Abstract
To enhance the efficacy of DNA malaria vaccines, we evaluated the effect on protection of immunizing with various combinations of DNA, recombinant vaccinia virus, and a synthetic peptide. Immunization of BALB/c mice with a plasmid expressing Plasmodium yoelii (Py) circumsporozoite protein (CSP) induces H-2Kd-restricted CD8+ cytotoxic T lymphocyte (CTL) responses and CD8+ T cell- and interferon (IFN)-γ-dependent protection of mice against challenge with Py sporozoites. Immunization with a multiple antigenic peptide, including the only reported H-2Kd-restricted CD8+ T cell epitope on the PyCSP (PyCSP CTL multiple antigenic peptide) and immunization with recombinant vaccinia expressing the PyCSP induced CTL but only modest to minimal protection. Mice were immunized with PyCSP DNA, PyCSP CTL multiple antigenic peptide, or recombinant vaccinia expressing PyCSP, were boosted 9 wk later with the same immunogen or one of the others, and were challenged. Only mice immunized with DNA and boosted with vaccinia PyCSP (D-V) (11/16: 69%) or DNA (D-D) (7/16: 44%) had greater protection (P < 0.0007) than controls. D-V mice had significantly higher individual levels of antibodies and class I-restricted CTL activity than did D-D mice; IFN-γ production by ELIspot also was higher in D-V than in D-D mice. In a second experiment, three different groups of D-V mice each had higher levels of protection than did D-D mice, and IFN-γ production was significantly greater in D-V than in D-D mice. The observation that priming with PyCSP DNA and boosting with vaccinia-PyCSP is more immunogenic and protective than immunizing with PyCSP DNA alone supports consideration of a similar sequential immunization approach in humans.
Keywords: Plasmodium, cytotoxic T lymphocytes, interferon-γ
After inoculation by mosquitoes, Plasmodium spp. sporozoites rapidly enter hepatocytes, where they develop to mature liver stage parasites during ≈2 days for the rodent parasite, Plasmodium yoelii (Py), and 5.5 days for the human pathogen, Plasmodium falciparum. This stage of the life cycle does not cause disease and is the only stage during which parasites reside in cells that consistently express major histocompatibility complex molecules.
One approach to malaria vaccine development is to induce protective CD8+ T cell responses to infected hepatocytes (1). This effort is in large part based on work in rodents that demonstrate that protective immunity induced by immunizing with irradiated sporozoites (2–4) or with DNA plasmids encoding Py proteins expressed in infected hepatocytes is eliminated by in vivo depletion of CD8+ T cells (5, 6).
The Py circumsporozoite protein (PyCSP) is present within Py-infected hepatocytes. In BALB/c mice immunized with PyCSP DNA vaccines, protection has varied from 22% to 75% (5, 6, 7) and is always dependent on CD8+ T cells. Protective immunity can be increased, and genetic restriction of the response to each individual protein can be circumvented by immunizing with mixtures of plasmids encoding liver stage proteins (6). Based on our work in mice, we have recently initiated a Phase I clinical trial of a P. falciparum CSP DNA vaccine designed to induce protective CD8+ T cells (8).
To improve the protection afforded by the PyCSP DNA vaccine, we assessed sequential immunization with DNA, recombinant vaccinia, and synthetic peptide PyCSP vaccines. These experiments indicated that priming with PyCSP DNA and boosting with recombinant vaccinia expressing PyCSP were associated with greater immunogenicity and protective immunity than priming and boosting with PyCSP DNA alone.
MATERIALS AND METHODS
Mice.
BALB/cByJ female mice (6–8 wk old) purchased from The Jackson Laboratory were used in all experiments.
Parasites.
P. yoelii (17XNL) clone 1.1 parasites were used. Sporozoites for challenges were obtained by hand dissection of infected mosquito glands in M199 medium containing 5% normal mouse serum.
Plasmid Constructions.
Two PyCSP DNA vaccines were used in this study. The plasmid designated 1012/tissue plasminogen activator protein leader peptide sequence (TPA)-PyCSP contained the DNA sequence encoding the full-length PyCSP, which included the native leader peptide sequence, fused in-frame with the leader peptide sequence from human tissue plasminogen activator protein. The 1012-PyCSP plasmid encoded no additional in-frame residues. The PyCSP encoding sequence was amplified by PCR from the plasmid nkCMVintPyCSP.1 (5). The resultant product was Pfu polymerase treated (Stratagene) and was gel-purified and ligated with the EcoRV-digested plasmid nkCMVintPolyLi. From this plasmid, the PyCSP encoding sequence was recovered on a SalI/XbaI fragment that was ligated with SalI/XbaI-digested VR1012 (9), which created the plasmid designated 1012-PyCSP. From the nkCMVintPolyLi/PyCSP plasmid, the PyCSP encoding sequence was recovered on a BamHI fragment and then was ligated with BamHI-digested pCMVintBL plasmid (10), which placed the coding sequence in-frame with the TPA. From this plasmid, the TPA-PyCSP encoding sequence was recovered on a SalI/BglII fragment that was ligated with SalI/BglII-digested VR1012, creating the plasmid designated 1012/TPA-PyCSP. The ability of these plasmids to express PyCSP was confirmed in vitro by using an anti-PyCSP mAb (11) and transiently transfected UM449 human melanoma cells (12). All DNA for injection was purified as described (8) by using cesium chloride-ethidium bromide density gradient centrifugation. DNA was solubilized in United States Pharmacopia saline for injection at ≈5.0 mg/ml and was stored at −20°C.
Recombinant Vaccinia Expressing PyCSP.
