Abstract
Patients with hematological malignancies can be successfully treated with HLA-matched T cell-depleted allogeneic stem cell transplantation (alloSCT) and subsequent donor lymphocyte infusions (DLIs). The efficacy of DLI is mediated by donor T cells recognizing minor histocompatibility antigens (mHags) on malignant recipient cells. Because HLA class II molecules are predominantly expressed on hematopoietic cells, mHag-specific CD4+ T cells may selectively mediate graft versus leukemia (GvL) reactivity without graft versus host disease (GvHD). In this study, we used a recombinant bacteria cDNA library for the identification of the first autosomal HLA class II (HLA-DQB1*0603)-restricted mHag LB-PI4K2B-1S encoded by the broadly expressed phosphatidylinositol 4-kinase type II β gene. A polyclonal CD4+ T cell response against LB-PI4K2B-1S was demonstrated in a patient with relapsed chronic myeloid leukemia (CML) who responded to DLI after HLA-matched alloSCT. LB-PI4K2B-1S-specific CD4+ T cells recognized and lysed the CD34+ CML cells of the patient and other leukemic cells as well as high HLA-DQ-expressing normal hematopoietic cells. HLA-DQ expression on normal cells of nonhematopoietic origin was moderately up-regulated by IFN-γ and not sufficient for T cell recognition. We hypothesize that LB-PI4K2B-1S-specific CD4+ T cells contributed to the antitumor response by both directly eliminating malignant cells as effector cells and stimulating CD8+ T cell immunity as helper cells.
Keywords: donor lymphocyte infusions, hematological malignancies, immunotherapy, stem cell transplantation, T lymphocytes
Patients with hematological malignancies can be successfully treated with HLA-matched T cell-depleted allogeneic stem cell transplantation (alloSCT) and subsequent donor lymphocyte infusions (DLIs) (1–3). The efficacy of DLI is mediated by donor T cells recognizing minor histocompatibility antigens (mHags) on malignant recipient cells. CD8+ T cells recognizing hematopoietic-restricted mHags may selectively mediate graft versus leukemia (GvL) reactivity, whereas CD8+ T cells directed against broadly expressed mHags may play a role in GvL as well as graft versus host disease (GvHD). Several HLA class I-restricted mHags, including HA-1 (4), HA-2 (5), LRH-1 (6), LB-ECGF-1H (7), and LB-ADIR-1F (8), have been identified as targets for CD8+ T cells induced in patients who developed strong GvL reactivity after treatment with DLI for relapsed hematological malignancies after alloSCT. In these patients, the appearance of circulating mHag-specific CD8+ T cells was immediately followed by complete remissions of their malignancies (6–9), indicating that mHag-specific CD8+ T cells can induce clinical responses.
HLA class I-restricted mHags have been identified by different strategies, including the biochemical characterization of eluted peptides (4, 5, 8), the screening of plasmid cDNA expression libraries (7, 10–12), and genetic linkage analysis (6, 13). In contrast to HLA class I molecules, which are expressed on all nucleated cells, HLA class II molecules are predominantly expressed on hematopoietic cells. Therefore, mHag-specific CD4+ T cells may selectively mediate GvL reactivity with no or limited GvHD (14–16). Because of technical limitations, only HLA class II-restricted H-Y antigens have been characterized by selecting and testing Y chromosome-specific genes for recognition by CD4+ T cells isolated from male patients after sex-mismatched alloSCT (17, 18). However, thus far, attempts to identify HLA class II mHags encoded by autosomal genes have been unsuccessful. The lack of identified HLA class II mHags greatly hampers detailed investigations of mHag-specific CD4+ T cells, and therefore their role in mediating GvL and GvHD remains largely unclarified. In contrast to endogenous antigens, which preferentially enter the HLA class I pathway, exogenous antigens are most efficiently processed and presented by the HLA class II pathway. Therefore, we developed recombinant bacteria cDNA expression libraries based on the delivery of exogenous antigens for the identification of HLA class II antigens. The feasibility of this rapid method, which allows the use of endogenous HLA class II molecules as expressed on EBV-transformed B lymphoblastoid cell lines (EBV-LCLs), was previously demonstrated by isolation of the DBY cDNA encoding an HLA-DQ5 H-Y antigen (19) and was used in the present study for identification of a human autosomal HLA class II mHag.
By screening a recombinant bacteria cDNA library, we identified an HLA-DQB1*0603-restricted mHag encoded by the broadly expressed phosphatidylinositol 4-kinase type II β gene (PI4K2B), designated as LB-PI4K2B-1S. A polyclonal CD4+ T cell response against LB-PI4K2B-1S was demonstrated in a patient successfully treated with DLI for relapsed chronic myeloid leukemia (CML) after HLA-matched alloSCT. The data also show that LB-PI4K2B-1S-specific CD4+ T cells were capable of mediating direct cytolyis of normal and malignant hematopoietic cells and therefore suggest involvement in antitumor immunity as effector as well as helper cells.
Results
Isolation and Characterization of CD4+ T Cell Clone ZRZ38.
