Abstract
RNA import into mitochondria is a widespread phenomenon. Studied in details for yeast, protists, and plants, it still awaits thorough investigation for human cells, in which the nuclear DNA-encoded 5S rRNA is imported. Only the general requirements for this pathway have been described, whereas specific protein factors needed for 5S rRNA delivery into mitochondria and its structural determinants of import remain unknown. In this study, a systematic analysis of the possible role of human 5S rRNA structural elements in import was performed. Our experiments in vitro and in vivo show that two distinct regions of the human 5S rRNA molecule are needed for its mitochondrial targeting. One of them is located in the proximal part of the helix I and contains a conserved uncompensated G:U pair. The second and most important one is associated with the loop E-helix IV region with several noncanonical structural features. Destruction or even destabilization of these sites leads to a significant decrease of the 5S rRNA import efficiency. On the contrary, the β-domain of the 5S rRNA was proven to be dispensable for import, and thus it can be deleted or substituted without affecting the 5S rRNA importability. This finding was used to demonstrate that the 5S rRNA can function as a vector for delivering heterologous RNA sequences into human mitochondria. 5S rRNA-based vectors containing a substitution of a part of the β-domain by a foreign RNA sequence were shown to be much more efficiently imported in vivo than the wild-type 5S rRNA.
Keywords: determinant, import, human, mitochondria, 5S ribosomal RNA
INTRODUCTION
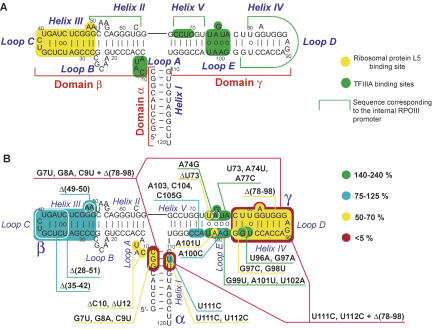
Among noncoding RNAs characterized to date, 5S ribosomal RNA is of particular interest for two reasons. First, despite its moderate size (∼120 nucleotides [nt]), 5S rRNA has quite a complicated structural organization, comprising plethora of noncanonical elements and motifs, as well as a sophisticated three-dimensional (3D) structure, which is maintained by various kinds of interactions (for review, see Szymanski et al. 2003). The molecule is composed of three quasi-independent structural domains (Fig. 1A). The α-domain encompasses helix I, while two other (major) domains, β and γ, are constituted by alternating helical and loop regions. A peculiar spatial organization of the internal loops B and E provides continuous stacking throughout the domains. All three “branches” of the molecule are arranged around a three-way junction (loop A), responsible for the higher-order geometry of the RNA molecule. Second, 5S rRNA seems to be one of the most interesting RNA species from the point of view of its intracellular traffic, which implicates a network of possible itineraries for various subcellular localizations. Being an integral part of large ribosomal subunit of almost all living organisms, it appears to perform a very important function in the translation process, as recent evidence suggests (Smith et al. 2001; Kiparisov et al. 2005; Kouvela et al. 2007). At the same time, it was shown that, at least in Xenopus laevis oocytes, to integrate a ribosome, a 5S rRNA molecule has to be first exported from the nucleus by the transcription factor IIIA (TFIIIA) (Pelham and Brown 1980; Guddat et al. 1990) and reimported into the nucleolus with the ribosomal protein L5; the 5S rRNA-L5 complex is then involved in the central protuberance formation (Steitz et al. 1988; Rudt and Pieler 1996). Although the 5S rRNA intracellular traffic in other eukaryotic systems has still to be studied, it is obvious that in most cases it should imply a complex pathway that operates through a consecutive exchange of carrier proteins.
FIGURE 1.
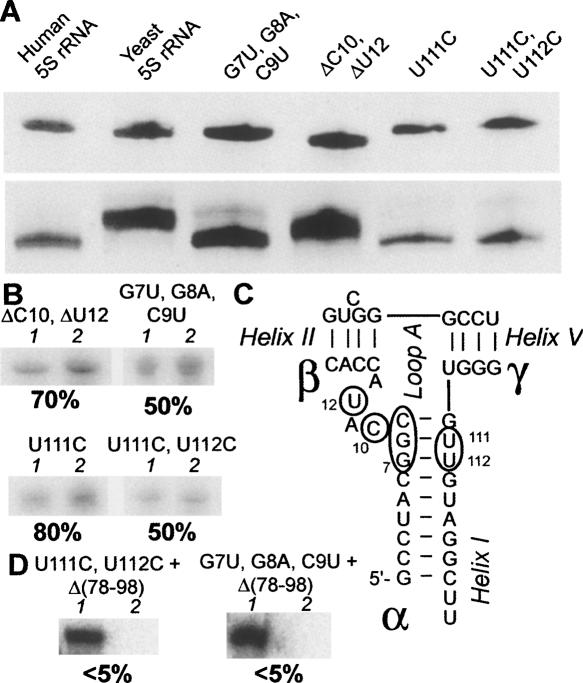
Human 5S rRNA secondary structure and mutation map. (A) Human 5S rRNA secondary structure (mfold, corrected manually according to structural studies published). Sites of protein binding are colored (Baudin et al. 1991; Chow et al. 1992; White et al. 1992; Allison et al. 1993; Wimberly et al. 1993; Szymanski et al. 2000; Huber et al. 2001; Lu et al. 2003; Zuker 2003). (B) The overall mutation map of the human 5S rRNA obtained in this work. Colors correspond to different import efficiencies in vitro.
As complicated as this pathway is, it became even more entangled after 5S rRNA import into mammalian mitochondria had been discovered (Yoshionari et al. 1994; Magalhaes et al. 1998). Being clearly demonstrated for several species, the mechanism by which 5S rRNA is imported into mitochondria and its functional significance remain poorly understood. Indeed, mammalian mitochondrial DNA is supposed to have lost the 5S rRNA gene in an early stage of evolution (Nierlich 1982). Furthermore, no attempts to find a 5S rRNA-like molecule in the mitoribosomal large particle were successful (Nagaike et al. 2001; Sharma et al. 2003). Still, 5S rRNA is one of the most abundant RNA species in mitochondria, which suggests its implication in some important processes inside the organelle (Magalhaes et al. 1998; Entelis et al. 2001a). Moreover, previously, it was shown that the number of 5S rRNA molecules in human mitochondria is sufficient to outfit all mitoribosomes (Entelis et al. 2001a). Thus, the 5S rRNA may be involved in mitochondrial translation or ribosomes biogenesis, although through a very unstable, if any, association with ribosomal particles.
