Abstract
Aminoacyl-tRNA synthetases are essential enzymes that help to ensure the fidelity of protein translation by accurately aminoacylating (or “charging”) specific tRNA substrates with cognate amino acids. Many synthetases have an additional catalytic activity to confer amino acid editing or proofreading. This activity relieves ambiguities during translation of the genetic code that result from one synthetase activating multiple amino acid substrates. In this review, we describe methods that have been developed for assaying both pre- and post-transfer editing activities. Pre-transfer editing is defined as hydrolysis of a misactivated aminoacyl-adenylate prior to transfer to the tRNA. This reaction has been reported to occur either in the aminoacylation active site or in a separate editing domain. Post-transfer editing refers to the hydrolysis reaction that cleaves the aminoacyl-ester linkage formed between the carbonyl carbon of the amino acid and the 2′ or 3′ hydroxyl group of the ribose on the terminal adenosine. Post-transfer editing takes place in a hydrolytic active site that is distinct from the site of amino acid activation. Here, we focus on methods for determination of steady-state reaction rates using editing assays developed for both classes of synthetases.
1. Introduction: Aminoacyl-tRNA Synthetase Fidelity
Aminoacyl-tRNA synthetases (aaRSs) catalyze the acylation of transfer RNAs (tRNAs) with cognate amino acids via a two-step mechanism (1-3). In the first step, an amino acid is activated with ATP to form an aminoacyl-adenylate (aa-AMP) intermediate. In the second step, the amino acid is transferred to the 3′ end of the cognate tRNA to form the aminoacylated tRNA product. Accurate aminoacylation is important to maintain the fidelity of protein biosynthesis (4-7).
The aaRSs are challenged during aminoacylation by intracellular pools of structurally related substrates that compete for binding. Discrimination of tRNA is based on identity elements encoded within the RNA sequence as well as unique structural motifs (1, 8). Negative or “anti-determinants” have also evolved to block charging of noncognate tRNAs.
About half of the aaRSs can easily distinguish their cognate amino acids based on structure or charge. These include aaRS that are responsible for activating tyrosine (9, 10), cysteine (11-14), asparagine (15, 16), and arginine (17, 18). However, many aaRSs are challenged in distinguishing the correct amino acid from other standard and non-standard amino acids that have steric and structural overlap (4, 6). For example, aliphatic amino acids such as isoleucine, methionine, leucine and valine and also non-standard amino acids including norvaline, norleucine, α-amino homocysteine, butyrate can be misactivated by LeuRS (19, 20).
Linus Pauling originally predicted that enzymes, in general, would not fully discriminate between isosteric substrates such as valine and isoleucine or alanine and glycine, which differ by a single methyl group (21). He proposed that protein misactivation rates would be as high as 1 out of 5 (21). Subsequently, the rate of valine misincorporation for isoleucine during protein synthesis was measured to be as low as 1 in 3000 (22). The increased fidelity is due to aaRS proofreading and editing mechanisms that clear their mistakes (23).
The aaRSs enhance amino acid discrimination, in part, by use of a “double sieve” mechanism (24-26). The first coarse sieve, an aminoacylation active site that is located in an ancient canonical core of the aaRS, is responsible for activating amino acids. A second fine sieve, a hydrolytic active site, has been clearly demonstrated to correct the enzyme's mistakes in a number of synthetase systems (27-35).
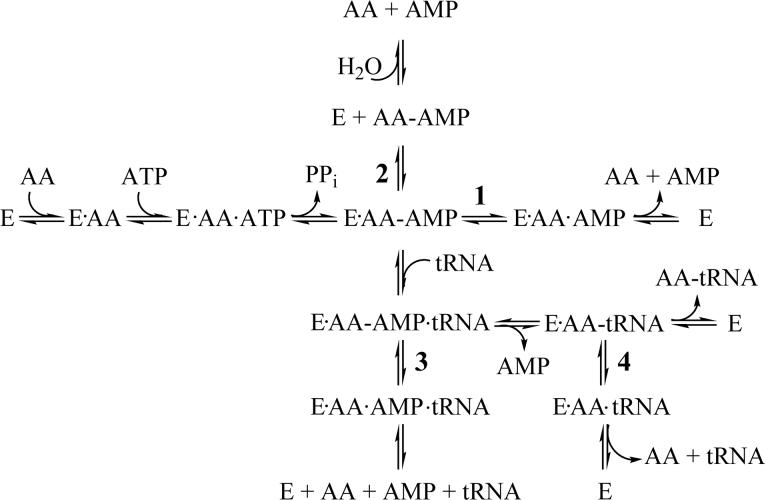
The aaRSs can target either misactivated amino acids or misaminoacylated tRNAs during editing (Figure 1). Post-transfer editing hydrolyzes mischarged tRNA (29, 36, 37), and requires translocation of the mischarged 3′-end of tRNA from the aminoacylation site to the distal hydrolytic active site on the aaRS (38, 39). In some cases, the mischarged tRNA is edited by a free-standing editing domain (40-44).
Figure 1.
Scheme showing possible pre-transfer (paths 1−3) and post-transfer (path 4) editing pathways for the aminoacyl-tRNA synthetases. The abbreviations are as follows: E, enzyme; AA, amino acid; AA-ATP, aminoacyl-adenylate; PPi, pyrophosphate; AA-tRNA, charged tRNA.
Pre-transfer editing is broadly defined as clearing misactivated aminoacyl-adenylates before the amino acid can be transferred to tRNA (6, 45, 46). Because the adenylate intermediate is transient, it is more difficult to investigate in a mechanistic and quantitative way compared to post-transfer editing. However, pre-transfer editing has been proposed to occur by a number of different mechanisms including translocation and hydrolysis in a distinct editing active site on the aaRS (47, 48), hydrolysis within the synthetic active site (49, 50), or by selective release into the cellular milieu (50, 51). In some cases, such as MetRS (52) and LeuRS (53), the misactivated adenylate intermediate cyclizes to form a lactone or thiolactone.
Since the original reports of proofreading by the aaRSs (36, 45), a number of techniques have been developed to characterize these editing functions. Herein, we describe a series of the most commonly used methods to investigate both pre- and post-transfer editing mechanisms in aaRSs. While several aspects of the assays are general and can be universally applied to all synthetases, in some cases, procedures must be adapted to the particular enzyme studied. Therefore, we have chosen to focus on examples from our own work centered on both Class I and Class II synthetases, and have added references for other synthetase examples (Table 1). The purpose of this article is to provide the reader with a practical description of the necessary protocols to characterize the hydrolytic editing activity of the aaRSs. For a comprehensive discussion of editing, the reader is encouraged to consult recent reviews (5, 6, 54).
Table 1.
Representative Editing Protocols for Aminoacyl-tRNA Synthetases
| Aminoacyl-tRNA Synthetase | Reference: |
|---|---|
| CLASS I |
|
| Leucyl-tRNA Synthetase | (30, 62, 70) |
| Isoleucyl-tRNA Synthetase | (65, 87, 94) |
| Valyl-tRNA Synthetase | (95, 96) |
| Methionyl-tRNA Synthetase |
(97, 98) |
| CLASS II |
|
| Alanyl-tRNA Synthetase | (33, 40, 99) |
| Phenylalanyl-tRNA Synthetase | (82, 100, 101) |
| Threonyl-tRNA Synthetase | (32, 102, 103) |
| Prolyl-tRNA Synthetase | (27, 28, 40, 42, 50, 51, 59, 66) |
| Lysyl-tRNA Synthetase | (104, 105) |
2. Pyrophosphate Exchange Assays
As a first step toward establishing whether editing is even required, pyrophosphate exchange assays may be carried out to determine which amino acids are misactivated by the aaRS (55, 56). In general, amino acid editing is predicted to be needed if the observed discrimination factor [1/(relative kcat/KM)] for the noncognate amino acid is less than ∼ 3,000, which is the observed overall error rate for protein synthesis (24). Perhaps more informative is the “effective discrimination factor”, which takes into account the relative cellular concentration of cognate and non-cognate amino acids (57, 58). For example, the in vitro discrimination factor for activation of alanine versus proline by E. coli prolyl-tRNA synthetase (ProRS) is 23,000 (59). However, upon accounting for the higher relative cellular concentration of alanine (148 μM alanine vs 9 μM proline) (60), the effective discrimination factor is only 1,200, which is well within the range where editing would be expected to be required (59).
