Abstract
A single ChIP sample does not provide enough DNA for hybridization to a genomic tiling array. A commonly used technique for amplifying the DNA obtained from ChIP assays is linker-mediated PCR (LMPCR). However, using this amplification method, we could not identify Oct4 binding sites on genomic tiling arrays representing 1% of the human genome (ENCODE arrays). In contrast, hybridization of a pool of 10 ChIP samples to the arrays produced reproducible binding patterns and low background signals. However, the pooling method would greatly increase the number of ChIP reactions needed to analyze the entire human genome. Therefore, we have adapted the GenomePlex whole genome amplification method for use in ChIP-chip assays; detailed ChIP and amplification protocols used for these analyses are provided as Supplementary Methods. When applied to ENCODE arrays, the amplicons prepared using this new method resulted in an Oct4 binding pattern similar to that from the pooled Oct4 ChIP samples. Importantly, the signal to noise ratio using the GenomePlex whole gene amplification method is superior to the LMPCR amplification method.
Keywords: chromatin immunoprecipitation, genomic microarray, ChIP-chip, ENCODE, Oct4, amplicons
INTRODUCTION
The technique of chromatin immunoprecipitation (ChIP) has proven to be a powerful tool, allowing the detection of protein-DNA interactions in living cells. Although this technique was first adapted for use with mammalian cells less than 10 years ago (1,2), it is now the gold standard experiment for the identification of a target gene of a particular transcription factor. Over the last several years, great strides have been made in expanding the use of ChIP from a one gene-at-a-time approach to a global analysis tool through the hybridization of the samples to genomic microarrays (i.e. the ChIP-chip assay). Today, arrays representing promoter regions (3), CpG islands (4-6), or entire genomes (7) are used in combination with ChIP to identify binding sites for transcription factors and components of the transcriptional machinery and to define chromatin structure. However, a single ChIP sample does not provide enough DNA for labeling and hybridization to an array. A commonly used technique for amplifying the DNA obtained from ChIP assays is linker-mediated PCR (LMPCR). Unfortunately, we have found that this method often produces very high background when samples are analyzed on genomic tiling arrays. In this study, we have compared three ChIP sample preparation methods, which differ in the background noise and reproducibility of binding site identification.
MATERIALS AND METHODS
Cell Culture
Ntera2 cells were grown in Dulbecco's Modified Eagle Medium supplemented with 2mM glutamine, 100 units/ml of penicillin and streptomycin, and 10% fetal bovine serum. All cells were incubated at 37°C in a humidified 5% CO2 incubator.
ChIP-chip Assays
ChIP assays (1 × 107 cells/assay) were performed following the protocol provided in Supplementary Methods (updates can be found at http://genomics.ucdavis.edu/farnham/ and http://genomecenter.ucdavis.edu/expression_analysis/). The Oct4 antibody used in this study was purchased from Santa Cruz Biotechnology (cat# sc-8628X) and the rabbit anti-goat IgG was purchased from MP Biomedicals (cat# 55335). For PCR analysis of the ChIP samples prior to amplicon generation, QIAquick-purified immunoprecipitates were dissolved in 50 uls of water. Standard PCR reactions using 2 uls of the immunoprecipitated DNA were performed. PCR products were separated by electrophoresis through 1.5% agarose gels and visualized using ethidium bromide.
Three different preparation methods were used to obtain enough ChIP DNA for application to genomic microarrays; ChIP-chip experiments were performed using two independent cultures of crosslinked Ntera2 cells for each method. Method 1: Linker-mediated PCR (LMPCR): For this method, one half of a ChIP sample (from 1 × 107 cells) was used for linker ligation. Amplification of the linker-ligated DNA using LMPCR is described in detail at http://genomics.ucdavis.edu/farnham/; see also (8). Method 2: Pooling ChIP samples: For this method, 10 individual Oct4 ChIP assays were performed from each of two sets of 1 × 108 cross-linked cells (1 × 107 cells per ChIP assay). ChIP samples were processed separately following the standard protocol except that after preclearing the chromatin with StaphA cells, all 10 ChIP samples were pooled into one tube for the washing steps. Washes and elution of the pooled ChIPs were then carried out as described in the standard protocol. Method 3: Whole genome amplification (WGA): An adaptation of the standard protocol for whole genome amplification using the Sigma GenomePlex WGA kit was used. Briefly, the initial random fragmentation step was eliminated and an entire ChIP sample (from 1 × 107 cells) or 10 ng of input chromatin was amplified. This usually provides enough sample for one array. However, if additional amplicon is needed then a second round of amplification (using 10−20 ng of the first amplification sample) can be performed. A detailed protocol for the WGA method is provided in Supplementary Methods.