Concerning the mechanism 5S rRNA molecules exploit to penetrate mitochondria, several facts can be outlined: (1) import needs ATP hydrolysis and the electrochemical potential across the mitochondrial inner membrane; (2) it depends on proteinase-sensitive outer membrane receptors; (3) the preprotein import apparatus needs be intact and functional; and (4) soluble cytosolic protein factors are required to direct it (Entelis et al. 2001a). Interestingly, these requirements strongly resemble those for tRNA import into yeast and human mitochondria, thus making a common point in their import mechanisms. But as a rule, protein factors, regulatory systems, and structural determinants needed for mitochondrial localization are unique for each type of imported RNA and strongly vary among species (which is not the case of preprotein import mechanisms, which proved to be highly conserved among species) (for review, see Schneider and Marechal-Drouard 2000; Entelis et al. 2001b; Tarassov et al. 2007). For example, soluble proteins necessary for artificial tRNA import into human mitochondria were shown to be incapable of directing 5S rRNA import in the same conditions (Entelis et al. 2001a). Thus, protein factors recruited for 5S rRNA targeting into mitochondria remain yet unidentified. The same can be said about the determinants of import, i.e., structural elements of 5S rRNA molecule needed for its directing into the mitochondria, a kind of signal of mitochondrial localization of the RNA.
In this work, we performed extensive analysis of a set of 5S rRNA variants in order to determine which structural elements of the molecule are implicated in its targeting to the mitochondria. We show that both the α-domain and especially the γ-domain contain structural elements required for its import. On the other hand, the β-domain has been shown to be dispensable for mitochondrial targeting. These findings allow for the construction of a 5S rRNA-based vector for delivering heterologous RNA sequences into human mitochondria in vivo, which may serve as a new tool both for mitochondrial genetic mechanisms investigation and for mitochondrial malfunctions therapy.
RESULTS
The 5S rRNA import determinants search strategy
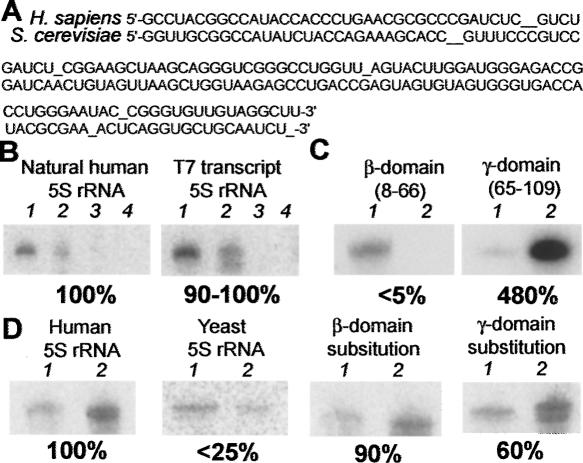
Previously, we have shown that radioactively labeled human 5S rRNA can be selectively imported into isolated human mitochondria in the presence of soluble protein fractions, ATP, and the ATP regeneration system (Entelis et al. 2001a). Since the human 5S rRNA bears no post-transcriptional modifications (Szymanski et al. 2003), we hypothesized that a T7 transcript of identical sequence would be as efficiently imported as the natural 5S rRNA. To choose among the numerous allelic variants of human 5S rRNAs reported to date (Kiparisov et al. 2005), we performed RT-PCR and sequenced 5S rRNAs isolated from HeLa cells or from purified mitochondria. In both cases, the same 5S rRNA sequence was found to be the most abundant and was thus chosen for further experiments (Fig. 2A). The T7 transcript with the corresponding sequence was shown to be imported into isolated mitochondria with an efficiency of 90%–100% with respect to that of the natural 5S rRNA, thus proving to be an adequate 5S rRNA substitute for further search of import determinants (Fig. 2B).
FIGURE 2.
Distinct role of 5S rRNA domains in import into isolated HepG2 mitochondria. (A) 5S rRNA sequences from human and yeast (manual alignment). (B) Import of the human 5S rRNA and a T7 transcript of identical sequence into mitochondria (autoradiograph of imported labeled RNAs). 1 indicates 5% of input RNA; 2, import assay; 3, import assay in absence of mitochondria; 4, import assay in absence of protein fractions directing import in vitro. Here and forth, relative import efficiencies (human wild-type 5S rRNA being 100%) are provided below the import pictures. (C) Import of isolated human 5S rRNA domains: 1 indicates 1% of input RNA; 2, import assay. (D) Import of the yeast 5S rRNA and “hybrid” 5S rRNA molecules: 1 indicates 1% of input RNA; 2, import assay.
To identify the structural elements needed for 5S rRNA import, the following strategy was adopted. First, to obtain crude results on determinants location in the molecule, import capacities of isolated major domains (β and γ) of the human 5S rRNA were checked in vitro (Fig. 2C). The β-domain was shown to be almost completely incapable of entering into mitochondria, while the γ-domain demonstrated even a higher import efficiency than the full-size human 5S rRNA.
Although these results are indicative, one cannot be sure that the isolated domains, devoid of their natural context, still preserve their original structural organization. This is why we used another approach. It was previously shown that Saccharomyces cerevisiae 5S rRNA can be very weakly internalized by isolated human mitochondria (Entelis et al. 2001a). This suggests that some differences between human and yeast 5S rRNA structures are responsible for their differential import. Indeed, sequences of both 5S rRNA species diverge in nearly 50 positions (depending on the variant) out of 120 (Fig. 2A), which is also clearly seen from their secondary structures. We modeled and constructed “hybrid” molecules where the β- or γ-domain of the human 5S rRNA was substituted by cognate yeast 5S rRNA domains. Thus, the general structural context of 5S rRNA was preserved while the domain structures changed. Interestingly, both mutant RNAs were shown to be imported into isolated mitochondria, but with different efficiencies, the γ-domain substitution being less active (Fig. 2D). Taken together, these data suggest that determinants of 5S rRNA import into human mitochondria are at least partially associated with the γ-domain, the β-domain being of little importance.
Next we performed a more focused analysis of 5S rRNA structural elements. A set of 5S rRNA variants, bearing mutations in various regions throughout the molecule, was modeled. The basic assumption, when choosing sites and kinds of mutations, is a possible correspondence between the determinants of 5S rRNA import into mitochondria and potential sites of import factor(s) binding. Thus, the elements that could be potentially involved in protein binding were of particular interest. All 5S rRNA variants genes were T7-transcribed in vitro, and the resulting transcripts were used to evaluate their import efficiency into isolated human mitochondria (Figs. 1B, 3–5; Table 1).
FIGURE 3.
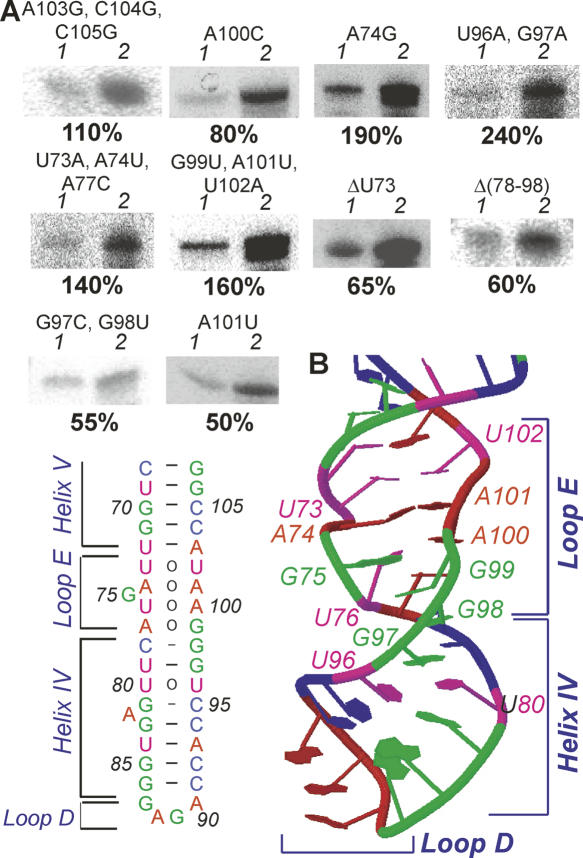
Import in vitro of 5S rRNA variants with mutations in the γ-domain. (A) Import of the γ-domain mutant variants: 1 indicates 1% of input RNA; 2, import assay. (B) Secondary and tertiary structures of the loop E-helix IV-loop D region from the Xenopus laevis 5S rRNA (JenaLib, 1un6) (Lu et al. 2003).