The P. yoelii 17X NL CSP gene (nucleotides 1–1757) (13) was cloned into the multiple cloning site COPAK H6 donor plasmid. The expression plasmid pMK4 was used in vivo to generate a recombinant virus (vP 1258) by using a New York Vaccinia (NYVAC) rescuing virus (14). The COPAK donor plasmid contains multiple cloning sites, with the gene of interest being placed under the control of an early–late H6 promoter (15). The plasmid also carries the K1L ORF flanked on either side with ORFs of the A24R and A27L (16). The foreign gene and the K1L gene are inserted into the ATI site of the NYVAC genome between the A24R and A27L ORFs. NYVAC virus differs from NYVAC(K1L) virus only by the absence of the K1L insertion. A control virus called vP993, containing a K1L insert but no foreign gene, was used as a control.
Immunizations with DNA Vaccines.
Mice were injected i.m. in the tibialis anterior muscle with PyCSP DNA. Negative control mice were injected with 1012/TPA DNA lacking the PyCSP gene. A 0.3-ml insulin syringe with a 29½G inch needle was used for all injections, and each single dose consisted of 100 μg that was delivered in a total volume of 100 μl and split between both legs. Either 9 wk (in Experiment 1) or 6 wk (in Experiment 2) after the first immunization, some groups of mice were boosted with the same dose of the PyCSP DNA (D-D) whereas some received either 107 plaque-forming units NYVAC(K1L)-PyCSP virus (vP1258) i.p. or i.m. (D-V and D-Vim, respectively) or PyCSP [multiple antigenic peptide (MAP4)(280-99)p2p30] peptide (D-P) s.c. A second challenge experiment was carried out to evaluate 1012/PyCSP (D2). In this experiment, 6 wk after the first immunization with 1012/PyCSP DNA, some mice were boosted with either the same plasmid or with NYVAC(K1L) PyCSP.
Immunizations Using Recombinant NYVAC(K1L)-PyCSP.
Mice were injected i.p. or i.m. in the tibialis anterior muscle with 107 plaque-forming units of NYVAC(K1L)-PyCSP (vP1258), and control mice were injected with the parental control virus NYVAC(K1L) (vP993) lacking the PyCSP gene. Some groups were boosted later with the same dose of the recombinant virus, PyCSP DNA vaccine, or PyCSP peptide.
Immunizations Using PyCSP Peptide.
Mice were injected s.c. with 16 μg of PyCSP peptide, MAP4(280–99)p2p30 consisting of four branches of SYVPSAEQILEFVKQISSQL and p2p30 (tetanus toxin), and a glycine-lysine core (17) mixed with Lipofectin (Life Technologies, Gaithersburg, MD) at 15 μl per dose. Groups were boosted 9 wk later with the same dose of the peptide (P-P), PyCSP DNA (P-D), or recombinant vaccinia virus (P-V).
Antibody Response
Indirect fluorescent antibody test.
The indirect fluorescent antibody test was used to detect anti-P. yoelii sporozoite antibodies in sera of immunized mice as described (18). In brief, diluted sera were reacted on air-dried sporozoites, and antibodies were detected by using fluorescein isothiocyanate-labeled rabbit anti-mouse Ig.
T Cell Assays
Cytotoxic T lymphocyte (CTL) Assays.
51Chromium release assays using bulk cultures were performed on individual mice as described (5). In brief, spleen cells (5 × 106) were stimulated in vitro for 6–7 days with a 16-mer peptide, PyCSP (280–295) (SYVPSAEQILEFVKQI), which contains the H-2Kd-restricted CTL epitope PyCSP (280–288) (SYVPSAEQI), in 24-well plates at 2.5 μM. Culture medium was RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, l-glutamine, 50 units/ml each of penicillin and streptomycin, and 2-mercaptoehanol at 5 × 10−5 M. P815(H-2d) cells (American Type Culture Collection) labeled with 0.1 mCi of 51Cr (Dupont/NEN) and pulsed with the PyCSP (280–288) peptide at 0.025 μM were used as target cells. Varying ratios of effector cells were added to 5,000 target cells in a 96-well U-bottom plate, standard chromium release methods were followed, and the percentage of specific lysis was calculated. The results are expressed as mean net percentage of specific lysis. The net percentage of specific lysis was calculated by subtracting the percentage of specific lysis of unpulsed target cells from the percentage of specific lysis of target cells pulsed with the experimental peptide PyCSP (280–288). The mean net percentage of specific lysis was the average of net percentages of specific lysis at each effector-to-target ratio of cells from the two mice studied per group.
ELIspot Assay for the Detection of Interferon (IFN)-γ-Producing Spleen Cells.
By using methods described by Miyahira et al. (19), spleen cells from mice at 14 days after the second immunization were used. This was at the time when the others were being challenged. In Experiment 1, spleen cells from two individual mice were assayed, and in Experiment 2, pooled spleen cells from two mice were used. The number of H-2Kd-restricted CTL epitope-specific IFN-γ -producing cells was determined in freshly isolated, unstimulated spleen cells 24–28 hr after being incubated with P815 cells (express Class I but not Class II major histocompatibility complex molecules) pulsed with 1 μM of the H-2Kd-restricted PyCSP (280–288) peptide.