3. Post-transfer Editing
Of the two major editing pathways (pre- and post-), post-transfer editing is better understood and is relatively straightforward to assay. A typical assay involves presenting mischarged tRNA substrate to the synthetase in trans. Thus, post-transfer editing assays consist of two main steps: 1) formation of mischarged tRNA and 2) measurement of deacylation activity. Post-transfer editing activity was first directly measured for IleRS utilizing Val-tRNAIle as a substrate (36).
3.1. Preparation of mis-charged tRNA substrates: general considerations
Several methods have been used to generate mischarged tRNA. Early procedures involved the use of buffer containing organic components such as 20% DMSO. Introduction of the organic solvent lowered specificity of the enzyme, allowing mischarged tRNA products to be produced, albeit in low yields (20−40%) (23, 36). More recent methods use mutant editing-defective enzymes, heterologous synthetases, and/or mutant tRNA substrates. The yield for Ser-tRNAAla production using E. coli C666A/Q584H mutant AlaRS (33) has been reported to be 15−30% (61) and a similar yield is obtained when Ile-tRNALeu is prepared by mischarging with E. coli TT/VV mutant LeuRS (62). Production of Tyr-tRNAPhe is prepared in ∼ 35% yield upon reaction of tRNAPhe and tyrosine in the presence of the E. coli αA294G-βA356W PheRS double mutant (63).
In some cases, these more modern methods allow for higher levels of mischarged tRNA to be produced (33, 59, 64, 65). For example, the mutant T242P IleRS is deficient in post-transfer editing of Val-tRNAIle and can produce the mischarged product in nearly quantitative yields (5, 64, 65). As an alternative strategy, mutations that allow mischarging may also be incorporated into the tRNA substrate. The triple mutant G1:C72/U70-tRNAPro, which contains the critical acceptor stem recognition elements for aminoacylation with alanine, was shown to be an excellent substrate for E. coli AlaRS, providing a convenient route to mis-charged Ala-tRNAPro (59). Using this method, 94% yield of mischarged tRNAPro can routinely be obtained (B. So and K. Musier-Forsyth, unpublished observations). To avoid the use of a mutant substrate, Deinococcus radiodurans ProRS, which has been shown to mis-acylate E. coli tRNAPro with alanine, can also be used (66), albeit with reduced yields (∼10%) (S. Hati and K. Musier-Forsyth, unpublished observations).
Recently, Suga and co-workers have used in vitro selection or SELEX techniques to develop a ribozyme known as “flexizyme” that catalyzes tRNA aminoacylation (67-69). By evolving novel flexizyme mutants, it is possible to catalyze tRNA misaminoacylation with virtually any amino acid. Typical yields for the production of aminoacyl-tRNA via this method are reported to be 20−50 % (69).
Regardless of the synthetic procedure employed, the mischarged products are recovered via phenol extraction under acidic conditions to stabilize the labile aminoacyl linkage (36) followed by ethanol precipitation. Once the mischarged tRNA substrate has been prepared, deacylation activity is measured by combining the mischarged tRNA product with the editing aaRS. Subsequently, mischarged and uncharged tRNA are precipitated with trichloroacetic acid for quantitation via scintillation counting, as described in more detail below (62, 70) for the preparation of Ala-tRNAPro.
3.2. Production of Ala-tRNAPro
Class II E. coli ProRS has been shown to misactivate multiple amino acids. Several editing mechanisms have been identified that ensure the fidelity of the formation of cognate Pro-tRNAPro (27, 28, 40-42, 44, 50, 51, 59, 71). In particular, mischarged Ala-tRNAPro is hydrolytically edited by bacterial ProRSs at a region of the enzyme termed the insertion (INS) domain (27, 28). Described below is a detailed procedure for the assay of post-transfer editing by E. coli ProRS.
Isolation of mischarged tRNA products involves a standard aminoacylation reaction that is allowed to reach plateau-level charging. The resulting aminoacyl-ester bond is stabilized via acidification, followed by phenol extraction and precipitation. The length of time required to reach a charging plateau is dependent upon the specific system and must be determined empirically.
A typical reaction for mischarging of E. coli tRNAPro with alanine using D. radiodurans is set up as follows:
- 50 μL 2× cocktail:
- 100 mM HEPES, pH 7.5
- 40 mM potassium chloride
- 40 mM β-mercaptoethanol
- 0.2 mg/mL bovine serum albumin (BSA)
- 50 mM MgCl2
- 8 mM ATP
- 13 μM [3H]-alanine (Perkin Elmer, SA = 50 Ci/mmol)
20 μL in vitro transcribed E. coli tRNAPro (5−10 μM final concentration)
30 μL [3H] alanine (stock concentration = 21.3 μM)
5 μL enzyme (1−2 μM final concentration)
One disadvantage to using tritiated amino acids or amino acids that contain [14C] is the relatively low specific activity (e.g., typically <50 Ci/mmol for [3H]-alanine or 152 mCi/mmol for [14C]-alanine). Therefore, when possible, cold amino acid is omitted from these reactions and only radiolabeled alanine (maximum volume possible) is used. However, in some systems, such as in the preparation of Ile-tRNALeu, the concentration of isoleucine in [3H]-Ile is so low (∼1 μM) that unlabeled amino acid must be included to reach a final concentration that is sufficient to drive enzyme catalysis. It is also possible to take a higher volume of radiolabeled amino acid and use a vacuum concentrator to concentrate the aliquot to dryness. The dried sample can then be re-hydrated at a lower volume to attain a known higher concentration. For the reaction described above, the final concentration of [3H]-alanine is 12.5 μM, when amino acid from the 2× cocktail and remaining reaction mixture are combined. As a result, sub-saturating levels of amino acid substrate are present in the charging reaction (KmAla = 140 mM) (59), and long reaction times are required (2−4 hrs).
To ensure a good yield of mischarged product, high concentrations of tRNA (5−10 μM) and enzyme (1−2 μM) are employed. Inorganic pyrophosphatase (4 U/mL) may be included to drive the reaction. However, its effect on aminoacylation activity should be tested beforehand. In rare examples, pyrophosphatase can inhibit charging activity (K. Splan and K. Musier-Forsyth, unpublished observations). The reaction (100 μl) is initiated with enzyme (5 μl), allowed to proceed for an empirically determined amount of time, and quenched upon the addition of 10 % acetic acid (2 μl).
The synthetase is removed via two extractions with an equal volume of phenol that has been equilibrated against diethyl pyrocarbonate (DEPC)-treated water to inhibit RNases and to maintain an acidic pH. The product is precipitated upon addition of 0.1 volumes of 7.5 M ammonium acetate, pH 5.0, and three volumes of ice-cold 100% ethanol for at least four hours at −80 °C. The resulting pellets are dried in a vacuum concentrator (∼10 min) and dissolved in 50 μL of 50 mM potassium phosphate, pH 5.0. Aminoacyl-tRNA should be kept on ice at all times and stored at −80 °C. Its useful shelf life is about 1−2 months.