FIGURE 5.
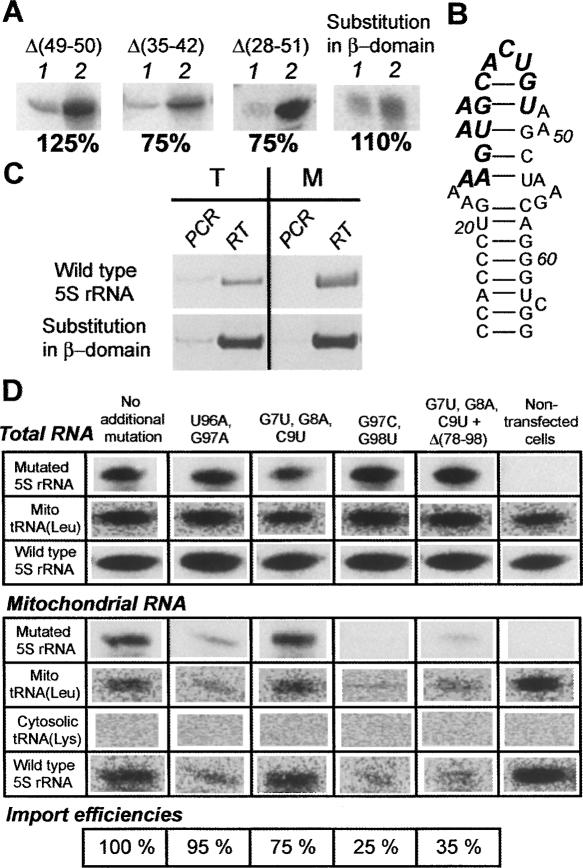
Import of 5S rRNA with mutations in the β-domain. (A) Import of 5S rRNA variants with mutations in the β-domain in vitro: 1 indicates 1% of input RNA; 2, import assay. (B) The secondary structure of the β-domain of the 5S rRNA variant with substitution of the helix III and the loop C for a heterologic sequence (bold italic). (C) RT-PCR analysis of presence of the 5S rRNA variant with substitution of the helix III and the loop C in total (T) and mitochondrial (M) RNA from stably transfected human cells. (D) Northern blot analysis of presence of the β-domain substitution 5S rRNA variants in total and mitochondrial RNA preparations two days after transient transfection. Relative import efficiencies are indicated below (the β-domain substitution 5S rRNA variant without any additional mutation being 100%). Probes used for hybridization are indicated in left column.
TABLE 1.
Mutant 5S rRNA variants descriptions and in vitro import efficiencies
Effects of structure changes in the γ-domain on the 5S rRNA import capacity
Since the γ-domain proved to be of greater importance for 5S rRNA mitochondrial targeting, a thorough dissection of this region was performed. Ten mutant variants were designed, constructed, and characterized (Fig. 3A,B). Depending on their import efficiencies, they could be divided into three classes.
Class I: Mutant variants with no significant import efficiency changes
Only two mutations in the 5S rRNA γ-domain fall into this class. One of them—A103G, C104G, C105G—leads to breaking the distal part of the helix V and destabilizing the loop E, suggesting that these regions are of little importance for 5S rRNA targeting to mitochondria. The second mutation—A100C—was previously shown to significantly affect the peculiar conformation of loop E, leading to its opening (Chow et al. 1992). These data suggest the particular 3D structure of loop E is dispensable for import.
Class II: Mutant variants with increased import efficiencies
Surprisingly, four mutant variants of the γ-domain group demonstrated higher level of import into isolated mitochondria than the wild-type human 5S rRNA. All of them were associated with the loop E and the proximal part of helix IV. A point mutation A74G was shown earlier to change the loop E conformation to a more closed one (Chow et al. 1992). Two other mutations—U73A, A74U, A77C and G99U, A101U; U102A—confer on loop E a quasi-regular A-form duplex structure with G75 bulged, which is significantly more closed and stable (Wimberly et al. 1993). Finally, the most prominent increase of import efficiency was observed when two consecutive unusual G-U and U-U pairs of helix IV were substituted by Watson–Crick A-U ones (U96A, G97A); the proximal part of the helix thus became a regular duplex. These mutations also led to closing of conformation and to general stabilization of the helix through a more efficient stacking (Szymanski et al. 2000). Taken together, these data clearly show that more closed conformation and stabilization of loop E and the adjacent part of helix IV lead to the 5S rRNA higher efficiency of import.
Class III: Mutant variants with decreased import efficiencies
Four mutations, all associated with loop E and helix IV, led to a moderate (to twofold) decrease of import capacity of the 5S rRNA. Interestingly, all of them are structural antipodes of the class II mutants, since they have significant destabilizing effect on loop E-helix IV structure. Thus, ΔU73 and A101U point mutations were previously reported to lead to profound changes in loop E conformation, its opening, and destabilization (Chow et al. 1992). Although a similar effect was demonstrated for A100C mutation, it did not significantly affect import of 5S rRNA, suggesting that another kind of reorganization took place. The G97C, G98U mutation disrupts two base pairs, which maintained both loop E and G-U/U-U pairs of helix IV, leading to dramatic disorganization of these structural modules. Finally, a deletion Δ(78–98) is removed from the molecule helix IV and loop D, making loop E terminal. These data suggest that the stability of loop E-helix IV region is very important for the efficient import of 5S rRNA. Still, no region of the γ-domain was shown to be critical for import, indicating that the α-domain and loop A may also contain import determinants.
Effects of mutations in the α-domain and loop A on the 5S rRNA import efficiency
Two highly conserved structural elements of the α-domain-loop A region were of particular interest to us, since both of them are distinguishable by their unusual conformations, most suitable for protein binding (Szymanski et al. 2000; Lu et al. 2003; Mokdad et al. 2006). The first one is loop A, a three-way junction, upon which the overall 3D structure of the molecule depends (Lescoute and Westhof 2006). The second comprises two consecutive G-U pairs, one compensated (i.e., with a purine residue at the position 5′ of the G) and another uncompensated (with a pyrimidine 5′ of the G) (Fig. 4C; White et al. 1992; Szymanski et al. 2000; Mokdad et al. 2006). In order to reveal possible implication of these elements in the 5S rRNA import process, four mutant 5S rRNA variants were designed.