Ninety-six-well nitrocellulose plates (Millipore) were coated with 75 μl of PBS containing 1 μg/ml of purified rat anti-mouse IFN-γ antibody (PharMingen). After overnight incubation at room temperature, the wells were washed repeatedly with culture medium and were incubated for 1 hr with 100 μl of culture medium containing 10% fetal calf serum. 100 μl of varying concentrations of effector cells together with 100 μl of 2.5 × 105 cells/ml irradiated P815 cells were placed in the antibody-coated wells. The starting concentration for the freshly isolated unstimulated effector cells was 30 × 106/ml, and doubling dilutions were assayed. One set of effector cells was cocultured with irradiated P815 cells that had been pulse-labeled with 1 μM PyCSP(280–288) peptide. The other set of cells was cocultured with irradiated P815 cells without peptide.
After incubation at 37°C and 5% CO2 for 24–28 hr, plates were washed extensively with PBS containing 0.05% Tween 20 (PBS/T). Wells then were incubated with 100 μl of a solution of 1 μg/ml biotinylated anti-mouse IFN-γ mAb (PharMingen) in PBS/T. After overnight incubation at 4°C, wells were washed with PBS/T and 100 μl of peroxidase-labeled streptavidin (Kirkegaard and Perry, Gaithersburg, MD) at a dilution of 1/1000 in PBS/T, was added to each well. After 1 hr incubation at room temperature, wells were washed extensively with PBS/T and twice with PBS. Spots were developed per instructions with the DAB Reagent set (Kirkegaard and Perry Laboratories). After 10–15 min, numbers of spots were determined by using a stereomicroscope. Spots corresponding to IFN-γ-producing cells in wells containing the different spleen cell dilutions were counted. Results were expressed as the number of IFN-γ-spot forming cells (SFCs) per 106 spleen cells.
Protection Against Challenge.
Two wk after the second immunization, mice were challenged by i.v. inoculation in the tail vein of 50 viable P. yoelii sporozoites. Mice were considered protected if malaria blood films taken from day 5 until day 14 after challenge were negative by microscopy.
Statistical Analysis.
The degree of protection afforded by the different immunization regimens was evaluated by using the χ2 test (epiinfo 6.04b) (Centers for Disease Control, Atlanta, GA). Other tests were performed in spss for Windows (version 6.1.4) (SPSS, Chicago, IL).
RESULTS AND DISCUSSION
Experiment 1
Protective Immunity.
Two wk after the second dose of vaccine, mice were challenged by i.v. injection of 50 Py sporozoites. Protection was highest among mice that received 1012/TPA-PyCSP DNA i.m. as the first dose and recombinant NYVAC(K1L) expressing PyCSP i.p. as the second dose (D-V) [11 of 16 mice (69%) were protected] (Table 1). Mice that received two doses of 1012/TPA-PyCSP DNA (D-D) had the next highest level of protection (7 of 16, 44%) (Table 1). Only these two groups had statistically significant protection on day 14 as compared with the pooled controls (P < 0.0001, D-V; P = 0.0006, D-D). All groups except the D-D group had significantly fewer protected mice than did the group primed with PyCSP DNA and boosted with NYVAC(K1L)-PyCSP (D-V) (Table 1, P < 0.02, χ2) and groups D-P, V-D, V-P, P-P, and P-D had significantly less protection (P < 0.05, χ2) than did the D-D group. However, some of the mice in three other groups did not develop parasitemia (Table 1). There was also a statistically significant delay in onset of parasitemia in four other groups: D-P, V-V, P-D, and P-V (P < 0.0001, χ2. Based on a 24-hr cycle in erythrocytes and a 10-fold increase in parasitemia each cycle (average of 15 nuclei per each mature erythrocytic stage parasite), a one-day delay in onset of parasitemia would represent a 90% reduction in parasite burden. This indicates that, in mice with a delay in onset of parasitemia, vaccine-induced immune responses had a significant effect on parasite development in the liver. All of the three groups primed with DNA, 2/3 of the groups primed with PyCSP CTL MAP, and only 1/3 of the groups primed with NYVAC(K1L)-PyCSP had statistically significant protection or delay in the onset of parasitemia as compared with controls (Table 1).
Table 1.
Experiment 1: Protection against sporozoite challenge after immunization with 1012/TPA-PyCSP (D), recombinant vaccinia NYVAC(K1L)-PyCSP (V), or PyCSP (MAP4(280-99)p2p30) peptide (P)
| Immunization
|
Protected/Challenged
|
% protected at day 14 | |||
|---|---|---|---|---|---|
| Prime | Boost | Day 5* | Day 6 | Day 14 | |
| D | D | 16/16 | 10/16 | 7/16 | 44 |
| D | V | 15/16 | 13/16 | 11/16 | 69 |
| D | P | 14/16 | 0/16 | 0/16 | 0 |
| V | V | 9/15 | 6/15 | 4/15 | 27 |
| V | D | 2/16 | 2/16 | 2/16 | 13 |
| V | P | 2/16 | 0/16 | 0/16 | 0 |
| P | P | 1/16 | 0/16 | 0/16 | 0 |
| P | D | 12/16 | 7/16 | 0/16 | 0 |
| P | V | 10/16 | 8/16 | 3/16 | 19 |
| VC1 | VC | 0/6 | 0 | ||
| PlC2 | PlC | 0/7 | 0 | ||
| Naive | — | 0/9 | 0 | ||
Days after sporozoite challenge.
Vaccincia control.
Plasmid control (VR1012/TPA).
Antibody Responses.
