Abstract
Modulation of the activity of the molecular chaperone HSP90 has been extensively discussed as a means to alter phenotype in many traits and organisms. Such changes can be due to the exposure of cryptic genetic variation, which in some instances may also be accomplished by mild environmental alteration. Should such polymorphisms be widespread, natural selection may be more effective at producing phenotypic change in suboptimal environments. However, the frequency and identity of buffered polymorphisms in natural populations are unknown. Here, we employ quantitative genetic dissection of an Arabidopsis thaliana developmental response, hypocotyl elongation in the dark, to detail the underpinnings of genetic variation responsive to HSP90 modulation. We demonstrate that HSP90-dependent alleles occur in continuously distributed, environmentally responsive traits and are amenable to quantitative genetic mapping techniques. Furthermore, such alleles are frequent in natural populations and can have significant effects on natural phenotypic variation. We also find that HSP90 modulation has both general and allele-specific effects on developmental stability; that is, developmental stability is a phenotypic trait that can be affected by natural variation. However, effects of revealed variation on trait means outweigh effects of decreased developmental stability, and the HSP90-dependent trait alterations could be acted on by natural selection. Thus, HSP90 may centrally influence canalization, assimilation, and the rapid evolutionary alteration of phenotype through the concealment and exposure of cryptic genetic variation.
Keywords: cryptic variation, morphological evolution
Wild-type phenotypes are remarkably robust despite genetic and environmental perturbations, yet evolutionary novelty continues to arise. Seminal studies by Waddington and others reported the expression of novel phenotypes under environmental stress in a variety of organisms (1–3). Such environmentally induced phenotypes, once exposed, can become independent of the inducing stress after several generations of selection (2). In an effort to explain the apparent inheritance of environmentally induced characters, Waddington (4) proposed that traits are buffered against minor environmental insults and genetic polymorphisms of small effect (“canalization”). Large perturbations may disrupt the canalized phenotype and reveal novel phenotypes, which may become “assimilated” through selection (2, 3). Both the existence of and potential molecular mechanisms leading to canalization and genetic assimilation have been much debated (5–7).
Several groups have found that reduction of HSP90 function dramatically affects a wide variety of morphological phenotypes in highly divergent multicellular eukarya (8–11). In the fruit fly Drosophila melanogaster (9) and the mustard plant Arabidopsis thaliana (8), the trait alterations revealed by reductions in HSP90 function differed among different genetic lines, suggesting that specific natural genetic variation had become differentially expressed upon HSP90 inhibition. Importantly, in both organisms, line-specific trait alterations observed with inhibited HSP90 could also be revealed by moderate environmental changes without any direct manipulation of chaperone function. Rutherford and Lindquist (9) demonstrated that selective breeding could increase the penetrance of traits dependent on HSP90 inhibition. Indeed, such traits could be driven to near fixation due to the enrichment of the population for normally cryptic natural variation predisposing the flies to the altered trait. During this breeding process, the traits became independent of the HSP90 mutation initially required to expose the altered phenotype. Thus, HSP90 appears to fulfill Waddington's concept of a developmental buffer or molecular canalization mechanism; one that can lead to the assimilation of novel traits.
A converse effect of HSP90 function has been observed in the yeast Saccharomyces cerevisiae and in mammalian cancer cell lines. In S. cerevisiae, HSP90 is required for new mutations to arise that mediate resistance to high concentrations of several antifungal agents (12). That is, high levels of HSP90 activity are necessary for the novel phenotype to be expressed, rather than a reduction of HSP90 activity leading to phenotype exposure as in D. melanogaster and A. thaliana. Similarly, oncogenic transformation in several mammalian cell lines is reversed upon HSP90 inhibition; HSP90 activity is required for the novel cancerous phenotype to be expressed (13). This effect occurs because of destabilization of certain oncogenic proteins by mutation, leading to a heightened requirement for HSP90 chaperone function (14). These two aspects of HSP90-dependent exposure and concealment of genetic variation have yet to be observed in the same organism.
