Abstract
Mutations in copper/zinc superoxide dismutase (SOD1) are causative for dominantly inherited amyotrophic lateral sclerosis (ALS). Despite high variability in biochemical properties among the disease-causing mutants, a proportion of both dismutase-active and -inactive mutants are stably bound to spinal cord mitochondria. This mitochondrial proportion floats with mitochondria rather than sedimenting to the much higher density of protein, thus eliminating coincidental cosedimentation of protein aggregates with mitochondria. Half of dismutase-active and ≈90% of dismutase-inactive mutant SOD1 is bound to mitochondrial membranes in an alkali- and salt-resistant manner. Sensitivity to proteolysis and immunoprecipitation with an antibody specific for misfolded SOD1 demonstrate that in all mutant SOD1 models, misfolded SOD1 is deposited onto the cytoplasmic face of the outer mitochondrial membrane, increasing antigenic accessibility of the normally structured electrostatic loop. Misfolded mutant SOD1 binding is both restricted to spinal cord and selective for mitochondrial membranes, implicating exposure to mitochondria of a misfolded mutant SOD1 conformer mediated by a unique, tissue-selective composition of cytoplasmic chaperones, components unique to the cytoplasmic face of spinal mitochondria to which misfolded SOD1 binds, or misfolded SOD1 conformers unique to spinal cord that have a selective affinity for mitochondrial membranes.
Keywords: degeneration, motor neuron disease, superoxide dismutase
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by the progressive and selective killing of motor neurons of the motor cortex, brainstem, and spinal cord (reviewed in ref. 1). Although most cases are of unknown etiology (referred to as sporadic ALS), ≈10% of all cases are genetically inherited. The most common form of adult-onset familial ALS is caused by mutations in the ubiquitously expressed housekeeping gene, superoxide dismutase 1 (SOD1). More than 115 disease-causing mutations, affecting all regions of the SOD1 gene product, have been identified. Study of rodent models expressing some of these SOD1 mutations has revealed that SOD1-mediated disease is not caused by a loss of its dismutase activity, but rather by the gain of one or more as yet unknown toxic property(ies) (2–4).
The first indications that mitochondria may contribute to disease arose from histopathological observations of disturbed mitochondrial ultrastructure in the motor neurons (and muscle) of both sporadic and familial ALS patients (5–8). Similar findings of vacuolated, dilated, and disorganized mitochondria were later confirmed in mutant SOD1 mouse models expressing dismutase-active (9–12), but not -inactive mutants (3). Initially shown for hSOD1G93A and hSOD1G37R (9, 12), these alterations in mitochondrial structure occur before any other pathological feature and any clinical disease feature (reviewed in ref. 13).
Although SOD1 is an abundant, ubiquitously expressed cytosolic protein, a proportion of SOD1 is located within the intermembrane space of mitochondria in yeast (14, 15) and rat liver (16). For spinal cord and brain mitochondria, the location(s) of mutant SOD1 is controversial. One report proposed that the majority of mitochondrial-associated, dismutase-inactive mutants were bound to the cytoplasmic face, whereas dismutase-active mutants were both bound at the surface and imported into the intermembrane space (17). Alternative proposals include mutant import into the matrix of mouse brain mitochondria (18) or that apparent mitochondrial association of unfolded, dismutase-inactive mutants represents simple contamination of mitochondrially enriched fractions with protein aggregates (19). We now test these competing proposals. Use of density gradient methods demonstrates that multiple mutant SOD1s float to the buoyant density of mitochondria, thereby eliminating the possibility that association reflects cosedimentation with much more dense protein aggregates. Furthermore, we uncover that the nature of both dismutase-active and -inactive mutant SOD1 association with spinal cord mitochondria involves very tight association with the outer mitochondrial membrane that is both alkali- and salt-resistant. The combination of sensitivity to proteolysis and immunoprecipitation with an antibody specific for misfolded SOD1 demonstrates that in all rodent models of mutant SOD1-mediated ALS, misfolded SOD1 is deposited onto the cytoplasmic face of the outer mitochondrial membrane, increasing antigenic accessibility of the normally structured electrostatic loop. Association of misfolded mutant SOD1 selectively onto spinal cord mitochondrial membranes documents that the nature of mutant SOD1 association with mitochondria is not the same in all tissues, implicating a unique tissue-selective composition of cytoplasmic chaperones, unique components on the cytoplasmic face of spinal mitochondria, or misfolded SOD1 conformers unique to spinal cord that have a selective affinity for mitochondrial membranes.