FIGURE 4.
Import of 5S rRNA variants with mutations in the 5S rRNA α-domain and with double (α/γ) mutations. (A) 5S rRNA variants mobility in gel. (Upper panel) Denaturing gel. (Lower panel) Native gel. (B) Import of the α-domain mutants: 1 indicates 1% of input RNA; 2, import assay. (C) The secondary structure of the helix I-loop A region of the human 5S rRNA. Nucleotides that were mutated in this work are framed. (D) Import of double (α/γ) mutants: 1 indicates 1% of input RNA; 2, import assay.
Since all these mutations are localized in or in close proximity to loop A, which is an element crucial for maintaining the general geometry of the molecule (Lescoute and Westhof 2006), their influence on the 5S rRNA tertiary structure was studied. Only one mutation of this group—deletion of two loop A nucleotides (ΔC10, ΔU12)—obviously affected the general conformation of the 5S rRNA, which is clearly seen from the shift of its mobility in native gel (Fig. 4A). Dramatic though these changes are, they only weakly influenced the 5S rRNA import efficiency, indicating that preservation of the particular overall conformation of the molecule is not necessary for import (Fig. 4B).
However, this is not the case of helix I mutations (Fig. 4B). Only one of them—U111C, changing the compensated G-U pair to the Watson–Crick G-C—did not significantly affect the import efficiency. But when both compensated and uncompensated G-U pairs were touched (U111C, U112C) or the whole proximal part of helix I was disrupted (G7U, G8A, C9U), the import efficiency of 5S rRNA was decreased twofold. These results suggest that the particular structure of this region (Fig. 4C) is, indeed, important for efficient 5S rRNA targeting to mitochondria.
Effects of double (γ+α-domains) mutations on import capacity of 5S rRNA
Although no single mutation was shown to drastically affect the import efficiency of 5S rRNA, two clear regions of the molecule could be distinguished, which, when destructed, significantly decrease 5S rRNA importability. The first one is associated with the loop E-helix IV region (γ-domain); the second, with the proximal part of helix I (α-domain). We hypothesized that one of these sites is sufficient for import in vitro, though simultaneous mutation of both of them should abolish 5S rRNA import. Indeed, when the deletion of helix IV and loop D was coupled with one of the helix I mutations (U111C, U112C or G7U, G8A, C9U), 5S rRNA import capacity was dramatically decreased, in perfect agreement with our expectations (Fig. 4D). This result clearly shows that both of these regions are responsible for the human 5S rRNA import capacity, though any one of them is sufficient for the 5S rRNA mitochondrial localization.
Effects of deletions in the β-domain on the 5S rRNA import efficiency
Although the isolated β-domain of the human 5S rRNA failed to be targeted to mitochondria, and its substitution by the yeast one did not significantly reduce the import efficiency, a series of deletions in this region were designed and tested in vitro (Fig. 5A). Deletion of a highly conserved A49-A50 two-nucleotide bulge in helix III as well as of loop C [Δ(35–42)], both needed for L18/eL5 family ribosomal proteins binding (see Fig. 1A; Huber and Wool 1984; Chow et al. 1992; Scripture and Huber 1995; Huber et al. 2001), led to no significant changes in 5S rRNA import efficiency. Even a more spacious deletion [Δ(28–51)], removing both helix III and loop C and destructing loop B structure, failed to substantially affect the import capacity of the 5S rRNA, in perfect agreement with the data presented above.
Import of 5S rRNA mutated versions into human mitochondria in vivo
Evaluation of import efficiencies of mutated 5S rRNA variants in vivo may appear problematic, because of an immense background of about 200 naturally present 5S rRNA genes, which makes chasing of a mutant 5S rRNA complicated. Fortunately, finding the β-domain dispensable for the 5S rRNA import machinery, enabled us to suggest that this region of the molecule can be used as a site to insert a tag-sequence. This would permit distinguishing mutated 5S rRNA molecules from the wild-type ones. For this, a mutant 5S rRNA variant was designed (Fig. 5B). It contained a substitution of helix III and loop C by a sequence of 13 unrelated nucleotides in a way that does not affect either the 5S rRNA gene promoter or the TFIIIA-binding sites, which are necessary for efficient expression and export of the RNA, respectively (see Fig. 1A). As expected, this mutant version efficiently penetrated into isolated human mitochondria (Fig. 5A). For in vivo expression, the corresponding mutant gene was cloned into pcDNA3.1(+) vector in a context optimal for correct transcript processing, and used for stable transfection of 143B human cells. RT-PCR analysis with the tag-specific primer proved the presence of the mutated RNA both in the cytosol and in purified mitochondria of the transfectants, indicating that it is expressed and imported, in agreement with our expectations (Fig. 5C).
On the other hand, due to the relatively low level of expression of the mutant 5S rRNA gene, it was impossible to correctly evaluate the mutant RNA import efficiency in vivo. Moreover, the approach used failed to deal with 5S rRNA variants that bear mutations in regions adjacent to loops A and E, which were shown to overlap with TFIIIA-binding sites (Fig. 1A). These variants would fail to bind TFIIIA, which presumably functions as the 5S rRNA exporter, and the mutated RNAs would be recognized by the Ro protein and then degraded (O'Brien and Wolin 1994; Shi et al. 1996). Indeed, all our attempts to express in vivo a 5S rRNA variant genes with mutations in import determinants regions were unsuccessful (data not shown).
In order to bypass this problem, we made use of an alternative approach. Human cells were transfected with T7 transcripts of mutated 5S rRNA variants that contained the β-domain substitution described above (Fig. 5B). One representative mutant 5S rRNA variant from each group described above was chosen for this experiment (Fig. 5D). In two days time, total and mitochondrial RNAs were isolated and analyzed by Northern blot hybridization. Since all 5S rRNA variants used in this experiment bear the characteristic 13-nt substitution in the β-domain (the tag), they could be easily traced by hybridization with a corresponding probe.
Interestingly, when one compares the total to mitochondria ratio of the mutated variants with that of the wild-type 5S rRNA (Fig. 5D), it can be clearly seen that in all cases RNAs with the substitution in the β-domain tend to localize inside the mitochondrial compartment. Indeed, when one considers the ratio between signals obtained from a mutated 5S rRNA in mitochondrial and total RNA preparations, it is always found to be 20 ± 7 times higher than that for the wild-type 5S rRNA. Thus, an apparent import efficiency increase was observed. A possible explanation for this effect implies the absence of competition between mitochondrial import factors and the ribosomal protein L5 for binding to a mutant 5S rRNA, since the site of recognition by the L5 (helix III and loop C) is demolished (Steitz et al. 1988; Rudt and Pieler 1996). Mitochondrial import—which apparently does not depend on the β-domain elements—remains the only possible itinerary for these mutant RNAs.
