Abstract
Class I MHC tetramers allow direct phenotypic identification of CD8+ T cell populations, but their production remains laborious. A peptide exchange strategy that employs class I MHC products loaded with conditional ligands (caged MHC molecules) provides a fast and straightforward method to obtain diverse arrays of class I MHC tetramers and facilitates CD8+ T cell epitope discovery. Here, we describe the development of photocleavable analogs of the FAPGNYPAL (SV9) epitope that bind H-2Kb and H-2Db with full retention of their structural and functional integrity. We ranked all possible H-2Kb octameric and H-2Db nonameric epitopes that span the genome of Chlamydia trachomatis and prepared MHC tetramers from ≈2,000 of the highest scoring peptides by replacement of the SV9 analog with the peptide of choice. The resulting 2,000-member class I MHC tetramer array allowed the discovery of two variants of an epitope derived from polymorphic membrane protein I (PmpI) and an assessment of the kinetics of emergence and the effector function of the corresponding CD8+ T cells.
Keywords: conditional ligands, crystal structure, consensus epitope prediction
Characterization of cytotoxic T lymphocyte (CTL) responses usually exploits their effector function. Techniques commonly used for epitope identification include the enzyme-linked immunospot (ELISPOT) assay (1), the chromium release cytotoxicity assay (2), or intracellular cytokine staining of CTL populations followed by flow cytometry (3). These methods require prolonged periods of ex vivo maintenance and expansion of CTLs before a response can be detected, and possibly bias the measured CTL response. Complementary methods, such as the quantitation of T cell receptor (TCR) transcripts, provide information on phenotype and clonality but not about antigen specificity or function (4, 5).
Direct visualization of antigen-specific T cells, independent of their cytotoxic capacity or ability to produce a particular cytokine, became possible with the development of MHC-tetramers (6–8). The production of a single peptide-MHC-complex is a time-consuming and labor-intensive task and yields a reagent capable of staining T cells specific only for that one MHC-peptide complex. This has limited the application of MHC tetramers in high-throughput screening formats. Schumacher and coworkers (9) reported a method in which class I MHC molecules are occupied transiently with a conditional ligand that self-cleaves into two fragments upon photolysis (9, 10). When caged MHC-tetramers are exposed to a large molar excess of a ligand of choice during photocleavage, tetramers of desired specificity are generated, provided the putative ligands can bind to the class I MHC in question. This strategy allows the use of a single batch of photocleavable MHC tetramer from which arrays of MHC tetramers of defined specificity can be rapidly generated for the purpose of high-throughput screening for CD8+ T cell epitopes. The method has been successfully used to identify new HLA-A2 epitopes from influenza virus (9). We sought to apply this technology to the murine H-2Kb and H-2Db molecules. The availability of a single conditional ligand for both these class I MHC products allows, in principle, the phenotypic analysis of all CD8+ T-lymphocytes that undergo clonal expansion after antigenic challenge in C57BL/6 mice.
To illustrate the use of H-2Kb and H-2Db tetramer arrays as epitope discovery tool, we produced a collection of ≈2,000 tetramers to explore the CD8+ T cell response against Chlamydia trachomatis in C57BL/6 mice. C. trachomatis is responsible for ocular, genital, and lymphatic infections and is the most common bacterial sexually transmitted disease in the United States (11). Adaptive immune effectors, including CD8+ T cells, are important for resolving infection and affording protection against C. trachomatis in both humans and mice (12–14). Thus far, only a single CD8+ T cell epitope—the H-2Db-restricted peptide ASFVNPIYL (CrpA63-71)—had been identified in the C57BL/6 strain (15).
Therefore, we scanned all ORFs from the genome of C. trachomatis for epitopes predicted to bind class I MHC products of the H-2b haplotype, using a recently developed consensus epitope prediction program (16), without any particular bias toward specific ORFs. H-2Kb and H-2Db tetramers for the 2,000 top-ranking epitopes were prepared by the peptide exchange strategy and used to screen for novel epitopes. This resulted in the identification of the H-2Db-restricted epitope polymorphic membrane protein I (PmpI)-L2612-620 (SAVSNLFYV) and the closely related PmpI-D612-620 (SAVSNLFYA), both of which elicit a CD8+ T cell response in C. trachomatis-infected C57BL/6 mice. This methodology can be readily extended to study the CD8+ T cell repertoire of any pathogen in this widely used mouse strain.
Results
Design and Properties of Photocleavable Ligands.