We have previously suggested that HSP90 may also affect microenvironmental canalization, or developmental stability in isogenic backgrounds (8). That is, modulation of HSP90 activity increases the variance in a phenotypic trait within an isogenic population. Other studies have either failed to observe similar effects in several traits in D. melanogaster (15, 16) or have found such effects to be quite restricted (17). However, the metrics used to assess developmental stability differ between studies. The experiments with D. melanogaster have usually measured developmental noise sensu Waddington, the accuracy with which a genotype produces repeated or symmetric characters within a particular individual (assessed by fluctuating asymmetry) (8). Instead, we measure the precision with which a genotype produces consistent trait values for a given phenotype across different individuals grown in the same environment. We thus examine developmental stability on the population level, rather than the individual level. The relationship between stability on the population and individual levels is unclear (8). It has also been recently suggested that natural variation may affect developmental stability (18); the interaction between such stabilizing polymorphisms and HSP90 is unknown.
As a highly connected node in genetic networks, HSP90 is ideally situated to affect many phenotypes. This molecular chaperone is essential for the proper function of a diverse set of key regulators of growth, development, and defense (19, 20). As a stress-inducible protein, HSP90 is at the intersection of organismic development and environmental responses. It has been previously suggested that HSP90's capacity to buffer genetic polymorphisms is diminished under stress, so such variation becomes phenotypically expressed (8, 9, 21). In addition, highly specific HSP90 inhibitors are secreted by some fungi and bacteria in natural environments, potentially affecting HSP90 buffering capacity in surrounding organisms (22).
The presence of cryptic polymorphisms whose expression depends on HSP90 activity (HSP90-dependent variation) has wide-ranging implications for phenotypic robustness and the ability of selection to effect phenotypic change (21, 23). To determine whether HSP90-dependent variation is globally important to phenotypic differences and evolutionary processes, the identity and frequency of HSP90-dependent variants must be ascertained in natural populations. Furthermore, the effects of HSP90 modulation on developmental stability must be compared with the magnitude of phenotypic change effected by revealed polymorphisms.
A. thaliana is a model system with several attributes that may greatly increase the probability of successful identification of HSP90-dependent polymorphisms. As a naturally inbreeding organism, A. thaliana allows any phenotype to be repeatedly measured on thousands of clonal siblings. Although heterozygosity is nearly absent in individual lines, a vast amount of genetic variation can be found among natural populations (24); some of these polymorphisms may be buffered by HSP90. Several studies have identified natural genetic variants affecting environmental responses in A. thaliana (24), making such traits particularly attractive for identifying HSP90-buffered genetic polymorphisms.
Our previous work demonstrated that HSP90 inhibition indeed affects an environmental response trait, seedling etiolation. In the dark, seedling hypocotyls (embryonic stems) elongate and cotyledon expansion is stalled; whereas, in the light, hypocotyl elongation is suppressed, cotyledons expand, and chlorophyll production ensues. The genetic circuits underlying this plastic and highly adaptive response (25) have been well characterized and are known to contain natural variation (26, 27). The investigation of seedling etiolation in the dark allows HSP90 activity to be modulated with geldanamycin (GDA), a highly specific HSP90 inhibitor that is light-sensitive (8, 13). Herein, we report the dissection of HSP90-dependent effects on seedling etiolation affecting both mean trait values and developmental stability.
Results
HSP90 Affects Genetic Variation in Hypocotyl and Root Elongation.
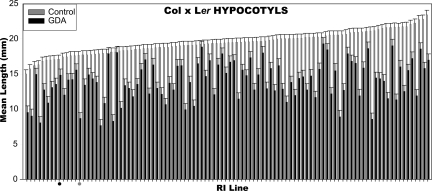
Previously, we have reported line-specific alterations in hypocotyl elongation in the dark in Cape Verde Island × Landsberg erecta (CVI × Ler) recombinant inbred lines (RILs) (8). We conducted quantitative genetic analysis of this trait for a core set of Columbia × Landsberg erecta (Col × Ler) RILs (28). Under control conditions, we observed a wide range of trait values, including many more extreme than either parent. This phenotypic transgression indicates the presence of multiple loci for which the parental alleles differentially contribute to the measured trait. Strikingly, this pattern of natural variation was completely altered when seedlings were grown in the presence of GDA (Fig. 1). Some lines with relatively long hypocotyls under control conditions were highly sensitive to HSP90 inhibition and exhibited relatively short hypocotyls, whereas others were insensitive to GDA and remained long (Fig. 1). None of the GDA lines produced hypocotyls as long as the controls, but roots were nearly always longer than controls.