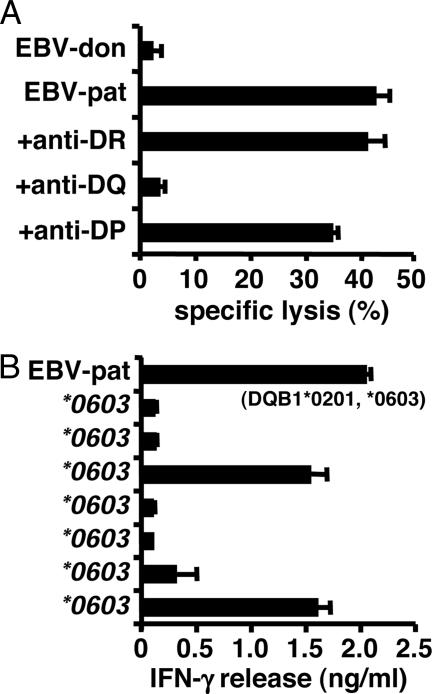
As described previously, CD4+ T cell clone ZRZ38 was isolated from patient RZ with relapsed CML who responded to DLI after HLA-matched alloSCT and developed only mild GvHD of the skin (20). Clone ZRZ38 differentially recognized and lysed patient and donor EBV-LCL in a standard 4-h 51Cr-release assay (Fig. 1). Lysis of patient EBV-LCL was inhibited after the addition of blocking antibodies against HLA-DQ, whereas antibodies against HLA-DR or HLA-DP had no effect (Fig. 1A). Screening of a panel of EBV-LCLs sharing one of the two HLA-DQ alleles of the patient indicated recognition of an HLA-DQB1*0603-restricted mHag (Fig. 1B). A haplotype consisting of HLA-DQA1*0103 and HLA-DQB1*0603 is present in ≈14% of the Caucasian population. Retroviral transductions with patient-derived HLA-DQ cDNAs confirmed the presentation of the mHag by an HLA class II heterodimer composed of the DQA1*0103 and DQB1*0603 subunits (data not shown).
Fig. 1.
Characterization of CD4+ T cell clone ZRZ38. (A) CD4+ T cell clone ZRZ38 was tested against donor and patient EBV-LCLs with and without blocking antibodies against HLA-DR, HLA-DQ, and HLA-DP in a 4-h 51Cr-release assay. Mean specific lysis of triplicate wells at an E/T ratio of 10:1 is shown. (B) A panel of EBV-LCLs sharing HLA-DQB1*0603 with the patient was tested for recognition by clone ZRZ38 in IFN-γ ELISA. The mean release of IFN-γ (ng/ml) in 50-μl supernatants of duplicate wells is shown. EBV-LCLs sharing HLA-DQB1*0201 with the patient were not recognized by clone ZRZ38 (data not shown).
Identification of LB-PI4K2B-1S as an HLA-DQB1*0603-Restricted mHag.
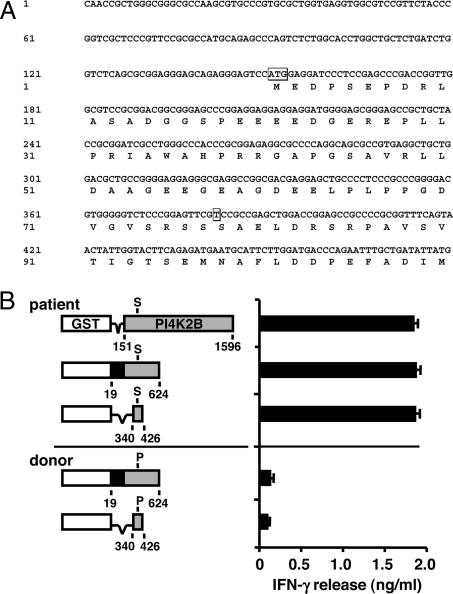
To identify the mHag recognized by CD4+ T cell clone ZRZ38, a recombinant bacterial cDNA library was constructed from EBV-LCL from patient RZ and screened for T cell recognition. Briefly, ≈4 × 106 random-primed cDNAs ranging from 400 bp to >3 kb were cloned into vector pKE-1, which encodes the glutathione-binding domain of GST under the control of an isopropyl β-d-thiogalactoside (IPTG)-inducible promoter (21). After induction of protein expression upon the addition of IPTG, bacteria were opsonized with complement and loaded onto donor EBV-LCLs, and pools of ≈50 different bacteria were screened for T cell recognition in IFN-γ ELISA. After a first screening of 960 pools, one pool strongly stimulated IFN-γ release by clone ZRZ38. Subcloning of this pool revealed a 3,295-bp cDNA, which was identical to the gene-encoding PI4K2B (GenBank accession nos. BC051749 and NM_018323), containing a known C-to-T SNP at 382 bp (SNP database ID no. rs313549) within the 151- to 1,596-bp protein-encoding region (Fig. 2A).
Fig. 2.
Identification of the PI4K2B cDNA region recognized by CD4+ T cell clone ZRZ38. (A) DNA sequence of the first 480 bp of the isolated 3,295-bp PI4K2B cDNA. Indicated are the ATG start at 151 bp and the SNP at 382 bp that leads to translation of a Pro (donor) or Ser (patient) at position 78 of the PI4K2B protein. (B) Different fragments of the isolated PI4K2B cDNA from patient RZ (151–1596, 19–624, and 340–426 bp) and donor Z (19–624 and 340–426 bp) were cloned in the same ORF as the GST sequence in pKE-1. The PI4K2B cDNA contains an ATG start at 151 bp and a TAG stop at 1,596 bp. Recombinant bacteria expressing truncated PI4K2B proteins were opsonized with complement, pulsed onto donor EBV-LCLs, and tested for recognition by CD4+ T cell clone ZRZ38 in IFN-γ ELISA. The mean release of IFN-γ (ng/ml) in 50-μl supernatants of duplicate wells is shown.