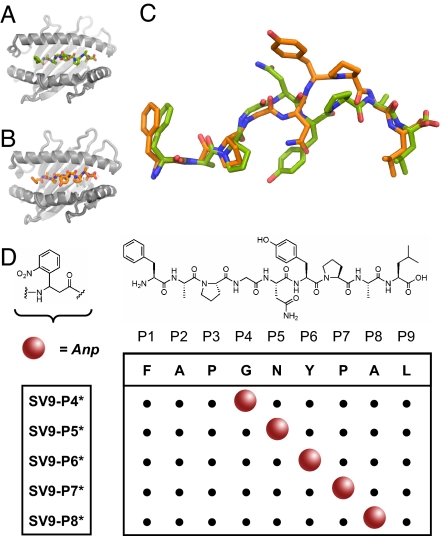
For the production of caged H-2Kb and H-2Db molecules, we took advantage of the capability of both MHC products to bind one and the same peptide—FAPGNYPAL (SV9324–332)—derived from Sendai virus nucleoprotein (17–20). Based on the reported crystal structures [Protein Data Bank (PDB) entries 2VAB for H-2Kb (19) (Fig. 1 A and C) and 1CE6 for H-2Db (20) (Fig. 1 B and C)] and our prior experience with photolabile MHC ligands (21), we systematically replaced five central amino acid residues in the primary peptide sequence of SV9 with the photocleavable 3-amino-3-(2-nitro)phenyl-propanoic acid (Anp) residue. The N- and C-termini were left unmodified, because these are known anchors of SV9 for binding the class I MHC molecules (22, 23). The five derivatives SV9-P4* to SV9-P8* (Fig. 1D) were assembled by standard solid-phase peptide synthesis. The introduction of racemic Anp resulted in the generation of two diastereoisomers for each SV9-derivative. The diastereoisomers were purified only for growing crystals of sufficient quality for x-ray crystallography, and crude peptide mixtures were used for the production of class I MHC tetramers (see below).
Fig. 1.
Design of photocleavable peptide ligands for H-2Kb and H-2Db. (A and B) The structures of H-2Kb (gray; PDB entry 2VAB) (A) and H-2Db (gray; PDB entry 1CE6) (B) complexed with the SV9 epitope. (C) The superimposed structures of SV9 as bound to H-2Kb (green) and H-2Db (orange). (D) In SV9, the positions P4 to P8 were substituted with the Anp-residue to produce photolabile derivatives SV9-P4* to SV9-P8*, respectively. Molecular graphics were produced with PyMOL software (http://pymol.org).
We examined the ability of the SV9 analogs to undergo UV-induced cleavage by electrospray ionization mass spectroscopy. The disappearance of the dominant singly charged ion species of the parent peptide, as shown for the representative example SV9-P7* [supporting information (SI) Fig. 6], correlated with the appearance of the N-terminal fragment as its major cleavage product (SI Fig. 7) upon exposure of the peptide to 365 nm UV-light. We observed that 10 min of UV-irradiation was adequate to drive the photocleavage reaction to near completion (SI Fig. 8) in agreement with previously reported data (10, 21). All remaining SV9 analogs performed identically in this assay (data not shown).
X-Ray Structure of H-2Db in Complex with SV9-P7*B.
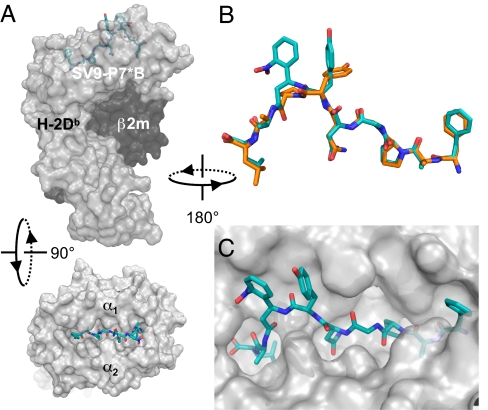
We performed x-ray crystallographic studies to determine the structural integrity of class I MHC molecules in complex with conditional ligands. Therefore, we first resolved the diastereoisomers of SV9-P7* by reversed-phase liquid chromatography (SI Fig. 9). The crude mixture, and purified peptides SV9-P7*A and SV9-P7*B, were refolded separately with the recombinantly expressed luminal portion of the class I MHC heavy chains for H-2Kb and H-2Db together with β2-microglobulin (β2m) light chain. When submitted to hanging-drop, vapor-diffusion crystallization conditions, only H-2Db in complex with peptide SV9-P7*B gave crystals that diffracted adequately, which allowed us to determine the three-dimensional structure to 2.65 Å resolution. The overall organization of this complex (Fig. 2A) shows the common tertiary structure for class I MHC molecules (23). It strongly resembles the reported structure of the H-2Db/SV9 complex (20), as is detailed in SI Materials and Methods. The main-chain conformations of both SV9-epitopes, when complexed to H-2Db, are essentially identical when superpositioned (Fig. 2B). Both complexes employ the N- and C- termini, and the deeply buried side-chains of the P2, P3, P5, and P9 residues as anchor residues (Fig. 2C). Furthermore, the crystal structure reveals a role for the tyrosine residue at P6 as an auxiliary anchoring residue (SI Fig. 11). Sufficient electron density for the photolabile peptide allowed the unambiguous assignment of the S-configuration to the Anp-residue of SV9-P7*B (SI Fig. 12). The nitro-group of this residue forms extensive hydrogen bonding networks with the H-2Db heavy chain, and makes crystal lattice contacts with adjacent molecules, which might explain why crystals were obtained only for a single peptide isomer.
