Abstract
Many alternative splice events result in subtle mRNA changes, and most of them occur at short-distance tandem donor and acceptor sites. The splicing mechanism of such tandem sites likely involves the stochastic selection of either splice site. While tandem splice events are frequent, it is unknown how many are functionally important. Here, we use phylogenetic conservation to address this question, focusing on tandems with a distance of 3–9 nucleotides. We show that previous contradicting results on whether alternative or constitutive tandem motifs are more conserved between species can be explained by a statistical paradox (Simpson's paradox). Applying methods that take biases into account, we found higher conservation of alternative tandems in mouse, dog, and even chicken, zebrafish, and Fugu genomes. We estimated a lower bound for the number of alternative sites that are under purifying (negative) selection. While the absolute number of conserved tandem motifs decreases with the evolutionary distance, the fraction under selection increases. Interestingly, a number of frameshifting tandems are under selection, suggesting a role in regulating mRNA and protein levels via nonsense-mediated decay (NMD). An analysis of the intronic flanks shows that purifying selection also acts on the intronic sequence. We propose that stochastic splice site selection can be an advantageous mechanism that allows constant splice variant ratios in situations where a deviation in this ratio is deleterious.
Keywords: purifying selection, subtle alternative splicing, tandem splice site, comparative genome analysis, Simpson's paradox
INTRODUCTION
Alternative splicing is a widespread mechanism to produce transcript and protein diversity in animals and plants (Campbell et al. 2006; Tress et al. 2007). Detailed studies revealed many examples where the existence and regulation of alternative splice variants are crucial for cellular functions. For example, alternative splice variants have important roles in the nervous (Ule et al. 2005; Licatalosi and Darnell 2006) and immune (Lynch 2004) systems and during sex determination in Drosophila (Black 2003). Moreover, human and mouse splicing factor genes extensively produce nonfunctional splice forms, which provides a potential mechanism for autoregulating the protein level (Stoilov et al. 2004; Lareau et al. 2007; Ni et al. 2007). Misregulation of alternative splicing is a frequent cause of disease (Pagani and Baralle 2004), and the human SFRS1 gene encoding the splicing factor ASF/SF2 was shown to be a proto-oncogene (Karni et al. 2007).
Despite these facts, the general extent of functional alternative splicing is unknown. Some splice forms such as the skipping of exon 12 of human CFTR were described to have no functional advantage (Raponi et al. 2007), and the tissue-specific inclusion of exon 8 of mouse Psap shows no phenotypic differences in a knockout mouse lacking this exon (Cohen et al. 2005). Furthermore, about one-third of the human alternative splice events lead to an early stop codon, thus yielding truncated proteins and/or subjecting the mRNA to the nonsense-mediated decay (NMD) pathway (Lewis et al. 2003). Apart from their potential to regulate the protein level by reducing the level of transcripts encoding the full-length protein, their function is often not obvious.
To assess function, one usually considers sequence conservation or the conservation of an event as an important criterion that implies purifying (negative) selection because deviations confer a disadvantage to the organism. Indeed, conserved exon skipping events have a tendency to preserve the protein reading frame (Resch et al. 2004; Sorek et al. 2004; Yeo et al. 2005). These exons and their intronic flanks also exhibit an increased sequence conservation (Sorek and Ast 2003). Furthermore, tissue-specific exon skipping is associated with conserved exons and with reading frame preservation (Xing and Lee 2005). However, genome-wide studies found only a small percentage (∼10%–20%) of exon skipping events to be conserved between human and mouse, with most alternative exons being either skipped in only one species or occurring only in one genome (Modrek and Lee 2003; Sorek and Ast 2003; Pan et al. 2004; Yeo et al. 2005). Thus, while alternative splicing is undoubtedly frequent, most of the splice events seem to have no functional role that is conserved in evolution.
Apart from exon skipping, numerous human and mouse alternative splice events occur at alternative donor and acceptor splice sites. The majority of these splice site pairs are in close proximity (Clark and Thanaraj 2002; Zavolan et al. 2003; Sugnet et al. 2004), thus leading to subtle mRNA changes. In this study, we analyze pairs of donor or acceptor sites that are 3–9 nucleotides (nt) apart (Δ3–Δ9 nt) and use the term “tandem sites” to denote these splice site pairs (Fig. 1). The most frequent of these subtle events is alternative splicing at NAGNAG acceptors (Zavolan et al. 2003; Hiller et al. 2004; Sugnet et al. 2004). At the donor site, Δ4 tandem splice sites are most prominent as dictated by the donor consensus sequence (Dou et al. 2006; Ermakova et al. 2007). For most tandem sites, it is likely that their underlying alternative splicing mechanism is based on a stochastic selection of either splice site, also called “noisy splicing” (Chern et al. 2006). A recent study showed that the region between the branch point and the acceptor has a strong influence on the splicing ratio of alternatively spliced NAGNAG sites (Tsai et al. 2007).
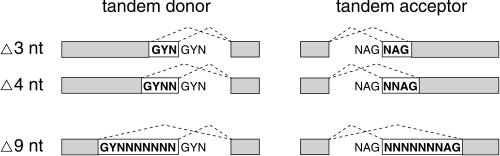
FIGURE 1.
Schematic representation of the tandem sites analyzed in this study. (Boxes) Exons; (dashed lines) splice events; (boldface) the variable exonic parts; (GYNGYN and NAGNAG) tandem sites with a distance of 3 nt; (Δ4–Δ9) tandem donors and acceptors that are 4–9 nt apart, respectively; (N) A, C, G, or T; (Y) C or T.
Targeted experimental studies have revealed functional roles for tandem splice events. For example, conserved tandem acceptors in human and mouse transcription factor genes (NAGNAG acceptors in PAX3 and PAX7, Δ6 acceptor in IRF2) result in protein isoforms that differ in the ability to activate transcription (Vogan et al. 1996; Koenig Merediz et al. 2000). Conserved Δ6 donors lead to protein variants of human ALDH18A1 that are insensitive to ornithine inhibition (Hu et al. 1999) and produce protein isoforms of mouse Fgfr1 that are unable to bind FRS2 and thus cannot activate the Ras/MAPK signaling pathway (Burgar et al. 2002). Furthermore, a splice event at a conserved Δ6 donor in human EDA tightly controls binding specificity by remodeling the properties of the receptor binding site, such that the longer protein binds only to the EDAR receptor, while the shorter variant binds only to the XEDAR receptor (Yan et al. 2000; Hymowitz et al. 2003). Another example is the Δ9 donor of human WT1 exon 9 that leads to the insertion of three amino acids (KTS). Both splice forms have distinct transcriptional regulation properties, hetero- and homozygous mouse mutants lacking one of the two splice forms show severe defects in kidney development and function (Hammes et al. 2001), and a mutation in this donor motif leads to Frasier syndrome in humans (Barbaux et al. 1997).
While these individual studies demonstrate that several of these subtle splice events are functionally important, the general extent remains unknown. Moreover, there is a discussion whether tandem sites that are alternatively spliced are better conserved in evolution than those that are constitutively spliced (Hiller et al. 2006c) since conflicting results were published for NAGNAG acceptors (Hiller et al. 2004; Chern et al. 2006). As alternative and constitutive NAGNAG sites have different preferences for specific NAGNAG motifs (Hiller et al. 2004; Akerman and Mandel-Gutfreund 2006), we considered the possibility that the comparison of two heterogeneous groups caused a statistical paradox, which is often called Simpson's paradox. This paradox is frequently encountered in biomedical studies (Julious and Mullee 1994) and describes a situation in which a trend observed between two groups is reversed when the two groups are split into several subgroups (Simpson 1951). A well-known example of Simpson's paradox is described in Bickel et al. (1975) and refers to university admission data. In this case, the overall admission rates indicated a significant bias against female applications, while investigating all departments individually provided evidence for the opposite—a bias in favor of female applicants. As described in Bickel et al. (1975), the explanation of this apparently paradox is: “The proportion of women applicants tends to be high in departments that are hard to get into and low in those that are easy to get into.”