Results
SOD1 Association with Mitochondria Demonstrated by Buoyant Density Centrifugation.
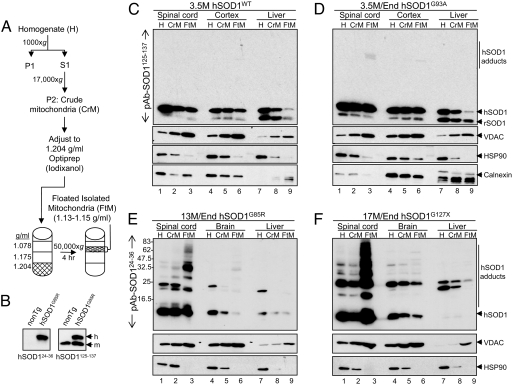
By using a single-step enrichment (a continuous iodixanol gradient) to fractionate initial tissue homogenates and in which both protein aggregates and mitochondria will sediment downward after sample loading at the top (19), mutant SOD1 association with mitochondria (especially for the dismutase-inactive mutant hSOD1G85R) has been proposed to represent coenrichment of SOD1 aggregates with mitochondria, rather than mutant association with mitochondria. To enrich for mitochondria while eliminating potential protein aggregates [which are known for all SOD1 mutants (3, 20, 21) but not wild-type (WT) SOD1 (12, 20)], differential sedimentation was initially used to enrich for mitochondria from tissue extracts. After this initial fractionation to remove most soluble components, samples were loaded at the bottom of a three-step iodixanol gradient and centrifuged. Under these conditions, mitochondria (and any proteins tightly associated with them) migrate upward (float) to their buoyant density (1.13–1.15 g/ml), banding at an interface between the 1.078-g/ml and 1.175-g/ml layers. Soluble or aggregated protein (density ≥1.26 g/ml), however, sediments downward.
Using multiple transgenic lines, we evaluated whether wild-type or mutant SOD1 protein is present in fractions containing floated mitochondria (Fig. 1A). Varying amounts of endogenous SOD1 [and the mitochondrial marker, voltage-dependent anion channel (VDAC)] were enriched in floated mitochondria from brain, spinal cord, and liver (Fig. 1 C and D, lanes 3, 6, and 9), in agreement with earlier work (16), and reinforcing the fact that mitochondria from different tissues have variable protein constituents [as most clearly demonstrated by mass spectrometry (22, 23)]. The abundant cytosolic marker Hsp90 was largely excluded, as expected. More hSOD1WT cofloated with mitochondria from spinal cord and cortex than those from liver (Fig. 1C, compare lanes 3, 6, and 9). As expected, mitochondrial association of hSOD1WT depended on its cytosolic level, as demonstrated by the comparison of three different hSOD1WT transgenic mouse lines accumulating varying SOD1 levels [supporting information (SI) Fig. 6].
Fig. 1.
A portion of SOD1 is localized to mitochondria enriched by buoyant density centrifugation. (A) Schematic outlining the enrichment of mitochondria according to buoyant density. (B) Demonstration of the selectivity of two SOD1 polyclonal antibodies used in this study. (C–F) SOD1 is enriched in floated mitochondria, compared with crude mitochondria and whole homogenates. SOD1 is enriched in spinal cord mitochondria and less so in cortical and liver mitochondria. VDAC, calnexin, and Hsp90 calnexin serve as markers for mitochondria, endoplasmic reticulum, and cytosol, respectively.
In each of four mutant SOD1 models (hSOD1G93A and hSOD1H46R rats; hSOD1G85R and hSOD1G127X mice), significantly higher levels of each mutant SOD1 were present in floated mitochondria from affected spinal cord tissues compared with liver, a tissue unaffected by disease (Fig. 1 D–F; compare lanes 3, 6, and 9; and SI Fig. 7). Furthermore, comparing spinal cord, brain, and cortical samples from the same animals, a higher proportion of dismutase-inactive mutants hSOD1G85R, hSOD1G127X, and hSOD1H46R floated with spinal cord mitochondria (Fig. 1 E and F, compare lanes 3 and 6; and SI Fig. 7). Almost equivalent levels of the dismutase-active mutant hSOD1G93A floated with mitochondria from both spinal cord and cortex (Fig. 1D, lanes 3 and 6). Low electrophoretic mobility adducts of mutant SOD1 that were detergent- and reducing agent-resistant (reminiscent of oxidized multimers described in ref. 24) also floated with mitochondria, indicating that they are tightly bound to floated mitochondria (Fig. 1 D–F, lanes 3). Thus, human SOD1 is a bona fide resident on or within spinal mitochondria, and there is preferential association of mutant SOD1 with mitochondria within affected tissues.