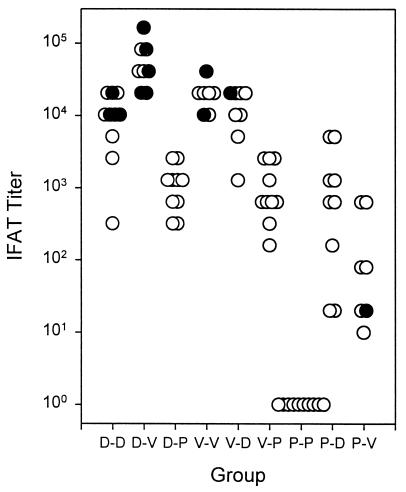
We assessed antibodies to native protein on sporozoites by indirect fluorescent antibody test in sera taken before sporozoite challenge in half of the mice in each group (Fig. 1). Mice in the D-V group, the group with the highest level of protection, had significantly higher antibody titers than did all other groups (Mann–Whitney U test, P = 0.0008). However, pre-challenge antibody levels in individual mice did not predict protection; three of the six mice with the highest levels of antibodies were not protected (Fig. 1). The D-D group, the only other group with significant protection, had antibody levels similar to mice in the V-D and V-V groups, groups without significant protection. Furthermore, in the D-D group, three of seven mice with the highest levels of antibodies were not protected. These data are consistent with our previous data and that of others (refs. 5 and 6; L. F. Scheller & S.L.H., unpublished material), which indicate that antibodies to sporozoites do not play a significant role in the protective immunity induced by the PyDNA vaccines, the Py irradiated sporozoite vaccine (2), the PyCSP CTL MAP or PyCSP recombinant vaccinia vaccines, or the Plasmodium berghei CSP recombinant vaccinia vaccines (20). The fact that the group with the highest level of protection (D-V) had the highest level of antibodies suggests that the antibody responses in this group may have been associated with other immune responses responsible for protection.
Figure 1.
End-point Indirect Fluorescent Antibody test titers to air-dried P. yoelii sporozoites, Experiment 1. Filled circles represent mice protected, and open circles represent those not protected on challenge. As a group, D-V had significantly higher titers than other groups (Mann–Whitney U test, P = 0.0008). D = 1012/TPA-PyCSP, V = NYVAC(K1L)-PyCSP, and P = PyCSP [MAP4(280–99)p2p30].
Cytotoxic T Cell Responses.
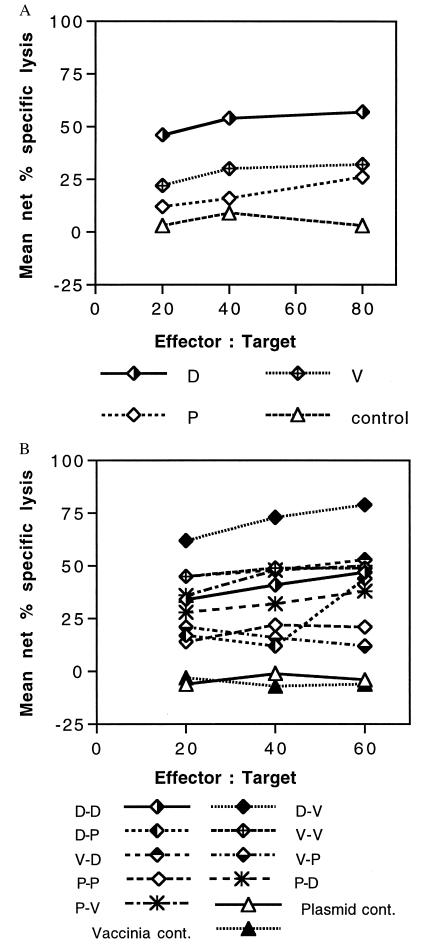
After a single injection in the two mice tested per group, both mice immunized with 1012/TPA-PyCSP had higher levels of CTL activity against the 9-aa, H-2Kd-restricted CTL epitope on the PyCSP than did mice immunized with the two other vaccines, and the mean net percentage of specific lysis of the two mice immunized with PyCSP DNA was the highest (Fig. 2A). Mice immunized with NYVAC(K1L)-PyCSP had the next highest level, and mice immunized with the PyCSP CTL MAP had the lowest level of cytolytic activity.
Figure 2.
Cytolytic activity induced by PyCSP vaccines after priming (A) and boosting (B). (A) CTL assay results of restimulated spleen cells taken from mice 2 wk after the first dose of 1012/TPA-PyCSP (D), NYVAC(K1L)-PyCSP (V), PyCSP (MAP4(280–99)p2p30) (P), or control mice. (B) CTL assay results of restimulated spleen cells taken 2 wk after the second dose of PyCSP vaccine, control plasmid, or controls (Experiment 1). The results are plotted as mean net percentage of specific lysis of P815 target cells (H-2d) pulsed with the H-2Kd-restricted, 9-aa PyCSP peptide 280–288 (see Materials and Methods). At all E/T ratios, combined D-V had higher net percentage of specific lysis than did any other group. (P = 0.013, Mann–Whitney U test.)
After two doses, the best responses were seen in the two D-V mice, the group best protected. Even at the lowest effector-to-target cell ratio (E/T) of 20:1, both mice in the D-V group had greater net percentages of specific lysis (63% and 60%) than all mice in all the other groups at any E/T ratio with the exception of one mouse at an E/T ratio of 60:1 (62%) in the V-D group and one mouse at an E/T ratio of 40:1 in the D-D group. The mean net percentage of specific lysis of the two D-V mice was higher than the mean net percentage of specific lysis of mice in all other groups (P = 0.013, Mann–Whitney U test) (Fig. 2B). Similar to the antibody data, mice from groups without significant protection had CTL activity comparable to mice in the D-D group that did have significant protection. These results must be interpreted with caution because there were only two mice tested per group and because only 69% of mice in the D-V group and 44% of the mice in the D-D group were protected and we do not know whether the mice that we assessed would have been protected. However, they clearly demonstrate that the D-V regimen induced the best H-2 Kd-restricted CTL activity.