Fig. 2.
Crystal structure of H-2Db in complex with the SV9-P7*B diastereoisomer. (A) Side and top views of the model with a transparent surface representation of the H-2Db heavy chain (gray), the β2m light chain (black), and stick representation of the peptide ligand (blue). (B) The conformation of SV9-P7*B (blue) superimposed on the previously reported structure of FAPGNYPAL (orange) shows their strong resemblance when complexed with H-2Db. (C) The N- and C- terminal residues of the ligand are firmly embedded within the peptide binding cleft, with P2, P3, P5, and P9 residue side-chains providing additional anchor motifs.
Validation of the Peptide Exchange Strategy.
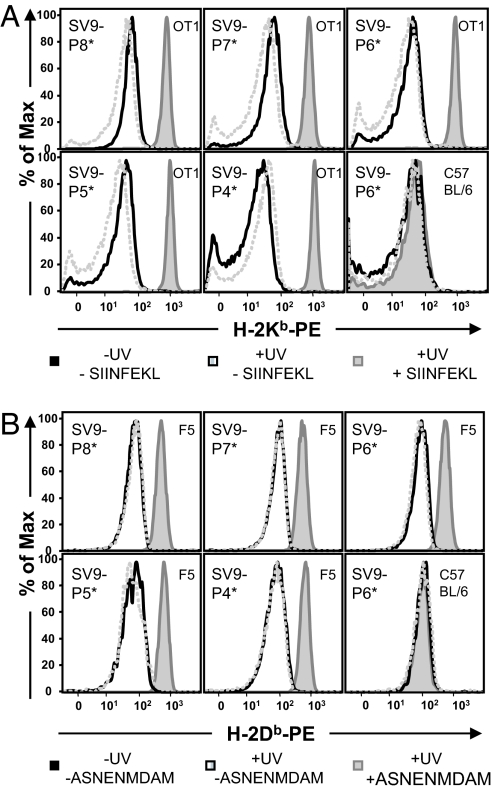
We refolded recombinantly expressed murine β2m with either H-2Kb or H-2Db heavy chain carrying a C-terminal 15-aa sequence for the E. coli-derived BirA biotin ligase (24, 25) in the presence of the photolabile derivatives SV9-P4* to SV9-P8*. The caged complexes were purified by size exclusion chromatography, biotinylated, and tetramerized on streptavidin R-phycoerythrin (SAPE) following well established procedures (26). As a proof of concept, UV-induced peptide exchange on the caged class I MHC tetramers was tested by staining monoclonal T cell populations from TCR transgenic mice, followed by flow cytometry. Caged H-2Kb tetramers could be reloaded with SIINFEKL index peptide and stained OT-I T cells from the ovalbumin-specific H-2Kb restricted TCR transgenic line of mice (Fig. 3A). T cells from F5 TCR transgenic mice recognized the H-2Db tetramers in complex with the influenza epitope ASNENMDAM, generated from caged MHC tetramers (Fig. 3B). Employing alternative index peptides during exchange resulted in class I MHC tetramers that did not stain either of these T cell populations (data not shown). Moreover, in the presence or absence of index peptide, both before and after UV irradiation, neither tetramer stained polyclonal T cells from unimmunized C57BL/6 mice. In conclusion, all five separate derivatives SV9-P4* to SV9-P8* can serve as a conditional ligand for H-2Kb and H-2Db and produce functional reagents that, upon exposure to mild peptide exchange conditions, enable T cell staining of the appropriate specificity. We arbitrarily selected SV9-P7* as the conditional ligand for the production of class I MHC tetramer arrays.
Fig. 3.
Photocleavage and peptide exchange on soluble class I MHC tetramers generates functional staining reagents. (A) Tetramers composed of H-2Kb and preloaded with any of the five different photocleavable ligands designed on SV9 stained CD8+ T cells from OT-1 TCR transgenic mice, only after long wave UV-irradiation in the presence of index peptide SIINFEKL. (B) H-2Db tetramers loaded with conditional ligands SV9-P4* to SV9-P8* stained CD8+ splenocytes from F5 TCR transgenic mice after irradiation at 365 nm in the presence of ASNENMDAM index peptide.
Epitope Prediction for C. trachomatis Serovar L2.