To determine the overall yield of mischarged tRNA substrate, the specific activity (SA; CPM/pmol) is first determined upon dilution of the 2× cocktail to 1× with the same volume of DEPC-H2O. Alternatively, a small aliquot of one or more of the final reaction mixtures can be tested. Aliquots (2−4 μL) of this solution are spotted onto filter pads in triplicate for quantitation via scintillation counting. Notably, these pads are not washed prior to scintillation counting.
At plateau levels of charging, an aliquot (1 μL) of the final mischarged tRNA is counted directly, and the yield of Ala-tRNAPro purification is then calculated as follows:
The tRNAPro amount is based on the reaction concentration. Loss of the labeled charged tRNA subsequent to purification can be further assessed via scintillation counting, once the tRNA is precipitated and re-hydrated. However, the total amount of tRNA (charged + uncharged) upon concentration and purification is difficult to measure directly. Thus, the overall reported yields of isolated mischarged tRNA should be considered as estimates and can vary, as noted above.
3.3. Assay to measure deacylation of Ala-tRNAPro
To measure ProRS-catalyzed deacylation of Ala-tRNAPro, the mischarged substrate is incubated with ProRS and acid-precipitable radioactivity (i.e. charged tRNA remaining) is measured as a function of time. In preparation for the assay, Whatman 3MM filter pads (Whatman catalog number 1003323) are labeled with pencil and pinned to a foil-covered styrofoam pad. One pad is included for each time point in addition to a zero time point. An aliquot (100 μL) of 5% trichloroacetic acid (TCA) is spotted onto each pad. Filters are dried under a heat lamp or overnight on the bench top. Reactions are set-up as follows:
- 35 μL 2× cocktail:
- 300 mM potassium phosphate, pH 7.0,
- 10 mM magnesium chloride
- 8 U/mL inorganic pyrophosphatase
- 0.2 mg/mL BSA
30 μL 3H-Ala-tRNAPro (1−2 μM final)
5 μL enzyme to initiate reaction
The best results are obtained utilizing at least 10,000 CPM of mischarged tRNA per reaction. Useful enzyme concentrations range from 0.005 − 1 μM, depending on the activity of the enzyme that is being investigated. To determine the rate of spontaneous deacylation, a reaction containing no enzyme that has been initiated with 150 mM KPi, pH 7.0, is also included. A positive control initiated with 0.2 M NaOH, pH 13, may also be performed. Under these conditions, complete deacylation should be observed within 15 min.
Before adding enzyme to initiate the reaction, a 10 μL aliquot is removed and spotted on the t = 0 filter pad, which is immediately dropped into a beaker of ice-cold 5% TCA (use ∼15 mL per pad). Subsequently, reactions are initiated with enzyme (or buffer as a control). Aliquots of 10 μL are removed at the desired time points, quenched by spotting on the appropriate filter pad that is pre-soaked in 100 μl of 5% TCA, and dropped into the 5% TCA wash. After all time points have been taken, the filter pads are washed 3 × 20 min in 5% TCA and 1 × 15 min in 95% ethanol to remove salt, dried under a heat lamp (10−15 min), and counted in 5 mL scintillation fluid (MP Biomedicals). For each reaction, the percent of Ala-tRNAPro remaining at each time point is determined and plotted as a function of time. Typical results are shown in Figure 2.
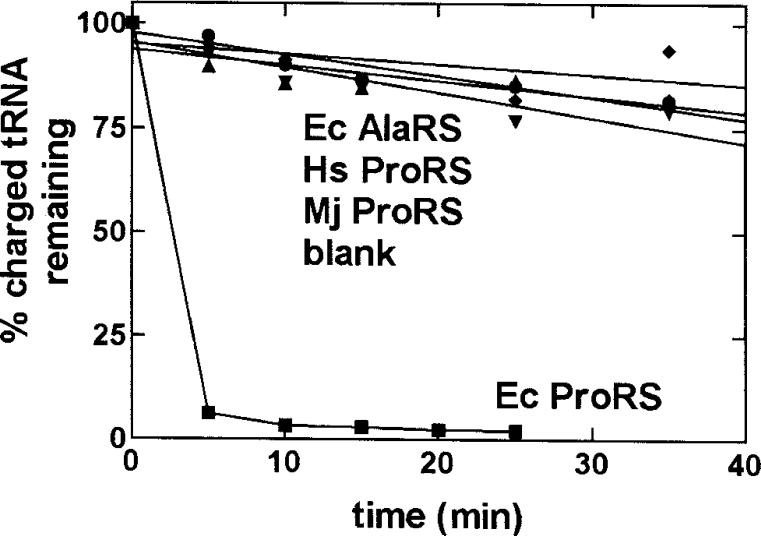
Figure 2.
Graph showing that the efficient deacylation of the Ala-tRNAPro variant is specific for E. coli (Ec) ProRS (0.5 μM) (■). E. coli AlaRS (1 μM) (◆), human (Hs) ProRS (0.6 μM) (•), and M. jannaschii (Mj) ProRS (0.4 μM) (▼) were unable to deacylate E. coli G1:C72/U70-Ala-tRNAPro. Also shown is a control reaction carried out in the absence of enzyme (▲). Figure reproduced with permission from reference (59).
3.4. Troubleshooting
3.4.1 Acid gel electrophoresis to confirm the location of the mischarged amino acid
Protein co-precipitates with tRNA during TCA precipitation (Section 3.3). Under conditions where the enzyme's charging activity is weak, such as during mischarging, it is possible for the enzyme to become self-labeled with radiolabeled amino acids (72-74). In addition, radiolabeled amino acid may be trapped in the protein's active site and also co-precipitated. In either case, quantitation based on TCA precipitable radioactivity would result in erroneously high measurements. To confirm that the amino acid is actually mischarged onto the tRNA, acid gel analysis of misaminoacylation reaction products can be performed (72, 75, 76). An example of a LeuRS misaminoacylation reaction contains:
- 2× cocktail:
- 120 mM TRIS-HCl, pH 7.5
- 20 mM MgCl2
- 2 mM DTT
- 8 μM tRNALeu
- 40 μM L-isoleucine
- 2 μM L-[14C]-isoleucine (Amersham Biosciences, 318 mCi/mmol)
4 mM ATP (final concentration)
1−2 μM of enzyme (final concentration)
Misaminoacylation is initiated by addition of 4 mM ATP to the reaction mixture. At a specific time point a 10 μl aliquot of the reaction is quenched with 10 μl 30 mM NaOAc pH 5.0 and 8 M urea. A 10 μl aliquot of this quenched reaction is separated by electrophoresis on a 10% acidic acrylamide gel prepared as described (72, 75, 76), that is pre-equlibrated with 25 mM NaOAc, pH 5.0. The acidic gel should be electrophoresed slowly at 5 mA and may also be carried out at 4 °C in a cold room to minimize overheating the gel apparatus. Upon completion, the acidic gel containing [14C]-labeled amino acids is dried and exposed to a phosphorimaging screen for about one month prior to analysis.
3.4.2. Varied deacylation plateau levels
Post-transfer editing activities for tRNA synthetases often plateau at levels that don't reflect the entire population of tRNA. This plateau level can vary widely between different experiments. Some tRNA synthetases, such as IleRS, ValRS, and LeuRS are known to be highly specific for either the 2′ or 3′ misaminoacylated ribose hydroxyl, but not both (47, 77). The amino acid is known to facilely migrate between the 2′ and 3′ position (78). Thus, we propose that the premature plateau likely reflects the changing population of 2′ versus 3′ aminoacylated tRNA. As a control, a final aliquot of the reaction can be quenched in base as described in Section 3.3 to fully hydrolyze the population of tRNA.
