Abstract
Helix 69 in 23S rRNA is a region in the ribosome that participates in a considerable number of RNA–RNA and RNA–protein interactions. Conformational flexibility is essential for such a region to interact and accommodate protein factors at different stages of protein biosynthesis. In this study, pH-dependent structural and stability changes were observed for helix 69 through a variety of spectroscopic techniques, such as circular dichroism spectroscopy, UV melting, and nuclear magnetic resonance spectroscopy. In Escherichia coli 23S rRNA, helix 69 contains pseudouridine residues at positions 1911, 1915, and 1917. The presence of these pseudouridines was found to be essential for the pH-induced conformational changes. Some of the pH-dependent changes appear to be localized to the loop region of helix 69, emphasizing the importance of the highly conserved nature of residues in this region.
Keywords: helix 69, 23S rRNA, pseudouridine, modified nucleotides
INTRODUCTION
The ribosome is an essential molecular machine composed of two subunits, which together carry out protein biosynthesis in all living cells. The ribosome is a large RNA–protein complex with more than 4000 nucleotides (nt) and over 50 different proteins. The large ribosomal subunit from bacteria sediments at 50S and consists of 23S and 5S rRNAs and 31 different proteins in Escherichia coli (Wilson and Nierhaus 2003). A number of high-resolution structures revealed that ribosomal proteins are mostly scattered at the surface of the ribosome and located away from the small and large subunit interface (Ban et al. 2000; Yusupov et al. 2001; Schuwirth et al. 2005). The 23S rRNA contains six large domains that form compact, intricate secondary structures and have unique tertiary structures. The region of interest in this study, helix 69 (H69), is located in domain IV of 23S rRNA, which forms part of a large diagonal flat surface at the interface of the two subunits. Region B2a comprises one of the most important ribosomal intersubunit bridges (Yusupov et al. 2001). A large part of the surface area of bridge B2a belongs to H69, which is flexible and partially disordered in X-ray crystal structures of the 50S subunit (Ban et al. 2000; Bashan et al. 2003). In 70S ribosomes, H69 stretches toward the decoding region (helix 44 or h44) of the small subunit rRNA and also interacts with the A- and P-site tRNAs through minor-groove contacts (Yusupov et al. 2001; Schuwirth et al. 2005).
Helix 69 of E. coli 23S rRNA has three pseudouridine (Ψ) residues at positions 1911, 1915, and 1917 (Fig. 1; Bakin and Ofengand 1993). The pseudouridine residue at position 1915 is methylated at the N3 position (Fig. 1; Kowalak et al. 1996). H69 also contains a G•U mismatch in the stem region. G•U mismatches often have different base-stacking properties than Watson–Crick base pairs, with greater base overlap on the 5′ side of U (Mizuno and Sundaralingam 1978). A recent crystal structure of the E. coli 70S ribosome at 3.5 Å revealed specific contacts with H69 that likely contribute to subunit association (Schuwirth et al. 2005). An A–A dinucleotide platform is formed with A1919 and A1918 due to the presence of a widened reversed-Hoogsteen base pair between residues Ψ1911 and A1919, bridged by the 2′-OH of A1918. Formation of this noncanonical base pair leads to projection of A1919 toward h44 of the small ribosomal subunit. Furthermore, the E. coli 70S crystal structure indicates that A1912 of H69 forms a distorted reversed-Hoogsteen base pair with Ψ1917, in which A1912 also projects toward the small ribosomal subunit (Schuwirth et al. 2005). In a 2.8-Å-resolution structure of Thermus thermophilus 70S ribosomes complexed with mRNA and tRNA, H69 is shifted toward the 30S subunit, and residue A1913 is flipped out and forms a hydrogen bond to the 2′ hydroxyl of ribose 37 of the A-site-bound tRNA anticodon stem–loop (Selmer et al. 2006). Thus, flexibility of H69 and its specific location in the ribosome at bridge B2a suggest that this helix may be involved in signal transmission between the two subunits at different stages of translation.
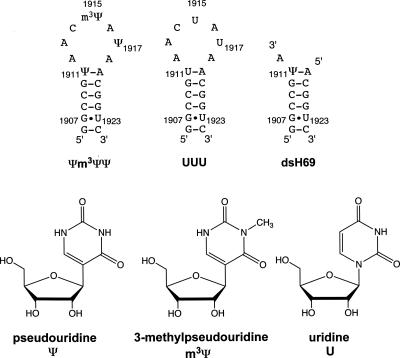
FIGURE 1.
The helix 69 (H69) RNA hairpins from E. coli are shown. The Ψm3ΨΨ and UUU RNA constructs are derived from positions 1906–1924 of E. coli 23S rRNA. The dsH69 RNA contains the stem-region of H69. The structures of pseudouridine (Ψ), 3-methylpseudouridine (m3Ψ), and uridine (U) are shown.
The ribosomal recycling factor (RRF) is a protein that is required for the final release step of translation (Hirashima and Kaji 1970; Subramanian and Davis 1973). This factor is believed to work in concert with the elongation factor G (EF-G) to promote dissociation of the ribosome subunits (Peske et al. 2005). A cryoelectron microscopy map of RRF bound to the E. coli 70S ribosome reveals that the recycling factor is bound close to bridge B2a, which involves H69 (Agrawal et al. 2004). A conformational change in the bridge B2a was observed upon binding of RRF. The same study produced a model in which the highly conserved amino acid residues E122 and V126 of RRF interact with m3Ψ1915 of H69, and V130 interacts with A1916. A more recent crystal structure (3.5 Å resolution) of E. coli RRF domain I bound to the Deinococcus radiodurans 50S subunit revealed a different set of contacts with H69 (Wilson et al. 2005). Together, these observed interactions with H69 emphasize its importance in proper functioning of the ribosome during protein synthesis. Thus, a study of the solution structure and dynamics of H69 may provide valuable insight into its biological functions.
RNA has considerable structural and functional diversity, perhaps in part due to the presence of a large number of modified bases (Limbach et al. 1994). Pseudouridine (Ψ) (Fig. 1), the most abundant base modification in rRNA, is found in the most functionally important regions of the ribosome (Ofengand and Bakin 1997). The physiochemical properties of Ψ are different from its isomer uridine due to several key variations in their structures. The first difference is a C–C glycosidic linkage in Ψ compared with a C–N linkage in uridine. Thus, there is a higher degree of rotational freedom for the base of Ψ, and more conformational flexibility of the nucleoside (Davis and Poulter 1991; Lane et al. 1995). The second difference is that Ψ contains an extra H-bond donor compared to uridine. The ΨN1H is available to participate in hydrogen-bonding interactions with neighboring groups (Davis and Poulter 1991). The crystal structure of tRNAGln complexed with Gln-tRNA synthetase revealed a unique hydrogen bond between ΨN1H and a water molecule coordinated to the 5′-phosphate group of Ψ and residue on the 5′ side (Arnez and Steitz 1994). The existence of a water-mediated hydrogen bond between ΨN1H and 5′-phosphate oxygen was further revealed in solution through novel NMR methods (Newby and Greenbaum 2002).
Pseudouridine residues often have altered sugar conformations and base-stacking interactions in RNA relative to the standard nucleotides. In modified RNAs studied to date, Ψ residues tend to have 3′-endo sugar puckers (N conformation) (Davis 1995). The Ψ residue in an AAΨA construct occurs in the N conformation (100%). When U replaces Ψ in the same construct, the level of N conformer is reduced to 53%. A higher percentage of N conformers is also observed in residues on the 5′ side of Ψ, and the stacking arrangement protects ΨN1H from exchange with solvent (Davis 1995).