A comparison of PI4K2B sequences between patient and donor revealed that the patient was C/T heterozygous at 382 bp, whereas the donor was C/C homozygous. This C-to-T transition created a Pro-to-Ser substitution at position 78 (P/S78) of the PI4K2B protein. To confirm that the SNP was differentially recognized by clone ZRZ38, different fragments of the PI4K2B cDNA (151–1,596, 19–624, and 340–426 bp) were amplified by PCR and cloned into pKE-1. Recombinant bacteria expressing truncated PI4K2B proteins were opsonized with complement, pulsed on donor EBV-LCLs, and tested for T cell recognition in IFN-γ ELISA (Fig. 2B). The data demonstrated that an 87-bp patient-derived PI4K2B cDNA (340–426 bp) with S78 (LB-PI4K2B-1S) stimulated IFN-γ release by clone ZRZ38, whereas no IFN-γ production was measured upon stimulation with a donor-derived 87-bp PI4K2B cDNA with P78 (LB-PI4K2B-1P).
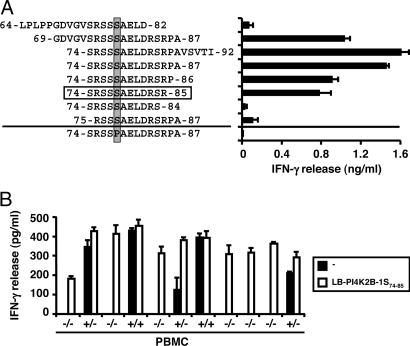
To identify the LB-PI4K2B-1S epitope recognized by clone ZRZ38, three overlapping 19-mer peptides (amino acids 64–82, 69–87, and 74–92) were synthesized and tested for T cell recognition (Fig. 3A). Two of the three peptides (amino acids 69–87 and 74–92) stimulated IFN-γ release by clone ZRZ38. An LB-PI4K2B-1S peptide (amino acids 74–87) containing the 14 aa shared by the two 19-mer peptides also stimulated IFN-γ release, in contrast to a 14-mer LB-PI4K2B-1P peptide. Serial truncation of the 14-mer LB-PI4K2B-1S peptide at the N and C termini revealed recognition of a minimal 12-mer SRSSSAELDRSR peptide (amino acids 74–85).
Fig. 3.
Identification of the LB-PI4K2B-1S epitope recognized by CD4+ T cell clone ZRZ38. (A) A series of peptides comprising the amino acids 64–92 region of LB-PI4K2B-1S were synthesized, pulsed onto donor EBV-LCLs, and tested for recognition by CD4+ T cell clone ZRZ38 in IFN-γ ELISA. Indicated is the S/P residue at position 78 (shaded) and the minimal 12-mer SRSSSAELDRSR peptide (amino acids 74–85, boxed) recognized by clone ZRZ38. The mean release of IFN-γ (ng/ml) in 50-μl supernatants of duplicate wells is shown. (B) PBMCs from HLA-DQB1*0603+ healthy individuals were genotyped for the LB-PI4K2B-1S/P polymorphism and tested for T cell recognition in IFN-γ ELISA (filled bars). As a control for cell viability and HLA-DQB1*0603 expression, PBMCs were tested for T cell recognition after 2 h of pulsing with the 12-mer LB-PI4K2B-1S peptide (open bars). Genotype data are shown as +/+ (S/S), +/− (S/P), and −/− (P/P). The mean release of IFN-γ (pg/ml) in 50-μl supernatants of duplicate wells is shown.
To confirm that LB-PI4K2B-1S is the mHag recognized by clone ZRZ38, peripheral blood mononuclear cells (PBMCs) from HLA-DQB1*0603+ healthy individuals were genotyped for the LB-PI4K2B-1S allele and tested for T cell recognition in IFN-γ ELISA. As a control for cell viability and HLA-DQB1*0603 expression, PBMCs also were tested after 2 h of pulsing with an LB-PI4K2B-1S peptide. Fig. 3B shows a population frequency of LB-PI4K2B-1S of 40–50% and a strict correlation between the presence of an LB-PI4K2B-1S allele and IFN-γ release by clone ZRZ38.
To determine the frequency of LB-PI4K2B-1S-specific CD4+ T cells in patient RZ, bone marrow mononuclear cells obtained 5 weeks after DLI were stained with antibodies against CD4 and HLA-DR, which is a marker for activated T cells. CD4+ and HLA-DR+ cells were sorted at 1 cell per well. Of the 21 CD4+ T cell clones with differential reactivity to patient and donor EBV-LCL, nine clones recognized donor EBV-LCL pulsed with an LB-PI4K2B-1S peptide (data not shown). These LB-PI4K2B-1S-reactive CD4+ T cell clones expressed different TCR BV chains with CDR3 regions variable in length and amino acid composition (data not shown), demonstrating the induction of a polyclonal CD4+ T cell response against LB-PI4K2B-1S.
Cell-Type Specificity of LB-PI4K2B-1S-Reactive CD4+ T Cells.