3.4.3. Re-charging assays to confirm editing activity
For a newly identified post-transfer editing activity, it is important to confirm that the ‘deacylation’ activity is not the result of non-specific nuclease activity present in the protein preparation. This can be readily accomplished by carrying out a re-charging assay (41). Briefly, following the editing reaction, the tRNA is recovered by phenol-chloroform extraction and ethanol precipitation. A standard charging assay is then carried out with the cognate amino acid using a known quantity of the recovered tRNA. To rule out nuclease activity, the extent of charging observed should correspond to the level expected based on the acceptor activity of the tRNA.
3.5. Alternative approach to monitor post-transfer editing: mischarging assays
While the above assays are described for the ProRS:Ala-tRNAPro system, they are based upon standard procedures and can be adapted for other synthetase systems, as long as appropriate mischarged substrates can be prepared. In addition to deacylation assays, mischarging assays can also be used to monitor post-transfer editing. These assays are especially useful once editing activity has been established, as they do not require the preparation of a mischarged tRNA substrate. Instead, the mischarged tRNA is formed in cis during the reaction. Under steady-state conditions, the misacylated species is not readily observed in the presence of the editing-active synthetase, but will be observed for variants that have significant defects in editing (70). Thus, mischarging assays are a rapid and straightforward way to screen a large number of mutants for the purpose of identifying active site residues. Indeed, post-transfer editing-deficient enzymes that can mischarge cognate tRNAs have been identified for several synthetases including IleRS (65, 79), ValRS (80), LeuRS (19, 62, 70), AlaRS (33, 81), ProRS (27, 50), ThrRS (32, 34) and both bacterial (63) and archaeal/eukaryal PheRS (82).
Mis-charging assays closely parallel cognate aminoacylation assays and will therefore be described only briefly. Certain assay parameters do need to be modified, particularly when working with weak editing-defective enzymes. Enzyme concentrations typically range from 0.5−5 μM, as compared to nM concentrations for charging of cognate amino acids. Mischarging plateaus can often be achieved within several min (65, 70). Despite these trends, both enzyme concentration and assay time course are best determined empirically for each synthetase variant.
The Km values for non-cognate amino acid substrates vary widely (0.35 mM – 150 mM), and in cases of weak amino acid affinity, only sub-saturating substrate levels can be achieved utilizing [3H]- and [14C]-based assays. As an alternative approach, mischarging activity of the aaRS can be monitored via an assay in which the 3′-terminal internucleotide linkage of the tRNA is labeled with [32P] (83). Upon completion of the assay, the tRNA is digested, generating a mixture of [32P]-labeled AMP and aminoacyl-AMP, which reflect the relative amounts of uncharged and charged tRNA, respectively. Importantly, use of this assay enables kinetic characterization of even weak mischarging activities for noncognate amino acids at saturating amino acid levels (50). In addition, this assay can be used to detect mischarging with amino acid analogs that are not readily available in [3H]- or [14C]-labeled form.
3.5.1. Potential problems with comparing post-transfer editing kinetics to mischarging activity
It should be noted that modest defects in editing activities that can vary for different enzymes may not necessarily lead to a detectable mischarging activity (70). Rather, a lowered threshold of editing activity may still be sufficient to maintain amino acid fidelity of the synthetase. In addition, it is possible that mischarging activity might occur in the presence of a robust editing activity that is similar to wild-type levels. This might occur when translocation mechanisms to move tRNA from the aminoacylation to the editing site are disrupted, which would prematurely release mischarged tRNA. These sorts of defects would not be detectable in standard post-transfer editing assays because translocation is bypassed to directly bind mischarged tRNA to the editing active site (84).
4. Pre-transfer Editing
4.1. General considerations
Although the concept of pre-transfer editing was first described in the 1960's (45), it has proven more difficult to characterize than post-transfer editing. In some systems, pre-transfer editing is stimulated only in the presence of cognate tRNA (5, 23, 45). Total editing activity can be assayed via the measurement of overall ATPase activity. However, since both pre- and post-transfer editing result in ATPase activity (Figure 1), the relative contributions of each pathway cannot be separated by this assay. One strategy that has been used successfully to isolate tRNA-dependent pre-transfer editing is to selectively inactivate post-transfer editing via mutation (65, 85). In another example, pre-transfer editing of Val-AMP by IleRS was observed in the presence of a DNA aptamer that was selected using the stable Val-AMP:IleRS complex as a target (86). In this experiment, the selected DNA aptamer specifically induced the hydrolysis of the enzyme-bound valyl-adenylate, but not the isoleucyl-adenylate. Furthermore, the rate of hydrolysis in the presence of saturating amounts of the aptamer coincided with the rate observed in the presence of saturating tRNAIle. This work demonstrated that pre-transfer editing was triggered by nucleic acid induced conformational changes and was not dependent on the ability of tRNA to serve as an amino acid acceptor.
In some aaRS systems, such as ProRS, pre-transfer editing is observed in the absence of tRNA (51, 59). Thus, ATPase assays performed in the absence of tRNA may be used as an initial evaluation of pre-transfer editing. These assays measure the amount of ATP consumed, which increases upon repeated cycles of misactivation and editing. However, it is important to remember that ATP hydrolysis is not a direct monitor of aminoacyl-adenylate hydrolysis and thus, these assays may not reflect the true rate of pre-transfer editing. For example, adenylate dissociation from the active site (Figure 1, path 2) represents one pathway that a weakly bound noncognate adenylate may be cleared. This path may trigger repeated cycles of ATPase activity, but does not correlate to enzymatic aminoacyl-adenylate hydrolysis. In contrast, direct measurement of adenylate hydrolysis may be achieved by monitoring AMP formation. Described below are two assays that are commonly used to monitor pre-transfer editing.
4.2. ATPase assays
4.2.1. Charcoal binding assay
Since ATP is consumed during charging and mischarging reactions, a charcoal-binding assay can be used to measure the release of [32P]-PPi from γ-[32P]-ATP. The latter binds to activated charcoal but inorganic pyrophosphate does not (87, 88). The quench solution (250 μl) containing 7% perchloric acid, 10 mM sodium pyrophosphate, and 3% activated charcoal (purchased from Sigma) is aliquoted into 1.5 mL microcentrifuge tubes (one per time point). A reaction is set up as follows:
- 20 μL 3× cocktail:
- 450 mM TRIS-HCl, pH 7.5
- 30 mM MgCl2
- 9 mM ATP
- 12 U/mL inorganic pyrophosphatase
- 0.03 μM γ-[32P]-ATP (Amersham Biosciences, 3000 Ci/mmol)
35 μL reaction mixture (amino acid, tRNA (15 μM), when applicable)
5 μL enzyme (1−4 μM final concentration)
The concentration of amino acid is at saturating levels (500 mM for alanine with ProRS), if possible, or is varied over a range of concentrations to obtain kcat and Km. Notably, some aliphatic and hydrophobic amino acid concentrations, such as leucine or tryptophan, are limited by solubility in aqueous conditions. A reaction without amino acid is also carried out to determine the background rate, which is presumably due to incomplete adsorption of ATP to charcoal.
Each reaction is initiated by the addition of enzyme. At the desired time point, a 10 μL aliquot is removed and injected into the acidic quench solution that contains charcoal. The quenched reaction is mixed vigorously by vortexing to ensure maximum adsorption by the charcoal. When all time points are completed, the tubes are centrifuged in a microfuge for 3 min at 10,000×g. ATP will be adsorbed by the charcoal, while released radioactive PPi will remain in the supernatant. Radioactivity of a portion of the supernatant (50 μL) is quantified in 5 mL of scintillation fluid (MP Biomedicals). Aliquots (4 × 10 μL) of 3× cocktail are also counted and the CPM measurements are averaged and used to calculate the specific activity of the reaction cocktail.