The structures and conformations of biomolecules are often dependent on solution conditions, such as salt concentration, pH, temperature, and the presence of specific metal ions (e.g., Mg2+). The overall thermodynamic stability and/or structural properties of some RNAs are influenced by pH (Durant and Davis 1999; Nixon and Giedroc 2000; Bayfield et al. 2004; Reiter et al. 2004). For example, formation of a protonated A+•C base pair in the anticodon stem–loop of tRNALys,3 stabilizes the hairpin structure (Durant and Davis 1999). NMR studies revealed that the structure of the RNA at pH 7 is much more dynamic than the structure at pH 5. A pH-dependent structural change was also predicted for an A2450+•C2063 mismatch in the peptidyl-transferase center (PTC) of the ribosome (Bayfield et al. 2004). A structure of the ribosomal 50S subunit crystallized at pH 5.8 confirmed the formation of the A2450+•C2063 mismatch pair (Ban et al. 2000). Of note, these RNAs are required for proper ribosomal function; furthermore, they contain Ψ nucleotides. Thus, given the structural and stability consequences of pseudouridine in these functionally important RNAs, it was of interest to investigate the effects of this highly conserved modified nucleotide on the structure and dynamics of H69. Studies to probe the changes in H69 structure under different solution conditions, such as varying pH, may give valuable insight into the role of this functionally important biomolecular system. Here, we report on the pH-dependent changes in H69 structure and stability determined through circular dichroism (CD) spectroscopy, one-dimensional (1D) imino proton NMR spectroscopy, and thermal melting. Furthermore, we have identified roles for specific pseudouridine residues in mediating these changes.
RESULTS
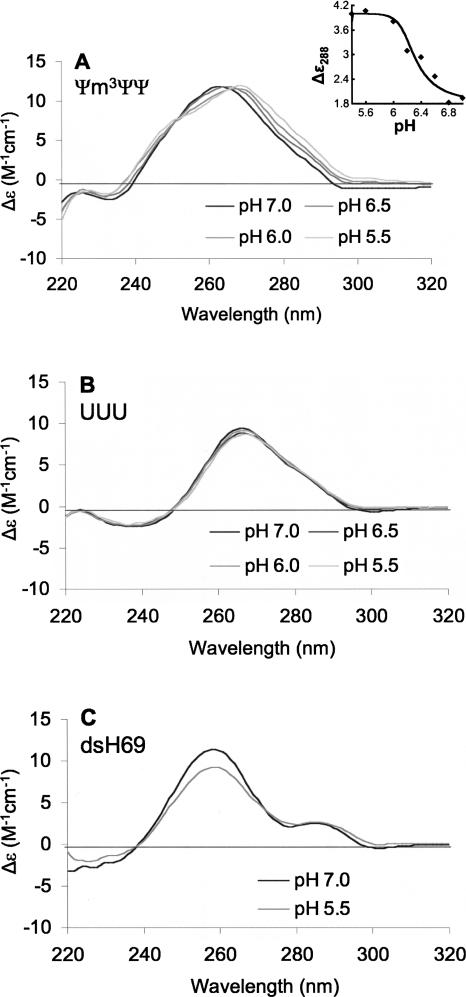
Circular dichroism spectroscopy is a useful technique to monitor conformational changes of nucleic acids (Ivanov et al. 1973). Historically, CD spectroscopy has been used to observe thermal or chemical denaturation, ligand binding, and conformational changes in nucleic acids as a function of salt concentration, level of hydration, and pH (Woody 1995). For these studies, we generated various RNA constructs representing H69 of E. coli 23S rRNA (Fig. 1). The construct referred to as Ψm3ΨΨ comprises residues 1906 through 1924 (E. coli numbering) and modified nucleotides Ψ1911, m3Ψ1915, and Ψ1917 of the wild-type, fully modified H69. The UUU construct is the unmodified variant with three uridine residues at positions 1911, 1915, and 1917. A third construct (dsH69) represents the modified stem region of H69. CD spectroscopy was used to monitor the pH sensitivity of the Ψm3ΨΨ and UUU structures (Fig. 2). At pH 7.0, the CD spectrum of Ψm3ΨΨ (Fig. 2A, black) shows a peak maximum at 264 nm and a peak minimum at 234 nm. The crossover point is at 240 nm. These features suggest that the H69 Ψm3ΨΨ construct forms an A-like structure. When the pH of the system is incrementally lowered from 7.0 to 5.5, the CD spectra of the H69 Ψm3ΨΨ construct change (Fig. 2A, dark gray to light gray). There are two distinct features of the CD spectra that are altered as the pH is lowered. The first change is a reduction of the molar ellipticity between 252 and 266 nm and development of a slight shoulder between 240 nm and 252 nm with an isosbestic point at 252 nm, suggestive of a two-state system. The second change is a red shift of the peak maximum to 268 nm and a slight blue shift of the crossover point to 238 nm at pH 5.5. Such changes are not observed when the pH is lowered in the unmodified construct (UUU) (Fig. 2B). These results reveal that the Ψ residues in H69 must be present in order for pH-dependent conformational changes to occur.
FIGURE 2.
CD spectra of modified (Ψm3ΨΨ, panel A) and unmodified (UUU, panel B) E. coli H69 RNAs in buffer A are shown. The CD spectra for dsH69 are in panel C. Each spectrum (room temperature) is an average of four scans and the RNA concentrations were 2.6 μM. The pH values varied from 5.5 (gray) to 7.0 (black). (Inset) Molar ellipticity at 288 nm plotted against pH and fit to an equation derived from the Henderson–Hasselbalch equation.
The molar ellipticity changes in the CD spectra were used to determine the pKa of the system. The molar ellipticity at 288 nm (Δε288) versus pH curve (Fig. 2A, inset) was fit to an equation derived from the Henderson–Hasselbalch equation (Bose et al. 2007), assuming that the change in Δε288 is directly proportional to the protonation of H69. Analysis at other wavelengths generated similar curves. This detailed analysis of the pH-dependent ellipticity change reveals an apparent pKa of 6.3 ± 0.2 for the structural change in modified H69.
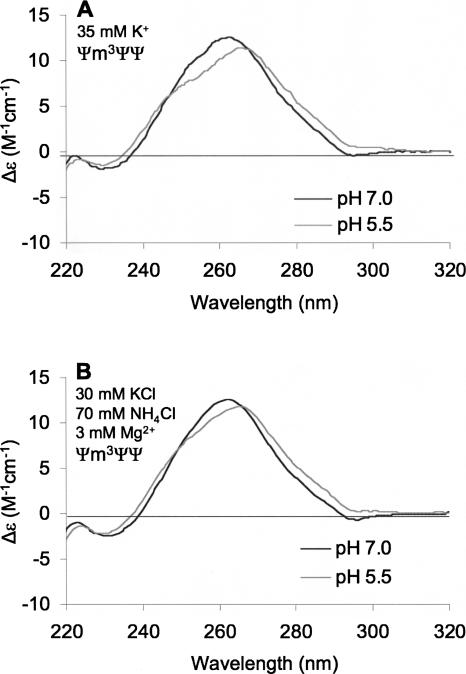
The CD experiments described above were carried out in low salt conditions (35 mM Na+), which are known to favor hairpin formation for H69 (Meroueh et al. 2000). The experiments were repeated in 35 mM K+ and salt concentrations that more closely mimic in vivo protein synthesis conditions (25 mM cacodylic acid, 25 mM Tris base, 70 mM NH4Cl, 30 mM KCl, 3 mM MgCl2, at pH 7.0 or 5.5) (Gromadski and Rodnina 2004), and similar results were obtained (Fig. 3) in which a 4-nm red shift in the CD spectra occurs as the pH is lowered from 7.0 to 5.5. Furthermore, identical results were obtained when the RNA samples were renatured by either slow or fast cooling (data not shown). At much higher Mg2+ and K+ concentrations (30 and 150 mM, respectively), the overall magnitude of the red shift is reduced slightly compared to that observed in the lower Mg2+ conditions, because the higher divalent metal ion conditions lead to a mixture of hairpin and duplex RNA structures, as determined by UV melting experiments (data not shown). This result reflects the limitation of using an RNA model system with a short hairpin stem (6 base pairs). Nonetheless, similar conformational changes of H69 are apparent under higher salt conditions.
