Abstract
Galileo is the only transposable element (TE) known to have generated natural chromosomal inversions in the genus Drosophila. It was discovered in Drosophila buzzatii and classified as a Foldback-like element because of its long, internally repetitive, terminal inverted repeats (TIRs) and lack of coding capacity. Here, we characterized a seemingly complete copy of Galileo from the D. buzzatii genome. It is 5,406 bp long, possesses 1,229-bp TIRs, and encodes a 912-aa transposase similar to those of the Drosophila melanogaster 1360 (Hoppel) and P elements. We also searched the recently available genome sequences of 12 Drosophila species for elements similar to Dbuz\Galileo by using bioinformatic tools. Galileo was found in six species (ananassae, willistoni, peudoobscura, persimilis, virilis, and mojavensis) from the two main lineages within the Drosophila genus. Our observations place Galileo within the P superfamily of cut-and-paste transposons and extend considerably its phylogenetic distribution. The interspecific distribution of Galileo indicates an ancient presence in the genus, but the phylogenetic tree built with the transposase amino acid sequences contrasts significantly with that of the species, indicating lineage sorting and/or horizontal transfer events. Our results also suggest that Foldback-like elements such as Galileo may evolve from DNA-based transposon ancestors by loss of the transposase gene and disproportionate elongation of TIRs.
Keywords: class II elements, transposase, terminal inverted repeats, 1360, inversions
Transposable elements (TEs) are intracellular parasites that populate most eukaryotic genomes and have a huge impact on their evolution (1). Their abundance and diversity are astonishing and a considerable effort is needed to put order in the increasing constellation of families being discovered. So far, two main classes are widely recognized, retrotransposons that transpose by an intermediate RNA molecule and transposons that move by using a single- or double-stranded DNA intermediate (2). Three subclasses of transposons have been defined based on the transposition mechanism: cut-and-paste, rolling-circle, and Mavericks (3). Cut-and-paste transposons possess TIRs, usually short, and encode a protein called transposase (TPase) that catalyzes their excision from the original location in the genome and promotes their reinsertion into a new site generating target site duplications (TSDs) in the process (4). The Drosophila elements P (5) and mariner (6) are among the best known families of cut-and-paste transposons but there are many more families classified in ten transposon superfamilies on the basis of similarity among the TPases: Tc1/mariner, hAT, P, MuDR, CACTA, PiggyBac, PIF/Harbinger, Merlin, Transib, and Banshee (3). Other elements are still unclassified, seemingly because only defective copies have been found. Defective (nonautonomous) copies coexist and often outnumber the canonical (autonomous) copies, and can move if there is a functional TPase provided by canonical copies present somewhere else in the same genome and if they conserve the signals required for TPase recognition (usually the TIR ends).
Foldback-like elements constitute a group of poorly known TEs with uncertain classification (2, 3). They take their name from the Foldback (FB) element of Drosophila melanogaster (7, 8) and are present in a diverse array of organisms (9–13). The unusual characteristics of Foldback-like elements include very long TIRs that make up almost the entire element and are separated by a middle domain with variable length and composition. No coding capacity has been found in many Foldback-like elements, and thus, their mechanism of transposition is uncertain. However, a small proportion (≈10%) of FB copies in D. melanogaster is associated with a 4-kb-long sequence called NOF encoding a 120-kDa protein of unknown function (14, 15). FB has been recently included in the MuDR superfamily (3) because of the similarity of the proteins encoded by both MuDR and NOF to that of Phantom, a transposon from Entamoeba (16). Besides, some copies of FARE, another Foldback-like transposon from Arabidopsis, harbor a large ORF with weak similarity to the MuDR TPase (13). The origin of many other Foldback-like elements is still uncertain.
Galileo was discovered in Drosophila buzzatii and is the only TE in the genus Drosophila that has been shown to have generated chromosomal inversions in nature (17–19). Other TEs, such as P, Hobo, or FB are known to induce chromosomal rearrangements in experimental populations of D. melanogaster (20), but there is no direct evidence of their implication in Drosophila chromosomal evolution. Galileo, together with two closely related elements, Kepler and Newton, were classified as Foldback-like elements because of their long, internally repetitive TIRs (18, 21). All copies of Galileo, Kepler, and Newton isolated so far from the genome of D. buzzatii lack any significant protein-coding capacity except for two Galileo copies bearing a short segment with weak similarity to the TPase of element 1360 (Hoppel) (21). An experimental search for Galileo sequences in other Drosophila species suggested that this TE has a rather restricted distribution, being only present in the closest relatives of D. buzzatii but not in more distantly related species within the repleta group (21). Here, we take advantage of the recently sequenced genomes of D. melanogaster (22), Drosophila pseudoobscura (23), and ten additional Drosophila species (24) to search for sequences similar to Galileo in these genomes by using bioinformatic tools. We found that Galileo has a much wider species distribution within the Drosophila genus than previously suspected. Furthermore, our results allow us to fully characterize the element Galileo and to classify it as a member of the P superfamily of cut-and-paste DNA transposons.