This highly efficient in vivo import system enabled us to representatively evaluate import capacities of the mutant RNAs in human cells. To this end, the ratio between the signal of the mutant RNA in the mitochondrial fraction and that of the naturally occurring mitochondrial tRNALeuUUR can be used. This ratio was normalized regarding differences between mutant RNA total levels in transfected cells (for details, see the Materials and Methods section). Resulting import efficiencies were expressed in form of percentage (the import efficiency of the β-substitution variant without any additional mutations chosen to be 100%) (Fig. 5D). It can be clearly seen that in vivo import efficiency of mutated 5S rRNAs correlates with those demonstrated in vitro. Indeed, mutations leading to destabilization or even destruction of the sites in the α-domain and especially in the γ-domain, which were shown to be critical for the import in vitro, also decrease import efficiency of the 5S rRNA in vivo, proving the importance of these structural elements for the molecule importability. On the other hand, the γ-domain site (loop E-helix IV) appears to be much more critical in vivo, indicating its paramount role in the 5S rRNA import system functioning.
DISCUSSION
A Plethora of mitochondrial import systems have been described to date in all kingdoms of eukaryotic organisms (for review, see Entelis et al. 2001b). Each of them is unique with regard to mechanisms exploited to target an RNA into mitochondria. Among them one can find systems with relatively low specificity for RNA transported (for example, tRNA import in trypanosomatids) (Schneider and Marechal-Drouard 2000), systems with several cytosolic RNA species specifically imported (like tRNA import into plant mitochondria) (Kumar et al. 1996), as well as very specific, selective systems, usually with but few RNAs imported (the best-studied case—tRNALysCUU import in yeast) (Tarassov et al. 1995; Entelis et al. 1998; Kazakova et al. 1999), one out of three imported yeast tRNA species (Rinehart et al. 2005). The pathway of directing the 5S rRNA into human mitochondria is obviously an example of highly selective targeting systems, since this is a single RNA species described to be efficiently targeted into the human organelle, while data on possible import of two other RNAs—RNA components of RNase P and MRP—are still contradictory and their import efficiency seems very low (Kiss and Filipowicz 1992; Li et al. 1994; Puranam and Attardi 2001). Although the selectivity of the 5S rRNA import has been demonstrated, no explanation for this specificity existed. In this work, we undertook a systematic analysis of human 5S rRNA structural elements in order to reveal those responsible for the import capacity. In in vitro experiments, two regions of human 5S rRNA were shown to be of utter importance. One of them is located in the proximal part of helix I (α-domain) and contains the conservative uncompensated G7-U112 pair (Fig. 4C). The second is associated with the loop E-helix IV region (γ-domain) with several noncanonical structural features (Fig. 3B). The 5S rRNA devoid of one of these sites demonstrates diminished efficiency of import into isolated human mitochondria, while destruction of both of them leads to irretrievable loss of import capacity.
We hypothesized that 5S rRNA import determinants should match import factors binding sites. Such an assumption is in good agreement with the “RNA zipcode” hypothesis (Jambhekar and Derisi 2007), suggesting that the subcellular localization of a given RNA molecule might be determined by particular structural motifs interacting with competing transport complexes. Indeed, the first region identified has a rather peculiar structure that significantly deviates from a regular A-form RNA-helix (White et al. 1992). The cause of this distortion is the displacement of the G residue of the uncompensated G-U pair toward the minor groove, resulting in a loss of stacking 5′ of guanosine and protruding of the C6-G113 pair toward the major groove (Mizuno and Sundaralingam 1978; White et al. 1992). This structural feature makes G-U pairs perfectly predisposed for protein binding in variety of RNAs. For example, the G3-U70 pair in tRNAAla is the major determinant for its recognition by the alanyl-tRNA synthetase (Hou and Schimmel 1988). A G-U pair in the ribosomal protein L32 pre-mRNA in yeast is recognized by the cognate protein (White and Li 1996). Numerous G-U pairs in ribosomal RNAs participate in ribosomal proteins binding. Moreover, the proximal part of helix I of prokaryotic 5S rRNAs was shown to bind to the ribosomal protein L18 (Mokdad et al. 2006). Thus, this region appears to be a likely binding site for an import factor.
The second site comprising the loop E-helix IV region (γ-domain), is even more distinguished with regard to structure peculiarities. Much studies were focused on the loop E conformation (Wimberly et al. 1993; Correll et al. 2003). Loop E-like motifs were many times shown to participate in protein binding; for example, the sarcin–ricin loop of 23S rRNA interacts with translation factors (Moazed et al. 1988), and eukaryotic 5S rRNA loop E is one of TFIIIA-binding sites (Lu et al. 2003). Helix IV is also remarkable by its unusual organization (Chow et al. 1992; Baeyens et al. 1995) and also was shown to participate in protein binding (to the ribosomal protein L30 of Haloarcula marismortui) (Mokdad et al. 2006). Still, our results clearly show that the particular structure of these elements is of no importance for 5S rRNA import into human mitochondria. The only parameter of this region that really matters seems to be its stability. Indeed, mutations that turn loop E or the proximal part of helix IV into regular duplexes, improve the 5S rRNA import efficiency, though destroy their original conformations. In contrast, destabilizing mutations not only affect the region structure but also decrease import efficiency.
In vivo, for both the G97C, G98U (γ-domain) and the double mutants where this region was subject to destabilizing mutations, the import efficiency was decreased even more substantially than in vitro. At the same time, disruption of the proximal part of helix I (G7U, G8A, C9U) (α-domain) only slightly affects the 5S rRNA importability. This suggests that the γ-domain site can, indeed, function as binding platform for the principal import factor, while the α-domain is an auxiliary binding site that may serve for more efficient positioning and/or binding of import factors. This hypothesis explains the importability of the isolated γ-domain even in absence of the α-domain site, as well as the fact that no single mutation in the γ-domain was able to prevent the 5S rRNA import in vitro if the α-domain site remained intact.
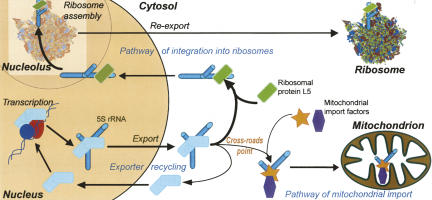
In this work, we also demonstrated that the 5S rRNA molecule with the β-domain mutated (and thus incapable to bind to the ribosomal protein L5) can be imported into human mitochondria with an apparent efficiency that ∼20 times exceeds that of the wild-type 5S rRNA. This finding enabled us to hypothesize that the 5S rRNA intracellular traffic is driven by a mechanism of competition/substitution between various carrier proteins (and respective targeting pathways) for their cargo (Fig. 6). Once a 5S rRNA molecule is exported from the nucleus, it is subject to two alternative import pathways. The first destination is the nucleolus where the 5S rRNA can become the part of a nascent large ribosomal particle. This pathway operates through the ribosomal protein L5 binding (Steitz et al. 1988; Rudt and Pieler 1996) and seems the most important and prevailing. Mitochondrial targeting appears to be the second destination (Magalhaes et al. 1998). 5S rRNA import into this organelle is ensured by import factors. Still, it appears much less prominent when compared with the first pathway. Indeed, it was shown that only ∼1% of the total 5S rRNA population in human cells is associated with mitochondria (Entelis et al. 2001a). What can be the cause of such a distribution pattern?