Biological replicates of LMPCR amplicons, pooled ChIP samples and WGA amplicons (a total of 6 samples) were applied to NimbleGen ENCODE oligonucleotide arrays containing ∼380,000 50mer probes per array, tiled every 38 bp. The regions included on the arrays encompassed the 30 MB of the repeat masked ENCODE sequences, representing approximately 1% of the human genome. The labeling of DNA samples for ChIP-chip analysis was performed by NimbleGen Systems, Inc. Briefly, each DNA sample (1 μg) was denatured in the presence of 5'-Cy3- or Cy5-labeled random nonamers (TriLink Biotechnologies, San Diego) and incubated with 100 units (exo-) Klenow fragment (NEB, Beverly, MA) and dNTP mix [6 mM each in TE buffer (10 mM Tris/1 mM EDTA, pH 7.4; Invitrogen)] for 2 h at 37°C. Reactions were terminated by addition of 0.5 M EDTA (pH 8.0), precipitated with isopropanol, and resuspended in water. Then, 13ug of the Cy5-labeled ChIP sample and 13ug of the Cy3-labeled total sample were mixed, dried down, and resuspended in 40 μl of NimbleGen Hybridization Buffer (NimbleGen Systems) plus 1.5 ug of human COT1 DNA. After denaturation, hybridization was carried out in a MAUI Hybridization System (BioMicro Systems, Salt Lake City) for 18 h at 42°C at the NimbleGen Service Laboratory. The arrays were washed using NimbleGen Wash Buffer System (NimbleGen Systems), dried by centrifugation, and scanned at 5-μm resolution using the GenePix 4000B scanner (Axon Instruments, Union City, CA). Fluorescence intensity raw data were obtained from scanned images of the oligonucleotide tiling arrays using NIMBLESCAN 2.0 extraction software (NimbleGen Systems). For each spot on the array, log2-ratios of the Cy5-labeled test sample versus the Cy3-labeled reference sample were calculated. Then, the biweight mean of this log2 ratio was subtracted from each point; this procedure is approximately equivalent to mean-normalization of each channel. Sites bound by Oct4 were identified using the peak calling algorithm described in Bieda et al. (9), with minor modifications (available upon request). The peaks called for both biological replicates of the LMPCR, pooling, and WGA methods are provided as Supplementary data. The array data has been deposited into GEO (series GSE5251)
RESULTS AND DISCUSSION
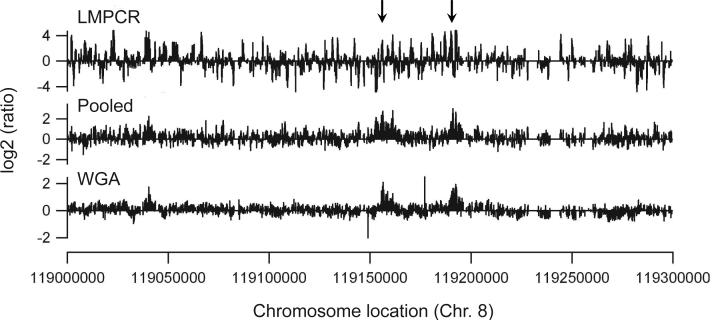
To identify Oct4 binding sites in the human genome, we first performed a ChIP experiment using an antibody to Oct4 and demonstrated that the Oct4 ChIP sample showed enrichment when primers specific to the NANOG and EVX1 promoters (known Oct4 binding sites) were used in PCR reactions, but no enrichment when negative control primers specific for the DHFR gene were used (data not shown). We then used LMPCR to amplify the Oct4 ChIP samples and hybridized the amplified samples to ENCODE arrays. Using ChIP samples amplified by LMPCR, we have previously identified binding sites for E2F family members using CpG island (4), promoter (9,10), and genomic tiling (9) arrays. However, using the LMPCR amplification method we found that Oct4 binding sites could not be distinguished from the background noise on the arrays (Figure 1, top panel). For example, although the Oct4 binding site in the EVX1 promoter is present on the array used in this study, it could not be identified above background noise. Also, two Oct4 binding sites (confirmed by PCR analysis of ChIP samples) within the EXT1 gene, indicated with arrows in Figure 1, do not show enhanced enrichment as compared to the surrounding DNA. Peak prediction analysis of two biologically independent ChIP-chip assays performed using the LMPCR method was carried out using a 98th percentile threshold of log2 oligomer ratios and a P-value P<0.0001 (9). Although hundreds of peaks were called for the two arrays using the LMPCR-derived amplicons, very few peaks were in common on both arrays (Table 1 and Supplementary data).