Here, we show that previous conflicting conclusions for the evolutionary conservation of NAGNAG acceptors (Hiller et al. 2004; Chern et al. 2006) arose from Simpson's paradox caused by substantial conservation differences between specific NAGNAG motifs. Controlling for biases, we found that alternatively spliced NAGNAG acceptors are significantly more conserved than those that are constitutively spliced. We extended the analysis to human tandem donor and acceptor sites that are up to 9 nt apart and estimated a lower bound for the fraction of tandem sites being under purifying selection, and thus expected to have an evolutionarily advantageous phenotype.
RESULTS
Conservation of human NAGNAG acceptors differs between the NAGNAG motifs
First, we analyzed the sequence conservation of human NAGNAG acceptor motifs that are located within the protein coding sequence (CDS). Our data set consists of 1597 confirmed (at least one mRNA/EST indicates splicing at the upstream and at least one mRNA/EST splicing at the downstream acceptor) and 7452 unconfirmed (currently available mRNA/EST data indicate no alternative splicing) NAGNAG acceptors (Hiller et al. 2007). We tested the pairwise conservation of human NAGNAG acceptors over different evolutionary distances: rhesus (∼23 million years ago [mya] since split of the common ancestor), mouse (∼90 mya), dog (∼92 mya), chicken (∼310 mya), and zebrafish and Fugu (∼450 mya) (Ureta-Vidal et al. 2003). Although the close distance human–rhesus might limit the power to detect conservation differences, we include rhesus to cover a large spectrum of evolutionary distances.
We used very stringent criteria to define conservation between two NAGNAG tandems to increase the likelihood that an orthologous tandem site, which is considered to be conserved, has the same splicing pattern (alternative or constitutive splicing) in the other species. Previous observations suggest that the tandem splice site motif is the strongest factor determining the splicing pattern (Chern et al. 2006; Hiller et al. 2006a). For this reason, we considered a human NAGNAG as conserved in another species if the orthologous acceptor pattern is identical to the human NAGNAG motif, except for the first N, where we allow variation between C and T. We allow this C/T variation since pyrimidines are the most frequent nucleotides at the −3 position of standard acceptors (Abril et al. 2005) and are not expected to affect the splicing efficiency significantly.
We first performed a global analysis and compared the conservation of all confirmed and all unconfirmed human NAGNAG acceptors in each of the other species. We found that unconfirmed NAGNAG acceptors are more conserved than confirmed ones in the pairwise comparisons (Fig. 2, left parts; Table 1), as previously reported for human and mouse (Chern et al. 2006). The differences are significant in a Fisher's exact test (P-values <0.01 for all pairwise comparisons). For this and the following tests, we also computed a standard measurement in biostatistics, the odds ratio (OR). The interpretation of an OR is as follows: an OR > 1 indicates higher conservation for confirmed NAGNAG tandems, an OR < 1 indicates higher conservation for unconfirmed tandems, and an OR = 1 indicates no differences between confirmed and unconfirmed tandems. In the global test, we observed ORs < 1 (Table 1), indicating higher conservation for unconfirmed ones.
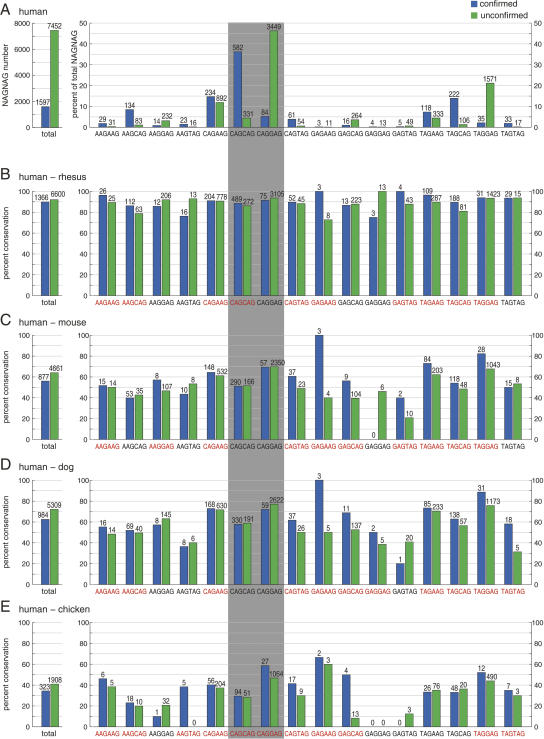
FIGURE 2.
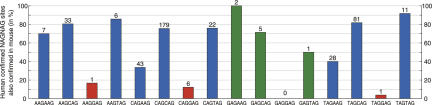
Overall and individual conservation of human confirmed and unconfirmed NAGNAG motifs. (A, left panel) Total number of confirmed and unconfirmed NAGNAG acceptors; (right panel) the fraction of individual motifs of the total number of confirmed and unconfirmed human NAGNAG acceptors. The numbers above the bars are absolute numbers of confirmed and unconfirmed NAGNAG acceptors. (B–E) The conservation of human NAGNAG acceptor motifs in (B) rhesus, (C) mouse, (D) dog, and (E) chicken was analyzed in a (left panel) global and (right panel) motif-specific comparison. As expected, the overall conservation drops with increased evolutionary distance from rhesus to chicken. A human NAGNAG acceptor is considered to be conserved if it is identical to the orthologous mouse acceptor motif except for an allowed variation between C and T at the first position. (Red) Motifs with a higher conservation for confirmed tandem acceptors. Note that all NAGNAG acceptors for which no pairwise alignment block with the respective species was found were discarded in the conservation analysis. The numbers above the bars are the absolute numbers of conserved NAGNAG sites.
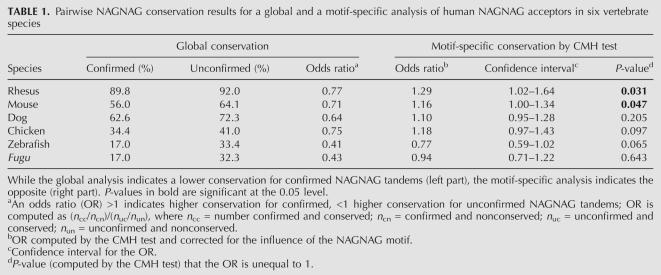
TABLE 1.
Pairwise NAGNAG conservation results for a global and a motif-specific analysis of human NAGNAG acceptors in six vertebrate species
Next, we compared the conservation between confirmed and unconfirmed tandems for each of the 16 NAGNAG motifs individually. Strikingly, this motif-specific comparison revealed for 10 of the 16 motifs a higher conservation level for confirmed NAGNAG acceptors in mouse (Fig. 2C). Similarly, confirmed NAGNAG acceptors are more conserved for 10 motifs in rhesus and for 11 in dog and chicken (Fig. 2B,D,E). This apparently contradicts the results of the global analysis. As evident from Figure 2, motifs differ considerably in their overall conservation levels. For example, 51% of all CAGCAG but 70% of all CAGGAG motifs are conserved in mouse.
We hypothesized that these substantial differences in the conservation levels are caused by constraints on the acceptor splice site consensus YAG | G (| indicates the intron–exon boundary; Y = C or T). While a G at the 5′ exon end conforms with the acceptor consensus sequence, a C at this position leads to a weaker acceptor. Thus, CAGCAG acceptors without functional importance are more likely to accept mutations of the unfavored C at position +4 in this motif, while CAGGAG acceptors are less likely to allow mutations of the preferred G at +4. To further test this, we grouped NAGNAG acceptors according to the nucleotide at the second N position and determined the overall conservation. We found that NAGGAG (68.1% conserved) and NAGAAG (62.3%) tandems are generally more conserved than NAGCAG (49.5%) and NAGTAG (56.6%) tandems, in agreement with the preferred 5′-most exon nucleotides, which are G and A followed by C and T (Abril et al. 2005). Moreover, GAG at the 5′ exon end might also be more constrained than CAG, since GAG is more often a core of the splicing enhancer motif identified in Stadler et al. (2006) than CAG (14% versus 12%). Thus, we identified the individual NAGNAG motif as a confounding variable that considerably affects the conservation levels. In such a situation, a global calculation can lead to wrong conclusions.