Results
Structure of Galileo in D. buzzatii.
By using as a query Galileo-3, a defective copy of Dbuz\Galileo (21), we carried out preliminary bioinformatic searches in the genome sequence of Drosophila mojavensis, another member of the repleta species group. Some of the hits, on close examination, bounded a protein-coding segment that might be the Galileo TPase. Several PCRs were then attempted to isolate longer Galileo copies from the D. buzzatii genome (see Methods). In each of them, one primer was anchored in the known Dbuz\Galileo TIRs and the other in the possible Dmoj\Galileo TPase. A putatively complete copy of Dbuz\Galileo could be assembled in this way (Fig. 1A). This copy is 5,406 bp long, possesses 1,229-bp TIRs and an intronless 2,738-bp ORF (nt 1348–4087) encoding a 912-aa protein (after fixing two STOP codons, and a 1-bp deletion that causes a frameshift mutation).
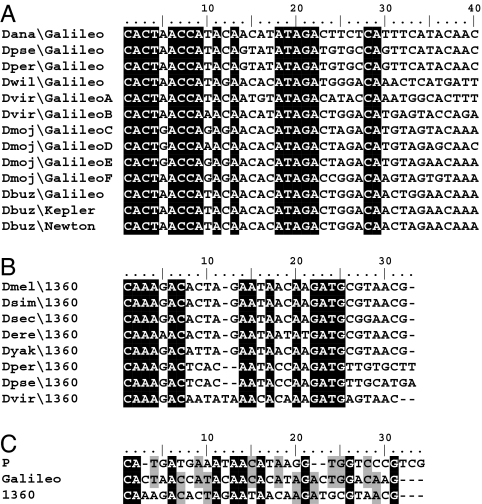
Fig. 1.
Most complete copies of Galileo and 1360 found in this work. (A) Putative complete Galileo copy from the D. buzzatii genome. (B) Most complete copies of Galileo found in the 12 sequenced genomes. (C) Most complete copies of 1360. TIRs are represented as arrows and TPases are represented as gray rectangles. The direct repeats of the TIRs in Dbuz\Galileo are indicated by striped patterns. Dmoj\Galileo internal inverted repeats are represented as little triangles. In D. mojavensis two Galileo copies representative of two subfamilies found in this species are depicted. See SI Table 4 for details.
A search using BLASTX revealed significant similarity of the Dbuz\Galileo TPase to those of the related D. melanogaster 1360 and P elements (25, 26) [AAN39288, E-value = 1e−95; Q7M3K2, E-value = 3e−25]. The Dbuz\Galileo TPase includes a THAP domain near the N terminus (amino acids 27–104) similar to the DNA binding domain of P element TPase (27–30). A copy of 1360 located in chromosome 4 of D. melanogaster (31) encodes a TPase (854 aa) longer than that in the National Center for Biotechnology Information database (25), including a THAP domain near the N terminus (after curation of a 1-bp frameshift mutation). A global alignment of the Dbuz\Galileo TPase with those of Dmel\1360 and Dmel\P yielded 34.5% and 27.6% identity, respectively. No significant similarity was found between the Dbuz\Galileo TPase and the proteins encoded by Dmel\FB (14, 15).
Distribution of Galileo and 1360 in the 12 Sequenced Drosophila Genomes.
Systematic bioinformatic searches using as queries the TPases and TIRs of Dbuz\Galileo and Dmel\1360 were carried out (see Methods). The results [supporting information (SI) Tables 1–3] suggested that elements similar to Galileo are present in D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. virilis, and D. mojavensis, whereas elements similar to 1360 are present in the five melanogaster subgroup species (melanogaster, simulans, sechellia, yakuba, and erecta) plus D. pseudoobscura, D. persimilis, and D. virilis. Therefore, none of the two TEs is seemingly present in D. grimshawi but both are found in D. pseudoobscura, D. persimilis, and D. virilis.
Characterization of Galileo Copies.
We characterized 46 relatively long copies of Galileo containing segments encoding a partial or full TPase from the six genomes where this TE is present (SI Table 4). All of them possess one or two long TIRs with similarity to those of Dbuz\Galileo (see below) and nine are flanked by perfect 7-bp TSDs. The structure of the longest, presumably most complete, copy in each species is depicted in Fig. 1B. These Galileo copies are 4,386 bp (D. willistoni) to 5,989 bp long (D. mojavensis) and exhibit TIRs of 684 bp (D. ananassae) to 813 bp (D. mojavensis). However, none of them contains a single ORF encoding a fully functional TPase (all bear STOP codons, frameshift mutations, and/or deletions). In D. mojavensis 16 long copies were characterized. Many of them include nearly complete TPase-coding segments and all but three contain one or more insertions of other TEs (SI Table 4). These 16 copies belong to two groups with distinctive structures (see Fig. 1B for representative copies) and encoding somewhat different TPases (see below).