IFN-γ Secreting Cells.
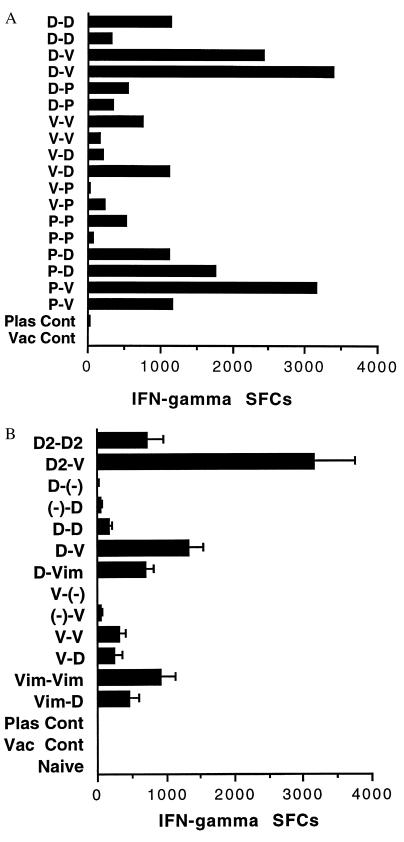
In BALB/c mice immunized with P. yoelii DNA vaccines, protection depends on CD8+ T cells, IFN-γ, and nitric oxide (6). One interpretation of these data is that the protection is not dependent on cytolytic CD8+ T cells but rather on CD8+ T cells that secrete IFN-γ, and that INF-γ induces the infected hepatocyte to produce nitric oxide, which kills the parasite (2, 6). We therefore assessed IFN-γ secretion by ELIspot in spleen cells taken 2 wk after the second immunization. Major histocompatibility complex-matched P815 cells (expressing Class I but not Class II molecules) pulsed with the 9-aa, H-2Kd-restricted PyCSP peptide were used as antigen-presenting cells. Four spleen cell concentrations (3, 1.5, 0.75, and 0.375 × 106 cells per well) were cocultured for 24–28 hr with P815 cells pulsed with the PyCSP (280–288) peptide or unpulsed P815 cells, and IFN-γ SFCs were counted. Because of the magnitude of the response in some of the groups, only wells with 0.375 × 106 cells could be counted. The number of spots in these wells was used to calculate the number of SFCs per million spleen cells. Coculture with unpulsed target cells always gave minimal to no spots. Data in Fig. 3A represent numbers obtained from two mice after subtracting spots in cultures with unpulsed P815 cells from spots in cultures with pulsed P815 cells. The best protected mice, mice primed with 1012/TPA-PyCSP and boosted with NYVAC(K1L)-PyCSP, had higher numbers of IFN-γ SFCs than did mice primed and boosted with DNA (D-D) (Fig. 3A). As with the antibody and CTL data, the only other group with significant protection, the D-D group, did not have higher numbers of IFN-γ SFCs than did mice from several other groups with no significant protection. Furthermore, one of the mice primed with peptide and boosted with NYVAC(K1L)-PyCSP had an excellent response. This group had only modest protection (3 of 16 mice completely protected during 14 days of follow-up). We cannot test cells from spleens from challenged mice at the time of challenge and therefore cannot determine whether a mouse with high activity actually would be protected.
Figure 3.
H-2Kd-restricted PyCSP-specific IFN-γ-SFCs by ELIspot. (A) IFN-γ SFCs in two individual mice in Experiment 1. (B) IFN-γ SFCs (mean plus SD) from triplicate cultures of pooled spleen cells from two mice in Experiment 2. D = 1012/TPA-PyCSP, D2 = 1012/PyCSP, V = NYVAC(K1L)-PyCSP given i.p., and Vim = NYVAC(K1L)-PyCSP given i.m. In all cases, mice primed with PyCSP DNA and boosted with NYVAC(K1L)-PyCSP had significantly more IFN-γ SFCs than did other groups primed and boosted with the same PyCSP DNA (t test, P < 0.001).
Experiment 2
Protective Immunity.
The results of the first experiment indicated that priming with 1012/TPA-PyCSP and boosting with recombinant NYVAC(K1L) expressing PyCSP (D-V) was significantly more protective than priming with NYVAC(K1L)-PyCSP and boosting with 1012/TPA-PyCSP DNA (V-D) or priming and boosting with NYVAC(K1L)-PyCSP (V-V) and was apparently more protective than priming and boosting with PyCSP DNA alone (D-D). Priming or boosting with the PyCSP CTL MAP did not provide protection comparable to that found in the D-V or D-D groups. The data also suggested that the ELIspot assay that measures IFN-γ SPCs was predictive of a difference in protection between the D-V and D-D groups. We therefore repeated the experiment but eliminated the PyCSP CTL MAP. In addition, we assessed i.m. administration of NYVAC(K1L)-PyCSP (Vim) and assessed a second PyCSP DNA vaccine, 1012/PyCSP (D2). This plasmid does not contain the TPA secretory leader peptide. We speculated that trafficking of the CSP would be different without this sequence, which would perhaps lead to enhanced intracellular processing of the CSP in the Class I pathway. However, comparison of in vitro expression of the two plasmids in mammalian cells did not reveal a difference in total expression or secretion of PyCSP (R.H., unpublished material).