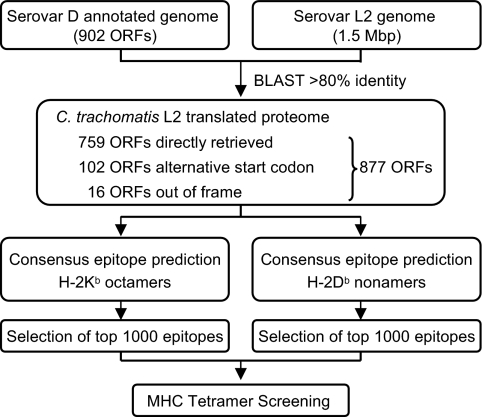
We targeted C. trachomatis, a bacterial pathogen with a relatively small genome, to demonstrate that the peptide exchange strategy lends itself to high-throughput screening for CD8+ T cell epitopes. Because immunization of C57BL/6 mice with C. trachomatis lymphogranuloma venereum (LGV) serovar L2 gives a robust CD8+ T cell response and an L2-specific epitope has been identified (15), C. trachomatis serovar L2 was selected as the infectious agent. Whereas the genome of C. trachomatis serovar D has been fully annotated (27), only preliminary genome sequence data are publicly available for serovar L2. To take advantage of the annotation data, we used the 902 ORFs identified in serovar D as a template to search for orthologous ORFs in serovar L2. Employing BLAST (28) to query the preliminary nucleotide sequence, 759 ORFs were identified as direct matches, 102 ORFs contained an alternative start codon, and 16 ORFs were found to be partially out of frame (Fig. 4). The resulting amino acid sequence data of these 877 ORFs of C. trachomatis L2 (SI Table 1) formed the basis for epitope prediction. We reconstructed a recently published consensus epitope prediction program (16), which combines four existing prediction routines, to identify candidate epitopes for H-2Kb and H-2Db. This program (available at http://jura.wi.mit.edu/bioc/grotenbreg), was used to rank the top 0.5% of all possible 8-mer H-2Kb epitopes (≈1,000 peptides) and 9-mer H-2Db epitopes (≈1,000 peptides) in the 877 ORFs of C. trachomatis L2 (SI Table 2). The cut-off limit was set based on the reported optimal performance of the consensus epitope prediction program (16), which still allowed the screening of a reasonable number of potential CD8+ T cell epitopes. The peptides corresponding to the predicted epitopes were subsequently produced synthetically on micromol scale.
Fig. 4.
Bioinformatic approach for the identification of candidate H-2Kb and H-2Db epitopes from C. trachomatis L2. The preliminary nucleotide sequence of serovar L2 was queried for ORFs, using the 902 ORFs identified in serovar D as template. The resulting 877 ORFs were searched for octameric H-2Kb epitopes and nonameric H-2Db epitopes, using the consensus epitope prediction program. The top-ranking 2,000 predicted epitopes were used for screening with class I MHC tetramer arrays.
Screening CD8+ T Cell Epitopes.
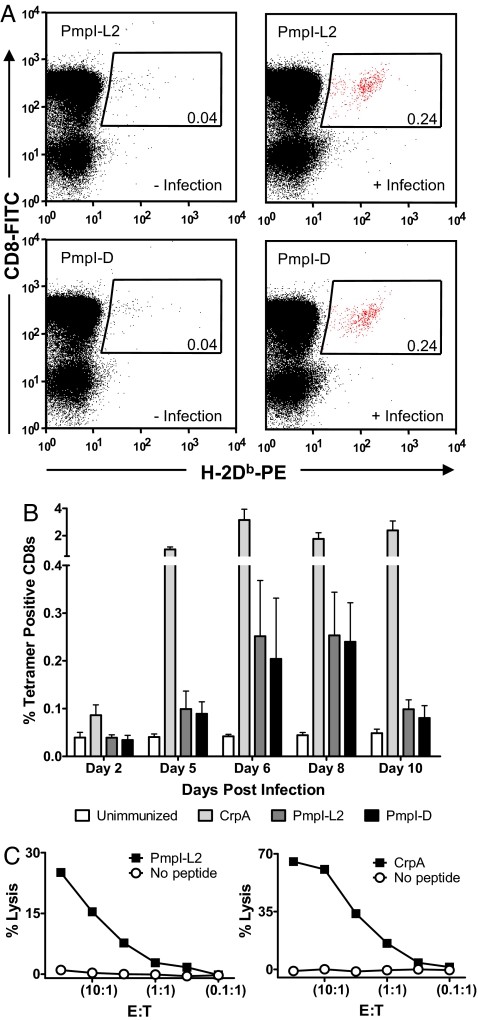
We prepared class I MHC tetramers from the 2,000 predicted and ranked C. trachomatis L2 epitopes and the photocleavable H-2Kb/SV9-P7* or H-2Db/SV9-P7* complexes. The tetramers were incubated with CD8+ splenocytes isolated from mice infected with C. trachomatis L2, and the T cell populations were analyzed by flow cytometry. In addition to the immunodominant H-2Db-restricted CrpA63-71 epitope, which also ranked in the top 0.5% of predicted epitopes, two variants of an epitope were identified that are derived from the polymorphic membrane protein I (PmpI). As shown in Fig. 5A, these epitopes PmpI-L2612-620 (SAVSNLFYV) and PmpI-D612-620 (SAVSNLFYA) are presented on H-2Db and recognized by CD8+ T cells as part of the immune response to C. trachomatis. Sequence identity of the peptides was confirmed by LC/MS/MS analysis (SI Figs. 13 and 14). Using the newly identified MHC tetramers, we analyzed the frequency and kinetics of the T cell response in infected mice with the CrpA epitope as a reference. Maximal expansion of PmpI-L2 and PmpI-D-specific CD8+ T cell populations occurs after 6 days with kinetics near-synchronous with the CrpA-specific population (Fig. 5B). The cytotoxic activity of the T cell populations specific for PmpI-L2 and PmpI-D in the context of H-2Db was confirmed by 51Cr-release assay of peptide-treated EL4 target cells followed by treatment with restimulated splenocytes from C. trachomatis immunized mice (Fig. 5C).