We also searched each of the six Drosophila genomes for short nonautonomous Galileo copies by using BLASTN and the most complete copy already found in the same genome (Fig. 1B) as query (see Methods). Galileo was rather abundant in the six genomes, the number of significant hits being >100 in all cases with a maximum of 495 in D. willistoni (SI Table 1). We identified and isolated 109 Galileo copies from the contigs producing significant hits in the six species. All of them possess two long TIRs separated by a relatively short middle segment and 97 show perfect 7-bp TSDs (SI Table 5). Thus, these copies are structurally similar to the copies of Galileo, Kepler, and Newton previously found in D. buzzatii (21). A summary of the characteristics of these relatively short nonautonomous copies is given in SI Table 6.
TSDs.
In D. buzzatii, Galileo generates on insertion 7-bp TSDs with the consensus GTAGTAC (21). Likewise, in the six Drosophila genomes analyzed here, 106 Galileo copies were flanked by identical 7-bp sequences (SI Tables 4 and 5). We calculated the frequency of the four nucleotides in each of the seven sites for each species separately. The frequency pattern observed in the six species was similar to that of Dbuz\Galileo and the 106 sequences were combined. All positions but the fourth show a significant departure from randomness, and the consensus is the palindrome GTANTAC.
Divergence Between Galileo Copies.
To estimate the time since the most recent transpositional activity of Galileo, we measured the average pairwise divergence between the short nonautonomous copies within each species (see Methods and SI Table 6). In D. ananassae, the average pairwise divergence among 20 copies was 2.8%, which implies a divergence time of ≈1.8 myr. However, evidence for more recent transpositional events was found because a subgroup of 13 copies shows an average divergence of 0.36% equivalent to a divergence time of only 0.225 myr. Similar observations were made in D. pseudoobscura, D. persimilis, and D. willistoni (SI Table 6). In each case, subgroups with ≈1% average divergence (implying divergence times ≈0.6 myr) were found. In D. virilis, analysis of 13 short nonautonomous copies uncovered two highly divergent groups that we named A and B (SI Fig. 5). Copies within each group were aligned and analyzed separately (SI Table 6). The average pairwise divergence within groups A and B was 4.6 and 5.7%, implying divergence times of 2.9 and 3.6 myr, respectively. Inclusion in the analysis of the longest copy found in the species (contig 16409) indicated unequivocally that it is a member of group A (SI Fig. 5). In D. mojavensis, analysis of 20 short nonautonomous copies revealed the presence of four well defined groups, here named C–F. We included in the analysis nine of the long copies containing the two TIRs and generated a phylogenetic tree with the 29 copies (Fig. 2). Groups C and D correspond to the two groups previously detected when the long, nearly complete, copies were analyzed. Copies within each group were separately aligned and analyzed. Average pairwise divergences within groups C through F were 2.2%, 2.3%, 2.4%, and 8.9%, respectively, indicating divergence times ranging from 1.4 to 5.5 myr (SI Table 6). The two and four Galileo groups or subfamilies found in D. virilis and D. mojavensis, respectively, seemingly represent relatively old tranposition bursts in these genomes. We suggest that the Newton and Kepler elements previously found in the D. buzzatii genome (18, 21) should likewise be considered only as different groups or subfamilies of Galileo in this species.
Fig. 2.
Neighbor-joining phylogenetic tree inferred from the analysis of 29 Galileo copies found in the D. mojavensis genome. The two TIRs of each copy were included in the tree as separate sequences to allow their comparison within and between copies. TIRa is the TIR located at 5′ from the TPase or the first TIR that appears in the contig if the copy could not be oriented. The complete deletion option was used leaving 269 informative sites. Bootstrap values at main nodes are shown. The average pairwise divergence between groups D and E is ≈25%, indicating a divergence time of ≈8 myr, and the average pairwise divergence between these two groups and groups C and F is ≈32%, implying a divergence time of ≈10 myr. The putative chimeric elements with highly divergent TIRs are marked with an arrow. Details of these Galileo copies are given in SI Tables 4 and 5.
One copy in D. pseudoobscura (contig 4355), one copy in D. willistoni (contig 10422), and three copies in D. mojavensis (contigs 11233, 10770.1, and 9832) are likely chimeric because they are flanked by dissimilar 7-bp sequences and show increased levels of divergence between the two TIRs (see for instance Fig. 2).
Characterization of 1360 Copies.
The longest and complete or nearly complete copies of element 1360 found in the eight genomes are shown in Fig. 1C (see also SI Table 7). The eight copies possess TPase-coding segments 2,428 bp (D. erecta) to 2,565 bp long (D. melanogaster), although only D. yakuba includes three different copies with 2,562-bp ORFs encoding a fully functional TPase. All of them bear 31- or 32-bp-long TIRs and total size for seemingly complete copies varies between 2,985 bp (D. persimilis) and 4,702 bp (D. virilis). The longest copies found in each species (Fig. 1C) were used as queries to interrogate the eight genomes by using BLASTN. The results showed that 1360 is very abundant in all genomes with a maximum number of 690 significant hits in D. sechellia (SI Table 1).