Dismutase-Inactive SOD1 Is Deposited on the Cytoplasmic-Facing Surface of Spinal Cord, but Not Brain, Mitochondria.
Multiple prior efforts have disagreed on how mutant SOD1 is associated with mitochondria. Efforts had argued that association was onto the cytoplasmic-facing mitochondrial surface [for hSOD1G85R (17)], in the intermembrane space (IMS) and the cytoplasmic surface [for hSOD1G37R (17)], in the IMS [for hSOD1G93A (14, 25–27)], and in the matrix [for hSOD1G93A and hSOD1G85R (18)]. To resolve mutant location on floated mitochondria that were free from protein aggregates, protease sensitivity was used to determine the proportion of mutant SOD1 accessible to protease by virtue of location on the cytoplasmic face of intact mitochondria. Because WT SOD1 and WT-like mutants (e.g., hSOD1G93A and hSOD1G37R) are known to be tightly folded, hyperstable proteins that are resistant to proteinase K digestion (28, 29) (Fig. 2A), this could only be done for the dismutase-inactive mutants hSOD1H46R, hSOD1G85R, and hSOD1G127X, which are known to be susceptible to protease digestion (28, 29), presumably because of the adoption of less well folded or misfolded conformations. By using a saturating amount of proteinase K (as demonstrated in SI Fig. 8), the majority of each of the three dismutase-inactive mutant SOD1s associated with intact spinal cord mitochondria was protease-sensitive even in the absence of detergent. Further, the remaining mutant protein (as well as Smac, a protein in the intermembrane space) was completely digested after detergent-mediated solubilization of mitochondrial membranes (Fig. 2A). In cortical mitochondria from the same animals, however, mutant SOD1 proteins remained protease-resistant when membranes were intact. These experiments indicate that these inactive mutant SOD1s are largely deposited onto the cytoplasmic face of the outer membrane of spinal cord mitochondria but are almost entirely sequestered within brain mitochondria. Additionally, by using anti-peptide antibodies predicted to detect each of the mutants with equal affinity (30), the amount of mutant SOD1 that is sequestered within mitochondria of both tissue types is almost identical.
Fig. 2.
SOD1 associates with the cytoplasmic face of the outer mitochondrial membrane and correlates with disease. (A) Mitochondria enriched via floatation from spinal cord, cortex, and liver were treated with 100 μg/ml proteinase K (PK) in the presence or absence of 0.5% Triton X-100 (Tx-100). The majority of mitochondrially associated hSOD1 mutants with a partially misfolded conformation including hSOD1H46R (A), hSOD1G85R (B), and hSOD1G127X (D), are found deposited on the cytoplasmic face of the outer mitochondrial membrane in spinal cord as evidenced by sensitivity to proteolysis. In cortical mitochondria, the same mutants are completely insensitive to protease until addition of detergent, consistent with an intramitochondrial localization. Note that the absolute amounts of mutant SOD1 localized inside mitochondria is comparable in spinal cord and cortex. (C and D) The accumulation of mutant SOD1 protein on the surface correlates with disease because it is much less apparent in asymptomatic hSOD1G85R (C) and hSOD1G127X (D) animals. As expected, (A) hSOD1G93A and endogenous rat SOD1 (rSOD1) are not protease-sensitive. In all cases, the IMS protein Smac is only susceptible to proteolysis in the presence of detergent. Age is indicated in months. Data shown are representative of three independent experiments.