Figure 1. Comparison of three different sample preparation methods for ChIP-chip assays.
The hybridization profile of a 300 kb region of chromosome 8 surrounding the EXT1 gene is shown for samples prepared by the LMPCR (top panel), pooled (middle panel), or WGA (bottom panel) methods. Oct4 binding sites confirmed by PCR are indicated with arrows.
Table 1.
_O'Geen
total number of peaks called on both arrays
if at least one of the ends of a peak region from one array overlapped a peak region from the other array, the peaks were considered to be overlapping
Because known Oct4 binding sites were enriched in the ChIP samples, it was likely that the inability to identify binding sites on the arrays was a result of the amplification method and not inefficient immunoprecipitation. To test this hypothesis, we performed 10 ChIP reactions for each of two biologically independent samples of cross-linked cells. The 10 ChIP samples from a given batch of cells were pooled, and the two pools were applied separately to genomic tiling arrays. We found that the pooling method greatly reduced the background noise on the array and produced reproducible binding patterns (Figure 1, middle panel). In fact, ∼70% of the peaks identified on one array were identified on the biological replicate array (Table 1 and Supplementary data).
Unfortunately, pooling ChIP samples is not always possible (e.g. if using specialized cell types or tumor tissues) and the need to pool 10 ChIP samples for every array would greatly increase the number of ChIP reactions needed to analyze the entire human genome. Therefore, we felt that a different method for amplifying ChIP samples was required. The method of whole genome amplification (WGA) has proven very useful for investigators performing comparative genomic hybridizations (see http://www.sigmaaldrich.com/sigma/bulletin/wga1bul.pdf.). The standard protocol for this technique is to first employ a random chemical fragmentation of the genome, producing a series of overlapping short templates averaging 400 base pairs. Next, the DNA fragments are efficiently primed to generate a library of DNA fragments with defined 3' and 5' termini. This library is then replicated using linear amplification in the initial stages, followed by a limited round of geometric amplifications. Because ChIP samples are obtained using sonicated chromatin that has an average size of 500 bp-1 kb, we reasoned that the chemical fragmentation step should not be necessary. Therefore, we used an entire ChIP sample (obtained from 1 × 107 cells) for the library generation and subsequent amplification. Using this protocol, we found that the predicted Oct4 peaks show a very similar pattern as in the pooled ChIP samples and the background noise was very low (Figure 1, bottom panel). Using the WGA method, we found that ∼63% of the peaks were detected on both arrays (Table 1 and Supplementary data). These results are very similar to those obtained by analysis of the arrays hybridized with the pooled samples. One reason why the overlap percentage was not higher than 63−70% when the pooled and WGA samples were analyzed is due to limitations of the peak-calling program. As shown in Supplementary Figure 1, very similar binding patterns of Oct4 on two arrays can lead to differences in the number and exact positions of called peaks.