FIGURE 3.
CD spectra of modified (Ψm3ΨΨ) E. coli H69 RNAs are shown. Each spectrum is an average of four scans and the RNA concentrations were 2.6 μM. The pH values were 5.5 (gray) or 7.0 (black). The buffers employed contained 35 mM K+ (buffer B, panel A) or 30 mM K+, 70 mM NH4Cl, 3 mM Mg2+ (buffer C, panel B).
For the next set of experiments, the goal was to determine which part of H69 is responsible for the observed pH-dependent CD changes of the Ψm3ΨΨ construct. A stem-only analog was generated by annealing two single strands obtained from chemical synthesis (Fig. 1, dsH69). The construct was made with an overhanging A residue on the 3′ end (corresponding to residue A1912) because the appropriate Ψ-containing support for solid-phase RNA synthesis was not available. In addition, the overhanging A residue allows the contribution of possible stacking or hydrogen-bonding interactions between Ψ1911 and the adjacent nucleotides to be considered. The dsH69 construct was investigated for pH-dependent CD changes. The CD spectrum of the stem at pH 7.0 shows a maximum at 258 nm and a small peak centered at ∼286 nm (Fig. 2C). The minimum at ∼230 nm is typical for A-form RNA, but not a predominant feature of the spectrum. When the pH of the dsH69 solution is at 5.5, the CD spectral features remain the same. The only difference between the CD spectra at different pH values is the magnitude of the peak maximum at 258 nm. The molar ellipticity at 258 nm is lower in the pH 5.5 spectrum compared to the pH 7.0 spectrum (Fig. 2C). We attribute this reduction of the ellipticity to partial denaturation of the H69 stem region. Together, these observations suggest that specific Ψ residues in the loop region, or cooperativity between these loop residues and the H69 stem, play a role in the pH sensitivity of the Ψm3ΨΨ construct.
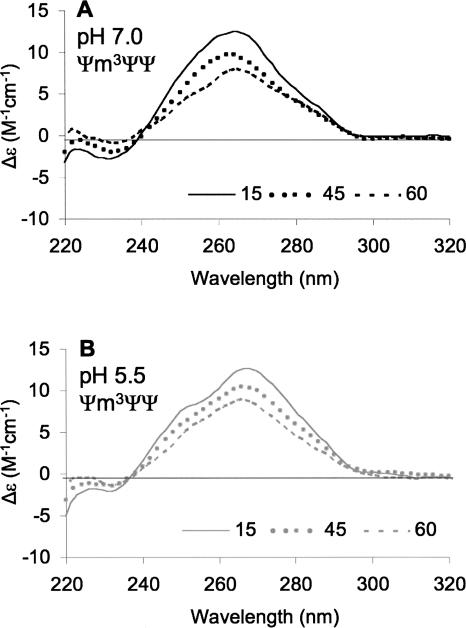
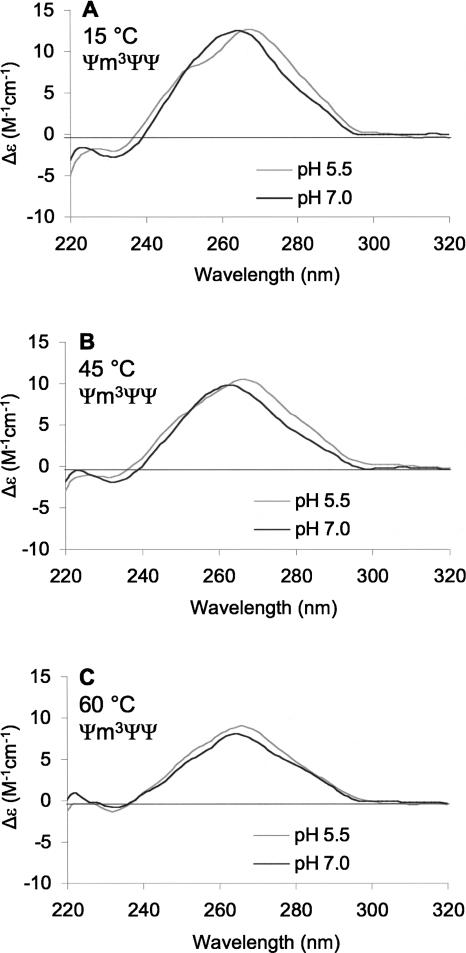
When the temperature of an RNA system is increased, weak interactions are completely disrupted and the hairpin loop structure denatures. If the pH sensitivity of modified H69 is due to weak interactions in the loop region, then those effects should diminish as the temperature of the system is increased. CD experiments (Fig. 4) at both pH 7.0 and 5.5 and different temperatures were carried out to understand the nature of the interactions that lead to the pH sensitivity of the overall structure of H69. In the buffer system employed (sodium cacodylate), the variation in pH with temperature over the range of 15°C – 60°C is minimal (ΔpH = 0.07) (Fukada and Takahashi 1998). As the temperature is raised from 15°C to 60°C for the Ψm3ΨΨ construct at pH 7.0, a clear reduction in the molar ellipticity at the peak maximum is observed (Fig. 4A). This change reflects Ψm3ΨΨ denaturation at pH 7.0. When the temperature is raised for the same RNA at pH 5.5, a similar reduction in the molar ellipticity at the peak maximum is observed (Fig. 4B). At pH 5.5, the small shoulder at 258 nm is less apparent as the temperature increased. If a direct comparison is made between the two systems (pH 7.0 and 5.5), distinct differences can be observed with increasing temperatures. At 15°C and pH 7.0, the Ψm3ΨΨ CD spectrum has a peak maximum at 264 nm, and a red shift of 4 nm occurs as the pH is lowered to 5.5 (Fig. 5A). At this lower temperature, a negligible difference in molar ellipticity at the peak maxima between the pH 7.0 and 5.5 spectra is observed. At a higher temperature of 45°C, the red shift in peak maximum is only 2 nm when the pH of the solution is lowered, but a small difference in the molar ellipticity is now apparent (Δε = 0.7 M−1 cm−1), suggesting a slight enhancement of stability of Ψm3ΨΨ at pH 5.5 compared to 7.0 (Fig. 5B). When the temperature is further elevated to 60°C, the difference in molar ellipticity at the two pH values is increased further (Δε = 1.0 M−1 cm−1), but the overall shapes of the CD spectra are more similar (Fig. 5C). This result indicates that the structure involving the protonated state of modified H69 is disrupted at higher temperatures, and the pH 7.0 and pH 5.5 states of Ψm3ΨΨ become conformationally equivalent, although the construct is slightly more stable at the lower pH.
