Abstract
Guanine nucleotide-exchange factors (GEFs) activate ADP-ribosylation factor (ARF) GTPases that recruit coat proteins to membranes to initiate transport vesicle formation. Three mammalian GEFs are inhibited by brefeldin A (BFA). GBF1, predominantly associated with cis-Golgi membranes, functions early in the secretory pathway, whereas BIG1 and BIG2 act in trans-Golgi or later sites. Perturbation of endoplasmic reticulum (ER) functions can result in accumulation of unfolded or misfolded proteins that causes ER stress and unfolded protein response (UPR), with accumulation of ER stress response element (ERSE) gene products. BFA treatment of cells causes accumulation of proteins in the ER, ER stress, and ultimately apoptosis. To assess involvement of BFA-sensitive GEFs in the damage resulting from prolonged BFA treatment, HepG2 cells were selectively depleted of BIG1, BIG2, or GBF1 by using specific siRNA. Only GBF1 siRNA dramatically slowed cell growth, led to cell-cycle arrest in G0/G1 phase, and caused dispersion of Golgi markers β-COP and GM130, whereas ER structure appeared intact. GBF1 depletion also significantly increased levels of ER proteins calreticulin and protein disulfide isomerase (PDI). Proteomic analysis identified ER chaperones involved in the UPR that were significantly increased in amounts in GBF1-depleted cells. Upon ER stress, transcription factor ATF6 translocates from the ER to Golgi, where it is sequentially cleaved by site 1 and site 2 proteases, S1P and S2P, to a 50-kDa form that activates transcription of ERSE genes. Depletion of GBF1, but not BIG1 or BIG2, induced relocation of S2P from Golgi to ER with proteolysis of ATF6 followed by up-regulation of ER chaperones, mimicking a UPR response.
Keywords: ADP-ribosylation factor, ER stress, chaperone, ATF6
Coat protein recruitment to membranes for transport vesicle generation at numerous intracellular sites between endoplasmic reticulum (ER) and plasma membrane is regulated by ADP-ribosylation factors (ARFs), 20-kDa GTPases that cycle between GDP-bound inactive and membrane-associated GTP-bound active forms. ARF activation requires a guanine nucleotide-exchange factor (GEF) to accelerate the replacement of GDP by GTP (1, 2). Mammalian cells contain diverse GEF proteins, three of which are inhibited by brefeldin A (BFA), a fungal fatty acid metabolite that reversibly causes disruption of Golgi structure (3). All GEFs contain a central Sec7 domain (≈200 aa) that catalyzes the release of ARF-bound GDP to permit GTP binding and is the site of its inhibition by BFA (4, 5). BFA binds the ARF–GDP–Sec7 domain complex of a sensitive GEF, preventing ARF activation (6, 7).
BFA-inhibited BIG1 and BIG2 were purified together with >670-kDa macromolecular complexes from bovine brain cytosol (8). Both seemed to activate preferentially class I ARFs (8) and localize to Golgi membranes (9). In contrast, GBF1 preferentially activated ARF5 in vitro and both ARF5 and ARF1 in vivo (10). Recently, GBF1 has been shown to coprecipitate with ARF1 and ARF4 from cells (11), suggesting that GBF1 interacts with both class II ARFs. GBF1, which promoted COPI recruitment to membranes both in vitro and in vivo (12, 13), was dynamically associated with elements termed vesicular tubular clusters (VTCs) and cis-Golgi membranes (14, 15). The GBF1 molecule contains a dimerization motif, first identified in its plant homolog GNOM (16), and likely exists as homodimer. GBF1 is the BFA-sensitive ARF GEF responsible for membrane association of COPI early in the secretory pathway (17), where it is essential to the translocation of pre-Golgi intermediates and the maintenance of Golgi integrity (18).
The complex system of ER membranes is the site of synthesis and maturation for membrane and secreted proteins. Perturbation of critical ER functions, such as Ca2+ homeostasis, protein glycosylation, or disulfide bond formation, can cause accumulation of unfolded or misfolded proteins that initiates ER stress. In the first of two major ER chaperone systems, lectin-like chaperones calnexin and calreticulin are critical to correct glycoprotein folding in the ER lumen (19, 20). In the second system, ER chaperone GRP78 (BiP) recognizes and interacts only with unfolded regions of proteins containing hydrophobic residues (21). Additionally, there are thio-oxidoreductases that catalyze folding, such as protein disulfide isomerase (PDI) and Erp72, known to bind some nascent ER proteins (22, 23). It later was reported that ER chaperones formed large ER-localized multiprotein complexes, which bound unfolded protein substrates (24). The unfolded protein response (UPR) in cells under going ER stress is characterized by up-regulation of ER chaperones, which can improve cell survival by facilitating correct folding or assembly of ER proteins and preventing their aggregation (25).