The results of this experiment were consistent with the previous experiment. Priming with 1012/TPA-PyCSP (D) or 1012-PyCSP (D2) and boosting 6 wk later with NYVAC(K1L)-PyCSP i.p. (V) or i.m. (Vim) was associated with the highest level of protection: 10 of 24 mice (42%) protected (Table 2). The next highest level of protection was with 1012/TPA-PyCSP administered as a single dose (−D) 8 wk before challenge or as two doses (D-D) at 6-wk intervals; 4 of 16 (25%) mice were protected. No other combination had more than one of eight mice protected, and none of the mice administered two doses of 1012-PyCSP (D2-D2) were protected (Table 2). We are not certain why the level of protection was lower in the D-D and D2-D2 groups as compared with our previously reported experiments with other PyCSP DNA plasmids (refs. 5 and 6; L. F. Scheller & S.L.H., unpublished material). However, the levels of protection are, in fact, consistent with other recently conducted work (7) and may be a reflection of waxing and waning of sporozoite infectivity over time.
Table 2.
Experiment 2: Protection against sporozoite challenge after immunization with 1012/TPA-PyCSP (D), 1012/PyCSP (D2), and recombinant vaccinia NYVAC(K1L)-PyCSP i.p. (V) or i.m. (Vim)
| Immunization
|
Protected/Challenged
|
% protected at day 14 | |||
|---|---|---|---|---|---|
| Prime | Boost | Day 6* | Day 9 | Day 14 | |
| D2 | D2 | 0/8 | 0 | ||
| D2 | V | 5/8 | 4/8 | 4/8 | 50 |
| D | — | 2/8 | 2/8 | 2/8 | 25 |
| — | D | 8/8 | 4/8 | 0/8 | 0 |
| D | D | 8/8 | 6/8 | 2/8 | 25 |
| D | V | 5/8 | 3/8 | 3/8 | 37.5 |
| D | Vim | 6/8 | 4/8 | 3/8 | 37.5 |
| V | — | 1/8 | 0 | ||
| — | V | 1/8 | 1/8 | 0/8 | 0 |
| V | V | 2/8 | 1/8 | 1/8 | 12.5 |
| Vim | Vim | 1/8 | 1/8 | 1/8 | 12.5 |
| V | D | 6/8 | 1/8 | 1/8 | 12.5 |
| Vim | D | 0/8 | 0 | ||
| VC1 | VC | 0/8 | 0 | ||
| PlC2 | PlC | 0/8 | 0 | ||
| Naive | — | 0/8 | 0 | ||
Days after sporozoite challenge.
Vaccinia control.
Plasmid control (VR1012/TPA).
IFN-γ-Secreting Cells.
Two wk after the second dose, spleens from two mice per group were pooled. Triplicate cultures of eight doubling dilutions beginning at 2 × 106 cells were assayed. Cell concentrations of 1.25, 0.625, and 0.3125 × 105 cells per well had easily countable spots and were used to calculate the number of spots per million spleen cells. At each of these concentrations, the number of spots per million spleen cells was extrapolated after subtracting the number of spots from cultures with unpulsed targets. The mean and SD then were calculated. Pooled cells from two mice primed with 1012/PyCSP DNA and boosted with NYVAC(K1L)-PyCSP i.p. (D2-V), the group with the highest level of protection (4 of 8, 50%), had the highest numbers of IFN-γ SFCs and significantly more INF-γ SFCs than mice immunized with 1012/PyCSP and boosted with 1012/PyCSP (D2-D2) (Fig. 3B). The next highest numbers of IFN-γ SFCs were found in mice primed with 1012/TPA-PyCSP and boosted with NYVAC(K1L)-PyCSP i.p. (D-V), and these mice had significantly greater numbers of INF-γ SFCs than did mice primed and boosted with 1012/TPA-PyCSP (D-D). As in the previous experiment, there was no absolute correlation between IFN-γ SFCs and protection. However, in all cases, mice primed with PyCSP DNA and boosted with NYVAC(K1L)-PyCSP had significantly greater IFN-γ SFCs and a higher percentage of protected animals than did the mice immunized and boosted with the same PyCSP DNA (D-V > D-D, D-Vim > D-D, and D2-V > D2-D2, t test, P < 0.001) (Fig. 3B).
Priming with PyCSP DNA and boosting with NYVAC(K1L)-PyCSP induced higher levels of antibodies, CTL, IFN-γ, SFCs, and protection than did priming and boosting with PyCSP DNA. There were four different comparisons in which mice primed with PyCSP DNA and boosted with NYVAC(K1L)-PyCSP were compared with mice primed and boosted with PyCSP DNA (Exp. 1: D-V vs. D-D; Exp. 2: D-V vs. D-D, D-Vim vs. D-D, and D2-V vs. D2-D2). In all comparisons, the group primed with PyCSP DNA and boosted with NYVAC(K1L)-PyCSP had a higher level of protection. The probability of this happening at random is 0.063. However, groups were small, and in only one of these comparisons were the differences statistically significant; D2-V vs. D2-D2 (P = 0.02, χ2). In the two experiments (Tables 1 and 2), 40 mice were primed with PyCSP DNA and were boosted with NYVAC(K1L)-PyCSP, and 32 mice were primed and boosted with PyCSP DNA. Fifty-three percent of the PyCSP DNA-primed NYVAC(K1L) PyCSP-boosted mice were protected, and 28% of the PyCSP DNA-primed and boosted mice were protected (P = 0.038, χ2).