FIGURE 4.
CD spectra of Ψm3ΨΨ in buffer A at pH 7.0 (A) or 5.5 (B) with varying temperature (15°C–60°C) are shown.
FIGURE 5.
CD spectra of Ψm3ΨΨ in buffer A at 15°C (A), 45°C (B), and 60°C (C) with varying pH (5.5 [gray] or 7.0 [black]) are shown.
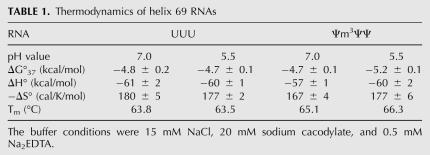
Thermal melting experiments enabled us to further probe the differences in H69 at pH 7.0 and 5.5. These experiments allow the thermodynamic parameters, ΔG°37, ΔH°, and ΔS°, to be determined. The UV melting profiles (absorbance vs. temperature) for the H69 Ψm3ΨΨ construct at different concentrations (8–80 μM) predict two-state equilibrium for both pH values (data not shown), consistent with previous studies (Meroueh et al. 2000; Chui et al. 2002). This two-state behavior is independent of the RNA concentration and reveals hairpin formation. The H69 Ψm3ΨΨ construct has a melting temperature (Tm) of 65.1°C at pH 7.0, but increases to 66.3°C at pH 5.5 (Table 1). When the thermodynamic stability for the Ψm3ΨΨ construct is considered, the ΔG°37 value is slightly more favorable by 0.5 kcal/mol as the pH is lowered (Table 1). In contrast, the difference in ΔG°37 value for the unmodified UUU construct is almost negligible at lower pH (ΔΔG°37 = 0.2 kcal/mol). For the Ψm3ΨΨ data, one might argue that the differences in ΔG°37 are insignificant due to the expected standard level of error (3%; ΔΔG°37 = 0.14 kcal/mol) (Allawi and SantaLucia 1997; SantaLucia and Turner 1997); however, each experiment was carried out in triplicate with five different RNA preparations, and the standard deviation in ΔG°37 between experiments was small (±0.1 kcal/mol, 2% error) (Table 1). Thus, with a more rigorous approach, higher precision in the ΔG°37 values can be achieved. For the Ψm3ΨΨ construct, the ΔH° component has a slightly more favorable contribution to the thermodynamic stability at lower pH. The −ΔS° value is slightly higher at pH 5.5 compared to pH 7.0, suggesting a less dynamic, or more ordered, system at the lower pH. For the UUU construct, the ΔΔS° value (ΔS°pH7.0 − ΔS°pH5.5) is almost negligible. These results suggest that Ψ and/or m3Ψ modifications lead to a slightly more stable and ordered system for H69 at lower pH. These results are also consistent with CD data discussed in the previous section.
TABLE 1.
Thermodynamics of helix 69 RNAs
As with the CD experiments, the UV melting studies were also carried out under higher salt conditions (150 mM K+/30 mM Mg2+). The overall trends are the same, in which H69 is slightly more stable at lower pH (data not shown); however, the melting curves reveal concentration-dependent behavior in the lower temperature range, suggesting the small model RNA exists in both duplex and hairpin conformations at the higher salt concentrations. Although the model system has limitations, as mentioned previously, the general trend in which H69 is stabilized at lower pH is consistent.
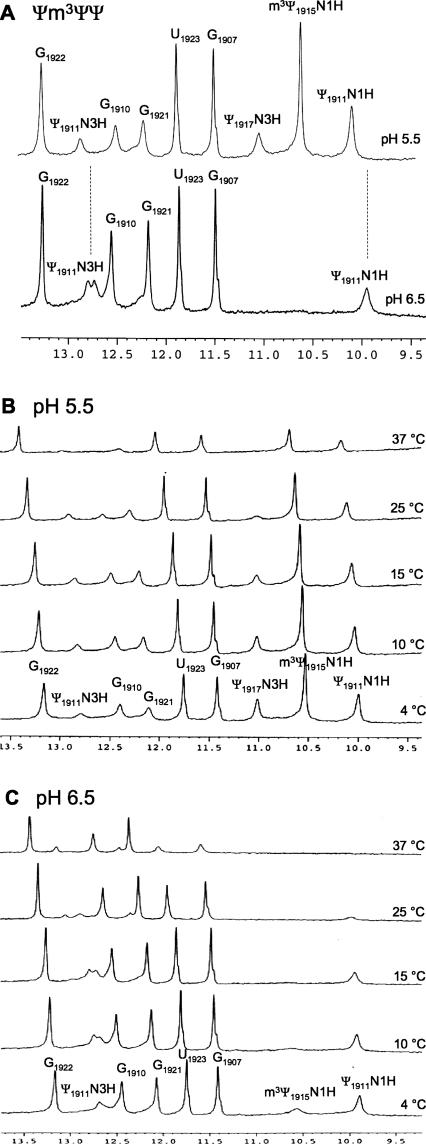
Although CD spectroscopy can reveal even small changes in nucleic acid conformations, this method cannot specifically identify the types of interactions present. Nuclear magnetic resonance (NMR) spectroscopy is a powerful technique that can be used to identify the molecular interactions within the nucleic acid (Fürtig et al. 2003). In our study, we used NMR experiments to understand the nature of the interactions in H69 and to identify the residues involved in those interactions. The hydrogen-bonding interactions will influence the overall conformation of H69. One-dimensional proton NMR experiments were carried out with the Ψm3ΨΨ and UUU constructs at pH 6.5 and pH 5.5. NMR spectral peaks in the 9.0–14.0 ppm region are indicative of hydrogen-bonding interactions of the imino protons in nucleic acids (Varani et al. 1996; Fürtig et al. 2003). The spectrum of modified H69 (Ψm3ΨΨ) at pH 6.5 at 15°C is shown in the lower half of Figure 6A. Seven spectral peaks are observed in the imino proton region. The peaks at 9.9 and 12.8 ppm correspond to the N1H and N3H, respectively, of the Ψ residue present at the loop-closing 1911 position (Meroueh et al. 2000; Chui et al. 2002). The Ψ1911N3 imino proton gives rise to two peaks with a 0.1-ppm separation. This result is likely an indication of conformational dynamics in the stem–loop junction of H69 or the existence of multiple conformations due to the fact that the pH of the solution is close to the apparent pKa of the structural transition. Spectral peaks at 11.5 and 11.9 ppm correspond to G1907 and U1923, which form the G•U mismatch present in the stem region. Imino protons that correspond to G1907 and U1923 show strong NOEs with each other, which is typical for a G•U mismatch (Varani et al. 1996). Spectral peaks at 12.2, 12.6, and 13.3 ppm correspond to G1921, G1910, and G1922, respectively, consistent with previous studies (Meroueh et al. 2000; Chui et al. 2002).
FIGURE 6.
The 1D imino proton (uridine H3, guanine H1, and pseudouridine N1H and N3H) NMR spectra of E. coli H69 RNA (Ψm3ΨΨ) are shown. The spectra in panel A were obtained at 15°C and pH 5.5 (upper) or pH 6.5 (lower). The spectra in panels B and C were obtained at 4°C, 10°C, 15°C, 25°C, and 37°C at pH 5.5 (B) or pH 6.5 (C). The buffer employed was 30 mM NaCl, 10 mM sodium phosphate, and 0.5 mM Na2EDTA in 90% H2O and 10% D2O. Nucleotides are assigned based on 1D NOE spectroscopy (Meroueh et al. 2000; Chui et al. 2002).