Transcriptional activation in the UPR depends on cis-acting ER stress response element (ERSE) in target gene promoters (26). The transcription factor ATF6 is an ER transmembrane protein that, in response to ER stress, translocates from the ER to Golgi. It is processed in the Golgi, via sequential cleavage by S1P and S2P, to a 50-kDa cytosolic form that activates transcription of ERSE genes (27). ER stress-induced movement of ATF6 to the Golgi is controlled by ER chaperone BiP. Dissociation of BiP from ATF6 unmasks an ER export signal, allowing its translocation to Golgi (28). When ER stress is not alleviated, an apoptotic pathway is activated (29).
Here, we report that cellular depletion of the ARF GEF GBF1, but not BIG1 or BIG2, induced UPR-like effects, with translocation of the enzyme S2P from the Golgi to ER, proteolytic activation of ATF6, and increased amounts of ER chaperone proteins.
Results
Effects of BIG1, BIG2, and GBF1 siRNA on Proliferation of HepG2 Cells.
Although acute effects of BFA treatment on vesicular trafficking and Golgi structure are apparently completely reversible, incubation of cells with BFA for >24 h resulted in cell death (30). To assess involvement of BFA-sensitive ARF-GEPs in these effects, HepG2 cells were selectively depleted of BIG1, BIG2, and GBF1 by using specific siRNA. Fig. 1A shows BIG1, BIG2, and GBF1 proteins in cells incubated with the vehicle alone (M) or with nontargeted (NT) or specific siRNAs against each of the three GEFs. Amounts of the three proteins from M and NT cells did not differ from those in untreated (C) cells, whereas the amount of each siRNA-targeted protein was clearly and selectively decreased after 48 h of transfection and was still lower after 72 h (Fig. 1A). The two bands of GBF1 diminished in parallel after GBF1 siRNA treatment, consistent with the existence of two forms of GBF1. Alternatively spliced variants of GBF1 with large in-frame deletions have been reported (31).
Fig. 1.
Effect of siRNA-induced depletion of BIG1, BIG2, or GBF1 on HepG2 cell proliferation. (A) Cells were incubated without additions (C), with vehicle alone (M), or with siRNA either nonspecific, NT, or specific for BIG1 (B1), BIG2 (B2), or GBF1 (G1) for 48 or 72 h before samples (30 μg) of total cell proteins were separated by SDS/PAGE and reacted with antibodies against BIG1, BIG2, GBF1, and α-tubulin. (B) Cells were treated as in A, and viable cells (identified by exclusion of 0.4% Trypan blue) were counted. Data are means ± SD of values from three experiments (*, P < 0.05 versus control).
After 24 h of incubation, numbers of viable cells in M, or NT, and BIG1, BIG2, or GBF1 siRNA-treated cells were similar to those of untreated cells (Fig. 1B). Depletion of BIG1, BIG2, and GBF1 for 48 h, however, resulted in numbers of cells (18–20 × 105) significantly lower than those of C and M-treated or NT-treated cells (28–30 × 105). After 72 h, differences in numbers of BIG1-, BIG2-, and GBF1-depleted cells were even larger. In particular, cells treated with GBF1 siRNA were fewer in number after 72 h than 48 h, consistent with extensive cell death (Fig. 1B).
Cell-Cycle Arrest in G0/G1 Phase and Cell Death After Depletion of GBF1.
After 24 h of incubation with BIG1, BIG2, or GBF1 siRNA, no significant effects on the cell-cycle distribution of HepG2 cells were detected (data not shown). After 48 h with GBF1 siRNA, the S-phase population was significantly less than that in untreated controls, i.e., 7.4 ± 0.8% versus 14.1 ± 0.7% (C), and cells in G0/G1 were 69.6 ± 1.6%, versus 61.9 ± 1.6% (C), with no significant change in percentages of G2/M cells [supporting information (SI) Table 1]. These differences were even larger after 72 h, when GBF1-depleted cells in G0/G1 were 82.9 ± 2.1% versus 65.4 ± 0.5% (C), and in S phase 2.2 ± 0.4% versus 10.1 ± 1.3% (C) (SI Table 1). The percentage of cells in G2/M also was significantly lower in GBF1-depleted (14.4 ± 1.1%) than in C (23.6 ± 0.7%) cells. There were no significant differences in phase distribution among cells incubated with NT, or BIG1, and BIG2 siRNA and control cells, suggesting that the specific depletion of cellular GBF1 induced a cell-cycle arrest with drastic reduction in the number of cells in the S phase and accumulation of surviving cells in G0/G1.