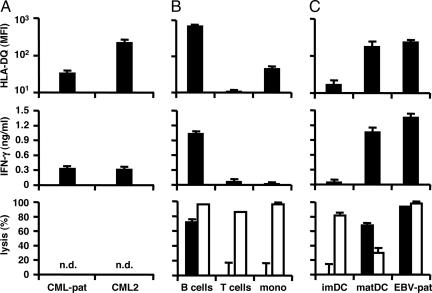
To determine the target cell-type specificity of LB-PI4K2B-1S-reactive CD4+ T cells, CD34+ CML cells isolated from bone marrow mononuclear cells from patient RZ upon relapse of the CML after alloSCT and another patient with chronic CML were analyzed for HLA-DQ surface expression and specific recognition by clone ZRZ38 in IFN-γ ELISA. In addition, various subsets of normal hematopoietic cells [B cells, T cells, monocytes, and immature and mature dendritic cells (DCs)] were isolated from PBMCs from an HLA-DQB1*0603+ and LB-PI4K2B-1S+ healthy individual and analyzed for HLA-DQ surface expression, as well as specific recognition and lysis by clone ZRZ38 in IFN-γ ELISA and CFSE-based cytotoxicity assays. LB-PI4K2B-1S-reactive CD4+ T cells recognized the CD34+ CML cells from patient RZ that moderately expressed HLA-DQ (Fig. 4A), as well as other high HLA-DQ-expressing CML (Fig. 4A). LB-PI4K2B-1S-reactive CD4+ T cells also recognized and lysed acute lymphoblastic leukemia (ALL) cells from an HLA-DQB1*0603+ and LB-PI4K2B-1S+ patient (data not shown). Flow-cytometric analysis demonstrated high HLA-DQ expression on normal B cells and EBV-LCLs, whereas monocytes displayed low HLA-DQ, and the majority of T cells and PHA-stimulated blasts completely lacked HLA-DQ expression (Fig. 4B) (data not shown). During differentiation of monocytes to immature DCs, HLA-DQ expression remained low, but HLA-DQ as well as CD80 and CD86 (data not shown) expression was significantly up-regulated upon DC maturation (Fig. 4C). Fig. 4 B and C shows a correlation between HLA-DQ expression and recognition and lysis of normal hematopoietic cells by clone ZRZ38, which is restricted to high HLA-DQ-expressing B cells, mature DCs, and EBV-LCLs.
Fig. 4.
Recognition and lysis of hematopoietic cells by LB-PI4K2B-1S-specific CD4+ T cells. (A) CD34+ CML cells isolated from bone marrow mononuclear cells from patient RZ (CML-pat) upon relapse of the malignancy after alloSCT and another HLA-DQB1*0603+ and LB-PI4K2B-1S+ patient with chronic CML (>75% purity, CML2). (B) B cells, T cells, and monocytes isolated from PBMCs from an HLA-DQB1*0603+ and LB-PI4K2B-1S+ healthy individual (>90% purity). (C) Immature and mature DCs and EBV-LCLs from patient RZ were stained with PE-labeled antibodies against HLA-DQ (Leinco Technologies) and analyzed by flow cytometry. (Top) The mean fluorescence intensity (MFI) of two stainings. These cell populations also were used as stimulator cells (3 × 104 cells per well) for CD4+ T cell clone ZRZ38 (5 × 103 cells per well) in IFN-γ ELISA. (Middle) The mean release of IFN-γ (ng/ml) in 50-μl supernatants of triplicate wells. In addition, these cell populations were stained with CFSE and used as target cells (1 × 104 cells per well) for CD4+ T cell clone ZRZ38 and alloreactive HLA-A*0201-specific CD8+ T cell clone MBM13 in CFSE-based cytotoxicity assays at an E/T ratio of 10:1. Numbers of isolated CD34+ CML cells were not sufficient to measure specific lysis. (Bottom) The mean percentage of lysis of CFSE+ cells of triplicate wells after coincubation with clone ZRZ38 (filled bars) and control anti-HLA-A*0201 CD8+ T cell clone MBM13 (open bars). n.d., not done.
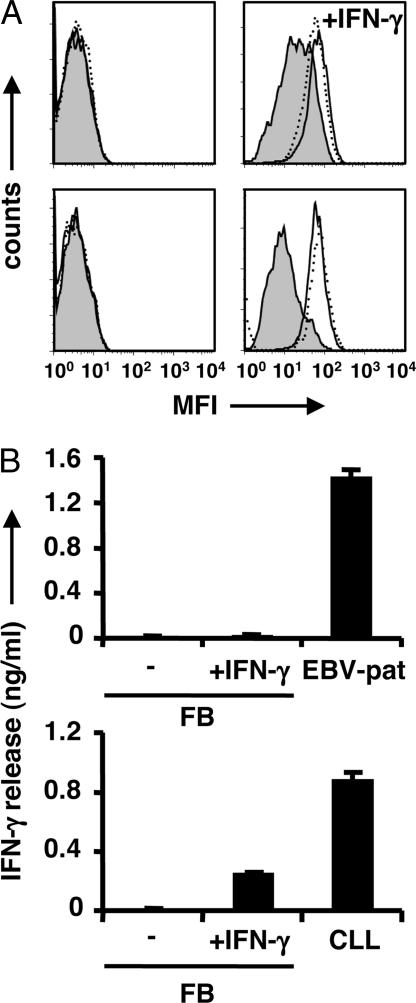
In addition to hematopoietic cells, the reactivity of LB-PI4K2B-1S-specific CD4+ T cells toward normal cells of nonhematopoietic origin, including dermal fibroblasts (FBs), keratinocytes (KCs), and biliary epithelial cells (BECs), was investigated. Flow-cytometric analysis demonstrated that the expression of HLA-DR and HLA-DP was significantly up-regulated on FBs, KCs, and BECs after treatment with IFN-γ, whereas HLA-DQ expression remained low and only measurable after long-term treatment with high doses of IFN-γ (Fig. 5A) (data not shown). Moreover, the IFN-γ-induced up-regulation of HLA-DQ expression on FBs was not sufficient for T cell recognition because no release of IFN-γ by CD4+ T cell clone ZRZ38 was measured after stimulation with IFN-γ-treated HLA-DQB1*0603+ and LB-PI4K2B-1S+ FBs, whereas alloreactive HLA-DPB1*0301-specific CD4+ T cells produced significant levels of IFN-γ after stimulation with IFN-γ-treated HLA-DPB1*0301+ FBs (Fig. 5B). These data suggest that CD4+ T cells recognizing LB-PI4K2B-1S in the context of HLA-DQB1*0603 are not reactive with normal cells of nonhematopoietic origin and therefore are not expected to cause GvHD.