When the pH of the system is lowered to 5.5, two additional peaks appear in the imino region of the 1D proton NMR spectrum of Ψm3ΨΨ at 10.6 and 11.0 ppm (Fig. 6A, upper spectrum). Specific 15N-labeling of Ψ in various H69 constructs enabled us to identify unambiguously these two peaks as m3Ψ1915N1H and Ψ1917N3H (Desaulniers 2005; data not shown). These data clearly demonstrate that Ψm3ΨΨ has an altered loop structure or altered loop dynamics at lower pH, such that the two additional Ψ imino peaks are observed. Another notable feature is the reduction in relative intensities of the peaks corresponding to G1921 and G1910, which reside in the stem region of H69. In addition, the chemical shift of the Ψ1911N3 imino proton changes from 12.8 to 12.9 ppm when the pH is lowered from 6.5 to 5.5, and that of Ψ1911N1H changes from 9.9 to 10.1 ppm. The changes in chemical shifts for these imino protons seem to be significant compared to the other protons, indicating that the protonation event may be occurring near Ψ1911 or causing a structural change that affects the loop-closing base pair.
The temperature of the system was changed to further probe the thermal stability of Ψm3ΨΨ at lower pH, and to compare with the CD and thermal melting data mentioned in the previous sections. When the temperature of the system is raised from 4°C to 37°C, the spectral intensities of all imino peaks are diminished as expected (Fig. 6B, pH 5.5); however, some of the imino proton peaks are less sensitive toward changes in temperature. Specifically, the peaks corresponding to Ψ1911N1H and m3Ψ1915N1H are still clearly observed at higher temperature (37°C). In contrast, the peak corresponding to Ψ1917N3H diminishes significantly at higher temperature (>25°C). The peaks corresponding to the imino protons of the G•U mismatch and the base stacked on the 5′ side of U1923 (G1922) are also less affected by the increase in temperature, which is consistent with an increased base-stacking interaction between the mismatch pair and adjacent Watson–Crick pair (Mizuno and Sundaralingam 1978). Overall, the NMR data support the hypothesis that the loop region of Ψm3ΨΨ has an altered structure at lower pH such that Ψ1911N1H and Ψ1915N1H are more protected from solvent.
When the temperature of the Ψm3ΨΨ construct at pH 6.5 is increased from 4°C to 37°C, the intensities of the imino proton peaks are diminished (Fig. 6C); however, in this case, the peaks corresponding to Ψ1911N1, G1907, and U1923 imino protons are more sensitive to the temperature increase. The imino proton peaks corresponding to stem residues G1922, G1921, and G1910 are affected less by the higher temperature. These results reflect differences in the melting behavior of Ψm3ΨΨ at pH 5.5 and 7.0. At lower pH, the G•U mismatch appears to be more stable than at higher pH. Similarly, the loop region of H69 seems to melt at a lower temperature at pH 6.5. These results support the CD and UV melting data, in which an altered conformation of modified H69 exists at lower pH, and the protonated form has slightly higher stability than the unprotonated form. The temperature-dependent NMR data reveal an increased stability of the loop region of H69 at lower pH and an increase in solvent exposure of the loop residues at higher pH.
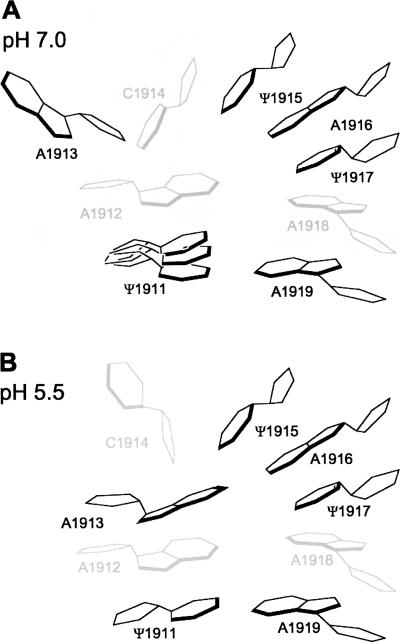
The bacterial 70S structure (Schuwirth et al. 2005) shows that residue A1913 of H69 is flipped out of the tip of the loop and exposed to solvent, whereas residues 1915–1917 are involved in stacking interactions. We present a model in Figure 7 based on our 1D NMR data and the available X-ray crystal structures, in which residues Ψ1915 and Ψ1917 stack with A1916, but are partially exposed to solvent due to the flipping out of A1913. The Ψ1911–A1919 base pair is dynamic and exists as a mixture of conformers. At lower pH, A1913 is oriented inside the loop and protects the remaining loop residues (Ψ1915 and Ψ1917) from solvent exchange. This alternative structure model resembles H69 in the 50S ribosome (Bashan et al. 2003). Overall, the H69 loop is more exposed to solvent and dynamic at higher pH, but stabilized at lower pH. These models may reflect the possible conformations of H69 at various stages of protein synthesis (e.g., 50S versus 70S ribosomes, in the presence or absence of the recycling factor).
FIGURE 7.
Schematic representations of the H69 loop region at (A) pH 7.0 and (B) pH 5.5 are shown.
DISCUSSION
Bashan and coworkers postulated that there could be more than one stable conformation of helix 69 in 23S rRNA during different stages of translation (Bashan et al. 2003). Ribosome movement is essential for protein synthesis to occur (Wilson and Nierhaus 2003); therefore, some regions of the ribosome are likely to be more dynamic than others. Some intriguing questions are (1) how does the RNA attain functionally important conformational states during the translation process, and (2) what are the roles of modified nucleotides that are concentrated in the functionally important regions of the ribosome?
The circular dichroism spectra presented here for the Ψm3ΨΨ RNA construct reveal that modified H69 undergoes a pH-dependent conformational change. In contrast, the structure of the unmodified H69 (UUU) is unaffected by pH. These results emphasize the importance of pseudouridine residues in influencing or facilitating the H69 structural change. Temperature-dependent CD studies also reveal a slight enhancement in H69 stability at lower pH, which is further supported by UV melting data (ΔG°37,pH5.5 − ΔG°37,pH7.0 = −0.5 kcal/mol). CD experiments with the stem-region RNA (dsH69) do not show any notable differences at pH 7.0 and 5.5, leading to the conclusion that the loop region plays a significant role in the pH-dependent structural change of H69. The reduction in molar ellipticity at the peak maximum suggests that stability of the H69 stem decreases at lower pH. These results are supported by 1D imino proton NMR spectra in which the H69 loop has an alternate conformation at lower pH such that the Ψ residues are protected from solvent, and base pairs in the upper stem region appear to denature more readily than the G•U mismatch or the loop region.
Our CD results reveal that a pH-dependent structural change in Ψm3ΨΨ occurs between pH 7.0 and 5.5. The apparent pKa of the protonation event is 6.3. Nucleic acid residues typically do not have pKa values near pH 6 (Saenger 1984). The pKas for imino protons of A and C residues are ∼4 and those for G and U nucleotides are ∼9. It has been shown, however, that the microenvironment of the RNA may allow for the base pKa to be shifted toward neutrality (Bevilacqua et al. 2004). In some cases, local regions of high electrostatic potential may drive the pKa shift. These changes in pKa may in turn affect the RNA function and contribute to enhanced catalytic activity (Wadkins et al. 2001; Kuzmin et al. 2005; Nakano and Bevilacqua 2007; Wilson et al. 2007). In most cases, it is challenging to assign a specific RNA functional group that undergoes protonation, and the exact contributions of a single nucleotide to catalytic function may be difficult to assess (Kuzmin et al. 2005). Experiments are currently in progress to identify the possible site of protonation on H69.
