Abstract
Two-component histidine kinases recently have been found in eukaryotic organisms including fungi, slime molds, and plants. We describe the identification of a gene, COS1, from the opportunistic pathogen Candida albicans by using a PCR-based screening strategy. The sequence of COS1 indicates that it encodes a homolog of the histidine kinase Nik-1 from the filamentous fungus Neurospora crassa. COS1 is also identical to a gene called CaNIK1 identified in C. albicans by low stringency hybridization using CaSLN1 as a probe [Nagahashi, S., Mio, T., Yamada-Okabe, T., Arisawa, M., Bussey, H. & Yamada-Okabe, H. (1998) Microbiol. 44, 425–432]. We assess the function of COS1/CaNIK1 by constructing a diploid deletion mutant. Mutants lacking both copies of COS1 appear normal when grown as yeast cells; however, they exhibit defective hyphal formation when placed on solid agar media, either in response to nutrient deprivation or serum. In constrast to the Δnik-1 mutant, the Δcos1/Δcos1 mutant does not demonstrate deleterious effects when grown in media of high osmolarity; however both Δnik-1 and Δcos1/Δcos1 mutants show defective hyphal formation. Thus, as predicted for Nik-1, Cos1p may be involved in some aspect of hyphal morphogenesis and may play a role in virulence properties of the organism.
One of the most common fungal pathogens in humans is Candida albicans. Although C. albicans is normally present in the gastrointestinal tract and does not cause disease, weakening of the host immune system through a variety of factors, such as administration of chemotherapeutic agents for cancer treatment or the presence of AIDS, often leads to colonization of tissues and resulting disease (1, 2). Systemic fungal infections are difficult to treat because most antifungal compounds exhibit unpleasant side effects and toxicities. Increasing resistance to antifungal compounds by common pathogens, as well as the increasing number of individuals who find themselves with compromised immunological function, warrants an increased understanding of the basic biology of fungi to identify potential antifungal targets. A number of factors have been implicated as being associated with the virulence properties of C. albicans, such as adherence to host cells and the ability to undergo the transition from yeast to hyphal growth known as dimorphism (3, 4). Recent studies from a number of laboratories have taken advantage of the fact that Saccharomyces cerevisiae can be induced to grow filamentously (pseudohyphal growth) (5) and that genes involved in the yeast-to-hyphal transition in C. albicans often can complement defects in genes in the S. cerevisiae pathway. This approach has led to the identification of a MAPK pathway involving the Candida genes CST20, HST7, and CPH1, which are homologs of the Saccharomyces STE20, STE7, and STE12 genes, respectively, that play a dual role in mating as well as in pseudohyphal growth (6–9). Deletion of the Candida genes results in organisms that are impaired in the ability to make hyphae under selected conditions, although not in response to serum (6–9). This result suggests that there is more than one pathway controlling hyphal growth. Previous work has demonstrated that, in S. cerevisiae, an additional mutation in the PHD1 gene is required to completely inhibit the filamentous transition of ste12 strains (10). EFG1, a PHD1 homolog, has been found in C. albicans, and its reduced expression causes loss of true hyhae formation (11). When double cph1/cph1 efg1/efg1 Candida mutants were made, they were unable to form hyphae under any condition tested and were avirulent in a mouse model (10). Additionally, a homolog of Saccharomyces cerevisiae Cla4p (a Ste20-like kinase) has been found in C. albicans (CaCla4p), and its deletion causes a complete abrogation of hyphal formation under all conditions (12). These mutants also demonstrate a loss of virulence in mice. This loss may be caused by the involvement of CaCla4p in polarized cell growth as demonstrated in S. cerevisiae. Recently, a gene encoding a protein with integrin-like properties was found in C. albicans. This protein, Int1p, is found on the cell surface and has been shown to affect the virulence properties of C. albicans. Deletion mutants of INT1 have reduced adherence to epithelial cells, and the deletion causes loss of hyphal formation (13). The above studies taken together demonstrate the importance of the transition from yeast-like to hyphal growth in Candida virulence.