Fig. 5.
Recognition of nonhematopoietic cells by LB-PI4K2B-1S-specific CD4+ T cells. (A) FBs from HLA-DQB1*0603+ and LB-PI4K2B-1S+ (Upper) or HLA-DPB1*0301+ (Lower) individuals were seeded at 3 × 103 cells per well and cultured for 7 days with or without 200 units/ml IFN-γ. FBs were analyzed by flow cytometry after staining with PE-labeled antibodies against HLA-DQ (filled histogram), HLA-DR (solid line), or HLA-DP (dotted line). Histograms show the MFI after culturing with (Right) or without (Left) IFN-γ. Nonstained FBs cultured with or without IFN-γ had similar MFI (data not shown). (B) (Upper) FBs from the HLA-DQB1*0603+ and LB-PI4K2B-1S+ individual treated with or without IFN-γ or patient EBV-LCLs were used as stimulator cells for CD4+ T cell clone ZRZ38 (5 × 103 cells per well) in IFN-γ ELISA. (Lower) FBs from the HLA-DPB1*0301+ individual cultured with or without IFN-γ or HLA-DPB1*0301+ CLL cells were used as stimulator cells for alloreactive HLA-DPB1*0301-specific CD4+ T cell clone CS2–1 (5 × 103 cells per well) in IFN-γ ELISA. The mean release of IFN-γ (ng/ml) in 50-μl supernatants of triplicate wells is shown.
Finally, expression of the PI4K2B gene in B cell [ALL, chronic lymphocytic leukemia (CLL), and multiple myeloma (MM)] and myeloid (acute myeloid leukemia and CML) malignancies and normal hematopoietic cells (PBMCs, B cells, T cells, and monocytes) and nonhematopoietic cells (mesenchymal stem cells and FBs) was investigated by quantitative real-time RT-PCR. The data showed that the PI4K2B gene was constitutively expressed in all normal and malignant hematopoietic and nonhematopoietic cells, confirming the broad tissue-distribution pattern of the PI4K2B gene as determined by public microarray databases (data not shown).
Discussion
CD4+ T cells are indispensable as helper cells in the induction and maintenance of CD8+ T cell immunity (22); they also have been described to exert direct cytolytic activity as effector cells (20, 23–25). CD4+ T cells recognize antigens in the context of HLA class II molecules, which are predominantly expressed on hematopoietic cells and may therefore selectively mediate GvL reactivity without GvHD. In this study, we demonstrated induction of a polyclonal CD4+ T cell response against an HLA-DQB1*0603-restricted mHag in a patient with relapsed CML who responded to DLI after HLA-matched alloSCT. Also, by screening a recombinant bacteria cDNA expression library, we identified an autosomal HLA class II-restricted mHag designated as LB-PI4K2B-1S.
The HLA-DQB1*0603-restricted mHag has a population frequency of 40–50% and is encoded by the PI4K2B gene, which is broadly expressed in human tissues. Our data show that recognition and lysis of normal hematopoietic cells by LB-PI4K2B-1S-specific CD4+ T cells critically depends on the number of HLA-DQ molecules expressed at the cell surface and is restricted to high HLA-DQ-expressing B cells, mature DCs, and EBV-LCLs. HLA-DQ expression on T cells, PHA-stimulated blasts, monocytes, and immature DCs is absent or low and not sufficient for T cell recognition. LB-PI4K2B-1S-specific CD4+ T cells also were demonstrated to recognize the CD34+ CML cells of the patient as well as other HLA-DQB1*0603+ and LB-PI4K2B-1S+ CML and ALL cells. Although tumor cells generally fail to induce immune responses due to lack or low expression of costimulatory and adhesion molecules (26), CD34+ CML precursor cells are capable of differentiating into functional antigen-presenting cells (APCs) (27, 28). Therefore, in the patient with relapsed CML in the chronic phase, LB-PI4K2B-1S-specific CD4+ T cells may have been induced by CD34+ CML cells or mature APCs originating from these malignant progenitor cells. Upon induction in vivo, LB-PI4K2B-1S-specific CD4+ T cells are expected to contribute to the antitumor response by directly eliminating malignant cells as effector cells and stimulating CD8+ T cell immunity as helper cells.
In several patients treated with alloSCT, high frequencies of mHag-specific CD4+ T cells have been shown to precede and closely correlate with the onset of clinical GvHD (23, 29). Although constitutive HLA class II expression is confined to hematopoietic cells, expression can be induced on a variety of nonhematopoietic cells after treatment with proinflammatory cytokines. Therefore, CD4+ T cells recognizing mHags in HLA class II may contribute to the development of GvHD when high amounts of proinflammatory cytokines are released as a consequence of tissue injury induced by conditioning regimens, chemotherapeutic drugs, or high pathogenic loads after transplantation (30).