Comparison of Galileo, 1360, and P Element TIRs.
With the exception of D. pseudoobscura and D. persimilis, the long Galileo TIRs show little similarity between the different species either in length or sequence composition. Conservation seems to be restricted to the terminus as revealed by the alignment of the first 40 bp of Galileo in D. buzzatii (including Kepler and Newton) and the six species analyzed here (including D. virilis groups A and B and D. mojavensis groups C–F). A total of 17 of the 40 terminal bp are conserved in the 13 sequences (Fig. 3A). Likewise, alignment of the 31 bp of 1360 TIRs in the longest copies described earlier (Fig. 1C) revealed 14 conserved bp (Fig. 3B). We generated the consensus sequences of the element terminus in Galileo and 1360 in the different species. Fifteen of 31 bp are identical, which provides further evidence of the evolutionary relationship between both TEs. In addition, the consensus Galileo terminus shares 17 bp with the 31-bp TIRs of Dmel\P (Fig. 3C).
Fig. 3.
Comparison of TIR ends. (A) Alignment of 40 bp of the TIR end of Galileo. A consensus sequence was constructed for Galileo TIRs in each TE subfamily and species. (B) Alignment of the 31-bp TIR of 1360. A representative TIR from a single copy of the TE is included. (C) Comparison of the Galileo TIR end with the TIRs of elements 1360 and P. Identical positions in all sequences are shown in black. Sites identical between Galileo and 1360 or P are shown in gray.
Comparison of Galileo, 1360, and P Element TPases.
We generated consensus amino acid sequences for the Galileo and 1360 TPases within each species (see Methods). For Dmoj\Galileo, the consensus sequences of the TPases encoded by copies in groups C and D are 937 and 936 aa long, respectively, and when aligned alone show a 87.2% identity and a 96.4% similarity.
A multiple alignment of the eight consensus Galileo TPases, the eight consensus 1360 TPases, and five TPases of representative P elements was carried out (SI Fig. 6). Besides, the human P-like THAP9 protein (32) was included in the analysis as outgroup. The Galileo TPases are 30–35% identical to those of 1360 and 20–25% identical to those of P elements (SI Table 8). Within the Galileo TPases, identity varies between 97.2% in the closely related pair D. pseudoobscura–D. persimilis, and 39.3% between D. persimilis and D. virilis. In addition, we examined the multiple alignment for conservation of several functional domains and motifs that have been identified in the Dmel\P TPase (5). The THAP domain is a zinc-dependent DNA binding domain evolutionarily conserved in an array of different proteins including the P TPase, cell-cycle regulators, proapoptotic factors, transcriptional repressors, and chromatin-associated proteins (28–30). It includes a metal-coordinating C2CH signature plus four other residues (P, W, F, and P) that are also required for DNA binding. These eight residues are fully conserved (with one exception) in positions C29, C34, P53, W63, C89, H92, F93, and P119 of the multiple alignment (SI Fig. 6). A leucine zipper coiled-coil motif involved in protein dimerization is located after the DNA binding domain (5). We predicted in silico a similar 22-aa-long coiled-coil motif after the THAP domain in the Galileo and 1360 TPases (SI Fig. 6). Finally, although the Dmel\P TPase does not contain the characteristic catalytic motif DD(35)E shared by many other TPases and integrases (4), the C-terminal portion of this protein contains numerous aspartic (D) or glutamic (E) residues and four of them seem to be critic for TPase function: D(83)D(2)E(13)D (see ref. 5). The first 3 aa are fully conserved in positions D677, D774, and E777 of the multiple alignment with one exception (SI Fig. 6), thus supporting this model (5). The conservation of the fourth amino acid is unclear.
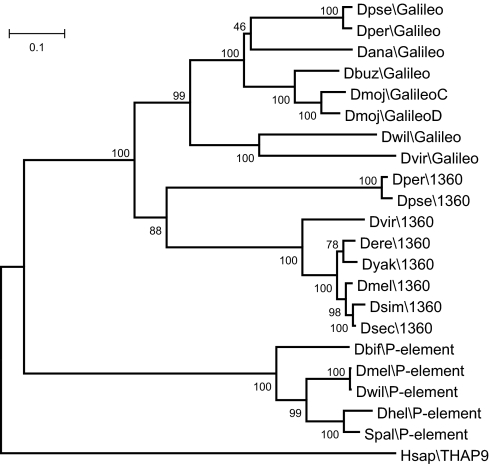
A phylogenetic tree was generated with the 21 Galileo, 1360, and P TPases and the human THAP9 protein (see Methods). The tree (Fig. 4) shows three clades corresponding to the Galileo, 1360, and P elements. Therefore, the three TEs seem monophyletic, although only the Galileo and P clades have very high statistical support. Galileo and 1360 are more closely related to each other than to the P element, which is connected to the other two by a deeper branch.