The involvement of homologous genes in controlling filamentous growth suggests that morphogenetic events involved in hyphal formation in various fungi may be conserved. We have shown previously that, in the filamentous fungus Neurospora crassa, a two-component histidine kinase, Nik-1, is involved in proper hyphal development and protection against increased environmental osmostress (14, 15). Unlike S. cerevisiae and C. albicans, N. crassa exhibits hyphal growth until it undergoes asexual spore formation, when it subsequently divides by budding (16). Deletion of nik-1+ causes multiple morphological defects such as misshapen hyphae that lyse and reduced conidiation. Under increased osmotic pressure, these defects become severe, resulting in loss of hyphal and spore formation capabilities. These characteristics are found for a number of osmotic mutants of N. crassa; os-1, os-2, os-4, os-5, and cut. (17). Osmotic mutants all demonstrate defective hyphal formation, reduced conidiation, and sensitivity to increased osmostress (17, 18). Osmotic mutants also are known to be insensitive to a number of dicarboximide fungicides (19) and have altered cell wall compositions (20, 21). Recently, the gene for one of the osmotic loci, os-1, was cloned by functional complementation of os-1 mutants and was shown to be identical to nik-1+ (15). The PCR-based screening method that led to the original identification of nik-1+ has been used subsequently to identify a homolog in C. albicans. In this report, we describe the isolation and characterization of COS1, whose predicted protein sequence shows it to be a member of the hybrid family of histidine kinases and a homolog of Nik-1 (Os-1) from N. crassa. As in N. crassa, Cos1p plays a role in hyphal development under certain growth conditions in C. albicans that will be discussed.
MATERIALS AND METHODS
Molecular Biological Manipulations.
All common molecular biological manipulations were performed by using standard methods or as described by the manufacturer. DNA sequencing was done by using dye terminator and dye primer chemistries on an automated sequencer (Applied Biosystems models 373 and 377) at the California Institute of Technology DNA Sequencing Facility. All DNA sequence data manipulation was performed with sequencher (version 2.1, Gene Codes, Ann Arbor, MI). Homology searches and alignments were done by using programs of the GCG package.
Strains and Culture Conditions.
The C. albicans strains used in this work are listed in Table 1 (22, 23). C. albicans strains were grown routinely in yeast extract/peptone/dextrose with uridine added to a final concentration of 50 μg/ml when necessary. Selection of URA3 transformants was done on agar plates of SD [0.67% (wt/vol) yeast nitrogen base without amino acids and 2% (wt/vol) glucose, final concentration]. Hyphal formation was monitored at 37°C by several methods: incubation of cells in liquid yeast extract/peptone/dextrose with 10% (vol/vol) bovine serum (Sigma), incubation in liquid Spider medium (6), incubation on solid Spider plates, and incubation on agar plates [Difco, 1.2% (wt/vol)] containing 10% (vol/vol) bovine serum.
Table 1.
C. albicans strains
| Strain | Genotype | Reference |
|---|---|---|
| 366 | wild-type isolate | ATCC |
| 36801 | wild-type isolate | ATCC |
| SC5314 | wild-type isolate | (22) |
| CAI4 | Δura3::imm434/Δura3::imm434 | (23) |
| LAC13 | Δura3::imm434/Δura3::imm434/Δcos1::hisG-URA3-hisG/COS1 | This work |
| LAC14 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/COS1 | This work |
| LAC15 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/COS1 | This work |
| LAC16 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/Δcos1::hisG-URA3-hisG | This work |
| LAC18 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/Δcos1::hisG-URA3-hisG | This work |
| LAC20 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/Δcos1::hisG | This work |
| LAC21 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/Δcos1::hisG | This work |
| LAC22 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/Δcos1::hisG (2 μ COS1) | This work |
| LAC23 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/Δcos1::hisG (2 μ COS1) | This work |
| LAC26 | Δura3::imm434/Δura3::imm434/Δcos1::hisG/COS1::URA3 | This work |
Identification of the COS1 Histidine Kinase Domain in C.