The ability of pH to influence RNA conformation has been known for some time (Bina-Stein and Crothers 1974). Recent examples of pH-dependent RNA tertiary structures include a retrovirus pseudoknot structure (Nixon and Giedroc 2000) and the U6 RNA of the nuclear pre-messenger RNA spliceosome complex (Reiter et al. 2004). The presence of a protonated base triple (C+•G–C; pKa = 6.8) contributes >3 kcal/mol in stability to an RNA stem–loop (Nixon and Giedroc 2000). Similarly, a protonated mismatch base pair (C•+A; pKa = 6.5) assists in stabilizing the flipped-out conformation of a neighboring U in the U6 RNA (Reiter et al. 2004). Durant and Davis (1999) showed similar stabilization (∼1.5 kcal/mol) at lower pH of the tRNALys,3 anticodon stem–loop by a protonated mismatch base pair (A+•C) (pKa ≈ 6), which further strengthened a neighboring A–Ψ pair and effectively reduced a 7-nt loop to a 3-nt loop. Our results with H69 provide further evidence for the roles of nucleotide modification and protonation in regulating RNA structure.
The question is whether pH-dependent conformational switching can occur at the ribosome level, which undergoes numerous structural changes during protein synthesis. The protonation events involved with catalysis (peptidyl transferase) must be distinguished from those required for other stages in protein synthesis, such as recycling and subunit dissociation. Several groups have proposed pH-dependent conformational changes in the peptidyl-transferase center (PTC) of the ribosome (Bayfield et al. 2001; Muth et al. 2001; Xiong et al. 2001; Hesslein et al. 2004), which was shown in one case to involve a A2453•C2499 wobble base pair (Bayfield et al. 2004). Chemical probing studies reveal different patterns of protection in the PTC of rRNAs from different organisms, suggesting that the pH-dependent conformational changes in the ribosome may involve more than one ionizing group (Beringer et al. 2005).
The pH-dependent conformational changes of E. coli helix 69 are of particular interest in this study. Helix 69 contains three highly conserved Ψ residues at positions 1911, 1915, and 1917. Residues in the loop region of H69 are also highly conserved. A1912 and U(Ψ)1917, which are both involved in a noncanonical reversed-Hoogsteen base pair (Schuwirth et al. 2005), were shown in genetic selection experiments to be essential in E. coli (Hirabayashi et al. 2006). Similarly, mutants A1912G, Ψ1917C, and A1919G show severe growth phenotypes (Liiv et al. 2005), and deletion of A1916 promotes misreading of the genetic code and is lethal in E. coli (O'Connor 2007). Absence of RluD, the pseudouridine synthase responsible for insertion of Ψ1911, Ψ1915, and Ψ1917 in E. coli 23S rRNA, leads to growth inhibition (Raychaudhuri et al. 1998). These data point to a critical biological role for the highly conserved Ψs in H69. Surprisingly, recent studies showed that H69 can be deleted and the ribosomes still maintain the ability to translate natural mRNA in vitro (Ali et al. 2006). Thus, the growth defects observed for the H69 mutants must be associated with another ribosome activity. Recent studies by Ejby et al. (2007) revealed that the helix 69 pseudouridines have a role in translation termination.
A major defect of H69 deletion is loss of requirement for RRF in ribosome recycling (Ali et al. 2006). RRF is found in close proximity of H69 in 70S ribosome cryo-EM structures, and causes H69 to undergo movement away from the 30S subunit (Agrawal et al. 2004; Gao et al. 2005). In comparing a number of high-resolution 70S and 50S ribosome structures, some key differences are observed. The 50S and 70S ribosome structures in the presence or absence of tRNA and mRNA components, various factors such as RRF or EF-G, or aminoglycosides vary in the relative positioning of H69, as well as the orientation of A1913. This residue has the ability to reside inside the loop or to flip out and make contacts with other ribosomal components (Gao et al. 2005; Petry et al. 2005; Schuwirth et al. 2005; Wilson et al. 2005; Selmer et al. 2006; Borovinskaya et al. 2007; Weixlbaumer et al. 2007). These conformational changes observed for H69 cause key bridge interactions with the 30S subunit to be affected (Wilson et al. 2005; Borovinskaya et al. 2007).
The changes that we observe in the isolated H69 structure with pH are consistent with the differences observed between the 50S and 70S ribosome structures. The CD and 1D NMR data for H69 at pH 7.0 are consistent with the bacterial 70S ribosome structure, in which A1913 is flipped out, whereas the low pH structure is predicted to be more constrained with a higher degree of base stacking, as observed in the 50S ribosome structure. The ability of H69 to undergo this conformational switching involving A1913 could be impaired in the absence of pseudouridine residues in the loop. Overall, our data provide support for the important biological roles of H69 and emphasize the importance of conformational flexibility in this highly modified, highly conserved region of the large subunit of the ribosome. This conformational change is dependent on the presence of Ψ in the loop region of H69. Although the exact site of protonation on H69 is unknown at this time, future structure and biochemical studies may provide clues toward understanding whether this conformational change via protonation occurs at the ribosome level during certain stages of protein synthesis.
MATERIALS AND METHODS
Preparation of RNAs
Two different 19-nt RNA analogs of helix 69 were used for pH-dependent experiments (residues 1906–1924). The first sequence contained two pseudouridine (Ψ) nucleotides (corresponding to positions 1911 and 1917 of E. coli 23S rRNA) and a 3-methylpseudouridine (m3Ψ) nucleotide (position 1915) (Ψm3ΨΨ), and the second sequence contained all uridines in place of the modified nucleotides (UUU). The third construct was composed of two separate strands (5′ half and 3′ half of dsH69 stem) that were annealed in an equimolar ratio. The sequences of the RNAs are as follows:
5′-GGCCGΨAAC(m3Ψ)AΨAACGGUC-3′ (Ψm3ΨΨ),
5′-GGCCGUAACUAUAACGGUC-3′ (UUU),
5′-GGCCGΨA-3′ (5′ half of dsH69 stem), and
5′-ACGGUC-3′ (3′ half of dsH69 stem).
These RNAs with the 2′-O-ACE protecting group were purchased from Dharmacon Research, where they were chemically synthesized on 0.2 or 1.0 μmol scales. The 3-methylpseudouridine phosphoramidite needed for generation of Ψm3ΨΨ was synthesized as described previously (Chui et al. 2002).
Deprotection of the RNAs was done by incubation in TEMED-acetate buffer (pH 3.8) for 30 min at 60°C (Scaringe 2001). The deprotected RNAs were purified by HPLC on an XTerra MS C18 column (2.5 μm × 50 mm; Waters). A 0.1 M TEAA (triethylamine acetate) buffer at pH 7.0 was used as the mobile phase with a 7%–11% linear gradient of CH3CN over 17 min. After HPLC purification, the RNAs were desalted by ethanol precipitation and dialysis for 3 d against RNase-free, double-deionized water. Authenticity and purity of the RNAs were confirmed by MALDI mass spectrometry and 20% denaturing polyacrylamide gel electrophoresis. The RNAs used for circular dichroism experiments and NMR experiments were renatured (Ψm3ΨΨ or UUU) or annealed (in the case of the double-stranded construct, dsH69) in buffers used for the respective experiment by heating to 90°C for 3 min and cooling slowly to room temperature. Alternatively, the RNA solution was cooled rapidly by placing on ice. RNA concentrations were calculated by using Beer's law and the single-stranded extinction coefficients (ε260nm = 189, 400 cm−1 M−1 for Ψm3ΨΨ and UUU, 69,500 cm−1M−1 for the 5′ half of dsH69 stem, and 56,100 cm−1M−1 for the 3′ half of dsH69 stem). Extinction coefficients were calculated as described previously using the value of 1 × 10−4 cm−1 M−1 for uridine, pseudouridine, and 3-methylpseudouridine (Meroueh et al. 2000; Richards 1975).