FIGURE 6.
A scheme of putative pathways of the human 5S rRNA intracellular localization (for details, see text).
Obviously, in both cases a carrier protein substitution has to take place. The exporter protein (TFIIIA, for instance) needs to be displaced by either the L5 or the mitochondrial import factors. The mechanism of the first substitution can be established from the data on the 5S rRNA-TFIIIA and 5S rRNA-L5 interactions. Binding of the 5S rRNA to the L5 proceeds through the “mutual induced fit” mechanism, when both molecules act as chaperones for each other. The resulting conformation of the 5S rRNA significantly differs from that in the complex with TFIIIA, ensuring the unidirectional displacement (DiNitto and Huber 2003). Although no data on the 5S rRNA-import factors interaction are available, from the results obtained in this work one can conclude that import factors and TFIIIA may exploit overlapping binding sites (especially in the domain γ). Thus, the mechanism of this substitution implies a competition for binding to the 5S rRNA between its nuclear exporter (TFIIIA) and its mitochondrial importer(s). Through consecutive exchange of protein partners, the 5S rRNA can make its way from the nucleus through the cytosol to the mitochondrion. It suggests that a 5S rRNA intracellular channeling mechanism may exist in vivo, by analogy with previously described tRNA channeling (for review, see Hopper and Phizicky 2003). But unlike the latter one, this channeling includes a cross-roads point where the final destination can be switched.
In this work, we designed human 5S rRNA variants for which no cross-road point exists (Fig. 5). Being as efficiently imported as the wild-type 5S rRNA in vitro, the 5S rRNA version with the substitution in the β-domain is free of the ambiguity in subcellular localization. This feature makes it move directly into mitochondria with apparent efficiency that significantly exceeds the natural one. Moreover, since the β-domain is clearly dispensable for import, it can be used for insertion of foreign RNA sequences in order to direct them into mitochondria in vivo. Here, we demonstrate for the first time that a 5S rRNA-based recombinant RNA, indeed, can be imported into human mitochondria in vivo, providing efficient delivery of a foreign RNA sequence of 13 nucleotides into the mitochondrial compartment. This tool may potentially become useful for suppression of mitochondrial malfunctions caused by mutations in mtDNA. Several strategies of mitochondrial disorders therapy based on artificially induced RNA import in human cells were proposed to date (Kolesnikova et al. 2000, 2004; Mahata et al. 2005, 2006). Still, all of them operate through complementation of mitochondrial tRNA mutations by substitute tRNAs import from the cytosol. On the other hand, the use of 5S rRNA–based vector systems exploiting the natural 5S rRNA import pathway can serve as a more universal tool for suppression of various kinds of mutations. As a possibility, oligonucleotides, complementary to mutated mitochondrial DNA sequences, inserted into the β-domain of such vectors may selectively suppress the mutated mitochondrial DNA replication, favoring wild-type DNA propagation. Such an approach was previously assayed with artificial PNA (peptide nucleic acids) oligonucleotides and proved to be efficient in vitro, but not in vivo (Taylor et al. 1997; Muratovska et al. 2001). The naturally occurring 5S rRNA import seems to provide us with a powerful tool to develop such an approach in vivo.
MATERIALS AND METHODS
Mutant 5S rRNA variants design and construction
All 5S rRNA variants secondary structures were modeled using the mfold secondary structure prediction algorithm (Zuker 2003) and corrected in accordance with data published on 5S rRNA structural elements (Chow et al. 1992; White et al. 1992; Wimberly et al. 1993; Szymanski et al. 2000; Huber et al. 2001). The variants genes were obtained by PCR-mutagenesis with primers bearing corresponding mutations, cloned into pUC119 vector with T7 promoter region, and sequenced. They were transcribed in vitro using the T7 RiboMAX Express Large Scale RNA Production System (Promega), and transcripts were purified by gel electrophoresis.
Import of 5S rRNA variants into isolated mitochondria
Human mitochondria and protein fractions directing import were isolated from HepG2 cells as described elsewhere (Gaines and Attardi 1984a; Entelis et al. 2001a). T7 transcripts were 32P-γ-ATP labeled by T4 polynucleotide kinase (Promega) and gel purified. The standard RNA import assay contained mitochondria (50 μg of mitochondrial protein), 3–5 pmol of a labeled RNA, and 10 μg of import directing protein fraction (Entelis et al. 2001a). After the import reaction and nonimported RNAs removal by RNase treatment and mitoplasts generation, mitochondrial RNAs were isolated, separated by denaturing gel electrophoresis, and analyzed with PhosphorImager (Fuji, Bas2000). As a standard for absolute import efficiency quantification, 1%–5% of input labeled RNA were deposed on gel. Assays in absence of either mitochondria or protein fractions were used as negative controls. All imports were performed at least twice in an independent way and presented values correspond to the average numbers with SDs less than 10%.
Native gel electrophoresis
For conformation studies, nonlabeled T7 transcripts were prefolded by thermal denaturing-renaturing in presence of Mg2+ (2.5 mM) and applied on a 10% PAAG gel. The gel was run at 10 V/cm in 1× TBE at 4°C; RNAs were visualized by ethidium bromide. To check RNA preparation homogeneity and size differences, standard denaturing gel electrophoresis (12% PAAG, 1× TBE, 8 M urea) was used.
RNA import in vivo
5S rRNA variants genes were constructed by PCR with primers including genomic 5′- and 3′-flanking sequences in order to ensure the proper processing of transcripts, and cloned into pcDNA3.1(+)Zeo vector (Invitrogen). The resulting construct was linearized and used for 143B cells stable transfection as described elsewhere (Mager-Heckel et al. 2007). Transfected cells were grown on DME medium (Sigma) with 100 μg/mL zeocine. Mitochondria were isolated and purified as described (Gaines and Attardi 1984b) and treated with RNase A and digitonin to get rid of nonspecifically attached RNAs. Total and mitochondrial RNAs were isolated with TRIZol reagent (Invitrogen) and analyzed by One Step RT-PCR (Qiagen) with primers to heterologous sequences inserted (or to the wild-type 5S rRNA as the positive control). The PCR products were separated by gel electrophoresis and visualized by ethidium bromide staining.
For transfection of HepG2 cells, 5 μg of T7 transcripts per 25 cm2 of confluent cells were used. The transfection was carried out by using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Two days after transfection, total and mitochondrial RNAs were isolated as described above, resolved in a denaturing gel, and analyzed by Northern blot hybridization with 32P-labeled oligonucleotides against the substitution region in the mutated 5S rRNAs (5′-GCTTACAGTGCTTACTT), mitochondrial tRNALeuUUR (5′-GAACCTCTGACTGTAAAG), cytosolic tRNALysUUU (5′-ACTTGAACCCTGGACC), and wild-type 5S rRNA (5′-CATCCAAGTACTACCAGGCCC).