Typical data is shown in Figure 3, plotted as nmol of PPi released versus time. The nmol PPi released (which corresponds to nmol ATP hydrolyzed) is obtained as follows: nmol PPi released = [CPM (time point) – CPM (background)]/specific activity (CPM/nmol) This value may then be divided by the total volume of the counted aliquot (50 μL) to obtain the concentration in the units of μM. As an alternative, adsorbed ATP can be separated upon filtration of the charcoal solution through glass fiber pads (Schleicher and Schuell) and counted directly. In this case, data is plotted as a decrease in ATP concentration over time (87).
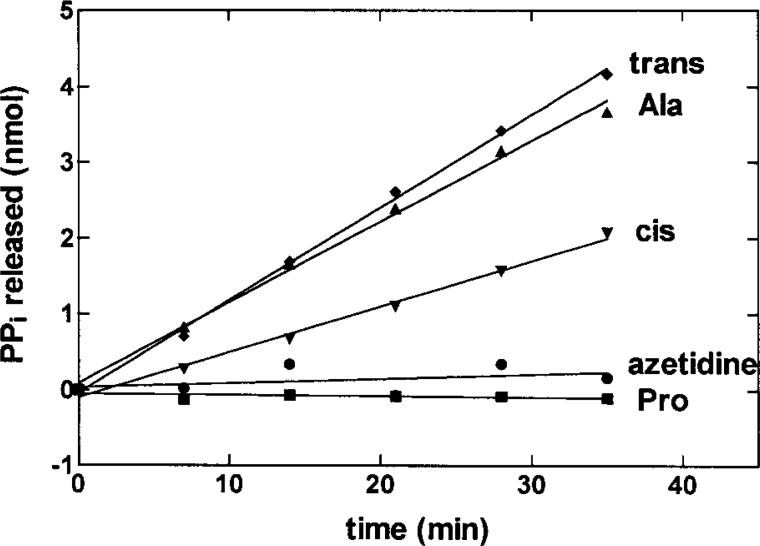
Figure 3.
Noncognate amino acids stimulate ATP hydrolysis by E. coli ProRS. Graph showing the ATP hydrolysis activity of ProRS (2 μM) in the presence of 250 mM trans-4-hydroxyproline (trans) (◆), 500 mM alanine (▲), 250 mM cis-4-hydroxyproline (cis) (▼), 250 mM azetidine-4-carboxylic acid (azetidine) (•), and 2 mM proline (■). Reproduced with permission from reference (59).
4.2.2. Direct measurement of formation and hydrolysis of aminoacyl-adenylate via thin-layer chromatography (TLC)
As described above, the ATPase assay measures the rate of ATP hydrolysis due to clearance of the noncognate aminoacyl-adenylate intermediate from the active site and repeated cycles of amino acid activation. However, this reaction rate may not necessarily be directly correlated to pre-transfer editing, which refers to adenylate hydrolysis. As an alternative, ATP consumption and its products can be monitored using ATPase assays that are resolved on nitrocellulose TLC plates. Assays of this type were recently used by Perona and co-workers to study a pre-transfer editing-like reaction catalyzed by Class I glutaminyl-tRNA synthetase (49). An additional mode of pre-transfer editing where misactivated homocysteine is converted to cyclic thiolactone by methionyl-tRNA synthetase (MetRS) can also be detected via TLC analysis (52).
Both γ-[32P]-ATP and α-[32P]-ATP can be incorporated to directly monitor pyrophosphate and AMP, respectively. Alternatively, [14C]-ATP can be incorporated into the reaction, which minimizes artifactual by-products and reactants that are common in [32P]-labeled nucleotides. The disadvantage is that phosphorimaging of TLC plates requires about a month for analysis.
A reaction is set up as follows:
- 20 μl 2× cocktail:
- 100 mM TRIS-HCl, pH 7.5
- 20 mM MgCl2
- 10 mM DTT (D,L-dithiothreitol)
- 36 μM ATP
- 0.06 μM γ - or α-[32P]-ATP (Perkin Elmer, 3000 Ci/mmol) or 3 mM [8-14C]-ATP (Amersham, 56 mCi/mmol)
20 μl reaction mixture ( amino acid, tRNA (10 μM), when applicable
0.5 − 1 μM enzyme (final concentration)
Each reaction is initiated by addition of enzyme. At specific time points, 2 μl aliquots are spotted onto TLC plates (J.T. Baker, Phillipsburg, NJ) that have been pre-run in water. The TLC plates are developed in 750 mM KH2PO4, pH 3.5 and air dried (89). Radiolabeled bands are detected and quantitated by phosphorimaging or autoradiography of the TLC plates. The ATP consumption is graphed as decreasing percentage of radioactivity (intensity/mm2) of ATP over time. The percentage of ATP is normalized based on the fraction of ATP consumed or AMP produced relative to all radioactive bands in the TLC lane. For example, the percentage of ATP is obtained as follows: % of ATP = [(band volume of ATP)/(volume of ATP + volume of AMP formed)] × 100, for each time point.
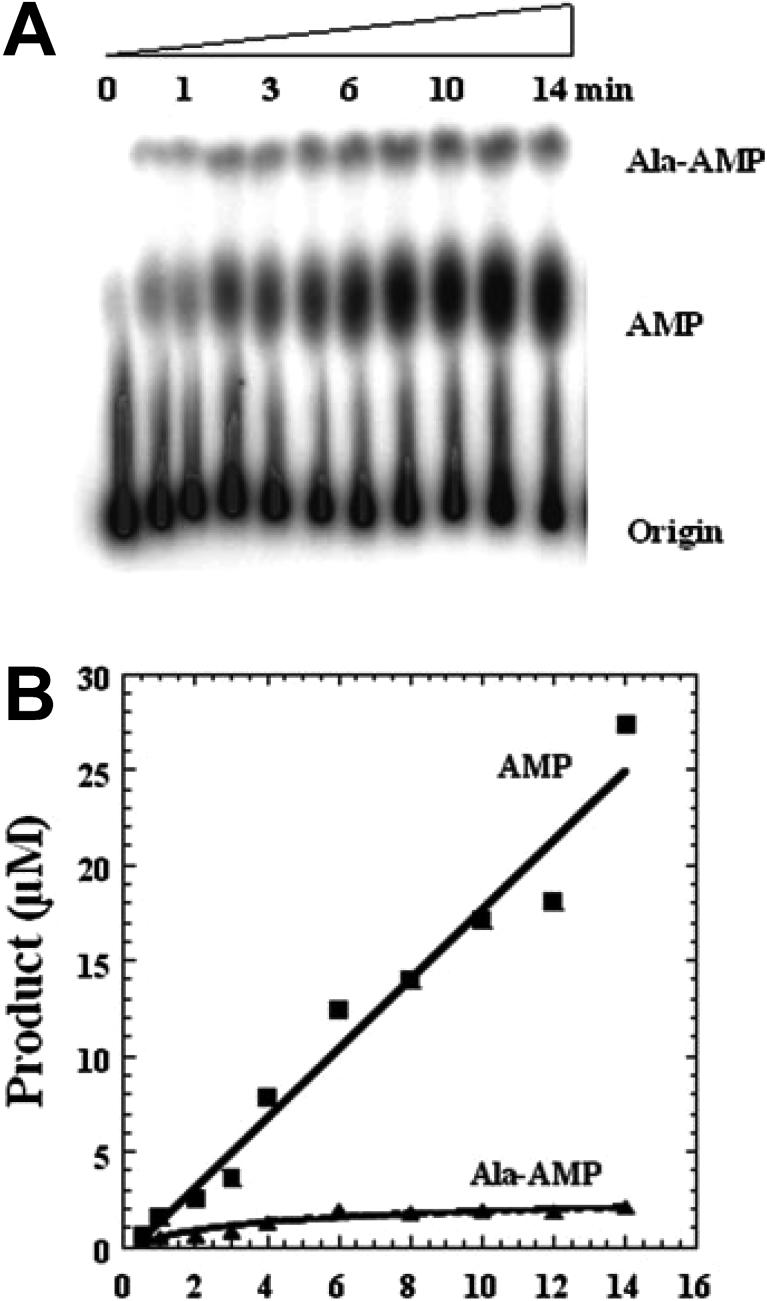
We have recently adapted the TLC based assay described above to study pre-transfer editing of alanine by E. coli ProRS (51). Since the [32P]-ATP is labeled at the α-phosphate, it is possible to visualize aa-AMP, AMP, and ATP by TLC analysis. A typical chromatography image for the formation and breakdown of Ala-AMP is shown in Figure 4A. The positions corresponding to elution of Ala-AMP and free AMP are confirmed by using chemically synthesized Ala-AMP (90) and commercially available AMP (Sigma). Non-hydrolyzed [α-32P]-ATP remains at the origin, while [32P]-AMP and Ala–[32P]-AMP migrate in the order indicated in Figure 4A. To account for spontaneous ATP hydrolysis activity, the no-enzyme time point is subtracted from the AMP band, which ensures that the remaining intensity is due to pre-transfer editing. To obtain initial rates, the intensity of each band is converted to concentration, multiplied by the dilution factor upon quenching, and plotted against time. Representative data is shown in Figure 4B for the formation and breakdown of Ala-AMP by E. coli ProRS.