Fig. 1.
Mean hypocotyl lengths in Col × Ler RILs. Mean hypocotyl lengths of 98 RILs (x axis) and the two parental accessions (Col, black dot; Ler, gray dot) are plotted, ordered by length on control medium. Black, GDA treatment; gray, control media treatment. Error bars denote standard error.
One potential explanation for these striking results is that genetic variation affecting drug resistance segregates between lines. We excluded this possibility by measuring root length of the same seedlings [supporting information (SI) Fig. 4]. Should the observed hypocotyl data be due to different genotypes having differential drug uptake or toxicity, roots of lines with strongly affected hypocotyls would also be strongly affected. However, the observed correlation is in the opposite direction; lines with highly HSP90-responsive hypocotyl length tend to have less responsive roots (R2 = 0.33; P = 1.3 × 10−4; Spearman's rank correlation).
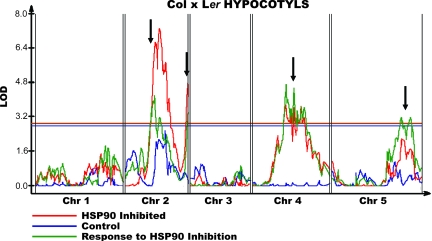
An alternative explanation is that HSP90-dependent variation has been revealed, changing which loci contribute to differences in phenotype. Indeed, QTL analysis of the hypocotyl response to HSP90 inhibition (defined in Materials and Methods) indicated the presence of at least five HSP90-responsive loci (Fig. 2), each accounting for 16–18% of the trait variance. No significant QTL that were not HSP90-dependent were detected. Several HSP90-responsive hypocotyl length QTL also significantly affected root length in an HSP90-responsive manner; however, not all affected both traits (SI Fig. 5). Details of map positions and additive effects are displayed in SI Table 3.
Fig. 2.
QTL affecting hypocotyl length in Col × Ler RILs. Red, HSP90-inhibited condition; blue, control; green, response to HSP90 inhibition. QTL (interval map) significance is in LOD; a horizontal line of the appropriate color denotes a genome-wide α = 0.05 significance threshold. Arrows point to significant HSP90-responsive QTL.
To assess whether the observed HSP90-dependent variation is specific to the Col × Ler RIL set, we performed a similar experiment with the more genetically divergent (29) CVI × Ler set (30). Again, HSP90 inhibition dramatically altered the pattern of natural phenotypic variation (SI Fig. 6). QTL analysis revealed a single HSP90-responsive QTL on chromosome 2, accounting for 30% of the trait variance. This QTL was highly reproducible over three experiments, with different incubators and seed batches used in different experiments. Notably, the chromosome 2 QTL is located in a comparable location to a QTL from the Col × Ler RI set, suggesting that CVI and Col may share an allele conferring reduced hypocotyl length responsiveness to inhibition of HSP90. No similar QTL was found for root response to GDA (SI Fig. 7); the hypocotyl response QTL appears to be trait-specific. Moreover, not all QTL are HSP90-responsive, as illustrated by the chromosome 5 QTL in the CVI × Ler set, which appears in both control and HSP90-inhibited conditions.
To ensure that our results were not due to pleiotropic effects of the erecta mutation, we also examined the Bay-0 × Shahdara RIL set (31), which does not segregate for erecta. Similar results regarding HSP90-responsive QTL were obtained (SI Figs. 8 and 9). Notably, one of these QTL is concealed rather than revealed upon HSP90 inhibition, reminiscent of results from S. cerevisiae (12) and cancer cell lines (13).
Thus, we firmly establish that HSP90-dependent variation can be readily mapped in environmental response traits across multiple populations by using a quantitative genetics approach. This observation confirms HSP90's proposed role in buffering specific genetic variants (8, 9) and establishes that such variation may be frequent, even among closely related populations.
Near-Isogenic Confirmation of HSP90-Dependent QTL.