albicans. The PCR primers H1A [CA(T/C)GAI(A/T/C)TI(C/A)GIACICC] and N2A [GC(A/G)TTIC(T/C)IACIA(G/A)(G/A)TT] (14) were used to amplify genomic DNA from wild-type C. albicans (ATCC 36801). C. albicans genomic DNA was prepared as described (24). PCRs contained 2.5 μM each primer, 1.5 μg genomic DNA, and 2.5 mM each dATP, dCTP, dGTP, and dTTP in 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 0.001% (wt/vol) Gelatin, 0.5 units of Amplitaq (Perkin–Elmer/Cetus) in a total volume of 100 μl. In a Perkin–Elmer thermocycler, all reaction components except the nucleotides and polymerase were incubated at 95°C (10 min) and then cooled to 4°C. Polymerase and nucleotides were added, and the reaction was cycled 30 times with the sequence at 94°C (1 min), 40°C (1 min), and 72°C (1 min) followed by extension at 72°C (10 min). PCR products were separated and purified by electrophoresis through a 1.5% (wt/vol) agarose gel. The purified products were ligated into a T-vector (Promega) and then sequenced.
Isolation of C. albicans COS1.
The PCR product generated from amplification of genomic C.albicans DNA was used to screen a size-selected C. albicans genomic library. The library was generated either by cloning size-selected HindIII/BamHI fragments of C. albicans ATCC 366 genomic DNA into pUC19 digested with the same restriction enzymes or by cloning 7- to 10-kb HindIII fragments of the same DNA into HindIII digested pUC19. Two clones were obtained that provided the templates for sequencing the COS1 gene. One is pCHK1, which has a 9.0-kb HindIII insert, and the other is pCA3.8, which has a 3.8-kb HindIII/BamHI overlapping insert of genomic DNA of pCHK1. The sequence of COS1 was obtained by primer walking on both strands of pCHK2 and pCA3.8.
Construction of COS1 Deletion Mutant.
Both alleles of COS1 were deleted by using the “URA blaster” technique (23). A 1.3-kb XhoI/NcoI fragment was removed from pCA3.8, and the NcoI site was blunted by treatment with Klenow fragment. To this was ligated a 3.6-kb SalI/BglII fragment from pBC19 in which the BglII site was blunted with Klenow. pBC19 is pUC19 where the BamHI site was converted to BglII and subsequently used to clone a BglII/SalI fragment from pCUB6 containing the hisG-URA3-hisG cassette (B. Cormack, Stanford University, unpublished results). The resulting plasmid is pKOCOS-1. Spheroplasts of C. albicans strain CAI4 (ura-) were transformed with pKOCOS-1 linearized with HindIII and ura+ transformants selected on SD plates containing 1 M sorbitol. Genomic DNA was isolated from several transformants and digested with BglII. DNA fragments were resolved by electrophoresis through an 0.8% (wt/vol) agarose gel and transferred to a nylon membrane. Blots were probed with a PCR fragment, which corresponds to nucleotides 3528–3996 of the genomic sequence, that was generated with the primers CA13 [TGGTACAGGTTTAGGGTTGTC] and CA15 [AACATGGCGTATTTGCATAGG]. Recombinants containing one wild-type allele and one disrupted allele (LAC13) were identified by the presence of the desired restriction fragments. One hisG repeat and the URA3 marker were removed by selection of LAC13 cells on media containing 5-fluoroorotic acid as described (25) except that uridine was substituted for uracil. The loss of hisG-URA3 was followed by Southern blotting with both the CA13/CA15 PCR product and the URA3 gene as probes. This yielded clones LAC14 and LAC15 (Δcos1∷hisG/COS1). These cells then were subjected to a second round of transformation and analysis as described for replacement of the first allele to give LAC16 and LAC18 (Δcos1∷hisG/Δcos1∷hisG-URA3-hisG) and finally LAC20 and LAC21 (Δcos1:hisG/Δcos1∷hisG).
Complementation of COS1 Deletion Mutant.