Higher conservation for confirmed versus unconfirmed human NAGNAG acceptors in rhesus, mouse, dog, and chicken
An unbiased analysis of the conservation level has to correct for the influence of the confounding variable NAGNAG motif. To this end, we used the Cochran–Mantel–Haenszel (CMH) test, which is an extension of the χ2 test and commonly used in such a situation. The CMH test estimates an OR that is corrected for the influence of the NAGNAG motif. As shown in the right part of Table 1, using the CMH test, we observed ORs > 1 for rhesus, mouse, dog, and chicken. This indicates a higher conservation for confirmed NAGNAG acceptors. However, in zebrafish and Fugu, confirmed NAGNAG tandems have a lower conservation even after correcting for the motif (Table 1), and this holds for the following analyses as well.
The contradictory results of the global and the motif-specific conservation analysis are an example of the above-described Simpson's paradox (Simpson 1951). Here, the paradox occurs since (1) the conservation level (Fig. 2B–E), and (2) the distribution of confirmed and unconfirmed NAGNAG acceptors (Fig. 2A) vary greatly among the different motifs. The most dramatic difference is caused by the weakly conserved CAGCAG motif (that makes up 36% of the confirmed but only 4% of the unconfirmed NAGNAG acceptors) and the strongly conserved motif CAGGAG (that makes up only 5% of the confirmed but 46% of the unconfirmed NAGNAG acceptors), shaded gray in Figure 2. Thus, confirmed NAGNAG acceptors are enriched in weakly conserved motifs, while unconfirmed ones are enriched in highly conserved motifs (analogous to the above example of Simpson's paradox). This bias causes the misleading result of a lower conservation of confirmed versus unconfirmed tandem acceptors in the global analysis. Moreover, this bias explains previous conflicting conclusions because the data set used in Hiller et al. (2004) (“intronic extra AGs”) (see Supplementary Note in Hiller et al. 2004) contains virtually none of the strongly conserved NAGGAG motifs, while NAGGAG motifs make up a large fraction of all unconfirmed NAGNAG sites that were analyzed in Chern et al. (2006).
The unequal NAGNAG motif distribution was observed in previous studies (Hiller et al. 2004; Akerman and Mandel-Gutfreund 2006) that showed that >90% of the alternative NAGNAG acceptors have an HAGHAG motif (H = A, C, T), while those tandems having a GAG are rarely alternatively spliced (Hiller et al. 2006b). Furthermore, standard acceptors are mostly CAG or TAG, with AAG and especially GAG being rare. This reflects the binding affinity of the U2AF35 splicing factor (Wu et al. 1999). Thus, the splicing machinery may select either acceptor in an HAGHAG motif, resulting in alternative splicing (Chern et al. 2006).
Estimating the number of human NAGNAG acceptors that are under purifying selection
Higher conservation of confirmed NAGNAG tandems indicates that a certain fraction is under purifying selection, which prevents the destruction of the NAGNAG motif in the course of evolution. Since the CMH test does not estimate how many confirmed tandem acceptors are under selection, we developed two simulations to answer this question. We used unconfirmed NAGNAG tandems to estimate the expected or background conservation that reflects evolutionary constraints to preserve a functional acceptor and the coding sequence that overlaps the NAGNAG motif. The number of confirmed and conserved tandem acceptors that exceed the expected conserved number is considered to be subject to purifying selection, which preserves the alternative splice event. In the following, we use fs for the fraction of confirmed and conserved tandem splice sites estimated to be under purifying selection.
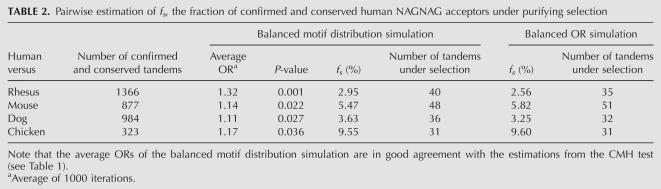
Applying the first simulation (called the “balanced motif distribution”; see Materials and Methods) to the rhesus, mouse, dog, and chicken conservation data, we estimate that between 2.95% (rhesus) and 9.55% (chicken) of the confirmed and conserved NAGNAG acceptors are under purifying selection (Table 2). Furthermore, the P-values are significant at the 0.05 level for all four comparisons. To further support this estimation, we applied another simulation (called the “balanced OR”; see Materials and Methods) and found highly similar results (Table 2).
TABLE 2.
Pairwise estimation of fs, the fraction of confirmed and conserved human NAGNAG acceptors under purifying selection
To extend the pairwise approach, we considered as a four-way conserved NAGNAG acceptor a human tandem that is conserved in rhesus, mouse, dog, and chicken (237 confirmed and 1360 unconfirmed four-way conserved sites) (Supplemental Table 1). NAGNAG sites that lack conservation in one or more species are considered as nonconserved in this test (1351 confirmed and 6101 unconfirmed sites). We found that confirmed NAGNAG acceptors have a significantly higher four-way conservation (CMH test: OR = 1.28, P = 0.014) than unconfirmed ones. The balanced motif distribution simulation estimates an fs of 20.3% (OR = 1.31, P < 0.0001), indicating that 48 of the 237 four-way conserved tandems are under selection. Thus, four-way conserved NAGNAG acceptors have a stronger tendency to be under purifying selection.
As pointed out above, CAGCAG is the motif with the highest number of confirmed human tandem acceptors. Confirmed CAGCAG acceptors show a slightly higher conservation than unconfirmed CAGCAG sites in rhesus and chicken but not in mouse and dog (Fig. 2B–E). To further investigate the conservation of this motif, we considered four-way conserved CAGCAG sites and found that human confirmed CAGCAG acceptors have a 3% higher four-way conservation level than unconfirmed ones, suggesting that 17 CAGCAG sites are under selection.
Finally, we analyzed conservation of NAGNAG tandems located in the untranslated region (UTR). In contrast to NAGNAG tandems in the CDS, we found no indication that UTR tandems are under selection (data not shown).
Conservation of human NAGNAG alternative splicing in mouse
Confirmed NAGNAG acceptor motifs are likely to be under purifying selection because the alternative splice event provides an advantageous phenotype. Therefore, we considered conservation of the alternative splice event in mouse. Of the human confirmed NAGNAG acceptors that are conserved in mouse, we found that 59% of the orthologous mouse NAGNAG acceptors are alternatively spliced in mouse. This shows that conservation of the NAGNAG motif is associated with conservation of the splice event. In particular, confirmed NAGNAG sites that have no GAG acceptor have a high chance to be confirmed in mouse (Fig. 3), presumably because their splice variant ratio is often rather balanced so that few ESTs can be sufficient to detect alternative splicing in mouse. As the mouse transcript coverage is only 62% of the human coverage (∼5 million mouse ESTs and mRNAs versus ∼8 million for human), our finding that 59% of the alternative splice events are conserved is a lower bound.
FIGURE 3.
Conservation of alternative NAGNAG splice events in human and mouse. Each bar is the percentage of human confirmed NAGNAG acceptors that is also confirmed in mouse, split into the 16 NAGNAG patterns. Absolute numbers are given above the bars. Only those human NAGNAG sites that are conserved in mouse are considered. (Blue) Tandem acceptors with the pattern HAGHAG (H = A, C, T); (red) acceptors with the pattern HAGGAG; (green) acceptors with the pattern GAGHAG. Note that there is no human confirmed GAGGAG acceptor that is conserved, hence none can be confirmed in mouse.