In conclusion, by screening a recombinant bacteria cDNA library, we identified an autosomal HLA class II (HLA-DQB1*0603)-restricted mHag that is recognized by CD4+ T cells induced in a patient with relapsed CML who showed a strong antitumor response after HLA-matched alloSCT and DLI. Because simultaneous CD8+ T cells recognizing HLA class I-restricted mHags, including HA-1 and HA-2, were demonstrated in this patient previously (9, 20), we hypothesize that LB-PI4K2B-1S-specific CD4+ T cells stimulated the induction and maintenance of mHag-specific CD8+ T cells, which subsequently played a dominant role in the direct elimination of the CML stem cells. Considering their crucial role as helper cells, local production of cytokines by mHag-specific CD4+ T cells may determine the numbers and overall reactivity of infiltrated CD8+ T cells and may therefore be a key aspect in the development of GvHD. We demonstrated that HLA-DQ expression on FBs, KCs, and BECs after extensive culturing with IFN-γ was moderately up-regulated, compared with HLA-DR and HLA-DP, and not sufficient for recognition by LB-PI4K2B-1S-specific CD4+ T cells. Therefore, CD4+ T cell recognition of mHags presented by HLA-DQ on nonhematopoietic cells may be limited, and HLA-DQ-restricted mHags may be appropriate targets for T cell therapies with the aim to selectively stimulate GvL without GvHD.
Materials and Methods
Cell Culture.
Peripheral blood, bone marrow, and skin biopsies were collected from healthy individuals and patients with hematological malignancies after approval by the Leiden University Medical Center Institutional Review Board and informed consent according to the Declaration of Helsinki. CD4+ T cell clone ZRZ38, alloreactive HLA-A*0201-specific CD8+ T cell clone MBM13, and alloreactive HLA-DPB1*0301-specific CD4+ T cell clone CS2–1 were cultured and stimulated every 10–20 days with irradiated allogeneic PBMCs and patient EBV-LCLs as described previously (7, 8). EBV-LCLs were cultured in IMDM (Cambrex) with 8% FBS. B cells, T cells, and monocytes were isolated from PBMCs and CD34+ CML cells from bone marrow mononuclear cells by using magnetic beads (Miltenyi Biotec). Primary hematopoietic cells were maintained in IMDM with 10% human ABO serum. Isolated monocytes were cultured to immature DCs in medium with 100 ng/ml GM-CSF (Novartis) and 500 units/ml IL-4 (Schering-Plough) for 7 days. Mature DCs were cultured from days 5–7 in medium with 100 ng/ml GM-CSF, 10 ng/ml TNFα, 10 ng/ml IL-1β, 10 ng/ml IL-6 (Cellgenix), 1 μg/ml prostaglandin E2 (Sigma–Aldrich), and 500 units/ml IFN-γ (Immukine/Boehringer Ingelheim). FBs were cultured from skin biopsies in DMEM with low glucose (Cambrex) and 10% FBS.
51Cr-Release Cytotoxicity Assay.
Target cells were labeled with 100 μCi (3.7 MBq) Na251CrO4 (Amersham Pharmacia Biotech) for 1 h at 37°C. After washing, target cells were preincubated with blocking antibodies against HLA-DR (B8.11.2), HLA-DP (B7.21), or HLA-DQ (SPV-L3) (kindly provided by A. Mulder, Department of Immunohematology and Blood Transfusion, Leiden University Medical Center) for 30 min at 37°C. Target cells (2 × 103 cells per well) were incubated with CD4+ T cell clone ZRZ38 at an E/T ratio of 10:1. After 4 h of incubation, 25 μl of supernatant was harvested for 51Cr analysis. The percentage of specific lysis was calculated according to the formula: [experimental release (cpm) − spontaneous release (cpm)]/[maximal release (cpm) − spontaneous release (cpm)] × 100%.
IFN-γ ELISA.
Hematopoietic cells were used as stimulator cells (3 × 104 cells per well) for CD4+ T cell clone ZRZ38 (5 × 103 cells per well) in U-bottom 96-well plates. Human FBs (3 × 103 cells per well) were cultured in flat-bottom 96-well plates with or without IFN-γ (200 units/ml) for 7 days and subsequently incubated with clone ZRZ38 (5 × 103 cells per well). After 24 h of incubation at 37°C, IFN-γ release in 50-μl supernatants was measured by ELISA (Sanguin).
CFSE-Based Cytotoxicity Assay.
CFSE-based cytotoxicity assays were performed as described previously (31). Target cells were stained with 2.5 μM CFSE (Molecular Probes) for 10 min at 37°C and incubated (1 × 104 cells per well) with CD4+ T cell clone ZRZ38 or CD8+ T cell clone MBM13 at an E/T ratio of 10:1. After overnight incubation at 37°C, cocultures were collected and stained with APC-labeled antibodies against markers for B cells (CD19), T cells (CD3), or myeloid cells (CD33) for 30 min on ice. Before analysis, propidium iodide (PI) was added at 1 μg/ml to exclude dead cells. Quantitative flow-cytometric analysis was performed by measuring absolute numbers of viable (PI−) CFSE+ and marker+ cells after the acquisition of a fixed number of Flow-Count Fluorospheres (Coulter). The mean percentage of lysis of CFSE+ and marker+ cells of triplicate wells after coincubation with clone ZRZ38 or MBM13 was calculated relative to the wells without T cells.
Construction of a Recombinant Bacteria cDNA Expression Library.
