Abstract
All organisms consist of cells that are enclosed by a cell membrane containing bipolar lipids and proteins. Glycerophospholipids are important not only as structural and functional components of cellular membrane but also as precursors of various lipid mediators. Polyunsaturated fatty acids comprising arachidonic acid or eicosapentaenoic acid are located at sn-2 position, but not at sn-1 position of glycerophospholipids in an asymmetrical manner. In addition to the asymmetry, the membrane diversity is important for membrane fluidity and curvature. To explain the asymmetrical distribution of fatty acids, the rapid turnover of sn-2 position was proposed in 1958 by Lands [Lands WE (1958) Metabolism of glycerolipides: A comparison of lecithin and triglyceride synthesis. J Biol Chem 231:883–888]. However, the molecular mechanisms and biological significance of the asymmetry remained unknown. Here, we describe a putative enzyme superfamily consisting mainly of three gene families, which catalyzes the transfer of acyl-CoAs to lysophospholipids to produce different classes of phospholipids. Among them, we characterized three important enzymes with different substrate specificities and tissue distributions; one, termed lysophosphatidylcholine acyltransferase-3 (a mammalian homologue of Drosophila nessy critical for embryogenesis), prefers arachidonoyl-CoA, and the other two enzymes incorporate oleoyl-CoAs to lysophosphatidylethanolamine and lysophosphatidylserine. Thus, we propose that the membrane diversity is produced by the concerted and overlapped reactions with multiple enzymes that recognize both the polar head group of glycerophospholipids and various acyl-CoAs. Our findings constitute a critical milestone for our understanding about how membrane diversity and asymmetry are established and their biological significance.
Keywords: glycerophospholipids, Lands' cycle, membrane remodeling, phospholipase A2, acyl-CoA
Glycerophospholipids are important structural and functional components of biological membranes and important constituents of serum lipoproteins and pulmonary surfactant (1). Additionally, phospholipids play important roles as precursors of lipid mediator such as platelet-activating factor (PAF) and eicosanoids (2). Tissues maintain distinct content and composition of various phospholipids such as phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), cardiolipin (CL), phosphatidylinositol (PI), and phosphatidylserine (PS) (3–5). Fatty acids are first activated to acyl-CoAs as described by Kornberg and Pricer (6). By the use of acyl-CoAs, phospholipids are formed from diacylglycerol by a de novo pathway, originally described by Kennedy in 1956 (7). However, the acyl groups in glycerophospholipids show high diversity and are distributed in an asymmetrical manner (3, 5). Saturated and monounsaturated fatty acids are usually esterified at the sn-1 position, whereas polyunsaturated acyl groups are located at the sn-2 position. These diversity and asymmetry are not fully explained by the Kennedy pathway. The rapid turnover of the sn-2 acyl moiety of glycerophospholipids was described by Lands as a remodeling pathway (Lands' cycle) (8) and is attributed to the concerted activation of phospholipase A2s (PLA2s) and lysophospholipid acyltransferases (LPLATs) (9, 10). Although these metabolic processes are carried out in a variety of tissues, information on the enzyme molecules involved in the phospholipid remodeling is limited.
We cloned lyso-PA acyltransferase α (LPAATα) and found that the enzyme used LPA as an acyl acceptor (11). Whereas the recombinant LPAATα was shown to use various fatty acyl-CoAs as acyl donors, no lysophospholipid other than LPA was found to serve as an acceptor. Several LPLATs, including LPAATα and β, lyso-PG acyltransferase (LPGAT), and lyso-CL acyltransferase (ALCAT), have been cloned in the last decade, and putative LPAATs (γ, δ, ε, ζ, and η) and tafazzin have been reported (12–19). Recently, our group identified two lyso-PC acyltansferases (LPCATs) functioning in the remodeling pathway, designated LPCAT1 and lyso-PAF acetyltransferase (LysoPAFAT)/LPCAT2, which are mainly expressed in lung and inflammatory cells, respectively (20, 21). These enzymes, which possess four well conserved domains, designated motifs 1–4 (LPAAT motifs) (22) and an endoplasmic reticulum (ER) retention sequence (23), were assigned to the LPAAT family (see Fig. 1A). Before the work presented here, there were no reports about mammalian lyso-PE acyltransferase (LPEAT), lyso-PS acyltransferase (LPSAT), or lyso-PI acyltransferase (LPIAT). Moreover, we believe that, in addition to LPCAT1 and LysoPAFAT/LPCAT2, a different class of LPCATs may exist for membrane biogenesis, because PC biosynthesis in the Lands' cycle occurs in a variety of tissues (1).
Fig. 1.
Phylogenetic tree of mouse LPLAT family and summary of characterized LPLATs. (A) A phylogenetic tree was drawn by using ClustalW (www.ebi.ac.uk/clustalw). Values show branch lengths that represent the evolutional distance between each pair of sequences. Putative LPLAT genes are indicated by asterisks. Sequence data of mouse acyltransferases are available in the DDBJ/European Molecular Biology Laboratory/GenBank databases. The accession numbers are shown in SI Table 4. (B) Functionally identified LPLATs were summarized. Darker colors indicate higher enzymatic activity for acyl-CoAs. The enzymes LPCAT3, LPCAT4, and LPEAT1 are marked as being shown in this study. Additional LPLATs that may exist are indicated with question marks. GPAT, glycerol-3-phosphate acyltransferase; MGAT, monoacylglycerol O-acyltransferase; ACAT, sterol O-acyltransferase; LRC4, leukocyte receptor cluster member 4.