To assess when mutant protein deposition onto the cytoplasmic surface occurs, we prepared floated mitochondria from asymptomatic hSOD1G85R (6.5 months old) and hSOD1G127X (12 months old) mice, time points at which there are neither clinical signs of disease nor detectable pathology. Most surprisingly, at these early time points, both mitochondrially bound hSOD1G85R and hSOD1G127X in both spinal cord and cortex were almost fully protected from proteinase K digestion in the absence of detergent (Fig. 2 C and D), indicative of location within those mitochondria. Thus, mutant SOD1 deposition on the cytoplasmic face of the outer mitochondrial membrane is not an intrinsic property of SOD1 but is acquired in an age-dependent manner consistent with a pathogenic role in development of age-dependent disease.
Dismutase-Inactive SOD1 Mutants Behave as Integral Membrane Proteins of Spinal Cord, but Not Brain, Mitochondria.
Divergent claims have been made concerning whether the association of mutant SOD1 with spinal cord mitochondria is as a loosely membrane-bound or soluble component (18) or is tightly associated with mitochondrial membranes whose linkage is resistant to alkali extraction (17). As an initial test of whether mutant SOD1 binding to the cytoplasmic face of spinal cord mitochondria was mediated by electrostatic interactions, floated mitochondria were repeatedly washed in a high-ionic strength buffer. However, just as was the case for the integral outer membrane protein VDAC, little to no mutant SOD1 was released (Fig. 3), demonstrating a salt-resistant linkage.
Fig. 3.
Mutant SOD1 is tightly bound to mitochondrial membranes in a salt-resistant manner. hSOD1H46R cannot be removed from isolated floated spinal cord mitochondria by extensive washing even with 1 M KCl. VDAC yields the expected pattern for an integral outer membrane protein. Data shown are representative of two experiments.
Next, although SOD1 protein does not contain any transmembrane segments nor a recognizable mitochondrial targeting signal (which we verified by using the bioinformatics tools DAS-TMfilter, TMPred, TMAP, HMMTOP, TMHMM, SPLIT, PRED-TMR2, and ConPredII), alkali extraction of isolated floated mitochondria from either spinal cord or cortex from a variety of transgenic lines was used to separate peripherally bound membrane components from those more tightly associated (Fig. 4A). This revealed that for spinal cord mitochondria, endogenous SOD1 was mostly released, as expected for a soluble component of the intermembrane space (or matrix), or a peripheral membrane component (Fig. 4B, lanes 2, 5, 8, and 11). For mutant SOD1, both alkali-sensitive and alkali-resistant pools were found in spinal cord mitochondria (Fig. 4B, lanes 2 and 3). In contrast, in the alkali-resistant fraction of identically treated cortical mitochondria isolated from the same animal, only a very small proportion of hSOD1G93A was recovered (Fig. 4B, lane 6), indicating that mutant SOD1s associated with cortical mitochondria are peripheral or freely soluble mitochondrial residents (Fig. 4 B, lane 5; C, lane 8; D, lanes 5 and 11; and E, lanes 5 and 11).
Fig. 4.
Mutant SOD1 is tightly bound to mitochondrial membranes in an alkali-resistant manner. (A) Schematic of alkali extraction. (B) Approximately one-third of hSOD1G93A, but almost no hSOD1WT protein, is tightly associated with spinal cord mitochondrial membranes in an alkali-resistant manner. In contrast, SOD1 is completely soluble in cortical mitochondria. (C) In contrast, hSOD1H46R, which adopts a partially unfolded conformation, is found in a higher proportion in spinal cord mitochondrial membranes compared with hSOD1G93A. (D and E) Time-dependent and disease-related integration of hSOD1G127X and hSOD1G85R into spinal cord mitochondrial membranes. VDAC and cytochrome c are used to identify behaviors of well defined mitochondrial components that are integral and peripheral membrane-associated, respectively. Data are representative of three experiments.
Alkali-resistant association with spinal cord, but not cortical, mitochondria was common to all three dismutase-inactive mutants examined (hSOD1H46R, hSOD1G85R, and hSOD1G127X) (Fig. 4 C–E). Cofloating with mitochondria were slow-mobility adducts of SOD1, and these were also found to behave as integral proteins of the outer membrane (Fig. 4D, lane 9, and SI Fig. 9). Importantly, consistent with a soluble intermembrane space component, comparable alkali sensitivity of mitochondrially associated hSOD1WT was found in both spinal cord and cortical mitochondria (Fig. 4B, lanes 8 and 11). Almost all mutant protein was restricted to the alkali-sensitive fraction of mitochondria derived from asymptomatic hSOD1G85R (Fig. 4E, lane 2) and hSOD1G127X mice (Fig. 4D, lane 2), demonstrating that association of mutant SOD1 proteins with the outer membrane of spinal cord mitochondria is acquired in an age-dependent manner.