Human tandem donors and acceptors with up to Δ9 nt under purifying selection
Next, we extended our conservation analysis to human tandem donors with Δ3–Δ9 nt and tandem acceptors with Δ4–Δ9 nt (Fig. 1) that are located within the CDS. As for NAGNAG acceptors, we found that constraints on the donor and acceptor consensus are one reason for the different conservation levels of individual tandem motifs (Materials and Methods). Furthermore, the motif distribution differs between confirmed and unconfirmed tandems, probably because some tandem motifs allow selection of either splice site by the spliceosome, while in other tandems the stronger splice site is used exclusively (Chern et al. 2006). To exclude potential biases, we used the balanced motif distribution simulation to assess fs in the following.
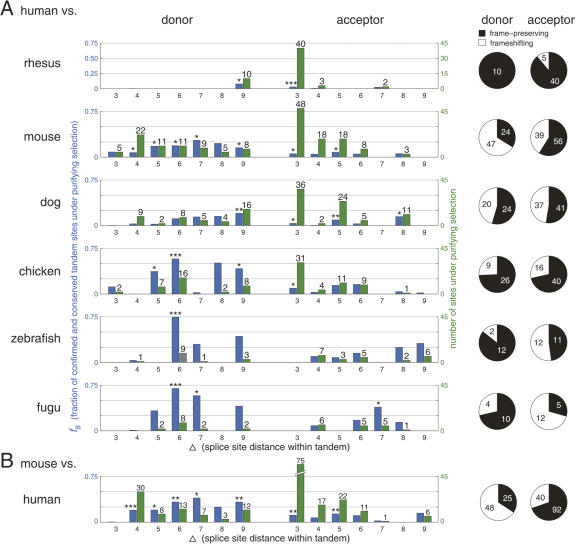
In contrast to NAGNAG acceptors, confirmed GYNGYN donors (Hiller et al. 2006b) are not conserved significantly more than unconfirmed ones. Only the mouse and chicken comparisons indicate that a few confirmed GYNGYN tandems might be under selection (Fig. 4A, left). However, conserved tandem donors with larger splice site distances contain more sites under purifying selection. We observed that fs increases with the evolutionary distance and that frame-preserving donor sites are preferentially under selection for large evolutionary distances (Fig. 4A, right). Strikingly, for the human–Fugu comparison, fs increases to 0.75 for Δ6 donors, indicating that three-quarters of the confirmed tandem donors that are conserved over ∼450 mya are under purifying selection. Indeed, these cases include the functionally important tandem donors in human EDA (Yan et al. 2000) and ALDH18A1 (Hu et al. 1999). Apart from frame-preserving sites, frameshifting tandem donors contain sites under selection, even in the human–fish comparison.
FIGURE 4.
Tandem donor and acceptor sites with Δ3–Δ9 nt under purifying selection. (A) Analyzing the conservation of confirmed human tandem sites in six vertebrate species. (Left chart, blue bars) fs; (green bars) the number of confirmed and conserved tandems under purifying selection (numbers >0 are given above the bars). (Right pie charts) The fraction of all frame-preserving and all frameshifting tandems that are under selection. (B) Analyzing the conservation of confirmed mouse tandem sites in human. P-values are determined by repeatedly testing the null hypothesis that confirmed tandems are conserved according to the motif-specific background conservation level (Materials and Methods). Significance is indicated as (***) P < 0.01; (**) P < 0.01; (*) P < 0.05.
While the fs estimations for individual Δ3–Δ9 acceptors are often lower than the respective donor classes and rarely significant, the absolute number under selection is mostly higher due to a larger number of confirmed tandem acceptors (Fig. 4A, left). In particular, NAGNAG sites contribute the biggest portion. As for tandem donors, fs increases for larger evolutionary distances. In general, Δ3–Δ6 tandems contain more sites under selection than Δ7–Δ9 tandems. The human–rhesus comparison reveals selection for only a few tandem site classes, which is presumably due to the close evolutionary distance that leads to a high background conservation rate.
Assessing purifying selection for mouse tandem splice sites
Up to now, we have assessed fs for human confirmed tandem sites by pairwise comparison with other species. Apart from human, only the mouse genome has a transcript coverage (∼5 million ESTs) that allows us to create sufficiently large sets of confirmed tandem sites. In contrast to mouse, many human ESTs are sampled from tumor tissue, and this might affect the above conclusions. To provide an independent estimation, we used the balanced motif distribution simulation to estimate fs for confirmed mouse tandem sites by analyzing the conservation in human. Noteworthy, a high fraction (70%) of the mouse confirmed and conserved NAGNAG sites is also alternatively spliced in human.
Consistently, the estimated number of mouse confirmed tandem sites under purifying selection is similar to the estimations for human confirmed sites (Fig. 4, cf. B and A). The mouse-based analysis estimates an even higher number of Δ4 donors, NAGNAG, and Δ9 acceptors to be under selection. It should be noted that mouse confirmed CAGCAG sites have a 3% higher conservation level than unconfirmed ones, suggesting that 13 confirmed mouse CAGCAG sites are under selection. This is in agreement with the estimation for four-way conserved human CAGCAG sites (see above).
Conservation of the intronic flanks
To provide further support that conservation of the tandem motif implicates conservation of the splicing pattern (alternative or constitutive splicing), we determined the conservation of the intronic flanking regions. Previous studies showed that exons, which are alternatively spliced in human and mouse, are flanked by highly conserved intronic regions (Sorek and Ast 2003; Yeo et al. 2005), and the same was observed for human and mouse confirmed GYNGYN and NAGNAG tandems (Akerman and Mandel-Gutfreund 2006; Hiller et al. 2006b). Thus, high intronic flank conservation is a hallmark of conserved alternative splice events. To abstract from pairwise conservation (often human–mouse), we used the PhastCons conservation scores, which are based on multiple genome sequences and a given phylogeny (Siepel et al. 2005).
Analyzing the average per-position conservation score for the 30-nt intronic flank of all tandems with Δ3–Δ9 nt, we found that confirmed and mouse-conserved human tandems have generally the highest intronic flank conservation, indicating that purifying selection also acts on the intronic context. In particular, Δ4, Δ6, Δ7, and Δ9 donors and Δ3 and Δ5 acceptors have significantly higher intron conservation (Supplemental Figs. 1,2), and this coincides with the tandem classes that have a significant fraction under purifying selection (Fig. 4A, mouse). These observations indicate that confirmed and conserved human tandem sites are associated with alternative splice events in other species.
DISCUSSION
Given the abundance of alternative splicing at tandem sites, it is of interest to find out what fraction of these events is biologically meaningful. Apart from experimental investigations (Condorelli et al. 1994; Vogan et al. 1996; Hu et al. 1999; Koenig Merediz et al. 2000; Yan et al. 2000; Joyce-Brady et al. 2001; Burgar et al. 2002; Tadokoro et al. 2005), another approach to address this question is to estimate the fraction of tandem sites under purifying selection. Here, we show that the sequence conservation level differs between tandem motifs due to constraints on the splice site consensus and possibly on splicing enhancer motifs as well as on the coding sequence. Together with differences in the tandem motif distribution, this bias seriously affects the conclusion whether confirmed tandems are more conserved than unconfirmed ones. Applying methods that control for this bias, we estimate the fraction of tandem sites under purifying selection.