In this study, we identified a family of LPLATs. The members of the family had been reported as uncharacterized proteins, termed membrane-bound O-acyltransferases (MBOATs) (24) that lack LPAAT motifs. Among them were three mammalian LPLATs, MBOAT1, -2, and -5. We found that mouse MBOAT1, -2, and -5 exhibited various LPLAT activities: (i) MBOAT1, LPEAT, and LPSAT; (ii) MBOAT2, LPCAT, and LPEAT; and (iii) MBOAT5, LPCAT, LPEAT, and LPSAT. To avoid confusion, we refer to mouse MBOAT1, -2, and -5, as mouse LPEAT1 (mLPEAT1), mLPCAT4, and mLPCAT3, respectively, based on their characteristic properties.
This is a documentation of cDNAs for mammalian LPEAT and LPSAT, and a finding of two additional LPCATs. Moreover, we functionally characterized MBOATs as a LPLAT family. MBOATs are critically important remodeling enzymes for membrane phospholipids biosynthesis in the Lands' cycle.
Results
Cloning of Mouse LPCAT3, LPCAT4, and LPEAT1.
To identify novel LPLATs, we focused on uncharacterized proteins in the MBOAT family. A phylogenetic tree was drawn by pairwise comparisons of amino acid sequences of LPAAT, diacylglycerol AT2 (DGAT2), and MBOAT family members by using ClustalW, European Bioinformatics Institute (http://www.ebi.ac.uk/) (25) (Fig. 1A). The cluster of MBOAT family proteins comprising LPLATs was different from that of the LPAAT or DGAT2 family. Mouse MBOAT5, -2, and -1 were designated mLPCAT3, mLPCAT4, and mLPEAT1, respectively, based on their activities characterized in this study. The functionally identified LPLATs are summarized in Fig. 1B. The higher enzymatic activities for acyl-CoAs are shown in darker color. Additional LPLATs may exist, especially LPIATs. The putative ORFs of mLPEAT1, mLPCAT4, and mLPCAT3 encoded proteins of 492 aa (56.1 kDa), 519 aa (58.9 kDa), and 487 aa (56.1 kDa), respectively [supporting information (SI) Fig. 6]. Boxes indicate that mLPCAT3, mLPCAT4, and mLPEAT1 were predicted by HMMTOP (26) to possess 10, 4, and 9 putative transmembrane domains, respectively. Although the enzyme sequences did not reveal LPAAT motifs as described above, LPCAT3 and LPEAT1 possess the C-terminal sequence motif KKXX (23), suggesting that the proteins are localized to the ER in the same way as LPCAT1 and LysoPAFAT/LPCAT2 (20, 21).
Subcellular Localization of mLPCAT3, mLPCAT4, and mLPEAT1.
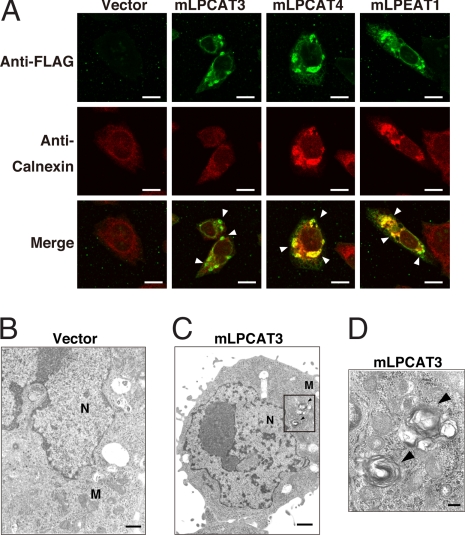
To facilitate Western blot and immunocytochemical analyses of mLPCAT3, mLPCAT4, and mLPEAT1, plasmids expressing N-terminal FLAG-tagged recombinant enzymes were constructed. Microsomal proteins (100,000 × g; 1-h pellet) of CHO-K1 cells transfected with empty vector or with each FLAG-tagged enzyme cDNA were subjected to Western blot analysis, which revealed apparent molecular masses of 40, 42, and 40 kDa for mLPCAT3, mLPCAT4, and mLPEAT1, respectively (SI Fig. 7A). These results are inconsistent with the molecular mass calculated from deduced amino acid sequences. The discrepancies could be due to the numerous predicted membrane spanning structures in these proteins (SI Fig. 6) that may affect their mobility during SDS/PAGE. The subcellular localization of these enzymes was examined by confocal microscopy. mLPCAT3, mLPCAT4, and mLPEAT1 colocalized with calnexin, an ER maker protein, suggesting that these enzymes are present mainly in ER (Fig. 2). Neither Golgi marker protein (GM130) nor mitochondria marker protein (cytochrome c oxidase) merged with the enzyme (SI Fig. 7 B and C). The distribution patterns were also confirmed by cell fractionation and Western blotting (SI Fig. 7D). Surprisingly, the staining of unidentified organelles colocalized with each enzyme and calnexin (Fig. 2A, arrowheads). To investigate these organelles, we analyzed CHO-K1 cells transfected with mLPCAT3 by transmission electron microscopy. Although ER structure in CHO-mLPCAT3 and CHO-Vector cells was the same, atypical multilayer membrane components (karmellae-like structures) (27) were observed. The number and size of these structures were greater in CHO-mLPCAT3 cells than in CHO-Vector cells (Fig. 2 B–D). The nature and origin of these structures remain to be clarified.
Fig. 2.