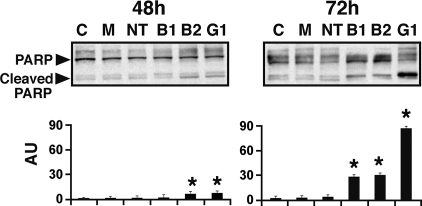
Poly(ADP-ribose) polymerase (PARP) and its cleavage products were assessed by Western blotting analysis of cell lysates (Fig. 2). After 48 h of incubation with siRNA, bands corresponding to a ≈90-kDa PARP cleavage product were significantly more prominent in samples from cells treated with BIG2 or GBF1 siRNA; after 72 h, the large accumulation of PARP cleavage product in GBF1-depleted cells was consistent with apoptosis. Amounts of 90-kDa PARP also were elevated in BIG1- and BIG2-depleted cells but much less dramatically (Fig. 2).
Fig. 2.
Effect of GBF1 depletion on PARP cleavage. HepG2 cells were incubated for 48 or 72 h as described in Fig. 1A, before separation of cell proteins (30 μg) by SDS/PAGE and reaction with antibodies against PARP. Representative blots and means ± SD of densitometric values (cleaved PARP) from three experiments are shown (*, P < 0.05 versus C).
Effects of BIG1, BIG2, and GBF1 siRNA on Golgi- and ER-Associated Proteins.
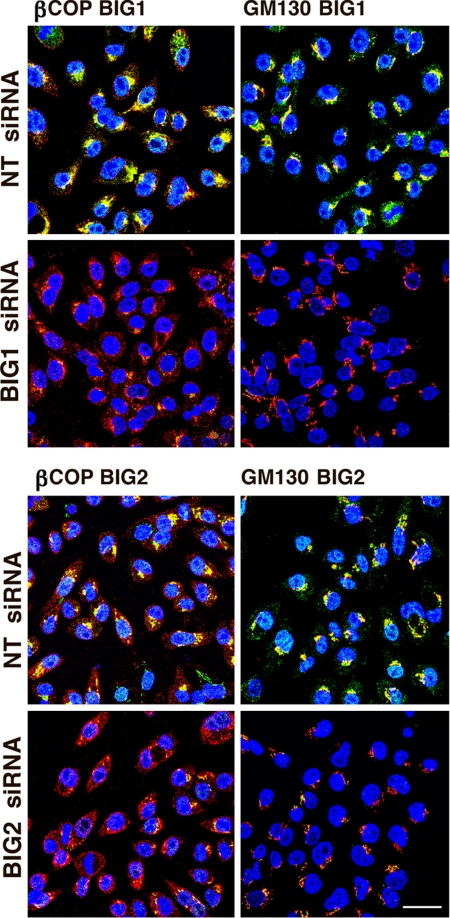
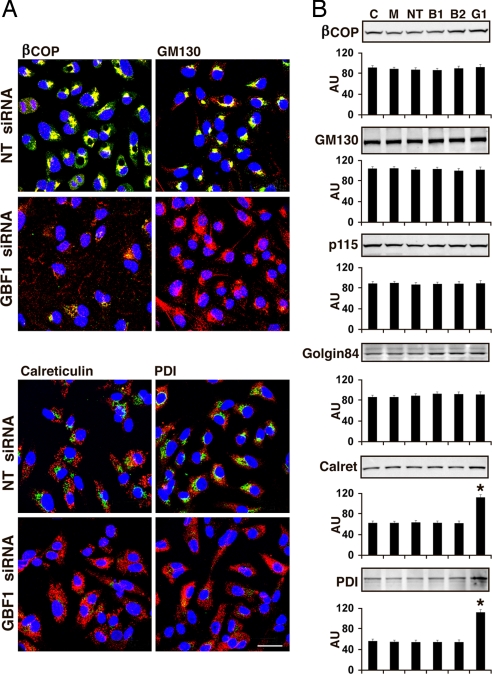
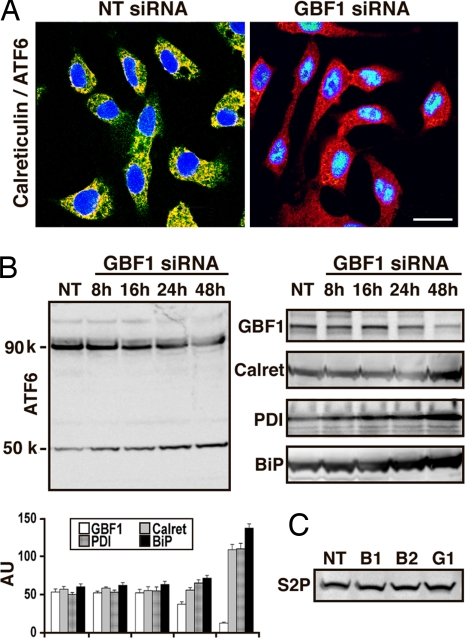
For additional studies, incubation of cells with siRNA was limited to 48 h to avoid the extensive cell death associated with longer exposure to GBF1 siRNA. In untreated cells, BIG1 and BIG2 (Fig. 3) and GBF1 (Fig. 4A) were seen largely in perinuclear structures, although none of these proteins colocalized with other two (10). In cells depleted of BIG1 or BIG2, β-COP, but not GM130, was dispersed somewhat from its perinuclear concentration in control cells (Fig. 3), whereas in GBF1-depleted cells, β-COP was redistributed and GM130 staining was widely scattered (Fig. 4A). The dissociation of β-COP and the extensive dispersion of the Golgi are analogous to the recently reported effects of GBF1 depletion in HeLa cells (11).