To make the template for homologous reintegration of the COS1 gene, the 9.0-kb HindIII fragment was removed from pCHK1 with HindIII and cloned into the HindIII site of pBluescript SK+ to give pBSCOS-1. A 1.3-kb fragment containing URA3 (generated from PCR amplification of pBC19 with U3BF [TGGATCCAGTACTAATAGGAATTGATTTGG] and U3BR [TGGATCCTCTAGAAGGACCACCTTTGATTG], cloned into a T-vector, and digested with BamHI, with the ends made blunt by treatment with Klenow fragment) was ligated into the EheI site downstream from COS1 in pBSCOS-1 to yield pBSCOS-1/URA3. The relevant insert was isolated after HindIII digestion and used to transform LAC20. Cells were selected on SD containing 1 M sorbitol. Genomic DNA was isolated from selected clones, digested with BglII, and blotted to nylon. The PCR product CA13/15 was used to probe for the reappearance of the desired band and yielded strains LAC26. Alternatively, the Δcos1/Δcos1 mutant LAC20 or LAC21 was transformed with pYPBCOS-1+, which is COS1 cloned into a 2-μ-based plasmid. The plasmid pYPBCOS-1+ was made by digesting pYPB-ADHpt (8) with NotI and SalI to remove the ADH promoter and ligating to this a NotI/SalI fragment from plasmid pT(CA32/33) containing COS1. PCR amplification (Taqplus, Stratagene) of pCHK1 with the primers CA32 [ATGCGGCCGCGGAGTTATGAAAGTTGTGGGC] and CA33 [ATGGTCGACAGTTCCTACACAACAATTTGGC] yielded a fragment spanning nucleotides 1060–4865 of the genomic sequence with an engineered NotI site at the 5′ end of the clone and a SalI site at the 3′ end, which was cloned into a T-vector (Promega) to give pT(CA32/33). The resulting ura+ transformants (LAC22, LAC23) were selected on SD plates containing 1 M sorbitol.
RESULTS
Sequence of COS1 from C. albicans.
PCR screening of C. albicans genomic DNA with degenerate primers that encode the sequence of the H and N boxes of two-component histidine kinases were used to clone a genomic DNA fragment that, upon sequencing, demonstrated homology to other histidine kinase family members (14). The fragment encoded a peptide that was most similar to the sequence of the Nik-1/Os-1 kinase from N. crassa (83% identity, data not shown). This PCR fragment was used as a probe to screen a genomic C. albicans library and led to the identifcation of a 9-kb HindIII and 3.8-kb HindIII/BamHI DNA fragment, which, upon subcloning, were sequenced.
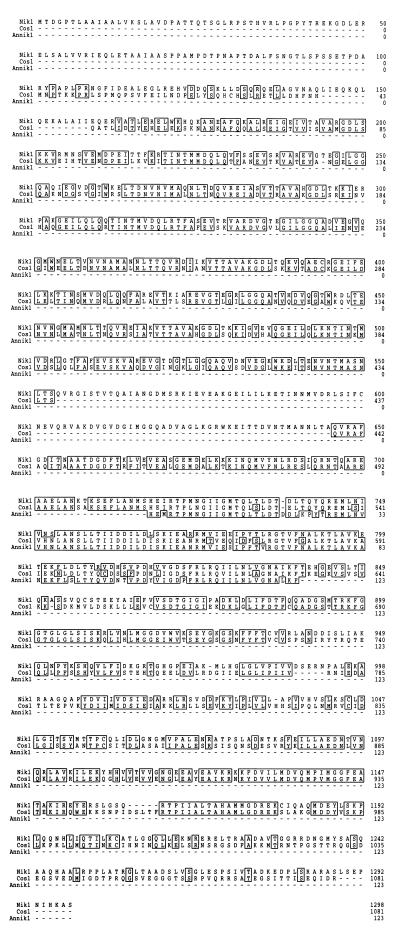
Translation of the COS1 gene from this fragment predicts a polypeptide of 1,081 amino acids (mw 118, 983) that, when aligned with other histidine kinases, demonstrated that, in fact, we had cloned a homolog of Nik-1(Os-1) (70% similarity overall); we call the gene “COS1” (Fig. 1) The sequence of the ORF predicted from COS1 is shown aligned with Nik-1 from N. crassa and the sequence of a translated DNA fragment that we obtained by PCR in Aspergillus nidulans (L.A.A., unpublished results) (Fig. 1). The sequence of Cos1p predicted from the gene indicates that this protein is a member of the hybrid family of histidine kinases, those that contain both a histidine kinase domain and a response regulator domain (26). Members of this class include the majority of histidine kinases described in eukaryotic organisms including fungi, yeasts, slime molds, and plants (reviewed in ref. 27). The overall domain organization is conserved between the Nik-1 and Cos1p, including the large N-terminal domain that contains tandem repeats of 90 amino acids, which is predicted to form a coiled-coil structure (14); however, Cos1p is missing one of the six repeats found in Nik-1. Of interest, coiled-coil regions have been implicated in the control of histidine kinase activity as in the case of the regulation of CheA, the kinase that controls bacterial chemotaxis (28). The major differences between Cos1p and Nik-1 occur at the very amino- and carboxy-terminal ends of the proteins. The N. crassa protein contains a longer amino-terminal domain (Fig. 1). Like Nik-1, Cos1p does not seem to contain regions that are likely to span the lipid bilayer, and hence this protein may be soluble. We note that there are two CTG codons that correspond to amino acids 463 and 899 of Cos-1p, which are translated as SER in C. albicans (29).