Mitochondrial-Associated SOD1 Is Misfolded.
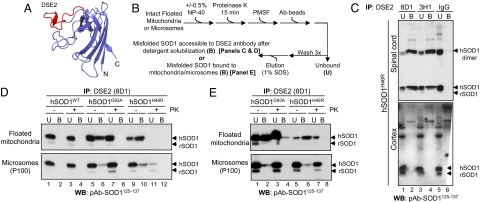
Antibodies that recognize epitopes typically unavailable within a stably folded protein conformation have been demonstrated to bind preferentially or exclusively to misfolded (or denatured) conformations (31–33), including misfolded mutant SOD1 in inherited ALS (34). To examine whether the age-dependent deposition of mutant SOD1 onto the cytoplasmic face only of spinal cord mitochondria reflected association of misfolded SOD1, we exploited monoclonal antibodies developed against the electrostatic loop, a structural element that is inaccessible in native well ordered SOD1 but can be accessible to antibody binding by mutant-mediated misfolding or zinc depletion (Fig. 5A). This surface represents one of several such domains proposed to represent disease-specific epitopes (DSE) exposed only on misfolded hSOD1 (34, 35).
Fig. 5.
Mutant SOD1 associated with floated spinal cord mitochondria is misfolded. (A) The DSE2 epitope mapped onto hSOD1G37R crystal structure (Protein Data Bank ID code 1AZV), as generated by PyMol. (B) Schematic of immunoprecipitation experiments. (C) Floated isolated mitochondria from hSOD1H46R spinal cord and cortex were immunoprecipitated with two independent antibodies to the DSE2 region. Each uniquely identified a pool of misfolded SOD1 associated with spinal cord but not cortical mitochondria. (D and E) DSE2 recognizes a protease-sensitive pool of hSOD1G93A and hSOD1H46R but not hSOD1WT in solubilized (D) or intact floated spinal cord mitochondria (E), but not microsomal membranes.
Two independent monoclonal antibodies were generated against the DSE2 region (amino acids 125–142). In detergent lysates of floated spinal cord mitochondria, both robustly detected misfolded SOD1 and covalently associated forms that migrated at positions consistent with cross-linked dimeric SOD1 (Fig. 5C, Upper, lanes 2 and 4). No similarly misfolded mutant SOD1 was associated with cortical mitochondria (Fig. 5C, Lower, lanes 2 and 4). Approximately 20% of hSOD1G93A and 50% of hSOD1H46R associated with floated spinal cord mitochondria were in a misfolded conformation that permitted DSE2 antibody binding (Fig. 5D). Protease sensitivity of the usually protease-resistant hSOD1G93A (28, 29) confirmed its misfolded conformation (Fig. 5D, Upper, compare lanes 6 and 8). Neither DSE2 antibody immunoprecipitated any misfolded SOD1 for either SOD1 mutant in the cytosol from either tissue (data not shown). Neither of the antibodies to DSE2 recognized hSOD1WT that had been imported into spinal mitochondria, even after detergent solubilization to liberate internal mitochondrial contents (Fig. 5D).
Recovery of intact, floated mitochondria (in the absence of detergent) by binding to a DSE2 antibody further demonstrated that for both dismutase-active and -inactive mutants, essentially all misfolded SOD1 was bound to spinal cord mitochondria that had antibody-accessible, misfolded SOD1 bound to the cytoplasmic face (Fig. 5E). This misfolded SOD1 was highly selectively enriched onto the outer mitochondria membrane, with a much smaller proportion (<10%) associated with microsomal fractions (100,000 × g pellet; Fig. 5 D, Lower, lanes 6 and 10, and E, Lower, lanes 2 and 6). Thus, mutant hSOD1 is tightly and selectively bound to the cytoplasmic-facing surface of spinal cord mitochondria in a misfolded conformation.