Fig. 3.
Effect of BIG1 or BIG2 depletion on Golgi complex. Cells depleted of BIG1 or BIG2 by incubation with specific siRNA were washed and fixed with 4% formaldehyde in PBS followed by immunostaining of BIG1 or BIG2 (green), β-COP, or GM130 (red). Findings were similar in three experiments. (Scale bar: 32 μm.)
Fig. 4.
Effect of GBF1 depletion on ER and Golgi proteins and structure. (A) Cells were incubated for 48 h with GBF1 siRNA, washed, and reacted with antibodies against GBF1 (green) and β-COP, GM130, calreticulin, or PDI (red). (Scale bar: 32 μm.) (B) Cells were incubated as described in Fig. 4A for 48 h before samples of cell proteins (30 μg) were separated by SDS/PAGE and reacted with antibodies against β-COP, GM130, p115, Golgin84 (Golgi markers), calreticulin, and PDI (ER markers). Representative blots and means ± SD of densitometric values from three experiments are shown (*, P < 0.05 versus C).
In control cells, calreticulin and PDI distributions were consistent with that of ER membranes, and no colocalization with GBF1 was evident (Fig. 4A). After 48 h of GBF1 depletion, localization of the two ER proteins appeared unchanged, although each appeared more abundant in some cells (Fig. 4A), and total amounts were significantly increased (Fig. 4B). Cell levels of β-COP, GM130, p115, and golgin84 were not detectably altered after 48 h of incubation with BIG1, BIG2, or GBF1 siRNA (Fig. 4B).
Effect of GBF1 Depletion with siRNA on ER Chaperones.
To show the effect of GBF1 depletion on protein composition, cells were incubated for 48 h with NT or GBF1 siRNA before proteomic analyses of whole extracts. MALDI-TOF identified five ER chaperones (BiP, ERp72, calreticulin, PDI, and ER-60) and mitochondrial chaperone Hsp60 as significantly increased (SI Table 2). Proliferating cell nuclear antigen (PCNA) was ≈80% decreased in GBF1-depleted cells, consistent with the limited proliferation, as was to a lesser extent Hsp90β (SI Table 2).
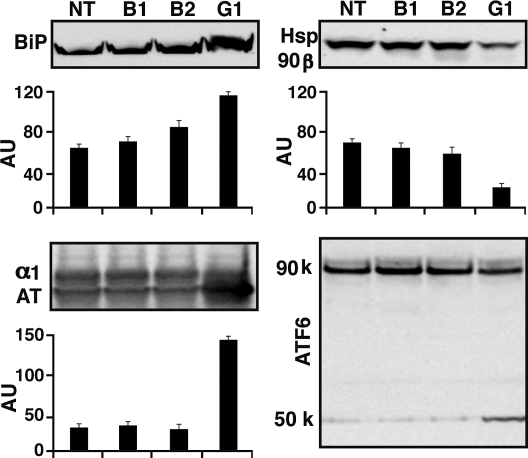
Elevated amounts of the ER chaperones BiP (Fig. 5), calreticulin, and PDI (Fig. 4B) were seen in cells incubated for 48 h with GBF1, but not BIG1 or BIG2 siRNA. Similarly, the amount of α1-antitrypsin protein was 4-fold that in control or BIG1- or BIG2-depleted cells (Fig. 5). Accumulation of α1-antitrypsin, associated with GBF1 depletion, resembled that reported after BFA treatment (32). Specificity of the decrease in Hsp90β content revealed by proteomic analyses was confirmed by the lack of similar changes in BIG1- or BIG2-depleted cells (Fig. 5).
Fig. 5.
Effect of GBF1 depletion on amounts of ER (BiP) and cytosolic (Hsp90β) chaperones, secreted protein α1-antitrypsin, and ER stress sensor ATF6. Cells were incubated for 48 h with NT, BIG1, BIG2, or GBF1 siRNA before samples of cell proteins (30 μg) were separated by SDS/PAGE and reacted with antibodies against BiP, HsP90β, α1-antitrypsin, or ATF6. Representative blots and means ± SD of densitometric values from three experiments are shown.
Effect of GBF1 Depletion on ATF6 Processing.
To assess potential involvement of ATF6 in effects of GBF1-depletion, we monitored its 50-kDa proteolytic cleavage product, not seen after BIG1 or BIG2 depletion (Fig. 5).