Figure 1.
Predicted amino acid sequence of Cos1p aligned with Nik-1(Os-1) of N. crassa and the homolog, AnNik-1, encoded by a PCR product identified from Aspergillus nidulans (L.A.A., unpublished results). Boxed residues are identical.
The POLIII gene is translated in the opposite direction to COS1, and its 5-prime end is just upstream of the COS1 gene (corresponding to nucleotide 960). Thus, the initiator ATGs of POLIII and COS1 are only 434 bp apart but are translated in opposite directions. There is a pyrimidine stretch (nucleotides 1,088–1,119) upstream from the predicted Cos-1p product as well as potential CAAT boxes (nucleotides 1,131–1,134 and 1,379–1,382) and a potential TATA box (nucleotides 1,382–1,385), which suggests that this region is the promoter for COS1. Unlike nik-1+, COS1 does not contain any introns. Following the stop codon (nucleotides 4,702–4,704), there are several AATAA sequences that may serve as Poly(A) addition signals (nucleotides 4,768–4,775, 4,813–4,821, 5,016–5,023, and 5,088–5,070). The genomic sequence of COS1 has been deposited in GenBank with the accession number U69886.
Cos1p Is Involved in Hyphal Development in C. albicans.
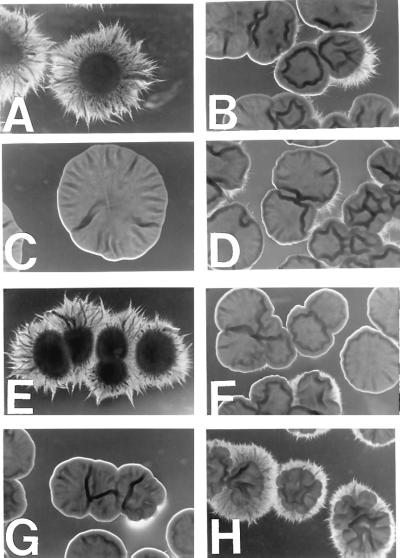
To ascertain a possible role for Cos1p in C. albicans, we constructed a diploid deleted for both copies of the gene (Fig. 2). The mutant and wild-type strains appeared similar when grown in a variety of liquid media at 30°C (data not shown). Upon deletion of only one copy (Δcos1/COS1), we noticed that the heterozygotes could not make hyphae as well as wild type when grown at 37°C on solid agar plates, either under conditions of nutrient deprivation (Fig. 3) or in response to serum (Fig. 4). This phenotype was more exaggerated in the Δcos1/Δcos1 mutants. Thus, like N. crassa Nik-1, C. albicans Cos1p is involved in hyphal development. One major difference is that the Δcos1 strains, when grown as yeasts, did not exhibit the osmosensitivity that N.crassa Δnik-1 strains demonstrate (data not shown). Increased osmolyte concentration inhibits hyphal development in the wild type such that differences between the wild type and mutant were difficult to assess. However, the effect on solid surfaces may be an osmotic-like effect because it could be that reduced water activity is the parameter actually being sensed. We were able to partially complement the defect in hyphal formation by the introduction of a wild-type copy of COS1 into Δcos1/Δcos1 strains either by homologously recombining it back into the genome or by expression from a 2 μ-containing plasmid (Figs. 3 and 4).
Figure 2.