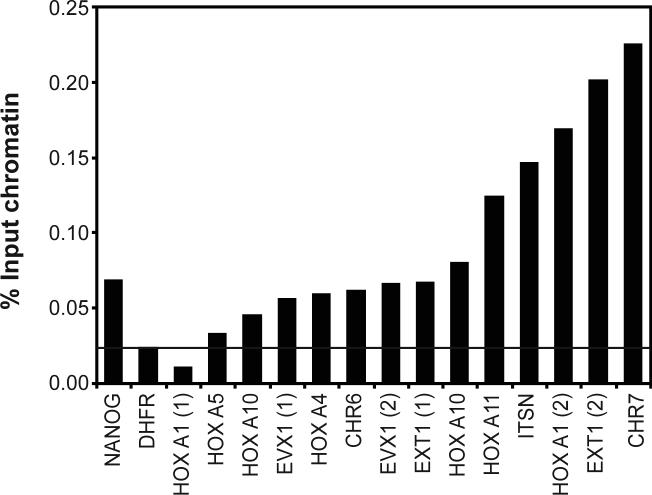
The Oct4 binding sites identified using the WGA method were tested by standard PCR analyses using a ChIP sample from a third independent culture of cells (Figure 3). After analyzing 14 predicted Oct4 binding sites, we obtained a 93% confirmation rate, indicating that the WGA amplification method results in an accurate representation of a ChIP sample obtained from a small number of cells.
Conclusions
We have shown that the method of LMPCR-mediated amplification does not work well for all ChIP samples, perhaps dependent upon the number of binding sites and the abundance of the factor. We have tested a different amplification method, originally developed to provide accurate representation of the genome for studies of copy number changes and SNP analyses in tumor samples. We found that the signal to noise ratio obtained from the hybridization of the WGA amplicons to genomic arrays is superior to the LMPCR method of amplification for ChIP samples, not only for Oct4 but also for a number of other human and mouse transcription factors (data not shown). Based on the low background, reproducibility, and the fact that a single ChIP sample provides sufficient material for several array hybridizations, we recommend the WGA protocol for ChIP-chip analyses.
Figure 2.
The results of PCR assays (using an independent ChIP sample) for 14 Oct4 binding sites predicted from the WGA array data are shown. Two different binding sites were assayed for the HoxA1, EVX1, and EXT1 genes. Primers specific to the NANOG promoter were used as a positive control and primers specific to the DHFR gene were used as a negative control. Enrichments above the negative control are considered confirmed Oct4 binding sites.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grant CA45250, HG003129, DK067889, and UCD FL69920. We thank the members of the Farnham lab for helpful discussion and data analysis and Heather Witt for preparation of the modified ChIP protocol.
Footnotes
COMPETING INTERESTS STATEMENT: Roland Green is an employee of NimbleGen Systems Inc; arrays from this company were used in the ChIP-chip studies. No other authors have competing interests.
Supplementary Material
REFERENCES
- 1.Boyd KE, Farnham PJ. Myc versus USF: Discrimination at the cad gene is determined by core promoter elements. Mol. Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 3.Squazzo SL, Komashko VM, O'Geen H, Krig S, Jin VX, Jang S-W, Green R, Margueron R, et al. Suz12 silences large regions of the genome in a cell type-specific manner. Genome Research in press. 2006 doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberley MJ, Inman D, Farnham PJ. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J Biol Chem. 2003;278:42466–42476. doi: 10.1074/jbc.M307733200. [DOI] [PubMed] [Google Scholar]
- 5.Wells J, Yan PS, Cechvala M, Huang T, Farnham PJ. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sties during S phase. Oncogene. 2003;22:1445–1460. doi: 10.1038/sj.onc.1206264. [DOI] [PubMed] [Google Scholar]
- 6.Weinmann AS, Yan PS, Oberley MJ, Huang TH-M, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oberley MJ, Tsao J, Yau P, Farnham PJ. High throughput screening of chromatin immunoprecipitates using CpG island microarrays. Methods in Enzymol. 2004;376:316–335. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- 9.Bieda M, Xu X, Singer M, Green R, Farnham PJ. Unbiased location analysis of E2F1 binding sites suggests a widespread role for E2F1 in the human genome. Genome Research. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin V, Rabinvich A, Squazzo SL, Green R, Farnham PJ. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data-a case study using E2F1. Genome Research in revision. 2006 doi: 10.1101/gr.5520206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.