Subcellular localization of FLAG-tagged LPLATs and transmission electron microscopy (TEM) of mLPCAT3 in CHO-K1 cells. (A) After transfection for 48 h, CHO-K1 cells were subjected to immunocytochemical analysis. ER and FLAG-tagged enzymes were visualized by using an anti-calnexin antibody (red) and an anti-FLAG antibody (green), respectively. The subcellular localizations of all enzymes were similar to the ER marker (merge staining patterns, yellow). (Scale bars, 20 μm.) Two independent experiments were performed with similar results. The arrowheads indicated karmellae-like membrane structures. (B–D) CHO-K1 cells transiently transfected with vector (B) or FLAG-tagged mLPCAT3 cDNA (C and D) were imaged by TEM. (D) The boxed area in C was magnified. M, mitochondria; N, nucleus. The arrowheads indicated the multilayer membrane components (karmellae-like structures). (Scale bars: B and C, 1 μm; D, 100 nm.) Two independent experiments were performed with similar results.
Tissue Distribution of mLPCAT3, mLPCAT4, and mLPEAT1.
Tissue distribution of mLPCAT3, mLPCAT4, and mLPEAT1 was analyzed by using quantitative real-time RT-PCR analyses. The mRNA of mLPCAT3 was detected ubiquitously with high expression levels in testis (SI Fig. 8A). However, mLPCAT4 mRNA was highly expressed in epididymis, brain, testis, and ovary; and mLPEAT1 mRNA in stomach, epididymis, and colon (SI Fig. 8 B and C). mRNA levels of β-actin were used to normalize the mRNA levels of each enzyme.
LPLAT Activities of FLAG-Tagged mLPCAT3, mLPCAT4, and mLPEAT1.
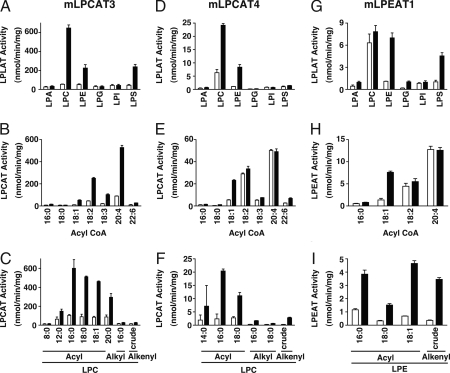
We next examined the LPLAT activities of FLAG-tagged mLPCAT3, mLPCAT4, and mLPEAT1 by using a variety of lysophospholipids as acceptors and acyl-CoAs as donors. mLPCAT3 exhibited detectable LPLAT activity for LPC, LPE, and LPS when arachidonoyl-CoA was used (Fig. 3A). mLPCAT4 possessed LPCAT and LPEAT activities (Fig. 3D), and mLPEAT1 showed LPEAT and LPSAT activities when oleoyl-CoA was used (Fig. 3G). mLPCAT3 recognized various polyunsaturated fatty acyl-CoAs, whereas mLPCAT4 and mLPEAT1 demonstrated preferential activity for oleoyl-CoA (Fig. 3 B, E, H; Table 1; and SI Fig. 9 A, C, E, and G).
Fig. 3.
Substrate selectivity of mLPCAT3, mLPCAT4, and mLPEAT1. (A) The LPLAT activities of mLPCAT3 were measured by using several 50 μM lysophospholipids, 25 μM [14C]arachidonoyl-CoA, and 0.5 μg of microsomal protein from mLPCAT3-CHO cells. LPCAT activities of mLPCAT3 using acyl-CoA (16:0, 18:0, 18:1, 18:2, 18:3, 20:4, and 22:6) with [14C]LPC (C16:0) (B) or several LPC (1-acyl LPC C8:0, 12:0, 16:0, 18:0, 18:1, 20:0, 1-alkyl LPC C16:0, and crude 1-alkenyl LPC) with [14C]arachidonoyl-CoA (C) were measured. Similarly, LPLAT assays were performed by using 100 μM lysophospholipids, 50 μM [14C]oleoyl-CoA, and microsomal protein (5 μg) of mLPCAT4- (D) or mLPEAT1-CHO (G) cells. LPCAT activities of mLPCAT4 using acyl-CoA (16:0, 18:0, 18:1, 18:2, 18:3, 20:4, and 22:6) with [14C]LPC (C16:0) (E) or several LPC (1-acyl LPC C14:0, 16:0, 18:0, 1-alkyl LPC C16:0, 18:0, and crude 1-alkenyl LPC) with [14C]oleoyl-CoA (F) were measured. LPEAT activities of mLPEAT1 were measured by using 14C-labeled acyl-CoA (16:0, 18:1, 18:2, and 20:4) with LPE (C18:1) (H) or [14C]oleoyl-CoA with several LPE (1-acyl LPE C16:0, 18:0, 18:1, and crude 1-alkenyl LPE) (I). Vector-transfected cells or each of LPLAT cDNA-transfected CHO-K1 cells are shown in open bars and closed bars, respectively. The data represent the mean + SD of triplicate measurements. Two independent experiments were performed with similar results.
Table 1.