Fig. 4.
Neighbor-joining phylogenetic tree constructed with the eight consensus Galileo TPases, eight consensus 1360 TPases, and five TPases from representative P elements. The human P-like THAP9 protein is included as an outgroup. The complete alignment without Gblocks filtering is shown in SI Fig. 6. The tree topology was identical when using maximum likelihood and parsimony methods.
Discussion
We characterized a seemingly complete copy of Galileo from the genome of D. buzzatii that contains a 2,738-bp ORF encoding a TPase. Three observations indicate that this is the true Galileo TPase instead of that of another TE accidentally associated with the long Galileo TIRs. (i) Two previously isolated Galileo copies bear a 141-bp portion of the same ORF in the right position and orientation (21), suggesting that all previously isolated Galileo copies are defective versions of the complete structure reported here. (ii) Our bioinformatic searches uncovered TEs structurally similar to Galileo in the genomes of six phylogenetically distant Drosophila species. These searches were carried out by using as queries the Dbuz\Galileo and Dmel\1360 TPases, and a careful scrutiny of the contigs producing significant hits led to the finding of the TIRs associated with the TPase segment and the characterization of the elements as either Galileo or 1360. No other TIRs besides those of these two TEs were found flanking the hits (but note that in Dmoj\Galileo 160-bp internal inverted repeats bound the TPase; Fig. 1B). The persistent association (over tens of myr) of this TPase with the same type of TIRs renders the possibility of an accidental association extremely unlikely. (iii) The presence of multiple Galileo copies comprising both TIRs and TPase-coding segments in seven Drosophila genomes suggests that these are integral components of the same elements, and these elements are (or have been) able to replicate and transpose within these genomes.
Further evidence leads us to infer that Galileo, previously considered a Foldback-like element, is in fact a transposon related to the D. melanogaster 1360 and P elements, and thus, it is probably a TE moving by a cut-and-paste reaction (3, 4). (iv) The Galileo TPase is 30–35% and 20–25% identical to those of 1360 and P elements, respectively, and the three proteins harbor similar functional domains such as a DNA binding THAP domain, a coiled-coil motif for protein dimerization, and a catalytic domain (5, 27–30). (v) Despite their dramatically different size (several hundred base pairs vs. 31 bp), the Galileo terminus includes sequences clearly related to the 1360 and P TIRs. Specifically, the consensus Galileo terminus shares 15 bp with the 1360 consensus TIR and 17 bp with the Dmel\P TIR. The three elements share identical 5′-CA…TG-3′ termini. (vi) Both Galileo and 1360 generate on insertion 7-bp TSDs that, in the case of Galileo, match the consensus sequence GTANTAC, a palindrome. The TSDs of Dmel\P are 8 bp long and the consensus also corresponds to a palindrome, GTCCGGAC, a fact related to the dimerization of the P TPase (5). This suggests that the functional Galileo TPase is also a dimer. We conclude that Galileo belongs to the P superfamily of cut-and-paste transposons.
A parsimonious interpretation of the phylogenetic tree relating Galileo with the 1360 and P elements (Fig. 4) suggests that Galileo arose from an ancestor with much shorter TIRs. Galileo long TIRs are variable in size both between and within species, suggesting a remarkable structural dynamism. For instance, in D. willistoni, the longest and putatively complete copy (contig 10048) has 765-bp TIRs, but another copy (contig 9452) has 959-bp-long TIRs. Similarly, TIRs of Galileo copies in D. mojavensis are 458 bp (contig 10940) to 1,260 bp (contig 10757.2) long. TIRs may accidentally shorten (e.g., by deletion) but very likely they may also be elongated by internal duplication, unequal recombination, and/or other mechanisms, such as long-tract gene conversion (33) or single-strand break and synthesis-repair (see figure 5B in ref. 34). We suggest that different Foldback-like elements might have originated from independent transposon lineages in a similar manner as the Drosophila element Galileo. In other words, TIR length and structure is not a reliable criterion for TE classification, and Foldback-like elements do not constitute a monophyletic group.
The phylogeny of the Galileo elements in the seven Drosophila species (Fig. 4) is clearly inconsistent with that of the species (cf. figure 1 in ref. 24). The elements of D. willistoni and D. virilis, pertaining to different subgenera (Sophophora and Drosophila, respectively) are each other's closest relative. Similarly, the Galileo elements of D. mojavensis and D. buzzatii (Drosophila subgenus) are more closely related to those of D. ananassae, D. pseudoobscura, and D. persimilis (Sophophora subgenus) than to those of D. virilis, a species from the same subgenus. Equally inconsistent with the species relationships is the phylogeny of the 1360 element (Fig. 4). There are two possible explanations for these topological disparities: lineage sorting and horizontal transfer (35). Lineage sorting refers to the vertical diversification of TE lineages and their differential loss along the branches of the species tree. Horizontal transfer is the process of invasion of a new genome by a TE, which is common for transposons and is considered as an integral phase of the transposon life cycle that allows long-term survival (6, 36). The strongest evidence for horizontal transfer is probably the detection of elements with a high degree of similarity in very divergent taxa, such as in the P element colonization of the D. melanogaster genome within the last century from the distantly related species D. willistoni (37). Many more events of horizontal transfer have occurred during the evolution of P elements in the genus Drosophila based on the available evidence (38). However, despite their close evolutionary relationship to P, the available evidence for horizontal transfer in Galileo and 1360 (Fig. 4) is not compelling and lineage sorting should be considered, at this time, as an equally likely explanation.