Discussion
Floatation of mutant SOD1 protein with spinal cord, but not cortical or liver, mitochondria unambiguously demonstrates that the presence of these mutant subunits cannot reflect contamination of protein aggregates with mitochondria, as has been proposed (19). Misfolded SOD1 is bound in a very tight, alkali-resistant linkage to the cytoplasmic face of the outer mitochondrial membrane (as shown by protease sensitivity of misfolded, dismutase-inactive mutants). These findings are further supported by immunoelectron microscopy of spinal cord motor neurons in situ in which hSOD1G93A is found both within mitochondria and in close proximity to the mitochondrial surface (25). Furthermore, similar studies in hSOD1G85R spinal motor neurons also demonstrate a preferential distribution at the mitochondrial surface (C.V.V. and D.W.C., unpublished data). This is common to multiple dismutase-active and -inactive mutants and is found only for mitochondria isolated from tissues at highest risk during disease, findings that support mutant-derived damage to spinal cord mitochondria as a central feature of pathogenesis from ubiquitously expressed SOD1 mutants. Deposition of mutant SOD1 onto the outer membrane may affect protein import, ionic homeostasis, mitochondrial motility, mitochondrial fission/fusion, or regulation of apoptosis. Indeed, an interaction between SOD1 and antiapoptotic Bcl-2 family members, which are residents of the outer mitochondrial membranes, has been proposed (36) but not yet confirmed (37).
A pool of mutant SOD1 is also found within mitochondria, most likely in the intermembrane space, as is known for the WT SOD1 protein. Both the physical properties and the dual localization of SOD1 (cytosol and mitochondria) are reminiscent of those known for yeast adenylate kinase, which is also known to fold rapidly and spontaneously into a thermal- and proteolytic-resistant hyperstable conformation (38). For adenylate kinase, partitioning between cytosol and mitochondria is caused by competition between rapid protein folding and inefficient mitochondrial targeting, i.e., once the protein folds, its cryptic mitochondrial targeting signal is inaccessible. In the context of mutant SOD1, slowed folding kinetics, which has been demonstrated in a cell-free system (39), would result in prolonged exposure of a cryptic targeting sequence.
Use of DSE2 antibodies directed to a normally buried domain of SOD1 has allowed us to determine that misfolded SOD1 conformers are associated with the cytoplasmic face of spinal cord mitochondria but not mitochondria from other tissues. These species have a high affinity for mitochondrial membranes, presumptively through exposure of a normally buried hydrophobic surface (40, 41). Although an inherently misfolded mutant SOD1 would be predicted to bind other intracellular membranes, which would be consistent with previous reports (29, 42), our data clearly demonstrate that misfolded SOD1 conformers are preferentially associated with mitochondria, with only small amounts detectable in microsomal fractions.
Two mechanisms, which are not mutually exclusive, are consistent with age-dependent binding of mutant SOD1 selectively to mitochondrial membranes only on spinal cord mitochondria. First, an enhanced level of a misfolded mutant SOD1 conformer in which a unique surface is exposed [perhaps because of fluctuations of the electrostatic loop (43–45)] may interact with unique, tissue-selective array of cytoplasmic chaperones to facilitate association/presentation to mitochondria. Second, there may be components unique to the cytoplasmic face of spinal mitochondria to which misfolded SOD1 binds. Indeed, it is recognized that mitochondria from different tissues (which perform different biological activities) obligatorily have different protein compositions (22, 23). Furthermore, the tissue type differences reported here call into question the pathological relevance of data collected on mitochondria from tissues that are not the primary target in disease. Combined with the very stable binding (resistance to alkali and high ionic strength), we propose that mutant SOD1 conformers bind to an integral mitochondrial component enriched in spinal mitochondria by exposure of one or more hydrophobic SOD1 surfaces.
In any case, a central determinant for mutant SOD1 association with mitochondria would be the steady-state proportion of misfolded SOD1. Thus, for mutants that adopt a well folded, native-like conformation (like hSOD1G37R or hSOD1G93A), high total accumulated levels would be required to drive mitochondrial membrane association because only a small proportion of the total protein is accumulated as a misfolded conformer. For dismutase-inactive mutants, accumulation at much lower levels would drive a comparable level of accumulation of misfolded mutant. For both mutant classes, SOD1 harboring ALS-linked mutations have significantly slower folding kinetics in vitro (38). Consistent with all of this, in mice, hSOD1G85R causes disease at levels up to 50 times lower than the dismutase-active mutants (3). In human disease as well, the least stable mutants (accumulating to the lowest levels) correlate with the most rapid disease progression (46).