Disruption of the COS1 gene from C. albicans. (A) Schematic showing the replacement strategy. Restriction sites are H (HindIII), Bg (BglII), X (XhoI), N (NcoI), and E (EcoRI). (B) Southern blot of genomic DNA to detect replacement of COS1 with hisG-URA3-hisG. After digestion of genomic DNA with BglII, the replacement event was detected by probing with a PCR product generated from CA13 and CA15 primers. Lanes: 1, CA I4 (wild-type parent); 2, LAC13 (Δcos1∷hisG-URA3-hisG/COS1); 3, LAC15 (Δcos1∷hisG/COS1); 4, LAC18 (Δcos1∷hisG/Δcos1∷hisG-URA3hisG); 5, CA I4 (wild type); 6, LAC13 (Δcos1∷hisG-URA3-hisG/ COS1); 7, LAC14 (Δcos1∷hisG/COS1); 8, LAC17 (Δcos1∷hisG/ Δcos1∷hisG-URA3-hisG); 9, LAC16 (Δcos1∷hisG/Δcos1∷hisG-URA3hisG).
Figure 3.
Morphology of C. albicans Δcos1/Δcos1 colonies grown on solid agar Spider media. Cells were grown at 37°C on solid Spider plates for 5 days (a-d) or 4 days (e-g). (a and e) SC5314 (wild- type COS1/COS1); (b and f) LAC13 (Δcos1/COS1); (c) LAC16 (Δcos1/Δcos1); (d) LAC26 (Δcos1/Δcos1 strain in which COS1∷URA3 has been recombined back at the cos1 locus, Δcos1/COS1∷URA3); (g) LAC18 (Δcos1/Δcos1); (h) LAC23 (Δcos1/Δcos1 with COS1 expressed from a 2-μ plasmid).
Figure 4.
Morphology of C. albicans Δcos1/Δcos1 colonies grown at 37°C for 3 days on agar plates containing 10% (vol/vol) fetal bovine serum. (a) SC5314 (wild-type COS1/COS1). (b) LAC13 (Δcos1/COS1). (c) LAC16 (Δcos1/Δcos1). (d) LAC 22 (Δcos1/Δcos1 with COS1 on 2-μ plasmid). (e) LAC 18 (Δcos1/Δcos1). (f) LAC 23 (Δcos1/Δcos1 with COS1 on 2-μ plasmid).
DISCUSSION
This work describes Cos1p, a eukaryotic two-component histidine kinase from C. albicans that is a homolog of Nik-1 from the filamentous fungus N. crassa. The same PCR-based strategy used to clone nik-1+ also allowed identification of COS1. From previous work, Nik-1 was implicated in hyphal morphogenesis because deletion mutants exhibited multiple morphological defects including reduced conidiation and aberrant hyphal structure with lysis. These defects were more dramatic when cells were exposed to growth conditions of high osmolarity such that they could no longer form spores or make hyphae (14). Staining with Calcofluor white showed that, when Δnik-1 mutants were grown in high salt, they displayed defective cell wall structure (L.A.A., unpublished results). Thus, the osmosensitivity observed is likely caused by altered cell wall structure. When grown as yeasts on solid or liquid media, Δcos1/Δcos1 mutants did not exhibit osmosensitivity and appeared like wild type (data not shown). However, when the Δcos1/Δcos1 mutants were placed on solid agar surfaces either on Spider medium (Fig. 3) or with 10% serum (Fig. 4), they showed a loss of hyphal formation compared with wild type. This phenotype is similar to C. albicans mutants in HST7, CST20, and CPH1, which are homologs of S. cerevisiae STE7, STE20, and STE12, respectively (6–9). The S. cerevisiae genes are involved in the MAPK pathway controlling pseudohyphal formation in response to nitrogen starvation and may have similar functions in C. albicans (30). hst7, ste20, and cph1 mutants only showed defects in hyphal formation when placed on solid Spider medium but responded normally on solid media with serum (6–9). Because it is known that, in S. cerevisiae, the histidine kinase Sln1p is coupled to the HOG1-MAPK pathway (31), deletion of the histidine kinase possibly could be rescued by overproduction of some downstream component. We were unable to rescue the cos1/cos1 mutant by overexpressing HST7 (a MEKK), suggesting COS1 may lie in a different pathway or interact with components downstream of HST7 (L.A.A., unpublished results).