The origin of the numerous chromosomal inversions in Drosophila and other Dipterans is still an open question and very few species have been investigated in this regard. Strong evidence implicating TE-mediated ectopic exchange has been found in four polymorphic inversions only, including the two D. buzzatii inversions generated by Galileo (39). In D. melanogaster and its close relatives, no TEs have been involved in the origin of three polymorphic inversions and only 2 of 29 fixed inversions contain repetitive sequences inverted with respect to each other at both breakpoints, pointing to a completely different mechanism for inversion generation (39). The fact that Galileo generated two independent inversions in D. buzzatii suggests that Galileo is not a passive substrate where ectopic recombination operates but may be actively generating inversions as a byproduct of its transposition mechanism. If this is correct, to create inversions, Galileo has to be active in a genome and a recent transpositional activity would be a necessary condition for Galileo to have any role in the generation of current inversions. We have not found any functional TPase in any of these species but only one genome was sequenced in each case, so they could still exist in unsequenced genomic regions, other genomes, and/or other natural populations. However, we have provided evidence of recent (<1 myr) transpositional activity of Galileo in D. ananassae, D. persimilis, D. pseudoobscura, and D. willistoni. These four are among the most polymorphic species of the genus with 24, 28, 13, and 50 inversions, respectively (40). In D. mojavensis, with fewer inversions (41), the most recent transpositional activity of Galileo seems somewhat older (≈1.5 myr). Finally, D. virilis with the oldest Galileo activity (≈3 myr) is chromosomally monomorphic (40). Therefore, there is a qualitative correlation between the number of inversions and the time of the most recent activity of Galileo in this small group of species. This correlation is suggestive but might be only coincidental. However, the detection of chimerical copies that may be the result of chromosomal rearrangements (19) indicates that, indeed, Galileo might have been involved in the origin of inversions, at least in some other species besides D. buzzatii.
Methods
PCR Amplification and DNA Sequencing.
Genomic DNA from D. buzzatii (strain st-1) and D. mojavensis (strain 15081-1352.22, Tucson Drosophila Stock Center) (as control) was used as template for PCR amplification of Galileo copies. Primers located in the TIRs were designed based on D. buzzatii known incomplete copies of Galileo (21), whereas primers inside the TPase were designed on the D. mojavensis putative complete TPases found in a preliminary bioinformatic search (SI Fig. 7). Primers in the TIRs were always used in combination with primers anchored in the TPase to avoid multiple bands generated by the highly repetitive primer alone or the amplification of defective copies without TPase. PCRs were carried out in a total volume of 25 μl including 100–200 ng of genomic DNA, 20 pmol of each primer, 200 μM dNTPs, 1.5 mM MgCl2, and 1–1.5 units of Taq DNA polymerase. PCR products were gel-purified by using QIAquick Gel Extraction kit (Qiagen) and sequenced directly with the amplification primers and sequencing primers designed over the end sequences to close gaps (SI Fig. 7). Sequences were aligned and assembled by using multialign software MUSCLE 3.6 (42).
Bioinformatic Searches.
BLAST searches were performed on the chromosome assemblies of D. melanogaster and D. simulans and the contig CAF1 assemblies of the other ten publicly available Drosophila genomes (http://rana.lbl.gov/drosophila). We used BLAST algorithm version 2.2.2 (43) implemented in the Drosophila Polymorphism Database server (http://bioinformatica.uab.es/dpdb) with default parameters. TBLASTN searches in the different species were performed by using as queries the TPases of Dbuz\Galileo and Dmel\1360 (SI Table 1). Hits with an E-value ≤ 10−20 (which in the conditions of our searches amounts roughly to ≈30% identity over a stretch of 200 aa) were considered significant. BLASTN searches were also carried out with the 40 terminal bp of Dbuz\Galileo and the 31 bp of the Dmel\1360 TIR (SI Table 1). The cutoff in this case was an E-value ≤ 10−3 (that requires ≈21–22 consecutive identical base pairs).
Contigs producing significant hits with the Dbuz\Galileo and Dmel\1360 TPases in each species were scrutinized to characterize the different copies of both TEs. TIRs and TSDs were searched around the putative TPases by using Dotlet 1.5 (44) to define the boundaries of each copy. Insertions of other TEs inside Galileo were identified by aligning the different Galileo copies found in the same species and further analyzing the sequences present in only one of them. Significant contigs <1 kb long and those that were found to contain complex clusters of several TE insertions (likely of heterochromatic origin) were not further investigated.