Total RNA was isolated from EBV-LCLs from patient RZ by using TRIzol (Invitrogen). After purification using the RNA cleanup protocol of the RNeasy kit (Qiagen), poly(A)+ mRNA was isolated by the PolyATract mRNA isolation system (Promega). Single-stranded cDNA was synthesized by using random primers 5′-GCTCGCCCTCGCGGCGCGCCNNNNNT-3′ with AscI restriction enzyme sites (italicized) as described previously by Davis and Benzer (21). After synthesis of double-stranded cDNA, BamHI-EcoRI adapters (Stratagene) were ligated, followed by digestion with AscI and size fractionation by column chromatography. Different cDNA fractions were ligated into the BamHI and AscI restriction enzyme sites of vector pKE-1 (21) and electroporated into Escherichia coli BL21(DE3) bacteria. Vector pKE-1 contains an isopropyl β-d-thiogalactoside (IPTG)-inducible tac promoter, an ampicillin-resistance gene, and a kanamycin-resistance gene. Recombinant bacteria were selected on agar plates with 50 μg/ml ampicillin.
Screening of a Recombinant Bacteria cDNA Expression Library.
The cDNA library was divided into pools of ≈50 different recombinant bacteria and screened for T cell recognition by the complement-opsonized bacteria assay (19). Briefly, bacteria were grown to OD600 of 0.5 in LB with 50 μg/ml ampicillin. Then 1 mM IPTG was added to induce protein expression, and expansion continued for 4 h. Bacteria were opsonized by adding human serum with 17% (vol/vol) of complement (Sigma–Aldrich), followed by additional expansion for 1 h. EBV-LCLs from donor Z (3 × 104 cells per well), which fully matched patient RZ at a high resolution level, were pulsed with complement-opsonized bacteria in IMDM with 10% FBS and 30 μg/ml gentamycin (Sigma–Aldrich) overnight at 37°C. CD4+ T cell clone ZRZ38 (5 × 103 cells per well) was added; after overnight incubation, supernatants were harvested for analysis of IFN-γ by ELISA.
SNP Genotyping.
Genomic DNA was isolated by using the PureGene genomic DNA isolation kit (Gentra Systems). Analysis for the SNP at 382 bp of the PI4K2B cDNA was performed by using the SNP genotyping assay (Applied Biosystems) containing forward and reverse primers for amplification of a 357- to 407-bp fragment and two TaqMan MGB probes labeled with VIC and FAM dyes to detect the C and T alleles, respectively. PCR amplification was performed according to the manufacturer's instructions by using 5 ng of genomic DNA and a real-time PCR ABI Prism 7700 sequence detector system (Applied Biosystems).
ACKNOWLEDGMENTS.
This work was supported by a European Union grant (6th Framework Program Allostem).
Footnotes
References
- 1.Kolb HJ, et al. Donor leukocyte transfusions for treatment of recurrent chronic myelogenous leukemia in marrow transplant patients. Blood. 1990;76:2462–2465. [PubMed] [Google Scholar]
- 2.Collins RH, et al. Donor leukocyte infusions in 140 patients with relapsed malignancy after allogeneic bone marrow transplantation. J Clin Oncol. 1997;15:433–444. doi: 10.1200/JCO.1997.15.2.433. [DOI] [PubMed] [Google Scholar]
- 3.Porter DL, et al. Treatment of relapsed leukemia after unrelated donor marrow transplantation with unrelated donor leukocyte infusions. Blood. 2000;95:1214–1221. [PubMed] [Google Scholar]
- 4.den Haan JM, et al. Identification of a graft versus host disease-associated human minor histocompatibility antigen. Science. 1995;268:1476–1480. doi: 10.1126/science.7539551. [DOI] [PubMed] [Google Scholar]
- 5.den Haan JM, et al. The minor histocompatibility antigen HA-1: A diallelic gene with a single amino acid polymorphism. Science. 1998;279:1054–1057. doi: 10.1126/science.279.5353.1054. [DOI] [PubMed] [Google Scholar]
- 6.de Rijke B, et al. A frameshift polymorphism in P2X5 elicits an allogeneic cytotoxic T lymphocyte response associated with remission of chronic myeloid leukemia. J Clin Invest. 2005;115:3506–3516. doi: 10.1172/JCI24832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slager EH, et al. Identification of the angiogenic endothelial-cell growth factor-1/thymidine phosphorylase as a potential target for immunotherapy of cancer. Blood. 2006;107:4954–4960. doi: 10.1182/blood-2005-09-3883. [DOI] [PubMed] [Google Scholar]
- 8.van Bergen C, et al. Multiple myeloma reactive T cells recognize an activation induced minor histocompatibility antigen encoded by the ATP dependent interferon responsive (ADIR) gene. Blood. 2007;109:4089–4096. doi: 10.1182/blood-2006-08-043935. [DOI] [PubMed] [Google Scholar]
- 9.Marijt WA, et al. Hematopoiesis-restricted minor histocompatibility antigens HA-1- or HA-2-specific T cells can induce complete remissions of relapsed leukemia. Proc Natl Acad Sci USA. 2003;100:2742–2747. doi: 10.1073/pnas.0530192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolstra H, et al. A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J Exp Med. 1999;189:301–308. doi: 10.1084/jem.189.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren EH, et al. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science. 2006;313:1444–1447. doi: 10.1126/science.1130660. [DOI] [PubMed] [Google Scholar]