Figure 4.
TLC-based assay to monitor AMP and aminoacyl-adenylate formation. A. Reaction time course showing chromatographic separation from top to bottom of Ala-[32P]AMP, [32P]AMP, and [32P]ATP. B. graphical representation of TLC data shown in panel A. Figure reproduced with permission from reference (51).
The assay described above directly measures the formation of both the aminoacyl-adenylate and the resulting AMP produced upon hydrolysis, allowing for mechanistic insights to be gained. As shown in Figure 1, tRNA-independent pre-transfer hydrolytic editing can be accomplished via one of two mechanisms: enzyme catalyzed hydrolysis of the adenylate (path 1) or “selective release” of the non-cognate adenylate followed by solution hydrolysis (path 2). To determine the dominant pathway, the rate of AMP formation measured via the above assay can be compared to the rate of solution hydrolysis. The latter may be measured via a so-called “chase” assay, where the adenylate is formed in the active site and chased into solution by addition of a vast excess of ATP or of an adenylate analog (49, 51, 91).
Pre-transfer editing of Ala-AMP by ProRS (50), Thr-AMP by SerRS (91), and Gln-AMP by GlnRS in the presence of a non-chargable tRNA substrate (49) has been shown to be enzyme-catalyzed, with the rate of AMP formation being significantly (17- to 35-fold) faster than the non-enzymatic rate of adenylate hydrolysis. Although in these examples enzyme-catalyzed hydrolysis is the dominant mechanism, the assays reveal an accumulation of aminoacyl-adenylate formation that exceeds the concentration of enzyme used, indicating that selective release contributes to total editing, albeit to a minor extent. In the case of bacterial ProRS, enzyme-catalyzed hydrolysis accounted for ∼80% of editing, with the remainder occurring via selective release (50).
Additional mechanistic insights can be obtained by performing the TLC-based pre-transfer editing assay under “burst” conditions (i.e., high enzyme concentrations (∼5 μM)). When AMP formation is plotted versus time, the presence or absence of the burst determines whether amino acid misactivation or hydrolysis of misactivated amino acid is rate-limiting. The presence of a burst at time zero indicates that aa-AMP was formed faster than it was hydrolyzed, suggesting that amino acid misactivation is not rate limiting relative to hydrolysis. Conversely, the absence of a burst indicates that misactivation of the amino acid is the rate-limiting step in this two-step process. Finally, it should be noted that a variation of the assay has recently been used to measure the pre-steady state kinetics of His-AMP formation by histidyl-tRNA synthease (92).
4.4. Fluorescence-based assay to monitor translocation of the aminoacyl-adenylate
In the IleRS system, non-cognate amino acid editing has been postulated to take place at a site distant from the synthetic active site (35, 38). To study the putative translocation step of misactivated substrate to the editing site, fluorescence-based techniques have been used (48, 79, 93). In this work, the fluorescent nucleotide N-methylanthraniloyl dATP (dATP†, where the dagger indicates the fluorescent analog) acts as an acceptor for tryptophan fluorescence when bound at the synthetic active site, allowing for active site occupancy to be conveniently monitored. Upon addition of either valyl-adenylate or isoleucyl-adenylate, or upon enzymatic formation of either aminoacyl-adenylate by IleRS, a fluorescence decrease is observed as dATP† is displaced from the synthetic active site. The addition of tRNAIle triggers hydrolytic editing, resulting in the emptying of the synthetic active site as misactivated amino acid is translocated to the editing site. In parallel, dATP† fluorescence is recovered as it rebinds to the synthetic active site. Significantly, fluorescence recovery is only observed in the presence of noncognate valine. Furthermore, a high concentration of dATP† is employed to ensure instantaneous binding, allowing for the translocation rate to be measured.
5. Conclusions
The editing activity of aaRSs has been actively studied for many years and considerable progress has been made on many levels. Decades of work to try to understand high fidelity mechanisms of tRNA synthetases have recently capitalized on more facile production of mischarged tRNA products to directly investigate amino acid editing mechanisms that have been identified in about half of this family of enzymes (Table 1). Mutational analysis and structural biology have localized editing active sites and their key determinants in each of the editing enzymes, although it has become clear that strategies for fidelity and their mechanisms can be completely different for each enzyme (6). Editing defects have also been linked to neurodegenerative disease (81). This has launched a new and important frontier in cell biology including the use of prokaryotic and eukaryotic systems to investigate the production and effects of statistical proteins in vivo that accumulate in the cell. The protocols detailed herein are representative of the most current methods utilized to characterize this important activity of the aminoacyl-tRNA synthetases.