Substrate selectivities of three acyltransferases
| Enzyme | Enzymatic activity | Acyl-CoA | Lysophospholipid |
|---|---|---|---|
| mLPCAT3 | LPCAT | 20:4 > 18:2 > 18:3 > 18:1 > 22:6 | 16:0 > 18:0 > 18:1 > 20:0 > 12:0 |
| LPEAT | 20:4 > 18:2 > 18:1 | 18:1 > 16:0 = alkenyl (crude) | |
| LPSAT | 20:4 > 18:2 > 18:1 | 18:1 | |
| mLPCAT4 | LPCAT | 18:1 | 16:0 > 18:0 |
| LPEAT | 18:1 | 16:0 > 18:1 > alkenyl (crude) > 18:0 | |
| mLPEAT1 | LPEAT | 18:1 | 18:1 > 16:0 > alkenyl (crude) > 18:0 |
| LPSAT | 18:1 | 18:1 |
mLPCAT3 and mLPCAT4 exhibited enzymatic activity for various acyl groups at the sn-1 position of LPC; higher activity was observed for 1-acyl-LPC as an acceptor than for 1-O-alkyl-LPC or 1-O-alkenyl-LPC (Fig. 3 C and F). Meanwhile, there were no clear differences in the activities of mLPCAT3, mLPCAT4, and mLPEAT1 for 1-acyl-LPEAT and 1-O-alkenyl-LPEAT (Fig. 3I and SI Fig. 9 B and F). mLPCAT3 and mLPEAT1 showed higher activity for 1-oleoyl-LPE (Fig. 3I and SI Fig. 9B), whereas mLPCAT4 showed a preference for 1-palmitoyl-LPE (SI Fig. 9F). Furthermore, mLPCAT3 and mLPEAT1 recognized both 1-oleoyl-LPS and a crude mixture of LPS (SI Fig. 9 D and H). The substrate selectivity and the apparent Km and Vmax values of the enzymes are summarized in Table 1 and SI Table 2, respectively. mLPCAT3 showed higher LPCAT activity when compared with LPEAT and LPSAT (Fig. 3A). Similarly, mLPCAT4 had higher LPCAT activity than LPEAT activity (Fig. 3D). mLPEAT1 showed similar activities for LPE and LPS (Fig. 3G and SI Table 2). The specific activities of mLPCAT3 were much higher than those of mLPCAT4 or mLPEAT1, partly because protein expression level of mLPCAT3 was extremely high (see SI Fig. 7A).
Reduction of Endogenous LPLAT Activities by mLPCAT3 siRNA.
We next investigated the role of endogenous mLPCAT3. To perform siRNA transfection, we chose B16 melanoma cells, because the cells exhibit high endogenous enzymatic activity and high mRNA expression of mLPCAT3 (data not shown). To investigate whether endogenous LPLAT activities are decreased by mLPCAT3-siRNA, we transfected B16 cells with siRNAs designed against three different regions of the mLPCAT3 gene or with a negative control (NC)-siRNA. mLPCAT3-siRNAs decreased the mRNA expression of mLPCAT3 20–30% compared with NC-siRNA without affecting the mRNA expression of mLPCAT4 and mLPEAT1 (SI Fig. 10B).
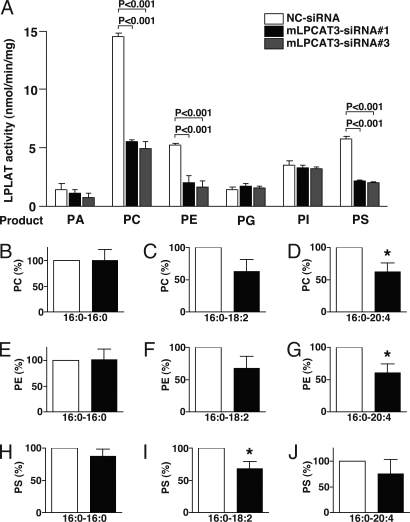
Endogenous LPCAT, LPEAT, and LPSAT activities, measured by using arachidonoyl-CoA as a donor, were shown to decrease in mLPCAT3-siRNA transfected cells (Fig. 4A). In cells transfected with siRNAs against mLPCAT4 and mLPEAT1, endogenous LPLAT activities for oleoyl-CoA remained unchanged (SI Fig. 10E), despite a reduction in the mRNA levels of mLPCAT4 and mLPEAT1 (SI Fig. 10 C and D). It is possible that mLPCAT3 compensates for LPEAT and LPSAT activities in B16 cells transfected with mLPCAT4- or mLPEAT1-siRNAs, and that the expression levels of these mRNAs were lower than that of mLPCAT3 (data not shown). Thus, mLPCAT3 appears to be a principal enzyme exhibiting LPCAT, LPEAT, and LPSAT activities in B16 cells. Transfection of B16 cells with mLPCAT3-, mLPCAT4-, or mLPEAT1-siRNAs did not affect LPAAT, LPGAT, or LPIAT activities (Fig. 4A and SI Fig. 10E).
Fig. 4.
Effect of mLPCAT3 knockdown on endogenous LPLAT activities and membrane phospholipid composition in B16 cells. (A) Endogenous LPLAT activities were measured by using 25 μM [14C]arachidonoyl-CoA with several lysophospholipids. The supernatants from 9,000 × g centrifugation were prepared from B16 cells transfected with mLPCAT3-siRNA or NC-siRNA and used for enzyme assays. Proteins were prepared from B16 cells transfected with mLPCAT3-siRNAs and NC-siRNA. The data represent the mean + SD of triplicate measurements. Statistical analyses were performed by ANOVA with Tukey's multiple comparison test (P < 0.001). Two independent experiments were performed with similar results. (B–J) The amounts of various phospholipids were measured by using a 4000 Q-TRAP, as described in Materials and Methods. The levels of PC (B–D), PE (E–G), and PS (H–J) in B16 cells were normalized to 17:0-LPC, which was added as a control in the lipid extraction step, and the values of NC-siRNA were considered to be 100%. Various lipids (sn-1-sn-2; 16:0–16:0, 16:0–18:2, and 16:0–20:4) were measured and shown. The data represent the mean + SD of three independent experiments. Statistical analyses were performed by one-sample t test (*, P < 0.05 vs. NC-siRNA).
The Effect of mLPCAT3 on Phospholipid Compositions as Determined by Liquid Chromatography (LC)–Mass Spectrometry.