Interestingly, we found that frame-preserving and frameshifting tandems are under selection. Frameshifting tandem splice events can have a functional role by creating truncated proteins as exemplified for a Δ4 acceptor in the last intron of the zebrafish pou5f1 gene (Takeda et al. 1994) or by subjecting the mRNA to the NMD pathway. In agreement with this, at least 21% of the human–mouse conserved exon skipping events lead to an NMD-inducing transcript, suggesting a potential role in regulating the protein level (Baek and Green 2005). Furthermore, NMD-inducing exon skipping and intron retention events in splicing factor genes are likely to be important because these alternative regions overlap highly or even ultraconserved elements (Lareau et al. 2007; Ni et al. 2007). It is noteworthy that experimental studies also revealed functional differences for tandem sites that lack deep evolutionary conservation. For example, the CAGCAG acceptor of human IGF1R exon 14, which leads to changes in the signaling activity and the internalization rate of the receptor (Condorelli et al. 1994), is not conserved in mouse, rat, dog, and chicken. Thus, similarly to the predicted functional roles of species-specific alternative splice events occurring at conserved exons (Pan et al. 2005), species- or lineage-specific alternative splice events at tandem sites may have functional consequences.
It is important to note that our estimation of the fraction of confirmed tandem sites under selection (Fig. 4) is a lower bound. A major reason is that our set of unconfirmed tandems is likely to be contaminated with sites that are alternatively spliced but currently lack transcript confirmation. To provide a rough estimate of how many unconfirmed NAGNAG acceptors might be alternatively spliced, we determined how many of those have a local context of 3 nt upstream and downstream (N3NAGNAGN3) that is identical to a confirmed NAGNAG. As the local sequence context primarily determines if a NAGNAG is alternatively spliced (Chern et al. 2006), these unconfirmed NAGNAG sites are expected to allow alternative splicing. We found that 10.5% of the unconfirmed human NAGNAG acceptors have a local context identical to a confirmed tandem. Remarkably, this fraction increases to 26% for those unconfirmed human NAGNAG acceptors with a C or T at the N-positions (YAGYAG), and these unconfirmed sites have a fivefold lower EST coverage than the confirmed ones. Requiring the identity of only 2 nt upstream and downstream (N2NAGNAGN2), 72% of the unconfirmed YAGYAG sites have a confirmed counterpart. Thus, a substantial fraction of the unconfirmed NAGNAG acceptors is likely to be alternatively spliced, although this is not indicated by current transcript data. Therefore, the background conservation level computed from unconfirmed tandems is likely to be overestimated, and consequently the real fraction under selection is underestimated. In particular, frameshifting tandem splice sites are expected to contain many unconfirmed but alternatively spliced cases, since NMD removes the alternative transcripts (Baek and Green 2005; Chern et al. 2006; Lareau et al. 2007; Ni et al. 2007). If the down-regulation of the mRNA encoding the full-length protein has functional relevance, unconfirmed but alternatively spliced tandem sites are probably conserved in evolution, which, in turn, leads to an overestimated background conservation level.
Two confirmed NAGNAG acceptors are located in ultraconserved elements (defined as at least 200-nt-long regions that are identical between human, mouse, and rat) (Bejerano et al. 2004). The first is the CAGCAG in PAX2 exon 2, which leads to a ProGly-to-Arg exchange immediately upstream of the Paired box domain. Interestingly, NAGNAG splice events within the Paired box domain in PAX3 and PAX7 affect DNA binding (Vogan et al. 1996). The second case is a CAGAAG in CLK4 exon 4 that leads to the insertion/deletion of a Lys upstream of the protein kinase domain. These two NAGNAG acceptors are also identical between human and chicken. Both ultraconserved elements overlap a large region of the intron–exon boundary; thus it is unknown if purifying selection on the NAGNAG acceptor and its context was the driving force for these ultraconserved elements.
Although tissue- or cell-line-specific splicing has been observed at tandem acceptors (Koenig Merediz et al. 2000; Hiller et al. 2004; Xu et al. 2004; Tadokoro et al. 2005) and tandem donors (Hu et al. 1999; Yan et al. 2000), stochastic selection of either of the two splice sites likely explains alternative splicing at most tandems (Chern et al. 2006). Stochastic splice events are expected to yield similar splice variant ratios in different tissues, and this was observed in many cases (Vogan et al. 1996; Hammes et al. 2001; Burgar et al. 2002; Tadokoro et al. 2005; Hiller et al. 2006b). Noteworthy, stochastic splicing does not preclude functional importance of the alternative splice event (Hiller et al. 2006c). Especially in a situation where both protein isoforms are required ubiquitously, stochastic splice site selection based only on spliceosomal core components offers the advantage of producing the two variants nearly independent of other conditions that regulate alternative splicing. This is likely to be the case for the functionally relevant tandem sites in mouse Fgfr1 and human PAX3 and PAX7 (see Introduction) that produce a constant ratio of the two splice variants (Vogan et al. 1996; Burgar et al. 2002). Another striking example is the Δ9 donor of human WT1 (see Introduction). This tandem donor site as well as its flanking regions is perfectly conserved between vertebrates, and the two splice variants have distinct functional roles. The splice variant ratio is constant in human tissues and cell lines (Barbaux et al. 1997; Davies et al. 2000) as well as in mouse (Hammes et al. 2001) and in zebrafish (C. Englert, pers. comm.). A deviation in this ratio is highly deleterious and leads to pronounced phenotypes (Hammes et al. 2001). In this case, stochastic donor selection by the ubiquitously expressed U1 snRNP would be a probable mechanistic basis of the constant ratio. Similar to NAGNAG acceptors (Tsai et al. 2007), sequences in the intronic flank might be important for the ratio of the two donor sites, which would explain the high intronic conservation. Apart from tandem sites, a stochastic mechanism that controls splicing of 48 mutually exclusive exons in Drosophila DSCAM is essential for axon guidance and is conserved over 300 mya in the insect lineage (Graveley 2005).
While we provided quantitative evidence that a fraction of tandem sites is under purifying selection and thus functional, their identity remains unknown. We found that NAGNAG acceptors with a strong minor splice site are more conserved than those with a weak one, suggesting that the frequency of the alternative splice event might be important. Furthermore, deep conservation in several species such as four-way conserved tandems (Supplemental Table 1), conservation over large evolutionary distances (Supplemental Table 2), and high intronic flank conservation (Supplemental Figs. 1,2) might be reasonable criteria to select promising candidates for further experimental studies.
MATERIALS AND METHODS
Data sets
We downloaded from the UCSC Genome Browser (Kuhn et al. 2007) the human genome assembly (hg17, May 2004) as well as RefSeq annotation (refFlat.txt.gz, November 2006). We screened all splice sites for the presence of a tandem donor and acceptor Δ3–Δ9 motif. Donor sites without GT or GC and acceptors without AG intron termini were omitted. The RefSeq annotation of the open reading frame was used to decide if a tandem site affects the CDS. A tandem site was considered as confirmed if there is at least one EST/mRNA that matches the short and at least one EST/mRNA that matches the long transcript. For NAGNAG and GYNGYN tandems, we downloaded EST information from TassDB (Hiller et al. 2007). For Δ4–Δ9 tandem sites, we used BLAST against all ESTs and mRNAs to obtain confirmation for the putative alternative splice event. BLAST was done as described in Hiller et al. (2006b). The total size of the obtained confirmed and unconfirmed data sets is as follows: GYNGYN: 116 confirmed and 8031 unconfirmed; Δ4 donors: 595 and 97,539; Δ5: 161 and 27,254; Δ6: 161 and 40,262; Δ7: 89 and 33,329; Δ8: 63 and 31,501; Δ9: 160 and 34,793; NAGNAG acceptors: 1597 confirmed and 7452 unconfirmed; Δ4 acceptors: 603 and 8093; Δ5: 364 and 7912; Δ6: 266 and 11,754; Δ7: 118 and 12,917; Δ8: 100 and 11,338; Δ9: 156 and 14,040.