Fig. 5.
CD8+ epitopes derived from PmpI are recognized in context of H-2Db after infection with C. trachomatis L2. (A) Staining of CD8+ T cells after photocleavage and peptide exchange with the two related peptides PmpI-L2612-620 (SAVSNLFYV) and PmpI-D612-620 (SAVSNLFYA) before and 6 days after infection with C. trachomatis L2. (B) H-2Db tetramer staining of the individual CD8+ populations for CrpA, PmpI-L2 and PmpI-D at indicated days after infection with C. trachomatis L2. Maximal expansion of PmpI-L2 and PmpI-D-specific CD8+ splenocytes occurs after 6 days. (C) Both CrpA- and PmpI-L2-specific CD8+ T cells are cytotoxic in a standard 51chromium release assay, using varying effector (E) to target (T) ratios. Experiments were performed at least in triplicate.
Discussion
Class I MHC tetramers of distinct specificity, yet accessible through a single precursor, enable the rapid generation of large tetramer arrays, which can be used as a screening tool for antigen-specific CD8+ T cell populations (7–9). We designed photocleavable ligands based on the SV9 (FAPGNYPAL) epitope for the H-2Kb and H-2Db MHC products. The conditional ligands SV9-P4* to SV9-P8* behave like typical H-2Kb and H-2Db ligands in all respects. The x-ray crystal structure of SV9-P7*B in complex with H-2Db was of sufficient resolution to determine the isomer configuration of the peptide and showed prominent roles for the Anp P7 residue in the formation of the crystal lattice and the Tyr P6 as auxiliary peptide anchor. Upon fragmentation of the photocleavable ligands by UV-irradiation, the uncaged MHC complexes could be reloaded with index peptides that had appreciable affinity for the complex. The functional integrity of the resulting H-2Kb and H-2Db complexes was confirmed by staining TCR transgenic T cell populations of known specificity.
We then applied a bioinformatic approach to focus our screening efforts on C. trachomatis serovar L2-derived epitopes most likely to elicit clonal expansion of specific CD8+ T cells, while taking into account all gene products of the pathogen in an unbiased manner. We identified ORFs in the recently sequenced genome of C. trachomatis L2, using the annotated proteome of C. trachomatis D as template. Only those ORFs that showed at least 80% identity were considered significant. Furthermore, those ORFs that are generated by alternative start codons, as apparent in this translation, were included, as alternative codon usage for translation initiation by C. trachomatis occurs (29–31).
We could readily track C. trachomatis L2 infection in C57BL/6 mice by staining the subpopulation of CD8+ T cells that recognize the known H-2Db-restricted CrpA epitope, using MHC tetramers generated by the peptide exchange strategy (data not shown). This immunodominant epitope is derived from the inclusion membrane-associated protein CrpA (15). The inclusion membrane sequesters the pathogen inside the host cell and provides an accommodating intracellular niche (11, 12). Similarly, a CD8+ T cell antigen that is presented in H-2d restricted fashion is derived from the Cap1 protein, which also localizes to this vacuolar membrane (29). The localization of these proteins, at the interface of the inclusion and the cytosol, gives them access to the class I MHC-processing pathways and makes them targets for CD8+ T cell recognition.
