-MRSA-II | + | + | + | − | − | − | − |
| C4000 | Like USA100/800 | t004 | ST225-MRSA-II | + | + | + | − | − | − | − |
| CMRSA3 | Like EMRSA1/4/11 | t037 | ST241-MRSA-III | + | + | + | − | − | − | − |
| CMRSA6 | Like EMRSA1/4/11 | t037 | ST239-MRSA-III | + | + | + | − | − | − | − |
| C1777 | Like EMRSA1/4/11 | t037 | ST239-MRSA-III | + | + | + | − | − | − | − |
| CMRSA4 | USA200/EMRSA16 | t018 | ST36-MRSA-II | + | + | + | − | − | − | − |
| C23374 | USA200/EMRSA 16 | t018 | ST36-MRSA-II | + | + | + | − | − | − | − |
| CMRSA5 | USA500 | t064 | ST8-MRSA-IVd | + | + | + | − | + | − | − |
| CMRSA8 | EMRSA 15 | t022 | ST22-MRSA-IV | + | + | + | − | − | − | − |
| CMRSA9 | t008 | ST8-MRSA | + | + | + | − | − | − | − | |
| C74 | t1154 | ST5-MRSA-IVb | + | + | + | − | − | − | − | |
| PVL+ MRSA | ||||||||||
| H435 | t311 | ST5-MRSA-II | + | + | + | + | − | − | − | |
| PMRSA-34 | t437 | ST59-MRSA-III | + | + | + | + | − | − | − | |
| MR37 | Like USA400 | t175 | ST1-MRSA-IVa | + | + | + | + | − | − | − |
| MR138 | t044 | ST80-MRSA-IVa | + | + | + | + | − | − | − | |
| H434 | Like USA1100 | t019 | ST30-MRSA IVc | + | + | + | + | − | − | − |
| PMRSA-29 | USA1100 | t019 | ST30-MRSA-IVc | + | + | + | + | − | − | − |
| C1538 | USA1100 | t019 | ST30-MRSA-IVc | + | + | + | + | − | − | − |
| PVL+ MSSA | ||||||||||
| MS02-W10 | USA800 | t015 | ST5-MSSA | + | + | − | + | − | − | − |
| SA5 | Unnamede | ST121-MSSA | + | + | − | + | − | − | − | |
| SA112 | t645 | ST121-MSSA | + | + | − | + | − | − | − | |
| SA125 | t436 | ST25-MSSA | + | + | − | + | − | − | − | |
| SA134 | t005 | ST22-MSSA | + | + | − | + | − | − | − | |
| SA28 | t437 | ST59-MSSA | + | + | − | + | − | − | − | |
| MS03-B1 | t437 | ST59-MSSA | + | + | − | + | − | − | − | |
| SA3 | Unnamede | ST30-MSSA | + | + | − | + | − | − | − | |
| H49 | t021 | ST30-MSSA | + | + | − | + | − | − | − | |
| SAF516 | t483 | ST30-MSSA | + | + | − | + | + | − | − | |
| PVL− MS-CoNS | ||||||||||
| S. epidermidis (ATCC12228) | ACME+ | + | − | − | − | − | − | + | ||
| PVL− MS-CoNS | ||||||||||
| S. epidermidis (CNS99-PF5) | ACME− | + | − | − | − | − | − | − | ||
| PVL− MS-CoNS | ||||||||||
| S. epidermidis (CNS99-PF7) | ACME+ | + | − | − | − | − | − | + | ||
| PVL− MR-CoNS | ||||||||||
| S. epidermidis (GISE 12333) | ACME+ | + | − | + | − | − | − | + | ||
| PVL− MR-CoNS | ||||||||||
| S. epidermidis (CNS99-PF6) | ACME− | + | − | + | − | − | − | − | ||
| PVL− MR-CoNS | ||||||||||
| S. epidermidis (CNS99-PF8) | ACME+ | + | − | + | − | − | − | + | ||
PFGE, PVL gene typing, SCCmec typing, spa typing, and MLST were used for strain characterization. The identification of MRSA isolates matching the USA300 and USA400 CA-MRSA strains was based on the similarity of PFGE patterns to those of the USA300 and USA400 control strains and the presence of PVL, SCCmec type IVa, spa type t008, and MLST type ST8 (for USA300) and of PVL, SCCmec type IVa, spa type t128, and MLST type ST1 (for USA400).
PFGE patterns A (indistinguishable from the USA300 control strain [CMRSA10]), B (an additional band around 150 kb in size), and C (the band at around 250 bp shifting to around 310 kb) for strain USA300 and patterns A, B, and C for strain SA400 (see reference 13 for details of the patterns) are indicated in brackets.