Evaluation of import efficiency
To estimate the apparent import efficiency increase of mutated 5S rRNAs, the following procedure was performed. Ratio I (for mutant 5S rRNA) was calculated between signals obtained after hybridization with an oligonucleotide against the substituted region in mitochondrial and total RNA preparations. Ratio II (for the wild-type 5S rRNA) was calculated between signals obtained after hybridization with an oligonucleotide against the wild-type 5S rRNA in the same preparations. The apparent import efficiency increase was calculated as a quadruple ratio between Ratios I and II.
To estimate the import efficiencies of mutated 5S rRNAs, the following approach was used. Ratio I was calculated between the signal obtained after hybridization with an oligonucleotide against the substituted region and that obtained after hybridization with an oligonucleotide against the mitochondrial tRNALeu UUR in the mitochondrial RNA preparation. Ratio II was calculated in the same way but in the total RNA preparation. Import efficiency of a mutated 5S rRNA was calculated as a quadruple ratio between Ratios I and II. In order to compare import efficiencies of different mutant 5S rRNAs, total mutant RNAs' levels in transfected cells should be taken into account and normalized. For this, the calculated import efficiencies were divided by Ratio II, which indicates the total level of mutated RNAs. Resulting normalized import efficiencies were expressed in the form of percentage of the efficiency thus obtained for the β-domain substitution 5S rRNA without any additional mutations.
ACKNOWLEDGMENTS
We thank other members of our laboratory: O. Kolesnikova, O. Karicheva, and C. Comte for helpful discussions; Y. Kharchenkov for the technical assistance; and C. Reibel for administrative help. This work was supported by the CNRS, Université Louis Pasteur, Moscow State University, AFM (Association Française contre les Myopathies), ANR (Agence Nationale de la Recherche), ARCUS (collaboration Alsace-Russia-Ukraine), RFBR (Russian Foundation for Basic Research), and GIS Institut de Maladies Rares.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.952208.
REFERENCES
- Allison, L.A., North, M.T., Murdoch, K.J., Romaniuk, P.J., Deschamps, S., le Maire, M. Structural requirements of 5S rRNA for nuclear transport, 7S ribonucleoprotein particle assembly, and 60S ribosomal subunit assembly in Xenopus oocytes. Mol. Cell. Biol. 1993;13:6819–6831. doi: 10.1128/mcb.13.11.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens, K.J., De Bondt, H.L., Holbrook, S.R. Structure of an RNA double helix including uracil-uracil base pairs in an internal loop. Nat. Struct. Biol. 1995;2:56–62. doi: 10.1038/nsb0195-56. [DOI] [PubMed] [Google Scholar]
- Baudin, F., Romaniuk, P.J., Romby, P., Brunel, C., Westhof, E., Ehresmann, B., Ehresmann, C. Involvement of “hinge” nucleotides of Xenopus laevis 5 S rRNA in the RNA structural organization and in the binding of transcription factor TFIIIA. J. Mol. Biol. 1991;218:69–81. doi: 10.1016/0022-2836(91)90874-6. [DOI] [PubMed] [Google Scholar]
- Chow, C.S., Hartmann, K.M., Rawlings, S.L., Huber, P.W., Barton, J.K. Delineation of structural domains in eukaryotic 5S rRNA with a rhodium probe. Biochemistry. 1992;31:3534–3542. doi: 10.1021/bi00128a030. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Beneken, J., Plantinga, M.J., Lubbers, M., Chan, Y.L. The common and the distinctive features of the bulged-G motif based on a 1.04 Å resolution RNA structure. Nucleic Acids Res. 2003;31:6806–6818. doi: 10.1093/nar/gkg908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNitto, J.P., Huber, P.W. Mutual induced fit binding of Xenopus ribosomal protein L5 to 5S rRNA. J. Mol. Biol. 2003;330:979–992. doi: 10.1016/s0022-2836(03)00685-5. [DOI] [PubMed] [Google Scholar]
- Entelis, N.S., Kieffer, S., Kolesnikova, O.A., Martin, R.P., Tarassov, I.A. Structural requirements of tRNALys for its import into yeast mitochondria. Proc. Natl. Acad. Sci. 1998;95:2838–2843. doi: 10.1073/pnas.95.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis, N.S., Kolesnikova, O.A., Dogan, S., Martin, R.P., Tarassov, I.A. 5 S rRNA and tRNA import into human mitochondria. Comparison of in vitro requirements. J. Biol. Chem. 2001a;276:45642–45653. doi: 10.1074/jbc.M103906200. [DOI] [PubMed] [Google Scholar]
- Entelis, N.S., Kolesnikova, O.A., Martin, R.P., Tarassov, I.A. RNA delivery into mitochondria. Adv. Drug Deliv. Rev. 2001b;49:199–215. doi: 10.1016/s0169-409x(01)00135-1. [DOI] [PubMed] [Google Scholar]
- Gaines, G., Attardi, G. Highly efficient RNA-synthesizing system that uses isolated human mitochondria: New initiation events and in vivo-like processing patterns. Mol. Cell. Biol. 1984a;4:1605–1617. doi: 10.1128/mcb.4.8.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines, G., Attardi, G. Intercalating drugs and low temperatures inhibit synthesis and processing of ribosomal RNA in isolated human mitochondria. J. Mol. Biol. 1984b;172:451–466. doi: 10.1016/s0022-2836(84)80017-0. [DOI] [PubMed] [Google Scholar]
- Guddat, U., Bakken, A.H., Pieler, T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K., Phizicky, E.M. tRNA transfers to the limelight. Genes & Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Hou, Y.M., Schimmel, P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Huber, P.W., Wool, I.G. Nuclease protection analysis of ribonucleoprotein complexes: use of the cytotoxic ribonuclease alpha-sarcin to determine the binding sites for Escherichia coli ribosomal proteins L5, L18, and L25 on 5S rRNA. Proc. Natl. Acad. Sci. 1984;81:322–326. doi: 10.1073/pnas.81.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, P.W., Rife, J.P., Moore, P.B. The structure of helix III in Xenopus oocyte 5 S rRNA: An RNA stem containing a two-nucleotide bulge. J. Mol. Biol. 2001;312:823–832. doi: 10.1006/jmbi.2001.4966. [DOI] [PubMed] [Google Scholar]
- Jambhekar, A., Derisi, J.L. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakova, H.A., Entelis, N.S., Martin, R.P., Tarassov, I.A. The aminoacceptor stem of the yeast tRNALys contains determinants of mitochondrial import selectivity. FEBS Lett. 1999;442:193–197. doi: 10.1016/s0014-5793(98)01653-6. [DOI] [PubMed] [Google Scholar]
- Kiparisov, S., Petrov, A., Meskauskas, A., Sergiev, P.V., Dontsova, O.A., Dinman, J.D. Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae . Mol. Genet. Genomics. 2005;274:235–247. doi: 10.1007/s00438-005-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T., Filipowicz, W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- Kolesnikova, O.A., Entelis, N.S., Mireau, H., Fox, T.D., Martin, R.P., Tarassov, I.A. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- Kolesnikova, O.A., Entelis, N.S., Jacquin-Becker, C., Goltzene, F., Chrzanowska-Lightowlers, Z.M., Lightowlers, R.N., Martin, R.P., Tarassov, I. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum. Mol. Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- Kouvela, E.C., Gerbanas, G.V., Xaplanteri, M.A., Petropoulos, A.D., Dinos, G.P., Kalpaxis, D.L. Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal nanomachine. Nucleic Acids Res. 2007;35:5108–5119. doi: 10.1093/nar/gkm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R., Marechal-Drouard, L., Akama, K., Small, I. Striking differences in mitochondrial tRNA import between different plant species. Mol. Gen. Genet. 1996;252:404–411. doi: 10.1007/BF02173005. [DOI] [PubMed] [Google Scholar]