Conservation was detected by analyzing the genome-wide pairwise alignments downloaded from the UCSC Genome Browser (assemblies: human hg17, rhesus rheMac2, mouse mm7, dog canFam2, chicken galGal2, zebrafish danRer3, fugu fr1) using the genomic locus of the human tandem sites to select the respective alignment chain. This approach allows a highly accurate detection of true orthologous splice sites, since the alignment of the individual exons and their splice sites is embedded in the syntenic context of the UCSC whole-genome alignment. Furthermore, coding exons are a class of functional elements that can be reliably aligned between distant genomes (Thomas et al. 2003). Tandem sites, for which no alignment chain was found, were excluded from the pairwise analysis as it is not clear if the entire exon is missing in the other species, if the tandem site is contained in two different alignment chains, or if these cases are due to wrong alignments. It should be noted that considering these tandem sites as nonconserved leads to an even higher conservation difference in favor of confirmed sites.
PhastCons scores for alignments of 16 vertebrate genomes with the human hg17 assembly (phastCons17way) were downloaded from the UCSC Genome Browser.
Statistics
The odds ratio is defined as (n cc/n cn)/(n uc/n un), where n cc is the number of confirmed and conserved tandems, n cn is the number of confirmed and nonconserved tandems, n uc is the number of unconfirmed and conserved tandems, and n un is the number of unconfirmed and nonconserved tandems. Statistical tests (Fisher's exact test, CMH test, Wilcoxon rank-sum test) were performed using the R software (http://www.r-project.org/).
Different filtering and conservation criteria for NAGNAG acceptors
Given two orthologous NAGNAG acceptor motifs, we define “conservation” as an identical NAGNAG motif allowing only a variation between C and T at the first position. We tested if the conservation results for NAGNAG tandems were affected by this definition of conservation. Higher conservation for confirmed NAGNAG acceptors was consistently found if we (1) consider NAGNAG conservation as the conservation of both AGs allowing both Ns to vary (CMH test OR: 1.26 for rhesus, 1.15 for mouse, 1.06 for dog, 1.16 for chicken) (Supplemental Fig. 3); (2) consider NAGNAG conservation as the identity of the entire hexamer; i.e., conservation of both AGs and both Ns (CMH test OR: 1.11 for rhesus, 1.1 for mouse, 1.01 for dog, 1.18 for chicken).
Higher conservation for confirmed NAGNAG tandems was also observed if we (1) exclude unconfirmed NAGNAG tandems from the analysis that have only single EST support and hence cannot be confirmed (CMH test OR: 1.24 for rhesus, 1.15 for mouse, 1.1 for dog, 1.18 for chicken); (2) exclude confirmed NAGNAG tandems where the minor acceptor is supported by only a single EST (CMH test OR: 1.35 for rhesus, 1.11 for mouse, 1.07 for dog, 1.25 for chicken). As confirmed NAGNAG acceptors have an approximately twofold higher EST coverage, we tested if the overall EST coverage affects our results. Splitting all confirmed and unconfirmed NAGNAG tandems into those with at most 10 and at least 10 ESTs, we found a higher conservation for confirmed NAGNAG sites in both groups except for dog (CMH test OR: 1.64 for at most 10 ESTs and 1.05 for at least 10 ESTs for rhesus, 1.23 and 1.04 for mouse, 1.08 and 0.97 for dog, 1.10 and 1.12 for chicken).
We also found higher conservation for confirmed NAGNAG acceptors, when we restrict the analysis only to those tandems that contain no GAG site (CMH test OR: 1.39 for rhesus, 1.12 for mouse, 1.1 for dog, 1.1 for chicken). Consistently, restricting the analysis only to those NAGNAG sites that have a C or T at both N positions, we also found higher conservation for confirmed ones (CMH test OR: 1.39 for rhesus, 1.07 for mouse, 1.11 for dog, 1.07 for chicken).
Balanced motif distribution simulation for NAGNAG acceptors
The basic idea for the balanced motif distribution simulation is that Simpson's paradox cannot occur if the distribution of the 16 motifs is equal between confirmed and unconfirmed NAGNAG tandems. To correct the unequal motif distribution, we did the following. For each NAGNAG motif, we constructed two lists containing the confirmed and unconfirmed tandems. From the list with the higher entry number, we randomly removed entries so that the entry number in this list equals the number in the other list. This procedure was repeated for all splice site motifs. Then, we combined all confirmed and unconfirmed lists, counted the total number of conserved confirmed and unconfirmed tandems, and determined the OR. Note that after correcting the unequal motif distribution (Supplemental Fig. 4), a global analysis provides a fair comparison; thus we can directly determine how many confirmed tandem acceptors are under purifying selection. The whole procedure was repeated 1000 times. Finally, we computed the average of the 1000 odds ratios and the difference between the average number of conserved confirmed and conserved unconfirmed tandems. This difference divided by the number of conserved and confirmed tandems is fs. We also tested bootstrapping (allowing a single tandem site to be selected more than once in one iteration) and found virtually identical results (data not shown).
Additionally, we computed a P-value by repeatedly testing the null hypothesis that confirmed tandems are conserved according to the motif-specific background conservation level. To this end, we used the motif-specific percentage p of conserved unconfirmed NAGNAG acceptors as the background conservation level. Let n be the number of confirmed NAGNAG acceptors with a given motif. Then, we generated n random numbers and counted as m the number of cases which are ≤p. The interval [0–p] represents the conserved part of the background, and the interval (p–1] represents the nonconserved part. For example, the background conservation level in mouse for AAGAAG is 50% (Fig. 2C). Since there are 29 confirmed AAGAAG acceptors in our data set, we generated 29 random numbers and counted how many of those are ≤0.5. We repeated that for all motifs and determined the total sum of motif-specific m. The P-value is the fraction of 10,000 performed iterations where this sum is equal to or higher than the actual number of confirmed and conserved tandems. This P-value is independent of the CMH test.
Balanced OR simulation
The rationale for the balanced OR simulation is that the CMH test should estimate an OR of 1 if there is no difference in the conservation. Thus, we determined which fraction of the confirmed and conserved tandems has to be artificially considered as nonconserved to get an OR of 1; this fraction is the estimation for fs. Specifically, for a given fraction f, we changed the conservation status of f · n randomly selected confirmed NAGNAG acceptors from conserved to nonconserved, where n is the total number of confirmed and conserved tandems. Then, we computed the OR using the CMH test. For a given f, this was repeated 1000 times, and we determined the average OR and the standard deviation. If f = fs, we expect that the OR = 1. Starting from f = 0, we increased f to obtain average ORs well below 1. The highest f for which the average OR is still >1 is taken as an estimate of fs (Supplemental Fig. 5).
The balanced OR simulation was only performed for NAGNAG acceptors as the number of motifs increases for GYNGYN and Δ4–Δ9 tandem sites, while the number of confirmed sites decreases. In a situation with many motifs mostly having a low case number, the CMH test cannot reliably estimate the OR.
Definition of conservation of two tandem sites
With increasing distance between the two acceptors of a confirmed tandem, the sequence between the two AGs has a tendency to contain pyrimidines (Dou et al. 2006), probably reflecting the requirement for a second polypyrimidine tract. Furthermore, the nucleotide downstream from the AGs, which is frequently a G for confirmed tandems, influences the splicing pattern (Dou et al. 2006). To account for these observations, we required identity of the +4 position (in the following, numbering starts at the first position in a Δ3–Δ9 acceptor or donor motif). All other positions between the first four and last three positions were required to be either a pyrimidine or a purine for Δ5–Δ9 motifs (for example, a CAGGCCAG is conserved to a TAGGTCAG but not to a TAGGACAG). To fulfill these constraints, two tandem acceptors have to be highly similar; indeed, tandem acceptors are often identical between species as the part downstream from the first AG overlaps with protein-coding sequence.