With depletion of GBF1, an accumulation of ATF6 in nuclei and absence from its previous perinuclear colocalization with calreticulin at the ER was seen (Fig. 6A). Fig. 6B shows a decline in 90-kDa ATF6 and concomitant increase in amounts of the 50-kDa cleavage product with time after addition of GBF1 siRNA to cells. Again, after 48 h with GBF1 siRNA, cell content of ER chaperones calreticulin, BiP, and PDI was significantly increased (Fig. 6B). ER stress initiates ATF6 translocation from ER to Golgi, where it is cleaved sequentially by S1P and S2P to the 50-kDa form that activates transcription of ERSE genes (30). No changes in amounts of S2P were detected in cells after incubation for 48 h with BIG1, BIG2, or GBF1 siRNA (Fig. 6C).
Fig. 6.
Proteolysis of ATF6 in cells incubated with GBF1 siRNA. (A) After incubation for 48 h with NT or GBF1 siRNA, cells were reacted with antibodies against ATF6 (green) and calreticulin (red). (Scale bar: 20 μm.) (B) Cells were incubated for the indicated time with GBF1 siRNA or for 48 h with NT siRNA before separation of proteins (30 μg) by SDS/PAGE and reaction with antibodies against ATF6, calreticulin, PDI, BiP, or GBF1. (Left) Representative blot showing full-length 90-kDa and activated 50-kDa ATF6. (Right) Representative blots are from the same experiment as that in Left. (Left Lower) Means ± SD of densitometric values from three experiments are shown. (C) Cells were incubated for 48 h with NT, BIG1, BIG2, or GBF1 siRNA before samples of cell proteins (30 μg) were separated by SDS/PAGE and reacted with antibodies against S2P. Findings were similar in three experiments.
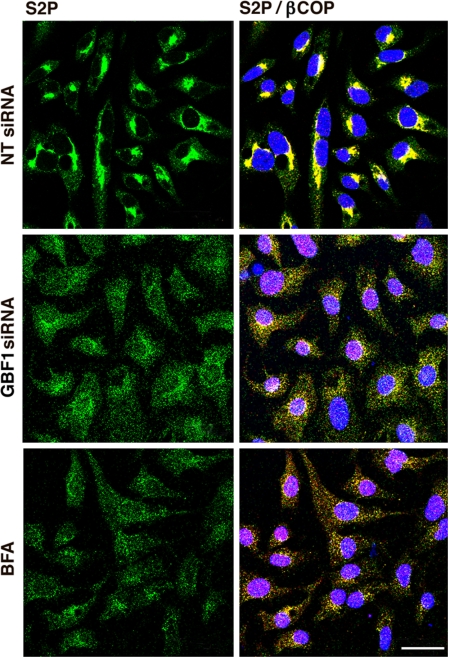
Redistribution of Enzyme S2P into the ER After GBF1 Depletion.
After 48 h of exposure to GBF1 siRNA, S2P was dispersed from its perinuclear colocalization with the Golgi marker β-COP (Fig. 7). The wide dispersion of S2P and β-COP in ER structures of GBF1-depleted cells resembles that reported in HeLa cells (33) and also seen in the HepG2 cells incubated for 1 h with BFA (Fig. 7). These data are consistent with cleavage of ATF6 at the ER to which the S2P enzyme responsible for ATF6 processing was relocated as a consequence of GBF1 depletion.
Fig. 7.
Effect of GBF1 depletion or BFA treatment on localization of S2P and β-COP. Cells were incubated with NT or GBF1 siRNA for 48 h or with 10 μg/μl BFA for 1 h before immunostaining for S2P (green) and β-COP (red). Findings were similar in three experiments. (Scale bar: 32 μm.)
Discussion
In mammalian cells, disregulation of proteins involved in vesicular membrane trafficking, such as GTP-binding proteins like ARFs that ensure coordinated fission–fusion or correct targeting of intracellular transport vesicles, may affect Golgi structural and functional integrity (34). In cells treated with BFA, Golgi membranes fuse with those of the ER, and anterograde vesicular traffic from ER exit sites to cis-Golgi rapidly ceases, without effect on retrograde transport from Golgi to ER (6). Three BFA-sensitive mammalian GEFs are known: BIG1, BIG2, and GBF1 (8, 14). GBF1 and the BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network (TGN), respectively (35). Most studies have been concerned with the reversible effects of BFA after relatively brief exposure to the drug, and alterations in Golgi morphology within 20 s are reported (3). Incubation of cells with BFA for >24 h, however, resulted in extensive swelling of the ER (36) and cell death (30). Treatment of cells with BFA causes an accumulation of proteins in the ER, which leads to ER stress and apoptosis (30), but BFA target proteins responsible for those effects have not been identified.