We next analyzed the components of membrane phospholipid and their amount in B16 cells transfected with mLPCAT3-siRNA or NC-siRNA. After supplementing with 10 nM arachidonic acid for 24 h, total lipids were extracted by the Bligh–Dyer method from washed cells. Lipids were analyzed by using a 4000 Q-TRAP quadrupole linear ion trap hybrid mass spectrometer with an ACQUITY Ultra Performance LC. Consistent with the substrate selectivity of the enzyme in vitro, the levels of PC, PE, and PS containing arachidonic acid (20:4) or linoleic acid (18:2) were decreased in mLPCAT3-siRNA transfected cells (Fig. 4 C, D, F, G, I, and J). Phospholipids possessing palmitic acid (16:0) were used as a negative control (Fig. 4 B, E, and H).
Discussion
Here, we have identified a LPLAT family, which is distinct from other LPLAT families because its members do not possess LPAAT motifs. Within this LPLAT family, we found key LPLAT enzymes, mLPCAT3, mLPCAT4, and mLPEAT1, which catalyze membrane phospholipid remodeling in the Lands' cycle. mLPCAT3 converted LPC, LPE, and LPS to PC, PE, and PS, respectively, by using polyunsaturated fatty acyl-CoAs (18:2 and 20:4). However, mLPCAT4 had LPCAT and LPEAT activities, and mLPEAT1 showed LPEAT and LPSAT activities. Both mLPCAT4 and mLPEAT1 demonstrated a clear preference for oleoyl-CoA (18:1) as an acyl donor.
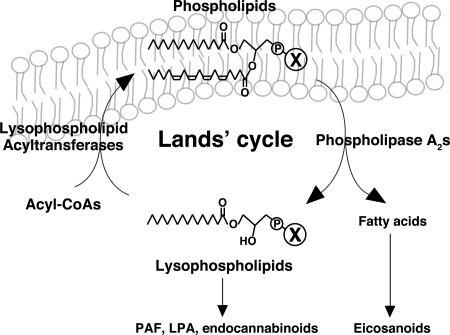
The diversity and asymmetry of glycerophospholipids are assumed to be regulated by several PLA2s and LPLATs that control lysophospholipids and phospholipid levels containing precursors of lipid mediators, such as arachidonic acid at the sn-2 position (2, 9). Arachidonic acid liberated by PLA2s is converted to eicosanoids (9, 28). Lysophospholipids are also precursors for a different class of lipid mediators including PAF, LPA, and endocannabinoids (29–35) (Fig. 5). Furthermore, membrane fluidity, curvature, and function may be attributed to phospholipid compositions (36–38). Although various types of PLA2s have been identified and their biological functions were uncovered, the properties of LPLATs and their number have not been clarified. To date, numerous studies have suggested the possible presence of polyunsaturated long chain fatty acyl-CoA specific LPCATs, LPEATs, and LPSATs (5, 39–42). Recently, we cloned and characterized two types of LPCAT: LPCAT1 and LysoPAFAT/LPCAT2. LPCAT1 may produce pulmonary surfactant phospholipids, a function postulated because of its high expression level in alveolar type II cells and its substrate selectivity (20, 43). LysoPAFAT/LPCAT2, which is mainly expressed in inflammatory cells, is up-regulated by endotoxin (a Toll-like receptor 4 agonist) stimulation in macrophages, and shows not only lysoPAF acetyltransferase activity (conversion of lysoPAF to PAF), but also LPCAT activity in the presence of Ca2+ (21). However, neither mLPCAT3, mLPCAT4, nor mLPEAT1 activity was Ca2+-dependent. Additionally, innate immune agonists (Toll-like receptor 3, 4, or 9 agonist) did not induce mRNA expression of these enzymes in macrophages (data not shown), indicating that mLPCAT3, mLPCAT4, and mLPEAT1 play roles in membrane biogenesis in a constitutive manner. Moreover, mLPCAT3 recognizes a wide range of acyl-CoAs, especially arachidonoyl-CoA, which is a major fatty acid of PC in several cell types (3, 5). Thus, it is possible that mLPCAT3 is a principal enzyme in the regulation of the remodeling pathway. Differences in phospholipid composition among various types of cells or tissues have been reported (5, 44). The substrate specificities and mRNA expression patterns of these enzymes may contribute to the production of the different varieties of membrane phospholipids seen in various tissues and determine their respective cell membrane characteristics.
Fig. 5.
Phospholipid metabolism in Lands' cycle. Fatty acids of phospholipid are liberated by PLA2s and converted to eicosanoids. Lysophospholipids are also precursors of a different class of lipid mediators including PAF, LPA, and endocannabinoids. Lysophospholipids are converted to phospholipids in the presence of acyl-CoA by LPLATs. X indicates several polar head groups of phospholipids. See text for details.
Until now, the LPAAT family has been characterized by LPLATs possessing LPAAT motifs. Among them, LPAATs are known to catalyze PA biosynthesis in the de novo pathway (Kennedy pathway), whereas LPGAT, ALCAT, LPCAT1, and LysoPAFAT/LPCAT2 exert their activities in the remodeling pathway (Lands' cycle) (Fig. 1). However, most LPAAT family genes have not yet been characterized, and many LPLAT cDNAs remain to be clarified (Fig. 1). In this study, we identified a LPLAT family denoted MBOAT, comprising reported proteins of unknown functions (24). Within the MBOAT family of enzymes, we identified additional LPCATs, LPEATs, and LPSATs. However, we did not identify LPIAT. Additional LPLATs having a preference for different acyl-CoAs may contribute to membrane composition diversity, which will be clarified in the future studies. The existence of multiple LPLATs may in some way be analogous to the 20 different aminoacyl-tRNA synthetases and multiple acyltransferases that act to incorporate amino acids into preexisting polypeptides (45).