Acknowledgements
This work was supported by grants from The National Institutes of Health (GM063789 to SAM and GM049928 to KM-F). We are also grateful to Ms. Byung-Ran So, Dr. Michael Ignatov, Dr. Sanchita Hati, and Dr. Joseph Chihade for helpful discussion and comments on the review and Dr. Michael Ignatov for design of Figure 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.First EA. In: The Aminoacyl-tRNA Synthetases. Ibba M, Francklyn C, Cusack S, editors. Landes Biosciences; Georgetown, TX: 2005. pp. 328–52. [Google Scholar]
- 2.Freist W. Biochemistry. 1989;28:6787–95. doi: 10.1021/bi00443a001. [DOI] [PubMed] [Google Scholar]
- 3.Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson TL, de Crécy-Lagard V, Schimmel P. Annu Rev Biochem. 2004;73:147–76. doi: 10.1146/annurev.biochem.73.012803.092429. [DOI] [PubMed] [Google Scholar]
- 5.Hendrickson TL, Schimmel P. In: Translation Mechanisms. Lapointe J, Braker-Gingras L, editors. Kluwer Academic/Plenum Publishers; 2003. pp. 34–64. [Google Scholar]
- 6.Mascarenhas A, Martinis SA, An S, Rosen AE, Musier-Forsyth K, Koehrer C. In: Protein Engineering. RajBhandary T, editor. Springer Verlag; (in press) [Google Scholar]
- 7.Jakubowski H, Goldman E. Microbiol Rev. 1992;56:412–29. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giegé R, Sissler M, Florentz C. Nucleic Acids Res. 1998;26:5017–35. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borgford TJ, Gray TE, Brand NJ, Fersht AR. Biochemistry. 1987;26:7246–50. doi: 10.1021/bi00397a008. [DOI] [PubMed] [Google Scholar]
- 10.Fersht AR. Biochemistry. 1987;26:8031–7. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- 11.Fersht AR, Dingwall C. Biochemistry. 1979;18:1245–9. doi: 10.1021/bi00574a020. [DOI] [PubMed] [Google Scholar]
- 12.Newberry KJ, Hou YM, Perona JJ. EMBO J. 2002;21:2778–87. doi: 10.1093/emboj/21.11.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CM, Christian T, Newberry KJ, Perona JJ, Hou YM. J Mol Biol. 2003;327:911–7. doi: 10.1016/s0022-2836(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 14.Zhang CM, Perona JJ, Hou YM. Biochemistry. 2003;42:10931–7. doi: 10.1021/bi034812u. [DOI] [PubMed] [Google Scholar]
- 15.Berthet-Colominas C, Seignovert L, Hartlein M, Grotli M, Cusack S, Leberman R. EMBO J. 1998;17:2947–60. doi: 10.1093/emboj/17.10.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki W, Sekine S, Kuroishi C, Kuramitsu S, Shirouzu M, Yokoyama S. J Mol Biol. 2006;360:329–42. doi: 10.1016/j.jmb.2006.04.068. [DOI] [PubMed] [Google Scholar]
- 17.Cavarelli J, Delagoutte B, Eriani G, Gangloff J, Moras D. EMBO J. 1998;17:5438–48. doi: 10.1093/emboj/17.18.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freist W, Sternbach H, Cramer F. Eur J Biochem. 1989;186:535–41. doi: 10.1111/j.1432-1033.1989.tb15239.x. [DOI] [PubMed] [Google Scholar]
- 19.Karkhanis VA, Mascarenhas AP, Martinis SA. J Bacteriol. 2007. in press. [DOI] [PMC free article] [PubMed]
- 20.Xu MG, Li J, Du X, Wang ED. Biochem Biophys Res Commun. 2004;318:11–16. doi: 10.1016/j.bbrc.2004.03.180. [DOI] [PubMed] [Google Scholar]
- 21.Pauling L. Festschrift für Arthur Stoll Siebzigsten Geburtstag. 1958:597–602. [Google Scholar]
- 22.Loftfield RB. Biochem. J. 1963;89:82–92. doi: 10.1042/bj0890082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fersht AR. Biochemistry. 1977;16:1025–30. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 24.Fersht AR. Proc. R. Soc. London Ser. B. 1981;212:351–79. doi: 10.1098/rspb.1981.0044. [DOI] [PubMed] [Google Scholar]
- 25.Fersht AR, Dingwall C. Biochemistry. 1979;18:2627–31. doi: 10.1021/bi00579a030. [DOI] [PubMed] [Google Scholar]
- 26.Fersht AR. Science. 1998;280:541. doi: 10.1126/science.280.5363.541. [DOI] [PubMed] [Google Scholar]
- 27.Wong FC, Beuning PJ, Nagan M, Shiba K, Musier-Forsyth K. Biochemistry. 2002;41:7108–15. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 28.Wong FC, Beuning PJ, Silvers C, Musier-Forsyth K. J Biol Chem. 2003;278:52857–64. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- 29.Lin L, Hale SP, Schimmel P. Nature. 1996;384:33–4. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 30.Betha AK, Williams AM, Martinis SA. Biochemistry. 2007;46:6258–67. doi: 10.1021/bi061965j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao MW, Zhu B, Hao R, Xu MG, Eriani G, Wang ED. EMBO J. 2005;24:1430–9. doi: 10.1038/sj.emboj.7600618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dock-Bregeon A, Sankaranarayanan R, Romby P, Caillet J, Springer M, Rees B, Francklyn CS, Ehresmann C, Moras D. Cell. 2000;103:877–84. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 33.Beebe K, Ribas De Pouplana L, Schimmel P. EMBO J. 2003;22:668–75. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dock-Bregeon AC, Rees B, Torres-Larios A, Bey G, Caillet J, Moras D. Mol Cell. 2004;16:375–86. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Science. 1998;280:578–82. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 36.Eldred EW, Schimmel PR. J Biol Chem. 1972;247:2961–64. [PubMed] [Google Scholar]
- 37.Mursinna RS, Lincecum TL, Jr., Martinis SA. Biochemistry. 2001;40:5376–81. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 38.Silvian LF, Wang J, Steitz TA. Science. 1999;285:1074–7. [PubMed] [Google Scholar]
- 39.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. Nat Struct Mol Biol. 2005;12:923–30. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 40.Ahel I, Korencic D, Ibba M, Söll D. Proc Natl Acad Sci U S A. 2003;100:15422–7. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An S, Musier-Forsyth K. J Biol Chem. 2004;279:42359–62. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 42.An S, Musier-Forsyth K. J Biol Chem. 2005;280:34465–72. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Huang K, Li Z, Banerjei L, Fisher KE, Grishin NV, Eisenstein E, Herzberg O. Proteins. 2000;40:86–97. doi: 10.1002/(sici)1097-0134(20000701)40:1<86::aid-prot100>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 44.Ruan B, Söll D. J Biol Chem. 2005;280:25887–91. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 45.Baldwin AN, Berg P. J Biol Chem. 1966;241:839–45. [PubMed] [Google Scholar]
- 46.Schimmel P, Schmidt E. Trends Biochem Sci. 1995;20:1–2. doi: 10.1016/s0968-0004(00)88937-9. [DOI] [PubMed] [Google Scholar]
- 47.Lincecum TL, Jr., Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grotli M, Martinis SA, Cusack S. Mol Cell. 2003;11:951–63. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 48.Nomanbhoy TK, Hendrickson TL, Schimmel P. Mol Cell. 1999;4:519–28. doi: 10.1016/s1097-2765(00)80203-8. [DOI] [PubMed] [Google Scholar]
- 49.Gruic-Sovulj I, Uter N, Bullock T, Perona JJ. J Biol Chem. 2005;280:23978–86. doi: 10.1074/jbc.M414260200. [DOI] [PubMed] [Google Scholar]
- 50.Splan KE, Ignatov M, Musier-Forsyth K. submitted. [DOI] [PubMed]