Finally, mitochondrial association is accompanied by accumulation of low-mobility species, the most prominent of which appears to be a covalently cross-linked dimer. A similar species has recently been shown to be a feature also common in sporadic disease (47), albeit in human disease it remains untested whether this dimer is selective for mitochondria. Determining this is now central to testing whether promiscuous mitochondrial–membrane association of SOD1 may contribute to pathogenesis in sporadic ALS.
Materials and Methods
Mice were deeply anesthetized by isoflurane inhalation and decapitated. Spinal cords were flushed out of the vertebral column with PBS and quickly placed in 5 volumes of ice-cold homogenization buffer (HB) [250 mM sucrose, 10 mM Hepes–NaOH (pH 7.4), 1 mM EDTA plus protease inhibitors). Cortices were quickly dissected from whole brain. Tissues were homogenized with seven strokes of a glass-pestle homogenizer at 4°C and on ice. Homogenates were centrifuged at 1,000 × g for 10 min. Supernatants were recovered, and pellets were washed with ½ volume HB. Supernatants were pooled and centrifuged at 17,000 × g for 15 min to yield S3 and a crude mitochondrial pellet. Crude mitochondria were gently resuspended in HB and then adjusted to 1.204 g/ml Optiprep (iodixanol) and loaded on the bottom of a polycarbonate tube. Mitochondria were overlaid with an equal volume of 1.175 g/ml and 1.078 g/ml Optiprep and centrifuged at 50,000 × g for 4 h (SW-55; Beckman). Mitochondria were collected at the 1.078/1.175 g/ml interface and washed once to remove the Optiprep. Optiprep stock solution (1.32 g/ml; Axis Bioshield) was diluted in 250 mM sucrose, 120 mM Hepes–NaOH (pH 7.4), 6 mM EDTA plus protease inhibitors.
Additional materials and methods are discussed in SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Sandrine Da Cruz and Philippe Parone for valuable discussions and Gregory Cox (Jackson Laboratory, Bar Harbor, ME), Stefan Marklund (Umeå University, Umeå, Sweden), Pak Chan (Stanford University, Palo Alto, CA), and Masashi Aoki (Tohoku University, Sendai, Japan) for generously sharing transgenic animals. This work was supported by National Institutes of Health Grant NS 27036 (to D.W.C.) C.V.V. is supported by a Development Grant from the Muscular Dystrophy Association. D.W.C. is supported by the Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712209105/DC1.
References
- 1.Boillee S, Vande Velde C, Cleveland DW. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:1–21. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Bruijn LI, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 3.Bruijn LI, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 4.Reaume AG, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 5.Hirano A, Donnenfeld H, Sasaki S, Nakano I. Fine structural observations of neurofilamentous changes in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 1984;43:461–470. doi: 10.1097/00005072-198409000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Hirano A, et al. Fine structural study of neurofibrillary changes in a family with amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 1984;43:471–480. doi: 10.1097/00005072-198409000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki S, Iwata M. Dendritic synapses of anterior horn neurons in amyotrophic lateral sclerosis: An ultrastructural study. Acta Neuropathol. 1996;91:278–283. doi: 10.1007/s004010050426. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki S, Iwata M. Mitochondrial alterations in the spinal cord of patients with sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2007;66:10–16. doi: 10.1097/nen.0b013e31802c396b. [DOI] [PubMed] [Google Scholar]
- 9.Dal Canto MC, Gurney ME. Development of central nervous system pathology in a murine transgenic model of human amyotrophic lateral sclerosis. Am J Pathol. 1994;145:1271–1279. [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins CM, Jung C, Xu Z. ALS-associated mutant SOD1G93A causes mitochondrial vacuolation by expansion of the intermembrane space and by involvement of SOD1 aggregation and peroxisomes. BMC Neurosci. 2003;4:16. doi: 10.1186/1471-2202-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong J, Xu Z. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J Neurosci. 1998;18:3241–3250. doi: 10.1523/JNEUROSCI.18-09-03241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong PC, et al. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 13.Bendotti C, Carri MT. Lessons from models of SOD1-linked familial ALS. Trends Mol Med. 2004;10:393–400. doi: 10.1016/j.molmed.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Field LS, Furukawa Y, O'Halloran TV, Culotta VC. Factors controlling the uptake of yeast copper/zinc superoxide dismutase into mitochondria. J Biol Chem. 2003;278:28052–28059. doi: 10.1074/jbc.M304296200. [DOI] [PubMed] [Google Scholar]