The MBOAT family is evolutionarily conserved from plants to mammals (24). Recently, four different research groups independently reported the identification of a yeast orthologue of this family as a LPLAT with most similarity to mLPCAT4 (MBOAT2) (46–49). Notably, the enzymatic properties of yeast MBOAT are different from those of mouse MBOAT. Differences in such enzymatic properties may account for the differences in membrane phospholipid composition between yeast and mammalian cells. Interestingly, the Drosophila orthologue of mLPCAT3 was reported as nessy, a gene controlled by Ultrabithorax (Ubx), homeotic (Hox), and other Hox proteins during Drosophila embryogenesis (50). This suggests that membrane phospholipid remodeling may be important in embryogenesis, and that mLPCAT3 may play a role in controlling the body plan by changing the phospholipid composition through the Lands' cycle. Furthermore, in a brachydactyly–syndactyly syndrome patient, human LPEAT1 located at chromosome 6 is disrupted (51), suggesting that human LPEAT1 contributes to the turnover of phospholipids for normal development and organogenesis.
In conclusion, we have identified three LPLATs belonging to a LPLAT family, MBOAT, that catalyze membrane biogenesis. Further studies are needed to elucidate the roles of mLPCAT3, mLPCAT4, and mLPEAT1 in vivo, and to determine their potential as therapeutic targets for various diseases. To date, the study of membrane biogenesis in the Kennedy-independent pathway has been delayed because of a lack of information about its key enzymes. Discovery of this LPLAT family paves the way for a better understanding of membrane asymmetry and diversity.
Materials and Methods
Materials.
Various phospholipids and acyl-CoAs were obtained from Avanti Polar Lipids. Linoleoyl-CoA (C18:2) was purchased from Sigma. 1-O-Alkyl-LPC (Lyso-PAF) was purchased from Cayman Chemical. 1-O-Alkenyl-LPC was from Doosan. [1-14C]Oleoyl-CoA (1.924 GBq/mmol), [1-14C]linoleoyl-CoA (2.035 GBq/mmol), and [1-14C]arachidonoyl-CoA (2.035 GBq/mmol) were purchased from Moravec Biochemicals. [1-14C]Palmitoyl-CoA (2.22 GBq/mmol) was from GE Healthcare.
Cloning of mLPCAT3, mLPCAT4, and mLPEAT1.
The entire coding region of mLPCAT3 [DNA Data Base in Japan (DDBJ) accession no. AB294194], mLPCAT4 (AB297383), and mLPEAT1 (AB297382) was amplified by PCR using DNA template from testis, stomach, and colon, respectively. For each gene, PCR was performed twice, and the FLAG epitope (DYKDDDDK) was attached to the N terminus by using a forward primer during the second PCR. The amplified DNA fragments were each cloned into pCXN2.1 vector (52) by using multiple cloning sites, and sequenced. The primers used for cloning these enzymes are shown in SI Table 3.
Expression of mLPCAT3, mLPCAT4, and mLPEAT1 in CHO-K1 Cells.
After 48 h of transfection with each FLAG-tagged LPLAT cDNA by using Lipofectamine 2000 (Invitrogen), cells from 10-cm dishes were scraped into 1 ml of ice-cold buffer containing 20 mM Tris·HCl (pH 7.4), 300 mM sucrose, and a proteinase inhibitor mixture, Complete (Roche Applied Science), and then sonicated three times on ice for 30 s. After centrifugation for 10 min at 800 × g (mLPCAT4 and mLPEAT1) or 9,000 × g (mLPCAT3), each supernatant was collected and centrifuged at 100,000 × g for 1 h. The resulting pellets were resuspended in buffer containing 20 mM Tris·HCl (pH 7.4), 300 mM sucrose, and 1 mM EDTA. Protein concentration was measured by the method of Bradford (53), by using a commercially prepared protein assay solution (Bio-Rad) and BSA (fraction V, fatty acid-free; Sigma) as a standard.
In Vitro LPLAT Assays.
The acyltransferase activity was measured in two ways: (i) by the transfer of [14C]acyl-CoA to lysophospholipids to form phospholipids or (ii) by conversion of [14C]palmitoyl-LPC to PC in the presence of acyl-CoA. Reaction mixtures contained 100 mM Tris·HCl (pH 7.4), 0.5–1 mM EDTA, indicated concentrations of acyl-CoA and lysophospholipid, and enzyme in a total volume of 0.1 ml. After incubation at 37°C for 10 min, reactions were stopped by the addition of 0.3 ml of chloroform:methanol (1:2, vol/vol). Total lipids were extracted by using the Bligh–Dyer method (54), and subsequently analyzed by TLC in chloroform:methanol:acetic acid:water (50:25:8:4, vol/vol/vol/vol). Bands at positions corresponding to the expected products were visualized by I2 vapor, cut off the plate, placed in Microscinti-O (PerkinElmer Life Sciences), and analyzed in a liquid scintillation counter LS6500 (Beckman).
siRNA Transfection.
B16 cells were grown in RPMI 1640 medium (Sigma) containing 10% FBS. Cells were transfected with 50 nM mLPCAT3-siRNAs (mLPCAT3-siRNA no. 1, ID no. 89055; no. 2, 89150; no. 3, 89240), mLPCAT4-siRNAs (mLPCAT4-siRNA no. 1, 79688; no. 2, 79779; no. 3, 179488), mLPEAT1-siRNAs (mLPEAT1-siRNA no. 1, 93126; no. 2, 93219; no. 3, 170568), or NC-siRNA (silencer negative control 1) by using Lipofectamine 2000 according to the manufacturer's protocol. siRNA were obtained from Ambion. The siRNA transfection was performed for 2–4 days in B16 cells.
Mice.