To confirm and further localize the HSP90-responsive QTL affecting hypocotyl length on chromosome 2 in the CVI × Ler RIL population, we created 50 stepped recombinant near-isogenic lines (NILs) with differing lengths of CVI genotype between 7.3 and 18.1 Mb on chromosome 2. The rest of the genome was Ler genotype. Measurement of hypocotyls and roots in these lines indeed confirmed a genotype-dependent HSP90-responsive effect on hypocotyl length (Table 1). This effect localizes to a region around 9.3 Mb, around 2 Mb toward the centromere from the ERECTA locus. Notably, ERECTA itself is not responsible for the HSP90-dependent QTL. However, the erecta phenotype shows an epistatic interaction with the HSP90-dependent polymorphism (Table 1 and SI Table 4). In a wild-type ERECTA background, the cryptic polymorphism is only revealed upon HSP90 inhibition. However, in a mutant erecta background, its effects are revealed both with and without HSP90 inhibition. That is, the highly pleiotropic erecta mutation may substitute for HSP90 inhibition to reveal a polymorphism at a second locus.
Table 1.
Confirmation of an HSP90-responsive QTL in near-isogenic lines
| Genotype at 9.35 Mb | Hypocotyl length, mm |
|||||
|---|---|---|---|---|---|---|
|
ERECTA |
erecta |
|||||
| Control | HSP90-inhibited | Response | Control | HSP90-inhibited | Response | |
| CVI | 22.7 | 17.0* | 5.7* | 22.1* | 16.2* | 5.9 |
| Ler | 22.7 | 14.5 | 8.2 | 20.7 | 14.0 | 6.7 |
Wilcoxon tests on line means were performed to test for allelic differences. *, P < 0.05.
The distal end of chromosome 5 displayed an HSP90-responsive effect on hypocotyl length in the Col × Ler RIL set. NILs were created to confirm the effect of genotype in this region in a Col genomic background. The HSP90-responsive hypocotyl length QTL was indeed confirmed (Table 2). This QTL appears to be trait specific because no similar effect was noted for root length (SI Table 5). Thus, the HSP90-responsive character of natural genetic variation may be recapitulated in lines differing by only a small portion of the A. thaliana genome.
Table 2.
HSP90 buffering affects both trait means and developmental stabilities
| Genotype at 23.3 Mb | Hypocotyl length, mm |
Hypocotyl developmental stability, LS |
||||
|---|---|---|---|---|---|---|
| Control | HSP90-inhibited | Response | Control | HSP90-inhibited | Response | |
| Col | 19.1 | 16.7** | 2.4* | 0.05 | 0.09** | 0.04* |
| Ler | 18.5 | 12.7 | 5.8 | 0.05 | 0.20 | 0.15 |
Near-isogenic analysis of a HSP90-responsive QTL on chromosome 5 in Col × Ler RILs confirms the effect and reveals HSP90-dependent buffering of developmental stability. *, P < 0.05; **, P < 0.01. Wilcoxon tests were performed to test for allelic differences in the HSP90-inhibited condition; the nonparametric test of epistasis from the companion article (32) was used to test the significance of the response.
HSP90 and Natural Variation Affect Developmental Stability.
We previously reported that reductions in HSP90 function can increase trait variability in genetically identical individuals because of effects on developmental stability (8). We observed that some RILs exhibited extreme variability in hypocotyl length between individuals upon HSP90 inhibition, whereas others displayed little variation. We took advantage of these differences to revisit the effect of HSP90 on phenotypic variation within a given genotype.
We use the median form of Levene's statistic (LS) (33) to measure developmental stability. This statistic affords an individual-based measure of within-genotype variability that is robust to differences in mean values. HSP90 inhibition dramatically decreases overall developmental stability in the Col × Ler RIL set for both hypocotyls and roots (P = 5.7 × 10−18 for hypocotyls, P = 1.0 × 10−8 for roots; Wilcoxon matched-pair test). Similar results were obtained for the CVI × Ler and Bay-0 × Shahdara RIL sets.