To assess involvement of BFA-sensitive ARF-GEPs in the injury resulting from prolonged BFA treatment, HepG2 cells were selectively depleted of BIG1, BIG2, or GBF1, by using specific siRNA. Only GBF1 siRNA dramatically slowed cell growth and led to cell-cycle arrest in G0/G1 phase. Growth arrest was evident after 72 h of GBF1 siRNA exposure with decreased cell content of cyclin D1, which is required for the progression from G1 to S phase (data not shown). Cells arrested in G0/G1 might proceed to apoptosis or could recover and then enter S phase. PARP and its cleavage products were quantified by Western blot analyses of lysates, revealing a large increase in amounts of ≈90-kDa proteolyzed PARP between 48 and 72 h of incubation with GBF1 siRNA, consistent with apoptosis.
To explore the ability of the Golgi and ER structures to maintain or organize themselves in the absence of individual GEFs, they were examined in cells treated with BIG1, BIG2, or GBF1 siRNA. Specific depletion of cellular GBF1 led to the dissociation of COPI coat complex from Golgi membranes, and redistribution of GM130, whereas ER structure appeared unaltered (Fig. 4A). All evidence indicated that the cell-cycle arrest and eventual apoptosis would be directly linked to the observed Golgi disintegration. COPI vesicles are tethered by a giantin–p115–GM130–GRASP65 complex that has important implications for both vesicle trafficking during interphase and Golgi breakdown at mitosis (37). p115 is required for membrane traffic from ER to and through the Golgi and for Golgi reassembly after mitosis (38). p115 binds to GM130 and giantin on Golgi membranes, and the interactions are important for COPI vesicle tethering in vitro (33). A novel interaction between p115 and GBF1 that is not required for targeting GBF1 or p115 to membranes has been described (39), although expression of the p115-binding region of GBF1 led to Golgi disruption, indicating that the p115-GBF1 interaction is functionally relevant.
At the onset of mitosis, dramatic breakdown of the Golgi structure results in clusters of small vesicles and tubules that are partitioned between two daughter cells. During telophase, clusters of vesicles and tubules fuse to generate stacked cisternae, which then reform the Golgi. Evidence indicates that the COPI complex is largely responsible for mitotic Golgi vesicle formation (40). Proteins implicated in COPI vesicle docking undergo alterations in phosphorylation at mitosis. Cdc2-cyclin B1-mediated phosphorylation of GM130 serine 25 inhibited its p115 interaction (41) so that COPI vesicles, unable to dock with Golgi cisternae, would accumulate followed by eventual cell-cycle arrest. No evidence of GM130 phosphorylation and no alteration in the interaction GM130 and p115 were detected in GBF1-depleted cells (data not shown), suggesting that COPI dissociation from Golgi membranes or GM130 redistribution induced by GBF1 siRNA was not directly responsible for the effects described.
Cell content of Golgi-associated β-COP, GM130, p115, and golgin84 was not detectably altered after 48 h of incubation with BIG1, BIG2, or GBF1 siRNA (Fig. 4B), but GBF1 depletion did significantly increase levels of ER proteins calreticulin and PDI (Fig. 4B; SI Table 2). Protein export from the ER takes place at ER exit sites, adjacent to clusters of elaborate tubule/vesicle membranes, and only when proteins are properly folded. Perturbation of ER functions can result in accumulation of unfolded or misfolded proteins that causes ER stress and the UPR. The last is characterized by up-regulation of ER chaperones that can be critical for cell survival by facilitating correct folding and assembly of ER proteins (25).
Induction of the mammalian UPR involves, in part, enhanced transcription of at least seven genes that encode ER chaperone molecules, such as BiP/GRP78 (23), which serves to correct protein misfolding. This response is concomitant with a marked decrease in the rate of overall protein synthesis and with arrest in the G1 phase of the cell division cycle (42). We examined the possibility that cell-cycle arrest and death resulting from GBF1 depletion were caused by an initial disruption of ER function attributable to retention and accumulation of proteins in this compartment. To identify proteins that were changed in amount after depletion of GBF1, proteomic analysis was used. Five ER chaperones (BiP, ERp72, calreticulin, PDI, and ER-60) and the mitochondrial chaperone Hsp60 were significantly increased in amounts.
Early in the UPR, proteolytic cleavage of the 90-kDa form of ATF6 releases the 50-kDa N-terminal portion (p50ATF6) containing the transcription activation region that moves into the nucleus and activates expression of the ER chaperone GRP78 and other proteins involved in protein folding (27). Amounts of p50ATF6 clearly increased with time of cell exposure to GBF1 siRNA but not to BIG1 or BIG2 siRNAs (Figs. 5 and 6B). Because translocation of p50ATF6 from ER to nucleus (Fig. 6A) is presumably part of the UPR, we investigated effects of GBF1 depletion on ATF6 and its cleavage enzymes S2P, which reside in ER and cis/medial-Golgi, respectively, of unstressed cells. GBF1 siRNA treatment specifically resulted in translocation of S2P from Golgi to ER, mimicking the effect of BFA described earlier.