C57BL/6J mice were obtained from Clea Japan. All animal studies were conducted in accordance with the guidelines for Animal Research at University of Tokyo and were approved by the University of Tokyo Ethics Committee for Animal Experiments. Also see SI Materials.
Supplementary Material
ACKNOWLEDGMENTS.
We are grateful to Dr. H. Aburatani (University of Tokyo) for gene expression analysis; Drs. M. Nakamura, T. Takahashi, Y. Kita (University of Tokyo), Mr. Y. Ihara, Y. Kihara, T. Harayama, and all of the members of our laboratory for valuable suggestions; and F. Hamano (University of Tokyo) for technical assistance. We also thank Dr. J.-i. Miyazaki (Osaka University, Osaka, Japan) for supplying pCXN2. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.S.). T.S. and H.S. were supported by the Center for NanoBio Integration at the University of Tokyo.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Base in Japan/European Molecular Biology Laboratory/GenBank databases [accession nos. AB297382 (mouse LPEAT1), AB294194 (mouse LPCAT3), and AB297383 (mouse LPCAT4)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0712245105/DC1.
References
- 1.MacDonald JI, Sprecher H. Phospholipid fatty acid remodeling in mammalian cells. Biochim Biophys Acta. 1991;1084:105–121. doi: 10.1016/0005-2760(91)90209-z. [DOI] [PubMed] [Google Scholar]
- 2.Shimizu T, Ohto T, Kita Y. Cytosolic phospholipase A2: Biochemical properties and physiological roles. IUBMB Life. 2006;58:328–333. doi: 10.1080/15216540600702289. [DOI] [PubMed] [Google Scholar]
- 3.Wood R, Harlow RD. Structural analyses of rat liver phosphoglycerides. Arch Biochem Biophys. 1969;135:272–281. doi: 10.1016/0003-9861(69)90540-2. [DOI] [PubMed] [Google Scholar]
- 4.Schlame M, Brody S, Hostetler KY. Mitochondrial cardiolipin in diverse eukaryotes. Comparison of biosynthetic reactions and molecular acyl species. Eur J Biochem. 1993;212:727–735. doi: 10.1111/j.1432-1033.1993.tb17711.x. [DOI] [PubMed] [Google Scholar]
- 5.Yamashita A, Sugiura T, Waku K. Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells. J Biochem (Tokyo) 1997;122:1–16. doi: 10.1093/oxfordjournals.jbchem.a021715. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg A, Pricer WE., Jr Enzymatic synthesis of the coenzyme A derivatives of long chain fatty acids. J Biol Chem. 1953;204:329–343. [PubMed] [Google Scholar]
- 7.Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipides. J Biol Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 8.Lands WE. Metabolism of glycerolipides: A comparison of lecithin and triglyceride synthesis. J Biol Chem. 1958;231:883–888. [PubMed] [Google Scholar]
- 9.Nakagawa Y, Waku K. The metabolism of glycerophospholipid and its regulation in monocytes and macrophages. Prog Lipid Res. 1989;28:205–243. doi: 10.1016/0163-7827(89)90013-1. [DOI] [PubMed] [Google Scholar]
- 10.Farooqui AA, Horrocks LA, Farooqui T. Glycerophospholipids in brain: Their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem Phys Lipids. 2000;106:1–29. doi: 10.1016/s0009-3084(00)00128-6. [DOI] [PubMed] [Google Scholar]
- 11.Kume K, Shimizu T. cDNA cloning and expression of murine 1-acyl-sn-glycerol-3-phosphate acyltransferase. Biochem Biophys Res Commun. 1997;237:663–666. doi: 10.1006/bbrc.1997.7214. [DOI] [PubMed] [Google Scholar]
- 12.Eberhardt C, Gray PW, Tjoelker LW. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J Biol Chem. 1997;272:20299–20305. doi: 10.1074/jbc.272.32.20299. [DOI] [PubMed] [Google Scholar]
- 13.Hollenback D, et al. Substrate specificity of lysophosphatidic acid acyltransferase beta—evidence from membrane and whole cell assays. J Lipid Res. 2006;47:593–604. doi: 10.1194/jlr.M500435-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Li D, et al. Cloning and identification of the human LPAAT-zeta gene, a novel member of the lysophosphatidic acid acyltransferase family. J Hum Genet. 2003;48:438–442. doi: 10.1007/s10038-003-0045-z. [DOI] [PubMed] [Google Scholar]
- 15.Cao J, Liu Y, Lockwood J, Burn P, Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J Biol Chem. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Cao J, Shi Y. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J Biol Chem. 2004;279:55866–55874. doi: 10.1074/jbc.M406710200. [DOI] [PubMed] [Google Scholar]
- 17.Lu B, et al. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem J. 2005;385:469–477. doi: 10.1042/BJ20041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye GM, et al. Cloning and characterization a novel human 1-acyl-sn-glycerol-3-phosphate acyltransferase gene AGPAT7. DNA Seq. 2005;16:386–390. doi: 10.1080/10425170500213712. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Malhotra A, Ren M, Schlame M. The enzymatic function of tafazzin. J Biol Chem. 2006;281:39217–39224. doi: 10.1074/jbc.M606100200. [DOI] [PubMed] [Google Scholar]
- 20.Nakanishi H, et al. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1): Expression in alveolar type II cells and possible involvement in surfactant production. J Biol Chem. 2006;281:20140–20147. doi: 10.1074/jbc.M600225200. [DOI] [PubMed] [Google Scholar]
- 21.Shindou H, et al. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:LYSO-PAF acetyltransferase. J Biol Chem. 2007;282:6532–6539. doi: 10.1074/jbc.M609641200. [DOI] [PubMed] [Google Scholar]