Because some RILs appeared to be far more variable than others, we next addressed whether differences in developmental stability might have a genetic basis. We estimated broad-sense heritabilities (H2) for both trait means and developmental stabilities (SI Table 6). Consistent with other authors' findings (18), estimated heritabilities for developmental stability tended to be far lower than those for trait means. Nonetheless, significant heritabilities were frequently observed, especially for hypocotyl elongation with HSP90 inhibited, suggesting that differences in developmental stability may have a genetic basis whose expression is influenced by HSP90.
To further explore natural genetic variation affecting developmental stability, we used QTL analysis of developmental stability across all three RIL populations. Specifically, we address three questions. First, are QTL for developmental stability amenable to genetic mapping? Second, are these QTL responsive to HSP90? Third, do these QTL exclusively affect developmental stability or do they also exert an influence on mean trait values?
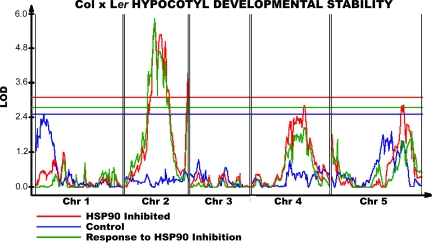
First, we did indeed observe significant QTL for developmental stability in all populations tested (Fig. 3 and SI Figs. 10–12). Second, HSP90 could dramatically affect such QTL. For example, a large-effect HSP90-responsive QTL affecting hypocotyl developmental stability was mapped to the middle of chromosome 2 in the Col-0 × Ler population; this QTL is only detected in HSP90-reduced conditions. This QTL colocalizes with a QTL for the trait mean of hypocotyl response to HSP90 inhibition. More generally, all of the three HSP90-dependent QTL affecting developmental stability colocalized with QTL affecting the trait mean; two colocalized with HSP90-responsive trait mean QTL. Remarkably, in these cases, the allele-dependent decrease in developmental stability did not compromise our ability to map the HSP90-responsive component of the trait mean.
Fig. 3.
QTL affecting hypocotyl and root length developmental stability in Col × Ler RILs. Red, HSP90-inhibited condition; blue, control; green, response to HSP90 inhibition. QTL (interval map) significance is in LOD; a horizontal line of the appropriate color denotes a genome-wide α = 0.05 significance threshold.
Third, we found QTL that appear to influence developmental stability exclusively, in addition to those that affect both trait mean and developmental stability. For example, a significant QTL affecting hypocotyl developmental stability in control conditions was localized to the top of chromosome 1 in the Col × Ler RIL set (Fig. 3). This QTL did not colocalize with any significant or nearly significant QTL affecting trait mean in this population. This failure to colocalize generalizes to all four QTL affecting developmental stability in control conditions, in contrast to our observations with HSP90-dependent developmental stability QTL. Because the estimated additive effects at most QTL affecting developmental stability observed on control media are similar to the effect estimated on GDA, these QTL are likely present in both conditions. The failure to observe most of these QTL on GDA is likely due to a lack of statistical power because HSP90-dependent polymorphisms have a greater overall effect on trait stability.
The Col × Ler RIL set displays a QTL affecting hypocotyl developmental stability on the distal end of chromosome 5 (Fig. 3). We attempted to confirm this predicted genotype dependence of developmental stability in the previously discussed Col/Ler NIL population with lines that differ in genotype only in this region. The QTL was indeed verified and depends on HSP90 activity levels (Table 2). Although the Col and Ler alleles had nearly identical developmental stabilities on control medium and developmental stability for both alleles increased upon HSP90 inhibition, the Ler allele had more than three times the response of the Col allele.
Other authors have suggested that mutations in ERECTA may lead to an increase in developmental stability in flowering time (18). We examined the CVI/Ler chromosome 2 NIL data discussed above to determine whether a mutant erecta affected the developmental stability of either hypocotyl or root length under control conditions. It did not (hypocotyl length P = 0.56, root length P = 0.55; Wilcoxon test). Instead, the HSP90-responsive QTL affecting hypocotyl developmental stability was verified with maximal probability of being located between 7.72 and 9.35 Mb (P = 0.003).