We are unaware of any studies in which antibodies, Class I-restricted CTL and INF-γ production, and protection against challenge with an infectious agent in animals primed with DNA and boosted with recombinant virus were shown to be higher than in animals primed and boosted with DNA. Primary immunization with a DNA vaccine and boosting with a recombinant fowl pox virus encoding the hemagglutinin antigen of influenza virus induced antibody levels in mice 30- to 120-fold higher than did either DNA or recombinant virus alone and induced protection against death in mice challenged with live influenza virus equivalent to that achieved with immunization with live virus alone or DNA alone (21). Immunization of sheep with DNA followed by recombinant adenovirus expressing Taenia ovis produced higher levels of antibody than did immunizing with protein/virus regimens and also produced high levels of protection (22). However, DNA/recombinant virus immunization was not compared with a DNA/DNA regimen. In another study, antibody titers induced in rabbits with 4–6 doses of DNA were increased by boosting with recombinant virus (23). In a mouse tumor model, immunization with a DNA vaccine and boosting with either a recombinant vaccinia or fowl pox induced antigen-specific CTL. Order of the immunization was also important; if the virus was given first and boosted with DNA, no lytic activity was noted. Boosting with an immunogen different from the one used for immunization (i.e., heterologous boosting) also produced increases in survival compared with animals receiving homologous boosts (24).
The immunogenicity of the D-V approach was greater than that of all other approaches we studied and, when coupled with the higher protection in these mice, suggested that these antibody and T cell assays predicted protection. However, the D-D group also was protected significantly in Experiment 1, but immune responses in these D-D mice were not significantly higher than in mice without significant protection. One explanation for this discrepancy is that we did not measure the appropriate immune responses or that our assays need refinement. The results in the D-V mice (the best immune responses and the best protection) could be a reflection of the global immune responses being greatest in these mice rather than any of the measured immune responses being critical for protection. The results also could have been influenced by the fact that we did not assay T cells from individual mice that then were challenged but rather took cells from spleens of mice that were not challenged. In these experiments, ≤69% of mice within a group were protected, and this variability in protection could explain why the results of testing cells from nonchallenged mice was not accurate in predicting protection.
These data demonstrate that priming with PyCSP DNA and boosting with recombinant vaccinia expressing PyCSP is more immunogenic and protective than is priming and boosting with PyCSP DNA or with vaccinia-PyCSP or priming with vaccinia-PyCSP and boosting with PyCSP DNA. Li and colleagues (24) reported that priming with recombinant influenza expressing the PyCSP H-2Kd-restricted CTL epitope and boosting with vaccinia-PyCSP was more protective than immunizing with either alone or with vaccinia-PyCSP first and the recombinant influenza second. We expected, therefore, to achieve good protection by priming with the PyCSP CTL MAP, which included the same epitope as the recombinant influenza. Priming with the CTL MAP and boosting with vaccinia PyCSP was associated with good CTL activity and IFN-γ production and a significant delay in onset of parasitemia but 19% complete protection (not significant).
Our results are consistent with those of Li (25) in regard to the need for delivering the recombinant vaccinia as the boosting rather than the priming dose. We do not have an explanation for either this requirement or the enhanced immunogenicity of the D-V approach as compared with the D-D approach. When using recombinant influenza or other viruses, it is always possible that the host develops immunity to the virus portion of the immunogen, thereby limiting the response to a booster dose. However, we have no evidence that this occurs with DNA and usually have seen good boosting with second and third doses of DNA (5, 26). It may be that the critical event in priming depends on the quality of the immunogen (DNA better than vaccinia) and that boosting is more dependent on the quantity of immunogen expressed (vaccinia better than DNA). Work is in progress to more clearly characterize how sequential immunization with PyCSP DNA and vaccinia-PyCSP improves the response to vaccination. Nonetheless, we think that these findings support assessing sequential immunization with plasmid DNA and recombinant vaccinia in humans.
Acknowledgments
The authors thank Hospital Corpsman Second Class A. Belmonte, Hospital Corpman Third Class R. Wallace, and Mr. S. Matheny for providing P. yoelii sporozoites and technical assistance. The assertions herein are the private ones of the authors and are not to be construed as official or as reflecting the views of the U.S. Navy or the Naval service at large. The experiments reported herein were conducted according to the principles set forth in the Guide for the Care and Use of Laboratory Animals, Institute of Laboratory Animal Research, National Research Council, National Academy Press (1996) and were approved under National Medical Research Institute Animal Care and Use Committee no. 9612. This work was supported by the Naval Medical Research and Development Command work units 1431 61102AA0101BFX and 1432 62787A00101EFX.
ABBREVIATIONS
- Py
Plasmodium yoelii
- CSP
circumsporozoite protein
- CTL
cytotoxic T lymphocyte
- IFN
interferon
- TPA
tissue plasminogen activator protein leader peptide sequence
- NYVAC
New York Vaccinia
- MAP
multiple antigenic peptide
- PBS/T
PBS containing 0.05% Tween 20
- SFC
spot-forming cells
A paper on rodent malaria reporting similar results has been published recently. See Schneider, J., Gilbert, S. C., Blanchard, T. J., Hanke, T., Robson, K. J., Hannan, C. M., Becker, M., Sinden, R., Smith, G. L. & Hill, A.V.S. (1998) Nature Med. 4, 397–402.
Footnotes
To whom reprint requests should be addressed at: Malaria Program, Naval Medical Research Institute, 12300 Washington Avenue, Rockville, MD 20852. e-mail: hoffmans@nmripo.nmri.nnmc.navy.mil.