- 22.Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 23.Shikano S, Li M. Membrane receptor trafficking: evidence of proximal and distal zones conferred by two independent endoplasmic reticulum localization signals. Proc Natl Acad Sci USA. 2003;100:5783–5788. doi: 10.1073/pnas.1031748100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 27.Snapp EL, et al. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narumiya S, Sugimoto Y, Ushikubi F. Prostanoid receptors: structures, properties, and functions. Physiol Rev. 1999;79:1193–1226. doi: 10.1152/physrev.1999.79.4.1193. [DOI] [PubMed] [Google Scholar]
- 29.Samuelsson B, Dahlen SE, Lindgren JA, Rouzer CA, Serhan CN. Leukotrienes and lipoxins: Structures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 30.Fukushima N, Kimura Y, Chun J. A single receptor encoded by vzg-1/lpA1/edg-2 couples to G proteins and mediates multiple cellular responses to lysophosphatidic acid. Proc Natl Acad Sci USA. 1998;95:6151–6156. doi: 10.1073/pnas.95.11.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii S, Shimizu T. Platelet-activating factor (PAF) receptor and genetically engineered PAF receptor mutant mice. Prog Lipid Res. 2000;39:41–82. doi: 10.1016/s0163-7827(99)00016-8. [DOI] [PubMed] [Google Scholar]
- 32.Hla T, Lee MJ, Ancellin N, Paik JH, Kluk MJ. Lysophospholipids—receptor revelations. Science. 2001;294:1875–1878. doi: 10.1126/science.1065323. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Liu J, Harvey-White J, Zimmer A, Kunos G. Endocannabinoid signaling via cannabinoid receptor 1 is involved in ethanol preference and its age-dependent decline in mice. Proc Natl Acad Sci USA. 2003;100:1393–1398. doi: 10.1073/pnas.0336351100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott SM, Zimmerman GA, Stafforini DM, McIntyre TM. Platelet-activating factor and related lipid mediators. Annu Rev Biochem. 2000;69:419–445. doi: 10.1146/annurev.biochem.69.1.419. [DOI] [PubMed] [Google Scholar]
- 35.Haeggstrom JZ, Wetterholm A. Enzymes and receptors in the leukotriene cascade. Cell Mol Life Sci. 2002;59:742–753. doi: 10.1007/s00018-002-8463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stubbs CD, Smith AD. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim Biophys Acta. 1984;779:89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 37.van Meer G, Vaz WL. Membrane curvature sorts lipids. Stabilized lipid rafts in membrane transport. EMBO Rep. 2005;6:418–419. doi: 10.1038/sj.embor.7400410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waku K, Lands WE. Control of lecithin biosynthesis in erythrocyte membranes. J Lipid Res. 1968;9:12–18. [PubMed] [Google Scholar]
- 40.Okuyama H, Eibl H, Lands WE. Acyl coenzyme A:2-acyl-sn-glycerol-3-phosphate acyltransferase activity in rat liver microsomes. Biochim Biophys Acta. 1971;248:263–273. doi: 10.1016/0005-2760(71)90014-2. [DOI] [PubMed] [Google Scholar]
- 41.Holub BJ. The biosynthesis of phosphatidylserines by acylation of 1-acyl-sn-glycero-3-phosphoserine in rat liver. Biochim Biophys Acta. 1980;618:255–262. doi: 10.1016/0005-2760(80)90031-4. [DOI] [PubMed] [Google Scholar]
- 42.Masuzawa Y, Sugiura T, Sprecher H, Waku K. Selective acyl transfer in the reacylation of brain glycerophospholipids. Comparison of three acylation systems for 1-alk-1′-enylglycero-3-phosphoethanolamine, 1-acylglycero-3-phosphoethanolamine and 1-acylglycero-3-phosphocholine in rat brain microsomes. Biochim Biophys Acta. 1989;1005:1–12. doi: 10.1016/0005-2760(89)90024-6. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Hyatt BA, Mucenski ML, Mason RJ, Shannon JM. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc Natl Acad Sci USA. 2006;103:11724–11729. doi: 10.1073/pnas.0604946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuksis A, Breckenridge WC, Marai L, Stachnyk O. Molecular species of lecithins of rat heart, kidney, and plasma. J Lipid Res. 1969;10:25–32. [PubMed] [Google Scholar]
- 45.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 46.Riekhof WR, Wu J, Jones JL, Voelker DR. Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2007;282:28344–28352. doi: 10.1074/jbc.M705256200. [DOI] [PubMed] [Google Scholar]
- 47.Jain S, et al. Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2007;282:30562–30569. doi: 10.1074/jbc.M706326200. [DOI] [PubMed] [Google Scholar]
- 48.Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J Biol Chem. 2007;282:30845–30855. doi: 10.1074/jbc.M702719200. [DOI] [PubMed] [Google Scholar]
- 49.Tamaki H, et al. LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae. J Biol Chem. 2007;282:34288–34298. doi: 10.1074/jbc.M704509200. [DOI] [PubMed] [Google Scholar]
- 50.Maurel-Zaffran C, et al. nessy, an evolutionary conserved gene controlled by Hox proteins during Drosophila embryogenesis. Mech Dev. 1999;86:159–163. doi: 10.1016/s0925-4773(99)00105-7. [DOI] [PubMed] [Google Scholar]
- 51.Dauwerse JG, et al. A t(4;6)(q12;p23) translocation disrupts a membrane-associated O-acetyl transferase gene (MBOAT1) in a patient with a novel brachydactyly-syndactyly syndrome. Eur J Hum Genet. 2007;15:743–751. doi: 10.1038/sj.ejhg.5201833. [DOI] [PubMed] [Google Scholar]
- 52.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 53.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 54.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.