Discussion
We have used three different RIL populations of A. thaliana to identify four different HSP90-responsive QTL affecting hypocotyl elongation and four affecting root length of seedlings grown in a dark environment. These results resolve several debated points regarding HSP90 buffering and its potential impact on evolutionary processes. First, HSP90-dependent alleles are amenable to genetic mapping techniques and are unlikely to be purely epigenetic. Second, continuous, environmentally responsive traits can be affected by HSP90-dependent polymorphisms, contrary to suggestions that such alleles only affect highly canalized traits (34). Third, HSP90-dependent alleles are frequent in natural populations and can have significant effects on natural phenotypic variation. Fourth, the effects of these polymorphisms are not immediately deleterious and could reasonably be advantageous under certain conditions. Finally, HSP90 modulation can both reveal and conceal the phenotypic effects of natural variation.
The HSP90-responsive nature of two QTL affecting hypocotyl length has been confirmed in near-isogenic settings. With one of these NIL systems, we observed that the pleiotropic erecta mutation could reveal cryptic HSP90-dependent genetic variation in the absence of HSP90 inhibition, thus experimentally supporting the suggestion (35) that the perturbation of highly connected nodes in genetic networks may represent a general method for uncovering hidden genetic variation.
We also demonstrated that developmental stability, the ability of a genotype to produce the same phenotypic outcome across isogenic individuals, has a genetic basis and can be affected by natural variation. HSP90-responsive QTL affecting trait means usually also affect developmental stability in an HSP90-dependent manner; one such case was confirmed in a NIL. Importantly, the effects of these QTL on trait means outweigh their effects on developmental stability. That is, if the polymorphisms are revealed, selection could act to increase their frequency in the population. Moreover, as recently observed by others (18), some QTL were detected that affect developmental stability but do not colocalize with a QTL affecting the trait mean. That is, some genetic variation may function solely to stabilize a phenotype.
These results suggest a model of the influence of HSP90 on canalization, assimilation, and the rapid evolutionary alteration of phenotype. Because HSP90 is a central node in many cellular networks, modulation of its activity, here achieved by inhibition but likely to naturally occur by environmental stress, will lead to increased instability of a variety of genetic pathways. This instability represents a reduction in canalization, leading to the exposure of the phenotypic effects of multiple cryptic polymorphisms. These phenotypic effects are initially poorly canalized; however, selection may enrich such polymorphisms or select for secondary stabilizing factors, increasing the developmental stability of the revealed alleles. Thus, the novel trait value is assimilated and may become HSP90-independent, as experimentally observed in other studies (9, 12). This study establishes the existence of all of the necessary prerequisites for this model: In multiple traits, HSP90 modulation reveals multiple natural genetic polymorphisms of significant but not extreme effect. The phenotypic effects of these alleles are initially unstable; however, the observed natural variation affecting developmental stability could be recruited to stabilize a new, beneficial trait value.
Materials and Methods
Hypocotyl and Root Assays.
All hypocotyl and root assays were performed according to the methods in ref. 8. The CVI × Ler RIL set was used in three independent experiments. In the first experiment, 10 seeds of each of the 50 RILs in the core set along with the parental lines were plated on each media treatment. Each RIL was plated on a single plate with seeds only from that RIL. The second experiment was of similar design, except that 50 seeds of each genotype were plated over five plates. This experiment required multiple days to plate. Twenty-three extra RILs were added to the third experiment to increase recombination frequency on chromosome 2. Nineteen seeds of each RIL were plated on each medium. Blocks of 10 genotypes were pseudorandomized such that one seed of each genotype was plated on each of 19 plates.
A similar pseudorandomized 10-genotype block design was used for the Col × Ler and Bay-0 × Shahdara RIL experiments. Nineteen seeds of 98 Col × Ler RILs and 15 seeds of 118 Bay-0 × Shahdara RILs, along with the parental genotypes of each set, were plated on each treatment. All RIL trait values are available in SI Table 7.