Elucidation of the mechanisms of these processes should tell us whether an unrecognized action(s) of GBF1, in addition to its unique function in activation of ARF for transport of newly synthesized proteins from ER exit sites to cis-Golgi, is responsible for cell-cycle arrest and death that result from its depletion or from prolonged inhibition of cells by BFA. Because BIG1 and BIG2, the only other BFA-inhibited GEFs in mammalian cells, are responsible for ARF activation at different sites of vesicular transport distal to that of introduction of new proteins to the trafficking system, their absence may not cause ER stress as quickly as does GBF1 depletion. The possibility that additional activities of the large, evolutionally conserved GBF1 molecule contribute to the drastic effects of its loss also must be considered.
Methods
Antibodies and Reagents.
Preparation and purification of antibodies against BIG1 and BIG2 are reported in refs. 13 and 43. Mouse monoclonal antibodies against GBF1, GM130, PARP, Golgin84, and BiP were purchased from BD, against ATF6 from Lifespan Biosciences, and against Hsp90β from Calbiochem. Rabbit antibodies against β-COP, GM130, and α1-antitrypsin were purchased from Abcam, p115 from Santa Cruz Biotechnology, S2P (MBTPS2) from Cell Signaling Technology, and calreticulin and PDI from Sigma. Horseradish peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG were purchased from Amersham Pharmacia.
Cell Culture.
HepG2 cells were grown (37°C, 5% CO2/95% air) on collagen I-coated 10-cm dishes (Becton Dickinson) in DMEM (GIBCO) with 10% FBS (GIBCO), penicillin (100 units/ml), and streptomycin (100 μg/ml). Cells (2 × 105) were grown for 24 h before incubation with vehicle alone (mock) or with NT, BIG1, BIG2, or GBF1 siRNA for 24, 48, or 72 h. Cell viability was assessed by 0.4% (wt/vol) Trypan blue (GIBCO) exclusion.
Cell-Cycle Analysis by Flow Cytometry.
Samples of cells (105-106) collected by centrifugation were dispersed in 0.5 ml of Vindelov's PI (10 mM Trizma base, 10 mM NaCl, 0.05 mg/ml propidium iodide, and 0.1% Nonidet P-40) containing 70 units of RNase for 2 h, before analysis using a FACScalibur flow cytometer (Becton Dickinson).
Experiments with siRNA.
siRNAs were designed and synthesized by Dharmacon Research. HepG2 cells were incubated with 100 nM siRNA or NT siRNA as negative control plus DharmaFECT 4 Transfection Reagent (Dharmacon Research) according to the manufacturer's directions for 24, 48, or 72 h before analysis. In three experiments, mean percentages of protein depletions with specific siRNA relative to control cells were 85%, 89%, and 90% for BIG1, BIG2, and GBF1, respectively, after 48 h of incubation and 95% after 72 h.
Western Blotting Analysis.
Cells collected by centrifugation after scraping were dispersed in lysis buffer (10 mM Tris·HCl, pH 8.0, 150 mM NaCl, and 1% Triton X-100) containing protease inhibitors. Proteins (30 μg) were separated by SDS/PAGE in 4–12% gel before transfer to nitrocellulose membranes for reaction with antibodies. Horseradish peroxidase-conjugated goat anti-rabbit and horse anti-mouse IgG secondary antibodies were detected by using SuperSignal Chemiluminescent substrate (Pierce Biotechnology). Densitometry was performed by using Fuji Image Gauge software (Fujifilm).
Confocal Immunofluorescence Microscopy.
Confocal immunofluorescence procedures are described in ref. 43.
2D-DIGE and MALDI TOF.
After treatment with NT or GBF1 siRNA for 48 h, cells (2 × 106) were washed three times in cold PBS and lysed in 250 μl of DIGE buffer (30 mM Tris·HCl, pH 8.5, 7 M urea, 2 M thiourea, and 4% CHAPS) containing protease inhibitors. Proteomic analysis was performed as described in ref. 44.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Christian Combs (National Heart, Lung, and Blood Institute Confocal Microscopy Core Facility), Dr. J. Philip McCoy and Ann Williams (National Heart, Lung, and Blood Institute Flow Cytometry Core Facility), and Dr. Rong-Fong Shen (National Heart, Lung, and Blood Institute Proteomics Core Facility). The Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute, supported this work.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712224105/DC1.