Investigators have reported the cloning of the S. cerevisiae SLN1 homolog from C. albicans CaSLN1 (32). A diploid knockout of CaSLN1 exhibited the osmosensitive phenotype seen for the S. cerevisiae mutant. The sequence of a second gene called CaNIK1 recently has been reported after its identification by using low stringency hybridization with CaSLN1 and is identical to COS1 (32). The phenotype of Casln1/Casln1 mutants could not be rescued by overexpression of CaNIK1/COS1 (32). This is consistent with our view that Cos1p probably is involved in the regulation of some aspect of cell wall maintenance during hyphal formation. Thus, mutations in this gene would give rise to defective hyphal formation and would have no effect on the yeast form. Because the yeast-to-hyphal transition is important for C. albicans virulence and Cos1p has a role in hyphal development, this protein may be a potential antifungal target, a possiblity currently under investigation.
Acknowledgments
We thank Brendan Cormack (Stanford University) for the gift of strains and plasmids as well as technical advice. We also thank Doreen Harcus (CNRC-NRC, Canada) for the gift of plamids and strains, Jacqueline Agnan for technical assistance in cloning COS1, and members of the Cal Tech Sequencing facility for their assistance in DNA sequencing. This work was funded by National Institutes of Health Grants AI 19296 (M.I.S.) and AI 33354 (C.P.S.).
Footnotes
Data deposition: The genomic DNA sequence encoding COS1 reported in this paper has been deposited in the GenBank database (accession no. U69886).
References
- 1.Odds F C. Candida and Candidiosis. London: Balliere Tindall; 1988. [Google Scholar]
- 2.Scherer S, Magee P T. Microbiol Rev. 1990;54:226–241. doi: 10.1128/mr.54.3.226-241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutler J E. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 4.Calderone R A, Braun P. Microbiol Rev. 1991;55:1–20. doi: 10.1128/mr.55.1.1-20.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Kohler J, Fink G R. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 7.Malathi K, Ganesan K, Datta A. J Biol Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- 8.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A, Brown A J P, Thomas D Y. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohler J R, Fink G R. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo H J, Kohler J R, Didomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 11.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas D Y. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- 13.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 14.Alex L A, Borkovich K A, Simon M I. Proc Natl Acad Sci USA. 1996;93:3416–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumacher M M, Enderlin C S, Selitrennikoff C P. Curr Microbiol. 1997;34:340–347. doi: 10.1007/s002849900193. [DOI] [PubMed] [Google Scholar]
- 16.Springer M L. BioEssays. 1993;15:365–374. doi: 10.1002/bies.950150602. [DOI] [PubMed] [Google Scholar]
- 17.Perkins D D, Radford A, Newmeyer D, Bjorkman M. Microbiol Rev. 1982;46:426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grindle M, Temple W. Trans Br Mycol Soc. 1985;84:396–372. [Google Scholar]
- 19.Grindle M, Temple W. Neurospora Newsl. 1982;29:16–17. [Google Scholar]
- 20.Livingston L R. J Bacteriol. 1969;99:85–90. doi: 10.1128/jb.99.1.85-90.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott W A. Annu Rev Microbiol. 1976;30:85–104. doi: 10.1146/annurev.mi.30.100176.000505. [DOI] [PubMed] [Google Scholar]
- 22.Gillum A M, Tsay E Y H, Kirsch D R. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 23.Fonzi W A, Irwin M Y. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 25.Boeke J, Lacroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 26.Alex L A, Simon M I. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 27.Loomis W F, Shaulsky G, Wang N. J Cell Sci. 1997;110:1141–1145. doi: 10.1242/jcs.110.10.1141. [DOI] [PubMed] [Google Scholar]
- 28.Falke J, Bass R B, Butler S, Chervitz S A, Danielson M A. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos M A S, Tuite M F. Nucl Acid Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H P, Styles C A, Fink G R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 31.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 32.Nagahashi S, Mio T, Ono N, Yamada-Okabe T, Arisawa M, Bussey H, Yamada-Okabe H. Microbiology. 1998;144:425–432. doi: 10.1099/00221287-144-2-425. [DOI] [PubMed] [Google Scholar]