Nonautonomous Copies.
BLASTN searches were carried out with the longest copies of Galileo and 1360 (Fig. 1 B and C) to estimate the abundance of the two TEs within each species (SI Table 1). Significant hits were those with E-value ≤ 10−20 (equivalent to ≈80% identity over a stretch of 200 bp). The number of significant contigs in these searches provides usually a minimum estimate for the number of TE copies because the searched databases were the CAF1 contig assemblies in most cases and each contig contains at least one copy but may actually contain two or more. For similarity analyses, only the TIRs were used as they produced the most reliable alignments. The two TIRs of each TE copy were analyzed separately to estimate the divergence between the two TIRs within each copy as well as the pairwise divergence between copies.
Consensus Sequences.
The consensus sequences for Galileo and 1360 TPases and Galileo TIRs were generated by using BioEdit 7.0.5 (45) after aligning the respective nucleotide sequences (SI Table 9) with MUSCLE 3.6 software (42). In the case of TPases, this consensus sequence was then translated into protein to allow the comparison among different species (SI Fig. 6). Conserved protein domains were detected by using InterProScan (46) and Conserved Domain Search (47). Coiled-coil regions were predicted by using the Coils server (48).
Phylogenetic Analyses.
TPase sequences were aligned with MUSCLE 3.6 (42) and the alignment was filtered with Gblocks version 0.91b (49) to remove the poorly aligned and highly divergent segments. Gblocks was used with the default parameters except for the maximum number of contiguous nonconserved positions = 15, the minimum length of a block = 6, and allowed gap position = half. These parameters were fixed so that the conserved THAP domain was included in the filtered alignment. All phylogenetic trees were constructed with MEGA 3.1 (50) by using the neighbor-joining method with complete deletion and 500 replicates to generate bootstrap values. Poisson correction and Kimura 2 parameters were used as substitution models for amino acid and nucleotide sequences, respectively. We dated the most recent transposition events within each species by dividing the average pairwise divergence between the elements in the same group or subgroup by the Drosophila synonymous substitution rate, 0.016 substitutions per nucleotide/myr (21). To date the divergence between different groups or subfamilies we calibrated the tree with the same substitution rate by using the appropriate option in MEGA (50). Time estimates for TEs should be taken with caution; if the synonymous substitution rate were an underestimate of the true mutation rate for TEs, our time estimates would provide an upper bound for the true values.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Margaret Kidwell, Cedric Feschotte, Dmitri Petrov, Mario Cáceres, Josefa González, and two anonymous referees for many constructive comments and Diana Garzón for help with the initial bioinformatic searches in D. mojavensis. This work was completed while A.R. was on sabbatical leave at Stanford University; he thanks Dmitri Petrov, Josefa González, James Cai, Yael Salzman, Ruth Hershberg, and Mike Macpherson for their warm hospitality and personal help. This work was supported by a Formación de Personal Investigador doctoral fellowship (to M.M.) and Secretaría de Estado de Universidades e Investigación (Ministerio de Educación y Ciencia, Spain) Grant BFU2005-022379 and mobility grant PR2006-0329 (to A.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Nucleotide sequences reported in this paper have been deposited in the DDBJ/EMBL/GenBank databases [accession nos. EU334682–EU334685 and BK006357–BK006363 (TPA section)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0712110105/DC1.