The H-2Db restricted epitopes that have been identified in this study stem from the polymorphic membrane protein I. The pmp family of genes consist of nine members (27, 31) that encode proteins homologous to autotransporters found in Gram-negative bacteria (32). These proteins are composed of a large surface-exposed functional domain at the N terminus, which carries several repeat motifs and is translocated through a β-barrel structure formed by the C-terminal membrane-associated domain. Their polymorphism suggests a role in diversifying exposed antigens to escape immune surveillance (33). Comparative expression profiling suggests that pmpI is the least abundantly expressed gene in the family, although its expression can be detected as early as 2 h after C. trachomatis L2 infection in cell culture (34). The PmpI protein also appears in proteomic analyses of complete protein extracts of C. trachomatis L2 (35). Pmp gene products contain numerous predicted MHC epitopes (36), but their verification was not reported until now. From the 877 ORFs identified for C. trachomatis L2 here and the ≈2,000 epitopes ranked to merit inclusion in our screen, 87 epitopes (37 for H-2Kb and 56 for H-2Db) mapped to the Pmp family of gene products (SI Table 3). Of the 12 H-2Db epitopes in the PmpI sequence that were screened here, only PmpI–L2612-620 elicited a CD8+ T cell response. The single amino acid difference between PmpI–L2612-620 and PmpI–D612-620 (Val620 versus Ala620, respectively) suggests that this epitope might map to a domain that is subject to immune selection.
The CD8+ T cell epitopes PmpI–L2612-620 and PmpI–D612-620 do, however, share eight of nine residues. Given the comparable staining patterns and the kinetics with which the CD8+ T cells specific for H-2Db with either PmpI-L2 or PmpI-D develop (Fig. 5), it appears that the alternative C terminus of these two epitopes does not influence TCR recognition. This C-terminal residue is deeply buried in the P9-pocket created by the α1- and α2-domains of the H-2Db heavy chain (Fig. 2C), and, consequently, CD8+ T cells are unlikely to be able to discriminate between the presented epitopes.
The screen performed in this report evaluated the top 0.5% of predicted octameric H-2Kb epitopes and nonameric H-2Db epitopes of the 877 ORFs identified in C. trachomatis L2. This screen is by no means exhaustive but is designed to demonstrate the feasibility of high-throughput class I MHC tetramer screening as a means to identify epitopes for organisms where only few such epitopes are known. The discovery of two related CD8+ T cell epitopes within a library of 2,000 peptides raises the question of why so few new epitopes were identified. The predictive method could have performed suboptimally because variables such as the pathogen's developmental lifecycle, protein expression rates, and ensuing antigen processing are not considered. Moreover, generation of unconventional epitopes, variations in epitope length or relaxed requirements for anchor positions further confound predictions (37, 38). Application of the consensus epitope prediction program in vaccinia virus infection, however, yielded a substantial number of epitopes with an apparent hierarchal distribution of dominance in which the H-2K and H-2Db restriction elements were represented equally (16). The confinement of C. trachomatis L2 to an intracellular vacuolar compartment limits access of pathogen-derived factors to the host's antigen processing machinery and provides an alternative explanation for the reduced breadth of the CD8+ T cell immune response. Furthermore, proteins that associate with the inclusion-membrane (such as CrpA) or that are surface-exposed (such as PmpI) and so interact with the host are polymorphic and may thus evade immune detection by sequence variation. Moreover, it has been demonstrated that class I MHC expression is inhibited in Chlamydia-infected cells (39) and that activated T cells in Chlamydia-infected individuals undergo macrophage-induced apoptosis (40).
In conclusion, we have shown that H-2Kb and H-2Db MHC tetramers can be effectively applied to screen arrays of ≈2,000 predicted epitopes and allow direct phenotypic identification of CD8+ T cells without relying on their effector function or proliferative capabilities. The number of CD8+ T cells that can be obtained from a mouse currently limits the number of peptides that can be screened to ≈50–100 candidate epitopes per animal, on the assumption that positive identification minimally requires ≈0.1–0.5% of tetramer-positive CD8+ T cells. The overall cost of a screen depends on the size of the pathogen's proteome and the desired sequence coverage and is currently mostly attributable to the cost of peptide synthesis. Collectively, these studies provide a framework for the identification of CD8+ T cell epitopes by class I MHC tetramer arrays in the C57BL/6-mouse model that are readily extendable to other pathogenic challenges.
Materials and Methods
Class I MHC Complex Production.
Peptides were constructed by standard Fmoc solid-phase peptide synthesis. Soluble H-2Kb and H-2Db complexes were produced following established protocols (26). Further experimental details, including crystallization, x-ray data collection, and structure refinement are provided in SI Materials and Methods. Structure coordinates and structure factors of H-2Db in complex with SV9-P7*B have been deposited in the Protein Data Bank (entry 2VE6).
Class I MHC Peptide Exchange.