For the confirmation of the QTL found on chromosome 2 in the CVI × Ler RIL set, 50 NILs were created from a cross of CS22095 with Ler. F2 individuals from a self-propagated F1 of this cross segregate for CVI genotype from 34 to 63 cM on chromosome 2 and from 84 to 107 cM on chromosome 5. F2 seedlings were selected for homozygous Ler genotype at a marker at 24.1 Mb on chromosome 5 and for a recombination between markers at 7.35 and 18.1 Mb on chromosome 2. Recombinants were self-propagated, and homozygotes for the recombined region on chromosome 2 were selected by PCR. These lines were then genotyped at insertion/deletion markers at 7.73, 9.35, 10.9, 12.9, 15.6, and 16.3 Mb on chromosome 2. See ref. 36 for marker information. All NILs were propagated at the same time, randomized with respect to genotype. Twenty-five seeds per condition of each NIL were used to measure hypocotyl and root length. This experiment also included two lines derived from the same cross that were selected to be Ler at all markers as well as 50 seeds per condition of the parental Ler accession (CS8581). This experiment was randomized with blocks of 10 genotypes as described previously. This experiment was automatically measured with HypocoTool (http://openwetware.org/wiki/HypocoTool), followed by manual quality control.
For the confirmation of the QTL observed on chromosome 5 in the Col × Ler RIL set, we obtained the STAIRS line CS9500 (37) from the Arabidopsis Biological Resource Center. This line has Col-0 genomic background except from 65.2 to 130 cM on chromosome 5, where it is Ler genotype. One hundred eight seeds per condition of CS9500 and Col-0 (CS60000), bulk propagated in the same flat, were used to measure hypocotyl and root length.
Statistical Analyses.
Least-squares means for the hypocotyl and root length of each RIL in the CVI × Ler experiments were derived from a linear analysis with each treatment examined separately. Genotype, experiment, plating date, and plate number were analyzed as fixed effects with plating date nested within experiment and plate number nested within experiment and plating date. For the chromosome 2 NIL experiment, least-squares means were derived from a linear regression model including treatment, block nested within treatment, plate number nested within block and treatment, and genotype nested within block and treatment. For the Col × Ler, Bay-0 × Shahdara, and chromosome 5 NIL experiments, simple traits means were used. For all traits, RIL means for the response to HSP90 inhibition are defined as the trait mean on control medium minus the trait mean on GDA for the same line. Because the interline variance on GDA was higher than on control medium, an alternative variance-dampening measure,
was also examined. No significant differences were observed from the presented results.
As a measure of developmental stability quantifiable for each individual, we used the log-median form of Levene's statistic (33):
where i represents each individual from line j. Simple means of LS were used for each line for all experiments except the CVI × Ler RIL experiments, where a multiple linear regression model with genotype and experiment as factors was used.
Broad-sense heritabilities were estimated by using variance components with the same models as described above, except that genotype was included as a random variable with restricted maximum likelihood statistical procedures. All statistical calculations were performed with JMP5.0 (SAS Institute).
QTL Mapping.
QTL mapping was performed with QTL Cartographer for Windows, v.2.5, using the interval map function. Least-squares trait means as calculated above were used. For the CVI × Ler RIL set, all AFLP markers from ref. 30 were used for QTL mapping. For the Col × Ler RIL set, the full set of markers compiled by the Nottingham Arabidopsis Stock Centre were used. For the Bay-0 × Sha RIL set, maps from ref. 31 were used. Significance thresholds for each trait were estimated by 1,000 permutations of the data relative to the genotypes.
Supplementary Material
ACKNOWLEDGMENTS.
We thank D. Weigel, J. Borevitz, and J. Maloof for discussions. T.A.S. was a Howard Hughes Medical Institute predoctoral fellow. T.A.S. and S.L. were supported by the Howard Hughes Medical Institute and the G. Harold and Leila Y. Mathers Foundation. N.S., S.U., K.S., and C.Q. were supported by National Institute of General Medical Sciences National Centers for Systems Biology Grant GM068763 and a Bauer Fellowship (to C.Q.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712200105/DC1.