References
- 1.Hoffman S L, Franke E D, Hollingdale M R, Druilhe P. In: Malaria Vaccine Development. Hoffman S L, editor. Washington, DC: Am. Soc. Microbiol.; 1996. [Google Scholar]
- 2.Doolan D L, Hoffman S L. Philos Trans R Soc London B Biol Sci. 1997;352:1361–1367. doi: 10.1098/rstb.1997.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schofield L, Villaquiran J, Ferreira A, Schellekens H R, Nussenzweig R S, Nussenzweig V. Nature (London) 1987;330:664–666. doi: 10.1038/330664a0. [DOI] [PubMed] [Google Scholar]
- 4.Weiss W R, Sedegah M, Beaudoin R L, Miller L H, Good M F. Proc Natl Acad Sci USA. 1988;85:573–576. doi: 10.1073/pnas.85.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedegah M, Hedstrom R, Hobart P, Hoffman S L. Proc Natl Acad Sci USA. 1994;91:9866–9870. doi: 10.1073/pnas.91.21.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doolan D L, Sedegah M, Hedstrom R C, Hobart P, Charoenvit Y, Hoffman S L. J Exp Med. 1996;183:1739–1746. doi: 10.1084/jem.183.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss. W. R., Ishii, K. J., Hedstrom R. C., Sedegah, M., Barnhart, K., Klinman, D. & Hoffman, S. L. (1998) J. Immunol., in press. [PubMed]
- 8.Hoffman S L, Doolan D L, Sedegah M, Wang R, Scheller L F, Kumar A, Weiss W R, Le T P, Klinman D M, Hobart P, et al. Immunol Cell Biol. 1997;75:376–381. doi: 10.1038/icb.1997.59. [DOI] [PubMed] [Google Scholar]
- 9.Hartikka J, Sawdey M, Cornefert-Jensen F, Margalith M, Barnhart K, Nolasco M, Vahlsing H L, Meek J, Marquet M, Hobart P, et al. Hum Gene Ther. 1996;7:1205–1217. doi: 10.1089/hum.1996.7.10-1205. [DOI] [PubMed] [Google Scholar]
- 10.Manthorpe M, Cornefert-Jensen F, Hartikka J, Felgner J, Rundell A, Margalith M, Dwarki V. Hum Gene Ther. 1993;4:419–431. doi: 10.1089/hum.1993.4.4-419. [DOI] [PubMed] [Google Scholar]
- 11.Charoenvit Y, Leef M F, Yuan L F, Sedegah M, Beaudoin R L. Infect Immun. 1987;55:604–608. doi: 10.1128/iai.55.3.604-608.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luke C J, Carner K, Liang X, Barbour A G. J Infect Dis. 1997;175:91–97. doi: 10.1093/infdis/175.1.91. [DOI] [PubMed] [Google Scholar]
- 13.De la Cruz V F, Lal A A, McCutchan T F. Mol Biochem Parasitol. 1988;28:31–38. doi: 10.1016/0166-6851(88)90176-4. [DOI] [PubMed] [Google Scholar]
- 14.Tartaglia J, Perkus M E, Taylor J, Norton E K, Audonnet J C, Cox W I, Davis S W, VanderHoeven J, Meignier B, Riviere M, et al. Virology. 1992;188:217–232. doi: 10.1016/0042-6822(92)90752-b. [DOI] [PubMed] [Google Scholar]
- 15.Perkus M E, Limbach K, Paoletti E. J Virol. 1989;63:3829–3836. doi: 10.1128/jvi.63.9.3829-3836.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 17.Franke E D, Corradin G P, Hoffman S L. J Immunol. 1997;159:3424–3433. [PubMed] [Google Scholar]
- 18.Charoenvit Y, Leef M F, Yuan L F, Sedegah M, Beaudoin R L. Infect Immun. 1987;55:604–608. doi: 10.1128/iai.55.3.604-608.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyahira Y, Murata K, Rodriguez D, Rodriguez J R, Esteban M, Rodrigues M M, Zavala F. J Immunol Methods. 1995;181:45–54. doi: 10.1016/0022-1759(94)00327-s. [DOI] [PubMed] [Google Scholar]
- 20.Lanar D E, Tine J A, de Taisne C, Seguin M C, Cox W I, Winslow J P, Ware L A, Kauffman E B, Gordon D, Ballou W R, et al. Infect Immun. 1996;64:1666–1671. doi: 10.1128/iai.64.5.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leong K H, Ramsey A J, Ramshaw I A, Morin M J, Robinson H L, Boyle D B. In: Vaccines 95. Chanock R M, Brown F, Ginsberg H S, Norrby E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 327–331. [Google Scholar]
- 22.Rothel J S, Boyle D B, Both G W, Pye A D, Waterkeyn J G, Wood P R, Lightowlers M W. Parasite Immunol (Oxf) 1997;19:221–227. doi: 10.1046/j.1365-3024.1997.d01-200.x. [DOI] [PubMed] [Google Scholar]
- 23.Richmond J F, Mustafa F, Lu S, Santoro J C, Weng J, O’Connell M, Fenyo E M, Hurwitz J L, Montefiori D C, Robinson H L. Virol. 1997;230:265–274. doi: 10.1006/viro.1997.8478. [DOI] [PubMed] [Google Scholar]
- 24.Irvine K R, Chamberlain R S, Shulman E P, Surman D R, Rosenberg S A, Restifo N P. J Natl Cancer Inst. 1997;89:1595–1601. doi: 10.1093/jnci/89.21.1595. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Rodrigues M, Rodriguez D, Rodriguez J R, Esteban M, Palese P, Nussenzweig R S, Zavala F. Proc Natl Acad Sci USA. 1993;90:5214–5218. doi: 10.1073/pnas.90.11.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman S L, Sedegah M, Hedstrom R C. Vaccine. 1994;2:1529–1533. doi: 10.1016/0264-410x(94)90078-7. [DOI] [PubMed] [Google Scholar]