Circular dichroism spectroscopy
Circular dichroism spectra for RNAs were obtained on a JASCO J600 or Chirascan circular dichroism spectropolarimeter equipped with a water bath to control the temperature. Experiments were carried out in buffer A (15 mM NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA, with pH adjusted by 1 M HCl), buffer B (20 mM cacodylic acid, 35–150 mM KCl, and 0–30 mM MgCl2 with pH adjusted by addition of KOH), or buffer C (25 mM cacodylic acid, 25 mM Tris [basic form], 30 mM KCl, 70 mM NH4Cl, and 3 mM MgCl2, with pH adjusted by addition of HCl). The RNAs were renatured by heating to 90°C for 3 min and cooling slowly to room temperature or by cooling rapidly on ice. The ΔA values (λ = 220 − 320 nm) were converted to molar ellipticity (Δε) values, with values expressed in moles of RNA molecules (Cantor and Schimmel 1980). The RNA concentrations were determined from the UV absorbance values (λ = 260 nm) at 95°C. All CD experiments were done in duplicate or triplicate to ensure consistent results.
The apparent pKa of the system was obtained by plotting ellipticity at various wavelengths (280–290 nm) versus pH and fitting to an equation derived using the Henderson and Hasselbalch equation (Bose et al. 2007, Equation 1). Equation 1 is derived considering that the change in ellipticity at a certain wavelength is proportional to the protonation. Δε288 nm,i is the molar ellipticity at the highest pH value, ΔΔε288 nm is the change in molar ellipticity, and pKa refers to the apparent pKa value for the transition:
Ellipticity values at 280–290 nm were obtained for H69 samples prepared in different buffer solutions with pH ranging from 5.4 to 7.0. Corrections were made to the ellipticity values to account for the changes in the concentration of H69. Data fitting for each of the six curves were carried out using Kaleidagraph v. 4.0 (Synergy Software).
Thermal melting experiments
UV-melting curves for RNAs with five different RNA concentrations (8–80 μM) in 15 mM NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA (pH 5.5 or 7.0) were obtained on an Aviv 14DS UV-visible spectrophotometer equipped with a five-cuvette thermoelectric controller. Five cuvettes with 1- and 2-mm pathlengths were employed, and the data were collected at 280 nm every 0.5°C between 0 and 90°C. Concentrations of the RNAs were calculated using the UV absorbance (λ = 260 nm) at 90°C and appropriate extinction coefficients. Thermodynamic parameters were derived using Meltwin 3.5 (McDowell and Turner 1996) assuming a two-state equilibrium for RNA melting. Melting experiments were carried out in triplicate, and average values for the thermodynamic parameters were obtained. Experiments were also carried out in 15 mM cacodylic acid, 150 mM KCl, and 0–30 mM MgCl2 with pH adjusted by addition of KOH.
NMR spectroscopy
NMR spectra for Ψm3ΨΨ were obtained on a Bruker AVANCE-AQS 700 MHz spectrometer equipped with a 5-mm triple-resonance cryoprobe. The concentration of the Ψm3ΨΨ RNA in each sample was 175 μM in buffer containing 10 mM Na2HPO4, 30 mM NaCl, and 0.5 mM Na2EDTA in 9:1 H2O/D2O (pH 6.5 or pH 5.5). The imino proton assignments of the H69 were compared and confirmed by earlier studies (Meroueh et al. 2000). Pseudouridine imino proton peaks were assigned using 15N-labeled samples (Desaulniers 2005). Watergate 5 with a gradient pulse sequence was used for the suppression of the water peak (Hwang and Shaka 1995). Digital Quadratic detection for at least 16,000 data points was carried out for acquisition of 1D-1H NMR spectra. Data were averaged for at least 128 scans to improve the signal-to-noise ratio.
ACKNOWLEDGMENTS
We thank Jay Herath and Yu-Cheng Chang for synthesis of the m3Ψ phosphoramidite and Mina Sumita, Larry Clos, Ravi Aduri, Andrew Feig, and John SantaLucia for instrument training and helpful discussions. This work was support by NIH GM54632.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.779908.
REFERENCES
- Agrawal, R.K., Sharma, M.R., Kiel, M.C., Hirokawa, G., Booth, T.M., Spahn, C.M.T., Grassucci, R.A., Kaji, A., Frank, J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc. Natl. Acad. Sci. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, I.K., Lancaster, L., Feinberg, J., Joseph, S., Noller, H.F. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Allawi, H.T., SantaLucia J., Jr Thermodynamics and NMR of internal G•T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- Arnez, J.G., Steitz, T.A. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- Bakin, A., Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bashan, A., Agmon, I., Zarivach, R., Schluenzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H.A.S., et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- Bayfield, M.A., Dahlberg, A.E., Schulmeister, U., Dorner, S., Barta, A. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc. Natl. Acad. Sci. 2001;98:10096–10101. doi: 10.1073/pnas.171319598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield, M.A., Thompson, J., Dahlberg, A.E. The A2453-C2499 wobble base pair in Escherichia coli 23S ribosomal RNA is responsible for pH sensitivity of the peptidyltransferase active site conformation. Nucleic Acids Res. 2004;32:5512–5518. doi: 10.1093/nar/gkh888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer, M., Bruell, C., Xiong, L., Pfister, P., Bieling, P., Katunin, V.I., Mankin, A.S., Böttger, E.C., Rodnina, M.V. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J. Biol. Chem. 2005;280:36065–36072. doi: 10.1074/jbc.M507961200. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., Brown, T.S., Nakano, S., Yajima, R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Bina-Stein, M., Crothers, D.M. Conformational changes of transfer ribonucleic acid. The pH phase diagram under acidic conditions. Biochemistry. 1974;13:2771–2775. doi: 10.1021/bi00710a017. [DOI] [PubMed] [Google Scholar]
- Borovinskaya, M.A., Pai, R.D., Zhang, W., Schuwirth, B.S., Holton, J.M., Hirokawa, G., Kaji, H., Kaji, A., Cate, J.H.D. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Bose, K., Yoder, N.C., Kumar, K., Baleja, J.D. The role of conserved histidines in the structure and stability of human papillomavirus type 16 E2 DNA-binding domain. Biochemistry. 2007;46:1402–1411. doi: 10.1021/bi0611255. [DOI] [PubMed] [Google Scholar]
- Cantor, C.R., Schimmel, P.R. Biophysical chemistry. Part II: Techniques for the study of biological structure and function. W.H. Freeman; San Francisco: 1980. [Google Scholar]
- Chui, H.M.-P., Desaulniers, J.-P., Scaringe, S.A., Chow, C.S. Synthesis of helix 69 of Escherichia coli 23S rRNA containing its natural modified nucleosides, m3Ψ and Ψ. J. Org. Chem. 2002;67:8847–8854. doi: 10.1021/jo026364m. [DOI] [PubMed] [Google Scholar]
- Davis, D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D.R., Poulter, C.D. 1H-15N NMR studies of Escherichia coli tRNAPhe from hisT mutants: A structural role for pseudouridine. Biochemistry. 1991;30:4223–4231. doi: 10.1021/bi00231a017. [DOI] [PubMed] [Google Scholar]
- Desaulniers, J.-P. Wayne State University; Detroit, MI: 2005. “Exploring nucleic acid modification in RNA hairpins and antisense oligonucleotides.”. Ph.D. thesis, [Google Scholar]