- Lescoute, A., Westhof, E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K., Smagula, C.S., Parsons, W.J., Richardson, J.A., Gonzalez, M., Hagler, H.K., Williams, R.S. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J. Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D., Searles, M.A., Klug, A. Crystal structure of a zinc-finger–RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- Magalhaes, P.J., Andreu, A.L., Schon, E.A. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol. Biol. Cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager-Heckel, A.-M., Brandina, I., Kamenski, P., Entelis, N., Martin, R.P., Tarassov, I. Methods of analysis of tRNA import into human mitochondria. Methods Mol. Biol. 2007;373:235–253. doi: 10.1007/978-1-59745-365-3_17. [DOI] [PubMed] [Google Scholar]
- Mahata, B., Bhattacharyya, S.N., Mukherjee, S., Adhya, S. Correction of translational defects in patient-derived mutant mitochondria by complex-mediated import of a cytoplasmic tRNA. J. Biol. Chem. 2005;280:5141–5144. doi: 10.1074/jbc.C400572200. [DOI] [PubMed] [Google Scholar]
- Mahata, B., Mukherjee, S., Mishra, S., Bandyopadhyay, A., Adhya, S. Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science. 2006;314:471–474. doi: 10.1126/science.1129754. [DOI] [PubMed] [Google Scholar]
- Mizuno, H., Sundaralingam, M. Stacking of Crick wobble pair and Watson–Crick pair: Stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon–anticodon wobble interaction. Nucleic Acids Res. 1978;5:4451–4461. doi: 10.1093/nar/5.11.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., Robertson, J.M., Noller, H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Mokdad, A., Krasovska, M.V., Sponer, J., Leontis, N.B. Structural and evolutionary classification of G/U wobble base pairs in the ribosome. Nucleic Acids Res. 2006;34:1326–1341. doi: 10.1093/nar/gkl025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratovska, A., Lightowlers, R.N., Taylor, R.W., Turnbull, D.M., Smith, R.A., Wilce, J.A., Martin, S.W., Murphy, M.P. Targeting peptide nucleic acid (PNA) oligomers to mitochondria within cells by conjugation to lipophilic cations: Implications for mitochondrial DNA replication, expression and disease. Nucleic Acids Res. 2001;29:1852–1863. doi: 10.1093/nar/29.9.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike, T., Suzuki, T., Tomari, Y., Takemoto-Hori, C., Negayama, F., Watanabe, K., Ueda, T. Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J. Biol. Chem. 2001;276:40041–40049. doi: 10.1074/jbc.M106202200. [DOI] [PubMed] [Google Scholar]
- Nierlich, D.P. Fragmentary 5S rRNA gene in the human mitochondrial genome. Mol. Cell. Biol. 1982;2:207–209. doi: 10.1128/mcb.2.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, C.A., Wolin, S.L. A possible role for the 60-kDa Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes & Dev. 1994;8:2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R., Brown, D.D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl. Acad. Sci. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam, R.S., Attardi, G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol. Cell. Biol. 2001;21:548–561. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart, J., Krett, B., Rubio, M.A., Alfonzo, J.D., Soll, D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes & Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudt, F., Pieler, T. Cytoplasmic retention and nuclear import of 5S ribosomal RNA containing RNPs. EMBO J. 1996;15:1383–1391. [PMC free article] [PubMed] [Google Scholar]
- Schneider, A., Marechal-Drouard, L. Mitochondrial tRNA import: Are there distinct mechanisms? Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- Scripture, J.B., Huber, P.W. Analysis of the binding of Xenopus ribosomal protein L5 to oocyte 5 S rRNA. The major determinants of recognition are located in helix III-loop C. J. Biol. Chem. 1995;270:27358–27365. doi: 10.1074/jbc.270.45.27358. [DOI] [PubMed] [Google Scholar]
- Sharma, M.R., Koc, E.C., Datta, P.P., Booth, T.M., Spremulli, L.L., Agrawal, R.K. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- Shi, H., O'Brien, C.A., Van Horn, D.J., Wolin, S.L. A misfolded form of 5S rRNA is complexed with the Ro and La autoantigens. RNA. 1996;2:769–784. [PMC free article] [PubMed] [Google Scholar]
- Smith, M.W., Meskauskas, A., Wang, P., Sergiev, P.V., Dinman, J.D. Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae . Mol. Cell. Biol. 2001;21:8264–8275. doi: 10.1128/MCB.21.24.8264-8275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, J.A., Berg, C., Hendrick, J.P., La Branche-Chabot, H., Metspalu, A., Rinke, J., Yario, T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, M., Barciszewska, M.Z., Erdmann, V.A., Barciszewski, J. An analysis of G-U base pair occurrence in eukaryotic 5S rRNAs. Mol. Biol. Evol. 2000;17:1194–1198. doi: 10.1093/oxfordjournals.molbev.a026402. [DOI] [PubMed] [Google Scholar]
- Szymanski, M., Barciszewska, M.Z., Erdmann, V.A., Barciszewski, J. 5 S rRNA: Structure and interactions. Biochem. J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov, I., Entelis, N., Martin, R. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov, I., Kamenski, P., Kolesnikova, O., Karicheva, O., Martin, R.P., Krasheninnikov, I.A., Entelis, N. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell Cycle. 2007;6:2473–2477. doi: 10.4161/cc.6.20.4783. [DOI] [PubMed] [Google Scholar]
- Taylor, R.W., Chinnery, P.F., Turnbull, D.M., Lightowlers, R.N. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nat. Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- White, S.A., Li, H. Yeast ribosomal protein L32 recognizes an RNA G:U juxtaposition. RNA. 1996;2:226–234. [PMC free article] [PubMed] [Google Scholar]
- White, S.A., Nilges, M., Huang, A., Brunger, A.T., Moore, P.B. NMR analysis of helix I from the 5S RNA of Escherichia coli . Biochemistry. 1992;31:1610–1621. doi: 10.1021/bi00121a005. [DOI] [PubMed] [Google Scholar]
- Wimberly, B., Varani, G., Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993;32:1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Yoshionari, S., Koike, T., Yokogawa, T., Nishikawa, K., Ueda, T., Miura, K., Watanabe, K. Existence of nuclear-encoded 5S-rRNA in bovine mitochondria. FEBS Lett. 1994;338:137–142. doi: 10.1016/0014-5793(94)80351-x. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]