Previously, we found that all GYNGYN donors that are confirmed in human and mouse are identical between both species and that the GTAGTT donor of STAT3 exon 21 even yields virtually identical splice variant ratios in human and mouse (Hiller et al. 2006b). Therefore, we required identity of the first and last three positions for Δ3–Δ9 donors. Analyzing the nucleotide preferences for the positions between the two GYNs, we found a preference for a purine at position +4 for Δ4–Δ9 donors, at +5 for Δ5–Δ9 donors, and at the position upstream of the second GYN for Δ6–Δ9 donors, which is in agreement with the general donor consensus. To account for this, we required either a purine or an identical nucleotide at these three positions.
Balanced motif distribution simulation for Δ4–Δ9 tandem sites
For tandem sites that are more than 6 nt apart, each motif basically becomes unique; thus it is no longer practical to compare in this simulation the conservation between confirmed and unconfirmed sites with equal motifs. Therefore, we modified the balanced motif distribution simulation to compare confirmed tandems with identical or highly similar unconfirmed tandems. To this end, we constructed for each confirmed tandem motif two lists: the first list contains all confirmed tandems with this motif, and the second list contains all unconfirmed tandems that are either identical or highly similar to this motif. Taking similar unconfirmed tandems into account makes the second list contain at least as many entries as the first one, so that this list can be used to sample a subset of unconfirmed tandems. Random sampling of unconfirmed tandems was repeated 1000 times.
For Δ4–Δ6 donors, we sampled only from identical unconfirmed tandems. Δ7–Δ9 donors were considered as similar if Δ7 motifs are identical in positions +1 to +5 and +7 to +10; Δ8 motifs are identical in positions +1 to +5 and +8 to +11; and there is at most one mismatch at positions +6 and +7; Δ9 motifs are identical in positions +1 to +5 and +9 to +12; and there are at most two mismatches at position +6, +7, and +8. Δ4–Δ9 acceptor motifs were considered as similar if they fulfill the conservation definition given above.
The reason to use this simulation is that the conservation differs between the motifs and the motif distribution differs between confirmed and unconfirmed ones. For example, the balanced motif distribution simulation estimates that 51% of the confirmed and 47.1% of the unconfirmed Δ4 donors are conserved; a difference of 3.9%. However, the global conservation is only 41.3% for unconfirmed Δ4 donors; a much higher difference of 9.7%. This indicates that unconfirmed Δ4 tandems are enriched in weakly conserved motifs that do not occur among the confirmed ones; for example, all of the 2226 GTAAGCA donors are unconfirmed, and this motif has an exceptionally low conservation level of 29.6% (660 of 2226). As the balanced motif distribution simulation compares either identical or highly similar motifs, it gives a fair estimation of a lower bound for fs.
Correlation between motif conservation and splice site consensus constraints
For NAGNAG acceptors, we found that constraints on the acceptor splice site consensus are one main reason for the motif-specific conservation differences. To further test this, we considered Δ4 acceptors. As most of these acceptors are predominantly spliced at the upstream acceptor, we focused on the +4 position, which is often the 5′ exon end. The conservation level is 62.2% for NAGGNAG sites, 61.5% for NAGANAG, 60.5% for NAGCNAG, and 59.4% for NAGTNAG. Thus, the order G > A > C > T exactly correlates with the preference of the +1 position in the acceptor consensus (Abril et al. 2005), even though the conservation differences are not as pronounced as observed for NAGNAG acceptors.
We also determined the overall conservation level of Δ4 donors with a GTNNGTN motif, focusing on the +4 position in the tandem motif. GTNAGTN donors have the highest overall conservation level with 45.4%, followed by GTNGGTN (40%), GTNTGTN (35.6%), and GTNCGTN (16.9%). Again, the order A > G > T > C correlates perfectly with the +4 position preference in the donor consensus (Abril et al. 2005). For donors with a GCNNGTN motif, the GTN donor is predominant in most cases; thus the +4 position in the tandem motif represents the −1 position in the donor consensus. At the −1 position, G is preferred over A, T, and C (Abril et al. 2005). Again, this order correlates with the conservation level: GCNGGTN, 55.6%; GCNAGTN, 54.8%; GCNTGTN, 46.2%; and GCNCGTN, 42.1%. Thus, constraints on the donor and acceptor consensus are likely to be a major reason for the observed differences in the overall conservation levels of tandem motifs.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Christoph Englert, Jürgen Schulte Mönting, and Anke Busch for helpful discussions; and three anonymous referees for useful comments and suggestions. This work was supported by grants from the German Ministry of Education and Research to S.S. (01GS0426) and M.P. (01GR0504, 0313652D) as well as from the Deutsche Forschungsgemeinschaft to M.P. (SFB604-02) and K.H. (Hu498/3-1).
Footnotes
Abbreviations: fs, fraction of confirmed and conserved tandem splice sites estimated to be under purifying selection; CMH test, Cochran–Mantel–Haenszel test.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.883908.
REFERENCES
- Abril, J.F., Castelo, R., Guigo, R. Comparison of splice sites in mammals and chicken. Genome Res. 2005;15:111–119. doi: 10.1101/gr.3108805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman, M., Mandel-Gutfreund, Y. Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res. 2006;34:23–31. doi: 10.1093/nar/gkj408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, D., Green, P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaux, S., Niaudet, P., Gubler, M.C., Grunfeld, J.P., Jaubert, F., Kuttenn, F., Fekete, C.N., Souleyreau-Therville, N., Thibaud, E., Fellous, M., et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat. Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., Haussler, D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bickel, P.J., Hammel, E.A., O'Connell, J.W. Sex bias in graduate admissions: Data from Berkeley. Science. 1975;187:398–404. doi: 10.1126/science.187.4175.398. [DOI] [PubMed] [Google Scholar]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Burgar, H.R., Burns, H.D., Elsden, J.L., Lalioti, M.D., Heath, J.K. Association of the signaling adaptor FRS2 with fibroblast growth factor receptor 1 (Fgfr1) is mediated by alternative splicing of the juxtamembrane domain. J. Biol. Chem. 2002;277:4018–4023. doi: 10.1074/jbc.M107785200. [DOI] [PubMed] [Google Scholar]
- Campbell, M.A., Haas, B.J., Hamilton, J.P., Mount, S.M., Buell, C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis . BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern, T.M., van Nimwegen, E., Kai, C., Kawai, J., Carninci, P., Hayashizaki, Y., Zavolan, M. A simple physical model predicts small exon length variations. PLoS Genet. 2006;2:e45. doi: 10.1371/journal.pgen.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, F., Thanaraj, T.A. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 2002;11:451–464. doi: 10.1093/hmg/11.4.451. [DOI] [PubMed] [Google Scholar]
- Cohen, T., Auerbach, W., Ravid, L., Bodennec, J., Fein, A., Futerman, A.H., Joyner, A.L., Horowitz, M. The exon 8-containing prosaposin gene splice variant is dispensable for mouse development, lysosomal function, and secretion. Mol. Cell. Biol. 2005;25:2431–2440. doi: 10.1128/MCB.25.6.2431-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli, G., Bueno, R., Smith, R.J. Two alternatively spliced forms of the human insulin-like growth factor I receptor have distinct biological activities and internalization kinetics. J. Biol. Chem. 1994;269:8510–8516. [PubMed] [Google Scholar]
- Davies, R.C., Bratt, E., Hastie, N.D. Did nucleotides or amino acids drive evolutionary conservation of the WT1 ± KTS alternative splice? Hum. Mol. Genet. 2000;9:1177–1183. doi: 10.1093/hmg/9.8.1177. [DOI] [PubMed] [Google Scholar]