+, present; −, absent.
New ST (MLST) profile: PMRSA-13 (3-3-1-4-4-4-3).
The spa type profiles are unnamed for SA5 (I2H2M) and SA3 (I2H2M).
Applicability and accuracy of M-PCR.
To address the applicability and accuracy of the M-PCR assay, we further applied our M-PCR assay to test a total of 1,133 local clinical MRSA isolates randomly selected from our Calgary frozen clinical isolate stock collection for the 18-year period from 1989 to 2006. We were able to accurately identify and classify all strains with available PFGE data, including 54 PVL+ USA300, 17 PVL+ USA400, 35 PVL− USA400, 40 PVL− CMRSA2, and 34 PVL− non-USA300/non-USA400/non-CMRSA2 MRSA strains. We were also able to clearly classify the remaining randomly chosen strains, including 514 S. aureus and 439 CoNS isolates. Once again, we noted that 10 (1.9%) of the isolates, including 5 PVL− MRSA and 5 PVL− MSSA isolates, were positive for the phage gene and yet did not belong to either USA300 or USA400. There was also one MRSA isolate that was positive for the PVL genes but negative for the φSa2mw/φSa2usa gene. Among 439 CoNS isolates tested, there was a 100% concordance with phenotypic susceptibility to methicillin, with 214 of the isolates being methicillin-susceptible CoNS (MS-CoNS) and 225 being methicillin-resistant CoNS (MR-CoNS). Of the MS-CoNS isolates, 75 (35.0%) of them were positive for arcA, while 93 (41.3%) of the MR-CoNS isolates were arcA positive. None of the CoNS isolates tested carried PVL, φSa2mw/usa phage, or USA400 marker MW0756 gene loci.
Our assay is capable not only of (i) accurately identifying and differentiating USA300 and USA400 strains but also of simultaneously detecting (ii) mecA to discriminate MRSA from MSSA and (iii) 16S rRNA and nuc to discriminate S. aureus from CoNS and of detecting (iv) PVL genes and a φSa2mw/usa-specific gene to determine whether the isolates/strains carry PVL genes and whether the PVL genes are carried by the φSa2mw/φSa2usa phage or other prophages in the population. Our M-PCR assay may also facilitate monitoring of the dynamic exchanges or evolution of genes among strains of MRSA, MSSA, and CoNS. This assay is based on the concept of certain strains carrying unique/specific genes/phages. However, since movement of these genes, phages, and other genetic elements is dynamic, the definition of a strain, as determined by the use of our assay for detection of gene targets uniquely present in individual CA-MRSA strains, may require reconsideration over time.
Acknowledgments
This work was partially supported by a grant from the Banting Research Foundation to K.Z. and by operating funds from the Centre for Antimicrobial Resistance, Calgary Health Region, Canada.
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.CDC. 2003. Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections—Los Angeles County, California, 2002-2003. MMWR Morb. Mortal. Wkly. Rep. 5288. [PubMed] [Google Scholar]
- 2.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis, J. S., M. C. Doherty, U. Lopatin, C. P. Johnston, G. Sinha, T. Ross, M. Cai, N. N. Hansel, T. Perl, J. R. Ticehurst, K. Carroll, D. L. Thomas, E. Nuermberger, and J. G. Bartlett. 2005. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin. Infect. Dis. 40100-107. [DOI] [PubMed] [Google Scholar]
- 5.Harmsen, D., H. Claus, W. Witte, J. Rothganger, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 415442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazakova, S. V., J. C. Hageman, M. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. K. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352468-475. [DOI] [PubMed] [Google Scholar]
- 7.McClure, J. A., J. M. Conly, V. Lau, S. Elsayed, T. Louie, W. Hutchins, and K. Zhang. 2006. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococci. J. Clin. Microbiol. 441141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulvey, M. R., L. Chui, J. Ismail, L. Louie, C. Murphy, N. Chang, and M. Alfa. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 393481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 373556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spratt, B. G., W. P. Hanage, B. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241129-134. [DOI] [PubMed] [Google Scholar]
- 12.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, K., J. McClure, S. Elsayed, J. Tan, and J. Conly. 2008. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J. Infect. Dis. 15195-204. [DOI] [PubMed] [Google Scholar]
- 14.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 435026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, K., J. Sparling, B. L. Chow, S. Elsayed, Z. Hussain, D. L. Church, D. B. Gregson, T. Louie, and J. M. Conly. 2004. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 424947-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]