References
- 1.Kidwell MG, Lisch DR. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: American Society for Microbiology; 2002. pp. 59–89. [Google Scholar]
- 2.Capy P, Bazin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Heidelberg: Springer; 1998. [Google Scholar]
- 3.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haren L, Ton-Hoang B, Chandler M. Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 5.Rio DC. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: American Society for Microbiology; 2002. pp. 484–518. [Google Scholar]
- 6.Hartl DL, Lohe AR, Lozovskaya ER. Modern thoughts on an ancient marinere: Function, evolution, regulation. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 7.Potter S, Truett M, Phillips M, Maher A. Eucaryotic transposable genetic elements with inverted terminal repeats. Cell. 1980;20:639–647. doi: 10.1016/0092-8674(80)90310-4. [DOI] [PubMed] [Google Scholar]
- 8.Truett MA, Jones RS, Potter SS. Unusual structure of the FB family of transposable elements in Drosophila. Cell. 1981;24:753–763. doi: 10.1016/0092-8674(81)90101-x. [DOI] [PubMed] [Google Scholar]
- 9.Liebermann D, et al. An unusual transposon with long terminal inverted repeats in the sea urchin Strongylocentrotus purpuratus. Nature. 1983;306:342–347. doi: 10.1038/306342a0. [DOI] [PubMed] [Google Scholar]
- 10.Rebatchouk D, Narita JO. Foldback transposable elements in plants. Plant Mol Biol. 1997;34:831–835. doi: 10.1023/a:1005855008823. [DOI] [PubMed] [Google Scholar]
- 11.Ade J, Belzile FJ. Hairpin elements, the first family of foldback transposons (FTs) in Arabidopsis thaliana. Plant J. 1999;19:591–597. doi: 10.1046/j.1365-313x.1999.00567.x. [DOI] [PubMed] [Google Scholar]
- 12.Simmen MW, Bird A. Sequence analysis of transposable elements in the sea squirt, Ciona intestinalis. Mol Biol Evol. 2000;17:1685–1694. doi: 10.1093/oxfordjournals.molbev.a026267. [DOI] [PubMed] [Google Scholar]
- 13.Windsor AJ, Waddell CS. FARE, a new family of foldback transposons in Arabidopsis. Genetics. 2000;156:1983–1995. doi: 10.1093/genetics/156.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Templeton NS, Potter SS. Complete foldback transposable elements encode a novel protein found in Drosophila melanogaster. EMBO J. 1989;8:1887–1894. doi: 10.1002/j.1460-2075.1989.tb03585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harden N, Ashburner M. Characterization of the FB-NOF transposable element of Drosophila melanogaster. Genetics. 1990;126:387–400. doi: 10.1093/genetics/126.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritham EJ, Feschotte C, Wessler SR. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol Biol Evol. 2005;22:1751–1763. doi: 10.1093/molbev/msi169. [DOI] [PubMed] [Google Scholar]
- 17.Cáceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- 18.Cáceres M, Puig M, Ruiz A. Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res. 2001;11:1353–1364. doi: 10.1101/gr.174001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casals F, Caceres M, Ruiz A. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol Biol Evol. 2003;20:674–685. doi: 10.1093/molbev/msg070. [DOI] [PubMed] [Google Scholar]
- 20.Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. BioEssays. 1994;16:269–275. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- 21.Casals F, Caceres M, Manfrin MH, Gonzalez J, Ruiz A. Molecular characterization and chromosomal distribution of Galileo, Kepler and Newton, three foldback transposable elements of the Drosophila buzzatii species complex. Genetics. 2005;169:2047–2059. doi: 10.1534/genetics.104.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 23.Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;480:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 25.Reiss D, Quesneville H, Nouaud D, Andrieu O, Anxolabehere D. Hoppel, a P-like element without introns: a P-element ancestral structure or a retrotranscription derivative? Mol Biol Evol. 2003;20:869–879. doi: 10.1093/molbev/msg090. [DOI] [PubMed] [Google Scholar]
- 26.Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 27.Lee CC, Beall EL, Rio DC. DNA binding by the KP repressor protein inhibits P-element transposase activity in vitro. EMBO J. 1998;17:4166–4174. doi: 10.1093/emboj/17.14.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clouaire T, et al. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci USA. 2005;102:6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roussigne M, et al. The THAP domain: A novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 2003;28:66–69. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 30.Quesneville H, Nouaud D, Anxolabehere D. Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol Biol Evol. 2005;22:741–746. doi: 10.1093/molbev/msi064. [DOI] [PubMed] [Google Scholar]
- 31.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagemann S, Pinsker W. Drosophila P transposons in the human genome? Mol Biol Evol. 2001;18:1979–1982. doi: 10.1093/oxfordjournals.molbev.a003739. [DOI] [PubMed] [Google Scholar]
- 33.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci USA. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page RD, Charleston MA. Trees within trees: Phylogeny and historical associations. Trends Ecol Evol. 1998;13:356–359. doi: 10.1016/s0169-5347(98)01438-4. [DOI] [PubMed] [Google Scholar]
- 36.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 37.Clark JB, Kidwell MG. A phylogenetic perspective on P transposable element evolution in Drosophila. Proc Natl Acad Sci USA. 1997;94:11428–11433. doi: 10.1073/pnas.94.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva JC, Kidwell MG. Horizontal transfer and selection in the evolution of P elements. Mol Biol Evol. 2000;17:1542–1557. doi: 10.1093/oxfordjournals.molbev.a026253. [DOI] [PubMed] [Google Scholar]
- 39.Ranz JM, et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sperlich D, Pfriem P. In: The Genetics and Biology of Drosophila. Ashburner M, Carson HL, Thompson JNJ, editors. London: Academic; 1986. pp. 257–309. [Google Scholar]
- 41.Ruiz A, Heed WB, Wasserman M. Evolution of the mojavensis cluster of cactophilic Drosophila with descriptions of two new species. J Hered. 1990;81:30–42. doi: 10.1093/oxfordjournals.jhered.a110922. [DOI] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Junier T, Pagni M. Dotlet: Diagonal plots in a web browser. Bioinformatics. 2000;16:178–179. doi: 10.1093/bioinformatics/16.2.178. [DOI] [PubMed] [Google Scholar]
- 45.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 46.Zdobnov EM, Apweiler R. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 47.Marchler-Bauer A, et al. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 49.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.