SAPE (Invitrogen) was added five times at 60 min intervals to biotinylated monomer—either H-2Kb or H-2Db with SV9-P7*—to a final molar ratio of 4:1 for monomer:SAPE. Tetramers were stored at 4°C and used within 1 month. Tetramers were diluted to 50 μg/ml in Dulbecco's modified Eagle's medium (DMEM) without phenol red and supplemented with 10% FCS, penicillin (100 units/ml), streptomycin (100 μg/ml), 2 mM glutamine, and 0.1% NaN3. The solution was deposited in a 96-well plate (V-bottom, 110 μl per well), and index peptide was added (2 μl per well). The plate was placed on ice and irradiated for 15 min in a Stratalinker 2400 UV cross-linker equipped with 365-nm UV-lamps at an ≈10- to 20-cm distance. After 1 h of incubation, the plate was centrifuged (3, 000 × g) for 20 min, and 50 μl of the supernatant was transferred into two new 96-well plates.
Flow Cytometry.
Splenocytes were enriched to >80% CD8+ T cells by negative selection, using Dynal beads (Invitrogen), and ≈1 × 107 cells were resuspended in RPMI medium 1640 supplemented as described above for DMEM. The CD8+ T cells were incubated with ethidium monoazide (15 min) while excluding light, washed, and irradiated (10 min) with incandescent light (100 W). Subsequently, the CD8+ T cells were distributed over the 96-well plates (≈1 × 105 cells in 50 μl per well) that contained saturating amounts of freshly prepared MHC tetramer and FITC-conjugated anti-CD8 mAb (Becton Dickinson) and were incubated for 45 min. The cells were washed with PBS and fixed with 4% formaldehyde in PBS. Cells were analyzed with a FACSCalibur flow cytometer. Data analysis was performed with FlowJo software (Tree Star).
Defining ORFs for C. trachomatis L2.
The 902 proteins encoded by C. trachomatis D/UW-3/CX were extracted from the National Center for Biotechnology Information GenBank database (www.ncbi.nlm.nih.gov) with accession number NC_000117. The sequence data for C. trachomatis serovar L2 were produced by the Sanger Institute and were obtained from www.sanger.ac.uk/Projects/C_trachomatis_L2.
Epitope Prediction.
The consensus epitope prediction program was reconstructed by using the scoring matrices published in ref. 16. The program, written as a Perl script, is available at http://jura.wi.mit.edu/bioc/grotenbreg, including documentation for its execution and sample data.
Mice Infection with C. trachomatis L2.
Elementary bodies of C. trachomatis serovar L2 434/Bu were propagated and purified as described in ref. 41. C57BL/6 mice were infected intravenously with 107 inclusion forming units of C. trachomatis serovar L2 and splenocytes were harvested six days later.
Chromium Release Assay.
Peptide-treated (100 nM) or untreated EL4 cells were loaded with 100 μCi sodium 51Cr for 1 h at 37°C, washed three times, and resuspended to 104 cells per well in 96-well plates. Splenocytes from immunized mice were restimulated five days with peptide, serially diluted, and added to a final volume of 200 μl per well. After 4 h incubation, the cytotoxic activity was assessed by the 51Cr in the supernatant, using a Wallac 1470 Wizard gamma counter. Maximum lysis was determined by the addition of 1% Triton X-100. Specific lysis was determined by using the following equation: [(release by CTL − spontaneous release)/(maximum release − spontaneous release)] × 100.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health grants (to M.N.S., J.-h.W., and H.L.P.), Novartis Vaccines, and a postdoctoral fellowship from the Netherlands Organization for Scientific Research (to G.M.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID code 2VE6 (H-2Db in complex with SV9-P7*B)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0711504105/DC1.