- 12.Kawase T, et al. Alternative splicing due to an intronic SNP in HMSD generates a novel minor histocompatibility antigen. Blood. 2007;110:1055–1063. doi: 10.1182/blood-2007-02-075911. [DOI] [PubMed] [Google Scholar]
- 13.Akatsuka Y, et al. Identification of a polymorphic gene, BCL2A1, encoding two novel hematopoietic lineage-specific minor histocompatibility antigens. J Exp Med. 2003;197:1489–1500. doi: 10.1084/jem.20021925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellucci R, et al. Immunologic effects of prophylactic donor lymphocyte infusion after allogeneic marrow transplantation for multiple myeloma. Blood. 2002;99:4610–4617. doi: 10.1182/blood.v99.12.4610. [DOI] [PubMed] [Google Scholar]
- 15.Soiffer RJ, et al. Randomized trial of CD8+ T-cell depletion in the prevention of graft-versus-host disease associated with donor lymphocyte infusion. Biol Blood Marrow Transplant. 2002;8:625–632. doi: 10.1053/bbmt.2002.v8.abbmt080625. [DOI] [PubMed] [Google Scholar]
- 16.Meyer RG, et al. Prophylactic transfer of CD8-depleted donor lymphocytes after T-cell-depleted reduced-intensity transplantation. Blood. 2007;109:374–382. doi: 10.1182/blood-2006-03-005769. [DOI] [PubMed] [Google Scholar]
- 17.Vogt MH, et al. The DBY gene codes for an HLA-DQ5-restricted human male-specific minor histocompatibility antigen involved in graft-versus-host disease. Blood. 2002;99:3027–3032. doi: 10.1182/blood.v99.8.3027. [DOI] [PubMed] [Google Scholar]
- 18.Spierings E, et al. Identification of HLA class II-restricted H-Y-specific T-helper epitope evoking CD4+ T-helper cells in H–Y-mismatched transplantation. Lancet. 2003;362:610–615. doi: 10.1016/S0140-6736(03)14191-8. [DOI] [PubMed] [Google Scholar]
- 19.van de Corput L, et al. A novel approach to identify antigens recognized by CD4 T cells using complement-opsonized bacteria expressing a cDNA library. Leukemia. 2005;19:279–285. doi: 10.1038/sj.leu.2403583. [DOI] [PubMed] [Google Scholar]
- 20.Kloosterboer FM, et al. Direct cloning of leukemia-reactive T cells from patients treated with donor lymphocyte infusion shows a relative dominance of hematopoiesis-restricted minor histocompatibility antigen HA-1 and HA-2 specific T cells. Leukemia. 2004;18:798–808. doi: 10.1038/sj.leu.2403297. [DOI] [PubMed] [Google Scholar]
- 21.Davis CA, Benzer S. Generation of cDNA expression libraries enriched for in-frame sequences. Proc Natl Acad Sci USA. 1997;94:2128–2132. doi: 10.1073/pnas.94.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toes RE, Schoenberger SP, van der Voort EI, Offringa R, Melief CJ. CD40-CD40Ligand interactions and their role in cytotoxic T lymphocyte priming and anti-tumor immunity. Semin Immunol. 1998;10:443–448. doi: 10.1006/smim.1998.0147. [DOI] [PubMed] [Google Scholar]
- 23.Faber LM, Luxemburg-Heijs SA, Veenhof WF, Willemze R, Falkenburg JH. Generation of CD4+ cytotoxic T-lymphocyte clones from a patient with severe graft-versus-host disease after allogeneic bone marrow transplantation: Implications for graft-versus-leukemia reactivity. Blood. 1995;86:2821–2828. [PubMed] [Google Scholar]
- 24.Dodi IA, et al. CD4(+) bias in T cells cloned from a CML patient with active graft versus leukemia effect. Cytotherapy. 2002;4:353–363. doi: 10.1080/146532402760271145. [DOI] [PubMed] [Google Scholar]
- 25.Michalek J, et al. Definitive separation of graft-versus-leukemia- and graft-versus-host-specific CD4+ T cells by virtue of their receptor beta loci sequences. Proc Natl Acad Sci USA. 2003;100:1180–1184. doi: 10.1073/pnas.0337543100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allison JP. CD28–B7 interactions in T-cell activation. Curr Opin Immunol. 1994;6:414–419. doi: 10.1016/0952-7915(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 27.Smit WM, et al. Generation of dendritic cells expressing bcr-abl from CD34-positive chronic myeloid leukemia precursor cells. Hum Immunol. 1997;53:216–223. doi: 10.1016/S0198-8859(96)00285-6. [DOI] [PubMed] [Google Scholar]
- 28.Choudhury A, et al. Use of leukemic dendritic cells for the generation of antileukemic cellular cytotoxicity against Philadelphia chromosome-positive chronic myelogenous leukemia. Blood. 1997;89:1133–1142. [PubMed] [Google Scholar]
- 29.Michalek J, Collins RH, Hill BJ, Brenchley JM, Douek DC. Identification and monitoring of graft-versus-host specific T-cell clone in stem cell transplantation. Lancet. 2003;361:1183–1185. doi: 10.1016/S0140-6736(03)12917-0. [DOI] [PubMed] [Google Scholar]
- 30.Ferrara JL. Cytokine dysregulation as a mechanism of graft versus host disease. Curr Opin Immunol. 1993;5:794–799. doi: 10.1016/0952-7915(93)90139-j. [DOI] [PubMed] [Google Scholar]
- 31.Jedema I, van der Werff NM, Barge RM, Willemze R, Falkenburg JH. New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood. 2004;103:2677–2682. doi: 10.1182/blood-2003-06-2070. [DOI] [PubMed] [Google Scholar]