- Dou, Y., Fox-Walsh, K.L., Baldi, P.F., Hertel, K.J. Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site. RNA. 2006;12:2047–2056. doi: 10.1261/rna.151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova, E.O., Nurtdinov, R.N., Gelfand, M.S. Overlapping alternative donor splice sites in the human genome. J. Bioinform. Comput. Biol. 2007;5:991–1004. doi: 10.1142/s0219720007003089. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes, A., Guo, J.K., Lutsch, G., Leheste, J.R., Landrock, D., Ziegler, U., Gubler, M.C., Schedl, A. Two splice variants of the Wilms' tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- Hiller, M., Huse, K., Szafranski, K., Jahn, N., Hampe, J., Schreiber, S., Backofen, R., Platzer, M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- Hiller, M., Huse, K., Szafranski, K., Jahn, N., Hampe, J., Schreiber, S., Backofen, R., Platzer, M. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am. J. Hum. Genet. 2006a;78:291–302. doi: 10.1086/500151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, M., Huse, K., Szafranski, K., Rosenstiel, P., Schreiber, S., Backofen, R., Platzer, M. Phylogenetically widespread alternative splicing at unusual GYNGYN donors. Genome Biol. 2006b;7:R65. doi: 10.1186/gb-2006-7-7-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, M., Szafranski, K., Backofen, R., Platzer, M. Alternative splicing at NAGNAG acceptors: Simply noise or noise and more? PLoS Genet. 2006c;2:e207. doi: 10.1371/journal.pgen.0020207. author reply e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, M., Nikolajewa, S., Huse, K., Szafranski, K., Rosenstiel, P., Schuster, S., Backofen, R., Platzer, M. TassDB: A database of alternative tandem splice sites. Nucleic Acids Res. 2007;35:D188–D192. doi: 10.1093/nar/gkl762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.A., Lin, W.W., Obie, C., Valle, D. Molecular enzymology of mammalian Δ1-pyrroline-5-carboxylate synthase. Alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. J. Biol. Chem. 1999;274:6754–6762. doi: 10.1074/jbc.274.10.6754. [DOI] [PubMed] [Google Scholar]
- Hymowitz, S.G., Compaan, D.M., Yan, M., Wallweber, H.J., Dixit, V.M., Starovasnik, M.A., de Vos, A.M. The crystal structures of EDA-A1 and EDA-A2: Splice variants with distinct receptor specificity. Structure. 2003;11:1513–1520. doi: 10.1016/j.str.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Joyce-Brady, M., Jean, J.C., Hughey, R.P. γ-Glutamyltransferase and its isoform mediate an endoplasmic reticulum stress response. J. Biol. Chem. 2001;276:9468–9477. doi: 10.1074/jbc.M004352200. [DOI] [PubMed] [Google Scholar]
- Julious, S.A., Mullee, M.A. Confounding and Simpson's paradox. BMJ. 1994;309:1480–1481. doi: 10.1136/bmj.309.6967.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni, R., de Stanchina, E., Lowe, S.W., Sinha, R., Mu, D., Krainer, A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig Merediz, S.A., Schmidt, M., Hoppe, G.J., Alfken, J., Meraro, D., Levi, B.Z., Neubauer, A., Wittig, B. Cloning of an interferon regulatory factor 2 isoform with different regulatory ability. Nucleic Acids Res. 2000;28:4219–4224. doi: 10.1093/nar/28.21.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, R.M., Karolchik, D., Zweig, A.S., Trumbower, H., Thomas, D.J., Thakkapallayil, A., Sugnet, C.W., Stanke, M., Smith, K.E., Siepel, A., et al. The UCSC Genome Browser Database: Update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau, L.F., Inada, M., Green, R.E., Wengrod, J.C., Brenner, S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Green, R.E., Brenner, S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi, D.D., Darnell, R.B. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Lynch, K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Lee, C.J. Alternative splicing in the human, mouse, and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Ni, J.Z., Grate, L., Donohue, J.P., Preston, C., Nobida, N., O'Brien, G., Shiue, L., Clark, T.A., Blume, J.E., Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani, F., Baralle, F.E. Genomic variants in exons and introns: Identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Shai, O., Misquitta, C., Zhang, W., Saltzman, A.L., Mohammad, N., Babak, T., Siu, H., Hughes, T.R., Morris, Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Bakowski, M.A., Morris, Q., Zhang, W., Frey, B.J., Hughes, T.R., Blencowe, B.J. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Raponi, M., Baralle, F.E., Pagani, F. Reduced splicing efficiency induced by synonymous substitutions may generate a substrate for natural selection of new splicing isoforms: The case of CFTR exon 12. Nucleic Acids Res. 2007;35:606–613. doi: 10.1093/nar/gkl1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch, A., Xing, Y., Alekseyenko, A., Modrek, B., Lee, C. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A., Bejerano, G., Pedersen, J.S., Hinrichs, A.S., Hou, M., Rosenbloom, K., Clawson, H., Spieth, J., Hillier, L.W., Richards, S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, E.H. The interpretation of interaction in contingency tables. J. R. Stat. Soc. [Ser A] 1951;13:238–241. [Google Scholar]
- Sorek, R., Ast, G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Shamir, R., Ast, G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Stadler, M.B., Shomron, N., Yeo, G.W., Schneider, A., Xiao, X., Burge, C.B. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2:e191. doi: 10.1371/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov, P., Daoud, R., Nayler, O., Stamm, S. Human tra2-β1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- Sugnet, C.W., Kent, W.J., Ares M., Jr, Haussler, D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004;2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- Tadokoro, K., Yamazaki-Inoue, M., Tachibana, M., Fujishiro, M., Nagao, K., Toyoda, M., Ozaki, M., Ono, M., Miki, N., Miyashita, T., et al. Frequent occurrence of protein isoforms with or without a single amino acid residue by subtle alternative splicing: The case of Gln in DRPLA affects subcellular localization of the products. J. Hum. Genet. 2005;50:382–394. doi: 10.1007/s10038-005-0261-9. [DOI] [PubMed] [Google Scholar]
- Takeda, H., Matsuzaki, T., Oki, T., Miyagawa, T., Amanuma, H. A novel POU domain gene, zebrafish pou2: Expression and roles of two alternatively spliced twin products in early development. Genes & Dev. 1994;8:45–59. doi: 10.1101/gad.8.1.45. [DOI] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- Tress, M.L., Martelli, P.L., Frankish, A., Reeves, G.A., Wesselink, J.J., Yeats, C., Olason, P.L., Albrecht, M., Hegyi, H., Giorgetti, A., et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl. Acad. Sci. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K.W., Tarn, W.Y., Lin, W.C. Wobble splicing reveals the role of the branch point sequence-to-NAGNAG region in 3′ tandem splice site selection. Mol. Cell. Biol. 2007;27:5835–5848. doi: 10.1128/MCB.00363-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule, J., Ule, A., Spencer, J., Williams, A., Hu, J.S., Cline, M., Wang, H., Clark, T., Fraser, C., Ruggiu, M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal, A., Ettwiller, L., Birney, E. Comparative genomics: Genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- Vogan, K.J., Underhill, D.A., Gros, P. An alternative splicing event in the Pax-3 paired domain identifies the linker region as a key determinant of paired domain DNA-binding activity. Mol. Cell. Biol. 1996;16:6677–6686. doi: 10.1128/mcb.16.12.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Romfo, C.M., Nilsen, T.W., Green, M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Xing, Y., Lee, C.J. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet. 2005;1:e34. doi: 10.1371/journal.pgen.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Belcastro, M.P., Villa, S.T., Dinkins, R.D., Clarke, S.G., Downie, A.B. A second protein L-isoaspartyl methyltransferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Plant Physiol. 2004;136:2652–2664. doi: 10.1104/pp.104.046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, M., Wang, L.C., Hymowitz, S.G., Schilbach, S., Lee, J., Goddard, A., de Vos, A.M., Gao, W.Q., Dixit, V.M. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- Yeo, G.W., Van Nostrand, E., Holste, D., Poggio, T., Burge, C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D.A., Hayashizaki, Y., Gaasterland, T. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]