- 51.Hati S, Ziervogel B, SternJohn J, Wong F, Nagan M, Rosen AE, Siliciano PG, Chihade JW, Musier-Forsyth K. J Biol Chem. 2006;281:27862–72. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- 52.Jakubowski H, Fersht AR. Nucleic Acids Res. 1981;9:3105–17. doi: 10.1093/nar/9.13.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Englisch S, Englisch U, von der Haar F, Cramer F. Nucleic Acids Res. 1986;14:7529–39. doi: 10.1093/nar/14.19.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jakubowski H. In: The aminoacyl-tRNA synthetases. Ibba M, Francklyn C, Cusack S, editors. Landes Bioscience; Georgetown, TX: 2005. pp. 384–96. [Google Scholar]
- 55.Fersht AR, Kaethner MM. Biochemistry. 1976;15:818–23. doi: 10.1021/bi00649a014. [DOI] [PubMed] [Google Scholar]
- 56.Heacock D, Forsyth CJ, Shiba K, Musier-Forsyth K. Bioorg Chem. 1996;24:273–89. [Google Scholar]
- 57.Freist W, Sternbach H, Pardowitz I, Cramer F. J Theor Biol. 1998;193:19–38. doi: 10.1006/jtbi.1998.0672. [DOI] [PubMed] [Google Scholar]
- 58.Ninio J. Biochimie. 1975;57:587–95. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 59.Beuning PJ, Musier-Forsyth K. Proc Natl Acad Sci U S A. 2000;97:8916–20. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raunio R, Rosenqvist H. Acta Chem Scand. 1970;24:2737–44. doi: 10.3891/acta.chem.scand.24-2737. [DOI] [PubMed] [Google Scholar]
- 61.Beebe K, Waas W, Druzina Z, Guo M, Schimmel P. Anal Biochem. 2007;368:111–21. doi: 10.1016/j.ab.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhai Y, Martinis SA. Biochemistry. 2005;44:15437–43. doi: 10.1021/bi0514461. [DOI] [PubMed] [Google Scholar]
- 63.Roy H, Ling J, Irnov M, Ibba M. EMBO J. 2004;23:4639–48. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hendrickson TL, Nomanbhoy TK, de Crécy-Lagard V, Fukai S, Nureki O, Yokoyama S, Schimmel P. Mol Cell. 2002;9:353–62. doi: 10.1016/s1097-2765(02)00449-5. [DOI] [PubMed] [Google Scholar]
- 65.Hendrickson TL, Nomanbhoy TK, Schimmel P. Biochemistry. 2000;39:8180–6. doi: 10.1021/bi0004798. [DOI] [PubMed] [Google Scholar]
- 66.Ahel I, Stathopoulos C, Ambrogelly A, Sauerwald A, Toogood H, Hartsch T, Söll D. J Biol Chem. 2002;277:34743–8. doi: 10.1074/jbc.M206928200. [DOI] [PubMed] [Google Scholar]
- 67.Kourouklis D, Murakami H, Suga H. Methods. 2005;36:239–44. doi: 10.1016/j.ymeth.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 68.Murakami H, Kourouklis D, Suga H. Chem Biol. 2003;10:1077–84. doi: 10.1016/j.chembiol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 69.Murakami H, Ohta A, Ashigai H, Suga H. Nat Methods. 2006;3:357–9. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 70.Mursinna RS, Martinis SA. J Am Chem Soc. 2002;124:7286–7. doi: 10.1021/ja025879s. [DOI] [PubMed] [Google Scholar]
- 71.Beuning PJ, Musier-Forsyth K. J Biol Chem. 2001;276:30779–85. doi: 10.1074/jbc.M104761200. [DOI] [PubMed] [Google Scholar]
- 72.Martinis SA, Schimmel P. Proc Natl Acad Sci U S A. 1992;89:65–9. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alexander RW, Schimmel P. Prog Nucleic Acid Res Mol Biol. 2001;69:317–49. doi: 10.1016/s0079-6603(01)69050-0. [DOI] [PubMed] [Google Scholar]
- 74.Gillet S, Hountondji C, Schmitter JM, Blanquet S. Protein Sci. 1997;6:2426–35. doi: 10.1002/pro.5560061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsu JL, Rho SB, Vannella KM, Martinis SA. J Biol Chem. 2006;281:23075–82. doi: 10.1074/jbc.M601606200. [DOI] [PubMed] [Google Scholar]
- 76.C U. Varshney,, Lee P, RajBhandary UL. J Biol Chem. 1991;266:24712–8. [PubMed] [Google Scholar]
- 77.Nordin BE, Schimmel P. Biochemistry. 2003;42:12989–97. doi: 10.1021/bi035052q. [DOI] [PubMed] [Google Scholar]
- 78.Taiji M, Yokoyama S, Miyazawa T. Biochemistry. 1983;22:3220–5. doi: 10.1021/bi00282a028. [DOI] [PubMed] [Google Scholar]
- 79.Bishop AC, Nomanbhoy TK, Schimmel P. Proc Natl Acad Sci U S A. 2002;99:585–90. doi: 10.1073/pnas.012611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Döring V, Mootz HD, Nangle LA, Hendrickson TL, de Crécy-Lagard V, Schimmel P, Marliere P. Science. 2001;292:501–4. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 81.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Nature. 2006;443:50–5. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 82.Sasaki HM, Sekine S, Sengoku T, Fukunaga R, Hattori M, Utsunomiya Y, Kuroishi C, Kuramitsu S, Shirouzu M, Yokoyama S. Proc Natl Acad Sci U S A. 2006;103:14744–9. doi: 10.1073/pnas.0603182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wolfson AD, Uhlenbeck OC. Proc Natl Acad Sci U S A. 2002;99:5965–70. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, Plattner JJ, Shapiro L, Martinis SA, Benkovic SJ, Cusack S, Alley MR. Science. 2007;316:1759–61. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 85.Williams AM, Martinis SA. Proc. Natl. Acad. Sci. USA. 2006;103:3586–91. doi: 10.1073/pnas.0507362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hale SP, Schimmel P. Proc Natl Acad Sci U S A. 1996;93:2755–8. doi: 10.1073/pnas.93.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt E, Schimmel P. Science. 1994;264:265–7. doi: 10.1126/science.8146659. [DOI] [PubMed] [Google Scholar]
- 88.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GL, Hartley BS. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 89.Martinis SA, Fox GE. Nucleic Acids Symp Ser. 1997;36:125–8. [PubMed] [Google Scholar]
- 90.Illangasekare M, Sanchez G, Nickles T, Yarus M. Science. 1995;267:643–7. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- 91.Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. FEBS Lett. 2007;581:5110–14. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 92.Guth EC, Francklyn CS. Mol Cell. 2007;25:531–42. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nomanbhoy TK, Schimmel PR. Proc Natl Acad Sci U S A. 2000;97:5119–22. doi: 10.1073/pnas.090102197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Farrow MA, Nordin BE, Schimmel P. Biochemistry. 1999;38:16898–903. doi: 10.1021/bi9920782. [DOI] [PubMed] [Google Scholar]
- 95.Hountondji C, Lazennec C, Beauvallet C, Dessen P, Pernollet JC, Plateau P, Blanquet S. Biochemistry. 2002;41:14856–65. doi: 10.1021/bi0205101. [DOI] [PubMed] [Google Scholar]
- 96.Tardif KD, Liu M, Vitseva O, Hou YM, Horowitz J. Biochemistry. 2001;40:8118–25. doi: 10.1021/bi0103213. [DOI] [PubMed] [Google Scholar]
- 97.Jakubowski H. J Biol Chem. 1993;268:6549–53. [PubMed] [Google Scholar]
- 98.Jakubowski H. Biochemistry. 1996;35:8252–9. doi: 10.1021/bi960344v. [DOI] [PubMed] [Google Scholar]
- 99.Tsui WC, Fersht AR. Nucleic Acids Res. 1981;9:4627–37. doi: 10.1093/nar/9.18.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roy H, Ling J, Alfonzo J, Ibba M. J Biol Chem. 2005;280:38186–92. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- 101.Ling J, Roy H, Ibba M. Proc Natl Acad Sci U S A. 2007;104:72–7. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beebe K, Merriman E, Ribas De Pouplana L, Schimmel P. Proc Natl Acad Sci U S A. 2004;101:5958–63. doi: 10.1073/pnas.0401530101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Korencic D, Ahel I, Schelert J, Sacher M, Ruan B, Stathopoulos C, Blum P, Ibba M, Söll D. Proc Natl Acad Sci U S A. 2004;101:10260–5. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jakubowski H. Biochemistry. 1997;36:11077–85. doi: 10.1021/bi970589n. [DOI] [PubMed] [Google Scholar]
- 105.Jakubowski H. Biochemistry. 1999;38:8088–93. doi: 10.1021/bi990629i. [DOI] [PubMed] [Google Scholar]