References
- 1.Miyahira Y, et al. Quantification of antigen specific CD8+ T cells using an ELISPOT assay. J Immunol Methods. 1995;181:45–54. doi: 10.1016/0022-1759(94)00327-s. [DOI] [PubMed] [Google Scholar]
- 2.Holden HT, Oldham RK, Ortaldo JR, Herberman RB. Standardization of the chromium-51 release, cell-mediated cytotoxicity assay: cryopreservation of mouse effector and target cells. J Natl Cancer Inst. 1977;58:611–622. doi: 10.1093/jnci/58.3.611. [DOI] [PubMed] [Google Scholar]
- 3.Kern F, et al. T-cell epitope mapping by flow cytometry. Nature Medicine. 1998;4:975–978. doi: 10.1038/nm0898-975. [DOI] [PubMed] [Google Scholar]
- 4.Moss PAH, et al. Persistent high-frequency of human immunodeficiency virus-specific cytotoxic T-cells in peripheral-blood of infected donors. Proc Natl Acad Sci USA. 1995;92:5773–5777. doi: 10.1073/pnas.92.13.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalams SA, et al. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J Exp Med. 1994;179:1261–1271. doi: 10.1084/jem.179.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 7.Klenerman P, Cerundolo V, Dunbar RR. Tracking T cells with tetramers: New tales from new tools. Nat Rev Immunol. 2002;2:263–272. doi: 10.1038/nri777. [DOI] [PubMed] [Google Scholar]
- 8.Bakker AH, Schumacher TNM. MHC multimer technology: Current status and future prospects. Curr Opin Immunol. 2005;17:428–433. doi: 10.1016/j.coi.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Toebes M, et al. Design and use of conditional MHC class I ligands. Nat Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 10.Rodenko B, et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat Protocols. 2006;1:1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- 11.Ward ME. The immunobiology and immunopathology of chlamydial infections. Apmis. 1995;103:769–796. doi: 10.1111/j.1699-0463.1995.tb01436.x. [DOI] [PubMed] [Google Scholar]
- 12.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: Implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 13.Ramsey KH, Rank RG. Resolution of chlamydial genital infection with antigen-specific T-lymphocyte lines. Infect Immun. 1991;59:925–931. doi: 10.1128/iai.59.3.925-931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez CM, et al. Relative importance of CD4+ and CD8+ T cells in the resolution of Chlamydophila abortus primary infection in mice. J Comp Pathol. 2006;134:297–307. doi: 10.1016/j.jcpa.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Starnbach MN, et al. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J Immunol. 2003;171:4742–4749. doi: 10.4049/jimmunol.171.9.4742. [DOI] [PubMed] [Google Scholar]
- 16.Moutaftsi M, et al. A consensus epitope prediction approach identifies the breadth of murine TCD8+-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher TNM, et al. Peptide selection by MHC class I molecules. Nature. 1991;350:703–706. doi: 10.1038/350703a0. [DOI] [PubMed] [Google Scholar]
- 18.Kast WM, et al. Protection against lethal Sendai virus infection by in vivo priming of virus-specific cytotoxic T lymphocytes with a free synthetic peptide. Proc Natl Acad Sci USA. 1991;88:2283–2287. doi: 10.1073/pnas.88.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 20.Glithero A, et al. Crystal structures of two H-2Db/glycopeptide complexes suggest a molecular basis for CTL cross-reactivity. Immunity. 1999;10:63–74. doi: 10.1016/s1074-7613(00)80007-2. [DOI] [PubMed] [Google Scholar]
- 21.Grotenbreg GM, et al. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J Biol Chem. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neefjes JJ, Dierx J, Ploegh HL. The effect of anchor residue modifications on the stability of major histocompatibility complex class I-peptide interactions. Eur J Immunol. 1993;23:840–845. doi: 10.1002/eji.1830230411. [DOI] [PubMed] [Google Scholar]
- 23.Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 24.Beckett D. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 26.Garboczi D, Hung D, Wiley D. HLA-A2-peptide complexes: Refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens RS, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Fling SP, et al. CD8(+) T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc Natl Acad Sci USA. 2001;98:1160–1165. doi: 10.1073/pnas.98.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero H, Zavala A, Musto H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 2000;28:2084–2090. doi: 10.1093/nar/28.10.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalman S, et al. Comparative genomes of Chlamydia pneumoniae and C-trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 32.Henderson IR, Lam AC. Polymorphic proteins of Chlamydia spp.—autotransporters beyond the Proteobacteria. Trends Microbiol. 2001;9:573–578. doi: 10.1016/s0966-842x(01)02234-x. [DOI] [PubMed] [Google Scholar]
- 33.Gomes JP, et al. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: Evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol. 2006;188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunes A, et al. Comparative Expression Profiling of the Chlamydia trachomatis pmp Gene Family for Clinical and Reference Strains. PLoS ONE. 2007;2:e878. doi: 10.1371/journal.pone.0000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skipp P, Robinson J, O'Connor CD, Clarke IN. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics. 2005;5:1558–1573. doi: 10.1002/pmic.200401044. [DOI] [PubMed] [Google Scholar]
- 36.Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun. 2005;73:6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 38.Shastri N, Schwab S, Serwold T. Producing nature's gene-chips: The generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 39.Zhong GM, Liu L, Fan T, Fan PY, Ji HZ. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in Chlamydia-infected cells. J Exp Med. 2000;191:1525–1534. doi: 10.1084/jem.191.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jendro MC, et al. Chlamydia trachomatis-infected macrophages induce apoptosis of activated T cells by secretion of tumor necrosis factor-a in vitro. Med Microbiol Immunol. 2004;193:45–52. doi: 10.1007/s00430-003-0182-1. [DOI] [PubMed] [Google Scholar]
- 41.Starnbach MN, Bevan MJ, Lampe MF. Protective cytotoxic T lymphocytes are induced during murine infection with Chlamydia trachomatis. J Immunol. 1994;153:5183–5189. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.