References
- 1.Schmalhausen II. Factors of Evolution: The Theory of Stabilizing Selection. Chicago: Univ of Chicago Press; 1949. [Google Scholar]
- 2.Waddington CH. Genetic assimilation of an acquired character. Evolution (Lawrence, Kans) 1953;7:118–126. [Google Scholar]
- 3.Waddington CH. Genetic assimilation of the bithorax phenotype. Evolution (Lawrence, Kans) 1956;10:1–13. [Google Scholar]
- 4.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 5.Meiklejohn CD, Hartl DL. A single mode of canalization. Trends Ecol Evol. 2002;17:468–473. [Google Scholar]
- 6.Dworkin I. A study of canalization and developmental stability in the sternopleural bristle system of Drosophila melanogaster. Evolution (Lawrence, Kans) 2005;59:1500–1509. [PubMed] [Google Scholar]
- 7.Debat V, David P. Mapping phenotypes: Canalization, plasticity and developmental stability. Trends Ecol Evol. 2001;16:555–561. [Google Scholar]
- 8.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 9.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 10.Sollars V, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 11.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: Drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 13.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: Essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Singer MA, Lindquist S. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc Natl Acad Sci USA. 1999;96:109–114. doi: 10.1073/pnas.96.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milton CC, Huynh B, Batterham P, Rutherford SL, Hoffmann AA. Quantitative trait symmetry independent of Hsp90 buffering: Distinct modes of genetic canalization and developmental stability. Proc Natl Acad Sci USA. 2003;100:13396–13401. doi: 10.1073/pnas.1835613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milton CC, Batterham P, McKenzie JA, Hoffmann AA. Effect of E (sev) and Su(Raf) Hsp83 mutants and trans-heterozygotes on bristle trait means and variation in Drosophila melanogaster. Genetics. 2005;171:119–130. doi: 10.1534/genetics.104.038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debat V, Milton CC, Rutherford S, Klingenberg CP, Hoffmann AA. Hsp90 and the quantitative variation of wing shape in Drosophila melanogaster. Evolution (Lawrence, Kans) 2006;60:2529–2538. [PubMed] [Google Scholar]
- 18.Hall MC, Dworkin I, Ungerer MC, Purugganan M. Genetics of microenvironmental canalization in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:13717–13722. doi: 10.1073/pnas.0701936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sangster TA, Queitsch C. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol. 2005;8:86–92. doi: 10.1016/j.pbi.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangster TA, Lindquist S, Queitsch C. Under cover: Causes, effects and implications of Hsp90-mediated genetic capacitance. BioEssays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 22.McLellan CA, et al. A rhizosphere fungus enhances Arabidopsis thermotolerance through production of an HSP90 inhibitor. Plant Physiol. 2007;145:174–182. doi: 10.1104/pp.107.101808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutherford SL. Between genotype and phenotype: Protein chaperones and evolvability. Nat Rev Genet. 2003;4:263–274. doi: 10.1038/nrg1041. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- 25.Dorn LA, Pyle EH, Schmitt J. Plasticity to light cues and resources in Arabidopsis thaliana: Testing for adaptive value and costs. Evolution (Lawrence, Kans) 2000;54:1982–1994. doi: 10.1111/j.0014-3820.2000.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 26.Borevitz JO, et al. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics. 2002;160:683–696. doi: 10.1093/genetics/160.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maloof JN, et al. Natural variation in light sensitivity of Arabidopsis. Nat Genet. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- 28.Lister C, Dean C. Recombinant inbred lines for mapping Rflp and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- 29.Aranzana MJ, et al. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 2005;1:531–539. doi: 10.1371/journal.pgen.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alonso-Blanco C, et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- 31.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred line population: A powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 32.Sangster TA, et al. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:2969–2974. doi: 10.1073/pnas.0712210105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz BB. Levene test for relative variation. Syst Zool. 1985;34:449–456. [Google Scholar]
- 34.Milton CC, Ulane CM, Rutherford S. Control of canalization and evolvability by hsp90. PLoS ONE. 2006;1:e75. doi: 10.1371/journal.pone.0000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 36.Salathia N, et al. Indel arrays: An affordable alternative for genotyping. Plant J. 2007 doi: 10.1111/j.1365-313X.2007.03194.x. in press. [DOI] [PubMed] [Google Scholar]
- 37.Koumproglou R, et al. STAIRS: a new genetic resource for functional genomic studies of Arabidopsis. Plant J. 2002;31:355–364. doi: 10.1046/j.1365-313x.2002.01353.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.