References
- 1.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 2.Jackson CL, Casanova JE. Turning on ARF: The Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 3.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: Insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson JD, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 5.Randazzo PA, Yang YC, Rulka C, Kahn RA. Activation of ADP-ribosylation factor by Golgi membranes: Evidence for a brefeldin A- and protease-sensitive activating factor on Golgi membranes. J Biol Chem. 1993;268:9555–9563. [PubMed] [Google Scholar]
- 6.Mansour SJ, et al. p200 ARF-GEP1: A Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc Natl Acad Sci USA. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peyroche A, et al. Brefeldin A acts to stabilize an abortive ARF–GDP–Sec7 domain protein complex: Involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 8.Morinaga N, Tsai S-C, Moss J, Vaughan M. Isolation of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP ribosylation factor (ARF) 1 and ARF3 that contains a Sec7-like domain. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaji R, et al. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc Natl Acad Sci USA. 2000;97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Lasell TK, Melancon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: Evidence for distinct functions in protein traffic. Mol Biol Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szul T, et al. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J Cell Sci. 2007;120:3929–3940. doi: 10.1242/jcs.010769. [DOI] [PubMed] [Google Scholar]
- 12.Claude A, et al. GBF 1, a novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamoto K, et al. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic. 2002;3:483–495. doi: 10.1034/j.1600-0854.2002.30705.x. [DOI] [PubMed] [Google Scholar]
- 14.Niu T-K, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol Biol Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szul T, et al. Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic. 2005;6:374–385. doi: 10.1111/j.1600-0854.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- 16.Grebe M, et al. A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, et al. GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J Cell Sci. 2006;119:3743–3753. doi: 10.1242/jcs.03173. [DOI] [PubMed] [Google Scholar]
- 18.García-Mata R, Szul T, Alvarez C, Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol Biol Cell. 2003;14:2250–2261. doi: 10.1091/mbc.E02-11-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Hendershot LM. ER chaperone functions during normal and stress conditions. J Chem Neuroanat. 2004;28:51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Schrag JD, Procopio DO, Cygler M, Thomas DY, Bergeron JJ. Lectin control of protein folding and sorting in the secretory pathway. Trends Biochem Sci. 2003;28:49–57. doi: 10.1016/s0968-0004(02)00004-x. [DOI] [PubMed] [Google Scholar]
- 21.Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 22.Bulleid NJ, Freedman RB. Defective co-translational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes. Nature. 1988;335:649–651. doi: 10.1038/335649a0. [DOI] [PubMed] [Google Scholar]
- 23.Spee P, Subjeck J, Neefjes J. Identification of novel peptide binding proteins in the endoplasmic reticulum: ERp72, calnexin, and grp170. Biochemistry. 1999;38:10559–10566. doi: 10.1021/bi990321r. [DOI] [PubMed] [Google Scholar]
- 24.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 26.Kokame K, Kato H, Miyata T. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J Biol Chem. 2001;276:9199–9205. doi: 10.1074/jbc.M010486200. [DOI] [PubMed] [Google Scholar]
- 27.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa T, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Gao X. Neuronal apoptosis induced by endoplasmic reticulum stress. Neurochem Res. 2002;27:891–898. doi: 10.1023/a:1020387414086. [DOI] [PubMed] [Google Scholar]
- 31.Claude A, Zhao BP, Melançon P. Characterization of alternatively spliced and truncated forms of the Arf guanine nucleotide exchange factor GBF1 defines regions important for activity. Biochem Biophys Res Commun. 2003;303:160–169. doi: 10.1016/s0006-291x(03)00316-4. [DOI] [PubMed] [Google Scholar]
- 32.Strous GJ, van Kerkhof P, van Meer G, Rijnboutt S, Stoorvogel W. Differential effects of brefeldin A on transport of secretory and lysosomal proteins. J Biol Chem. 1993;268:2341–2347. [PubMed] [Google Scholar]
- 33.Shen J, Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J Biol Chem. 2004;279:43046–43051. doi: 10.1074/jbc.M408466200. [DOI] [PubMed] [Google Scholar]
- 34.Pfeffer SR. GTP-binding proteins in intracellular transport. Trends Cell Biol. 1992;2:41–45. doi: 10.1016/0962-8924(92)90161-f. [DOI] [PubMed] [Google Scholar]
- 35.Manolea F, Claude A, Chun J, Rosas J, Melançon P. Distinct functions for Arf nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and TGN, respectively. Mol Biol Cell. 2007 Nov 14; doi: 10.1091/mbc.E07–04-0394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez C, Fujita H, Hubbard A, Sztul E. ER to Golgi transport: Requirement for p115 at a pre-Golgi VTC stage. J Cell Biol. 1999;147:1205–1222. doi: 10.1083/jcb.147.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sönnichsen B, et al. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shorter J, Warren G. A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García-Mata R, Sztul E. The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucleotide-exchange factor. EMBO Rep. 2003;4:320–325. doi: 10.1038/sj.embor.embor762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misteli T, Warren G. COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in cell-free system. J Cell Biol. 1994;125:269–282. doi: 10.1083/jcb.125.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowe M, et al. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 42.Brewer JW, Hendershot LM, Sherr CJ, Diehl JA. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc Natl Acad Sci USA. 1999;96:8505–8510. doi: 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu K-F, Shen X, Li H, Pacheco-Rodriguez G, Moss J, Vaughan M. Interaction of BIG2, a brefeldin A-inhibited guanine nucleotide-exchange protein, with exocyst protein Exo70. Proc Natl Acad Sci USA. 2005;102:2784–2789. doi: 10.1073/pnas.0409871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schieke SM, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.