- 15.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone CCS, localize to the intermembrane space of mitochondria: A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J Biol Chem. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 16.Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Vijayvergiya C, Beal MF, Buck J, Manfredi G. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J Neurosci. 2005;25:2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergemalm D, et al. Overloading of stable and exclusion of unstable human superoxide dismutase-1 variants in mitochondria of murine amyotrophic lateral sclerosis models. J Neurosci. 2006;26:4147–4154. doi: 10.1523/JNEUROSCI.5461-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Xu G, Borchelt DR. High molecular weight complexes of mutant superoxide dismutase 1: Age-dependent and tissue-specific accumulation. Neurobiol Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- 21.Jonsson PA, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2003;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 22.Bailey AO, et al. RCADiA: Simple automation platform for comparative multidimensional protein identification technology. Anal Chem. 2007;79:6410–6418. doi: 10.1021/ac070585g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 24.Ezzi SA, Urushitani M, Julien JP. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J Neurochem. 2007;102:170–178. doi: 10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- 25.Higgins CM, Jung C, Ding H, Xu Z. Mutant Cu,Zn superoxide dismutase that causes motoneuron degeneration is present in mitochondria in the CNS. J Neurosci. 2002;22:215. doi: 10.1523/JNEUROSCI.22-06-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaarsma D, et al. CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. 2001;102:293–305. doi: 10.1007/s004010100399. [DOI] [PubMed] [Google Scholar]
- 27.Okado-Matsumoto A, Fridovich I. Amyotrophic lateral sclerosis: A proposed mechanism. Proc Natl Acad Sci USA. 2002;99:9010–9014. doi: 10.1073/pnas.132260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratovitski T, et al. Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Hum Mol Genet. 1999;8:1451–1460. doi: 10.1093/hmg/8.8.1451. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi H, et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc Natl Acad Sci USA. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardo CA, et al. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc Natl Acad Sci USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paramithiotis E, et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat Med. 2003;9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 32.Cashman NR, Caughey B. Prion diseases: Close to effective therapy? Nat Rev Drug Discov. 2004;3:874–884. doi: 10.1038/nrd1525. [DOI] [PubMed] [Google Scholar]
- 33.Urushitani M, Ezzi SA, Julien JP. Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2007;104:2495–2500. doi: 10.1073/pnas.0606201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 35.Cashman NR, et al. Active and passive immunization of superoxide dismutase-1 disease specific epitopes in a mouse model of amyotrophic lateral sclerosis. Ann Neurol. 2007;62:542–543. [Google Scholar]
- 36.Pasinelli P, et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Gould TW, et al. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J Neurosci. 2006;26:8774–8786. doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnoys EJ, Wang JL. Dual localization: Proteins in extracellular and intracellular compartments. Acta Histochem. 2007;109:89–110. doi: 10.1016/j.acthis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Bruns CK, Kopito RR. Impaired post-translational folding of familial ALS-linked Cu,Zn superoxide dismutase mutants. EMBO J. 2007;26:855–866. doi: 10.1038/sj.emboj.7601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiwari A, Xu Z, Hayward LJ. Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J Biol Chem. 2005;280:29771–29779. doi: 10.1074/jbc.M504039200. [DOI] [PubMed] [Google Scholar]
- 41.Zetterstrom P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urushitani M, et al. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 43.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu,Zn superoxide dismutase mutants. Proc Natl Acad Sci USA. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: Parallels to precursors in amyloid disease. Proc Natl Acad Sci USA. 2006;103:10218–10223. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strange RW, Yong CW, Smith W, Hasnain SS. Molecular dynamics using atomic-resolution structure reveal structural fluctuations that may lead to polymerization of human Cu-Zn superoxide dismutase. Proc Natl Acad Sci USA. 2007;104:10040–10044. doi: 10.1073/pnas.0703857104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato T, et al. Rapid disease progression correlates with instability of mutant SOD1 in familial ALS. Neurology. 2005;65:1954–1957. doi: 10.1212/01.wnl.0000188760.53922.05. [DOI] [PubMed] [Google Scholar]
- 47.Gruzman A, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2007;104:12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.