- Durant, P.C., Davis, D.R. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- Ejby, M., Sørensen, M.A., Pedersen, S. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc. Natl. Acad. Sci. 2007;104:19410–19415. doi: 10.1073/pnas.0706558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada, H., Takahashi, K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Protein Struct. Funct. Genet. 1998;33:159–166. [PubMed] [Google Scholar]
- Fürtig, B., Richter, C., Wöhnert, J., Schwalbe, H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- Gao, N., Zavialov, A.V., Li, W., Sengupta, J., Valle, M., Gursky, R.P., Ehrenberg, M., Frank, J. Mechanism for the disassembly of the post-termination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B., Rodnina, M.V. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Hesslein, A.E., Katunin, V.I., Beringer, M., Kosek, A.B., Rodnina, M.V., Strobel, S.A. Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes. Nucleic Acids Res. 2004;32:3760–3770. doi: 10.1093/nar/gkh672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi, N., Sato, N.S., Suzuki, T. Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J. Biol. Chem. 2006;281:17203–17211. doi: 10.1074/jbc.M511728200. [DOI] [PubMed] [Google Scholar]
- Hirashima, A., Kaji, A. Factor dependent breakdown of polysomes. Biochem. Biophys. Res. Commun. 1970;41:877–883. doi: 10.1016/0006-291x(70)90165-8. [DOI] [PubMed] [Google Scholar]
- Hwang, T.-L., Shaka, A.J. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J. Magn. Reson. A. 1995;112:275–279. [Google Scholar]
- Ivanov, V.I., Minchenkova, L.E., Schyolkina, A.K., Poletayev, A.I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12:89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- Kowalak, J.A., Bruenger, E., Hashizume, T., Peltier, J.M., Ofengand, J., McCloskey, J.A. Structural characterization of U*-1915 in domain IV from Escherichia coli 23S ribosomal RNA as 3-methylpseudouridine. Nucleic Acids Res. 1996;24:688–693. doi: 10.1093/nar/24.4.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin, Y.I., Da Costa, C.P., Cottrell, J.W., Fedor, M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lane, B.G., Ofengand, J., Gray, M.W. Pseudouridine and O2′-methylated nucleosides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bonds in proteins. Biochimie. 1995;77:7–15. doi: 10.1016/0300-9084(96)88098-9. [DOI] [PubMed] [Google Scholar]
- Liiv, A., Karitkina, D., Maiväli, Ü., Remme, J. Analysis of the function of E. coli 23S rRNA helix-loop 69 by mutagenesis. BMC Mol. Biol. 2005;6:18. doi: 10.1186/1471-2199-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbach, P.A., Crain, P.F., McCloskey, J.A. Summary: The modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.A., Turner, D.H. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- Meroueh, M., Grohar, P.J., Qiu, J., Santalucia J., Jr, Scaringe, S.A., Chow, C.S. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res. 2000;28:2075–2083. doi: 10.1093/nar/28.10.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, H., Sundaralingam, M. Stacking of Crick Wobble pair and Watson–Crick pair: Stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon–anticodon Wobble interaction. Nucleic Acids Res. 1978;5:4451–4462. doi: 10.1093/nar/5.11.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth, G.W., Chen, L., Kosek, A.B., Strobel, S.A. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA. 2001;7:1403–1415. [PMC free article] [PubMed] [Google Scholar]
- Nakano, S., Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: A mutant of the C41 motif provides insight into the positioning and thermodynamic linkage of metal ions and protons. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- Newby, M.I., Greenbaum, N.L. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. 2002;99:12697–12702. doi: 10.1073/pnas.202477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, P.L., Giedroc, D.P. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- O'Connor, M. Interaction between the ribosomal subunits: 16S rRNA suppressors of the lethal ΔA1916 mutation in the 23S rRNA of Escherichia coli . Mol. Genet. Genomics. 2007;278:307–315. doi: 10.1007/s00438-007-0252-y. [DOI] [PubMed] [Google Scholar]
- Ofengand, J., Bakin, A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria, and chloroplasts. J. Mol. Biol. 1997;266:246–268. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- Peske, F., Rodnina, M.V., Wintermeyer, W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Petry, S., Brodersen, D.E., Murphy F.V., IV, Dunham, C.M., Selmer, M., Tarry, M.J., Kelley, A.C., Ramakrishnan, V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri, S., Conrad, J., Hall, B.G., Ofengand, J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli . RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, N.J., Blad, H., Abildgaard, F., Butcher, S.E. Dynamics in the U6 RNA intramolecular stem–loop: A base flipping conformational change. Biochemistry. 2004;43:13739–13747. doi: 10.1021/bi048815y. [DOI] [PubMed] [Google Scholar]
- Richards, E.G. Handbook of biochemistry and molecular biology. CRC Press; Cleveland, OH: 1975. Use of tables in calculation of absorption, optical rotary dispersion, and circular dichroism of polyribonucleotides; pp. 596–599. [Google Scholar]
- Saenger, W. Principles of nucleic acid structure. Springer-Verlag; New York: 1984. [Google Scholar]
- SantaLucia J., Jr, Turner, D.H. Measuring the thermodynamics of RNA secondary structure formation. Biopolymers. 1997;44:309–319. doi: 10.1002/(SICI)1097-0282(1997)44:3<309::AID-BIP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Scaringe, S.A. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods. 2001;23:206–217. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H.D. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy F.V., IV, Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Subramanian, A.R., Davis, B.D. Release of 70S ribosomes from polysomes in Escherichia coli . J. Mol. Biol. 1973;74:45–56. doi: 10.1016/0022-2836(73)90353-7. [DOI] [PubMed] [Google Scholar]
- Varani, G., Aboul-ela, F., Allain, F.H.-T. NMR investigation of RNA structure. Progress in NMR Spectroscopy. 1996;29:51–127. [Google Scholar]
- Wadkins, T.S., Shih, I.-h., Perrotta, A.T., Been, M.D. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- Weixlbaumer, A., Petry, S., Dunham, C.M., Selmer, M., Kelley, A.C., Ramakrishnan, V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- Wilson, D.N., Nierhaus, K.H. The ribosome through the looking glass. Angew. Chem. Int. Ed. Engl. 2003;42:3464–3486. doi: 10.1002/anie.200200544. [DOI] [PubMed] [Google Scholar]
- Wilson, D.N., Schluenzen, F., Harms, J.M., Yoshida, T., Ohkubo, T., Albrecht, R., Buerger, J., Kobayashi, Y., Fucini, P. X-ray crystallography study on ribosome recycling: The mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.J., Ouellet, J., Zhao, Z.-Y., Harusawa, S., Araki, L., Kurihara, T., Lilley, D.M.J. Nucleobase catalysis in the hairpin ribozyme. RNA. 2007;12:980–987. doi: 10.1261/rna.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody, R.W. Circular dichroism. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Polacek, N., Sander, P., Böttger, E.C., Mankin, A. pKa of adenine 2451 in the ribosomal peptidyl transferase center remains elusive. RNA. 2001;7:1365–1369. [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Zh., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]