Abstract
The ability to visualize mRNA in single living cells and monitor in real-time the changes of mRNA level and localization can provide unprecedented opportunities for biological and disease studies. However, the mRNA detection specificity and sensitivity are critically dependent on the selection of target sequences and their accessibility. We carried out an extensive study of the target accessibility of BMP-4 mRNA using 10 different designs of molecular beacons (MBs), and identified the optimal beacon design. Specifically, for MB design 1 and 8 (MB1 and MB8), the fluorescent intensities from BMP-4 mRNA correlated well with the GFP signal after upregulating BMP-4 and co-expressing GFP using adenovirus, and the knockdown of BMP-4 mRNA using siRNA significantly reduced the beacon signals, demonstrating detection specificity. The beacon specificity was further confirmed using blocking RNA and in situ hybridization. We found that fluorescence signal from MBs depends critically on target sequences; the target sequences corresponding to siRNA sites may not be good sites for beacon-based mRNA detection, and vice versa. Possible beacon design rules are identified and approaches for enhancing target accessibility are discussed. This has significant implications to MB design for live cell mRNA detection.
INTRODUCTION
The ability to detect, quantify and monitor the expression of specific genes in living cells, tissues and animals will offer unprecedented opportunities for the early detection of disease, clinical diagnosis, prognosis and treatment monitoring based on molecular markers of the disease. Although in vitro assays such as DNA microarrays, RT–PCR and northern blotting can quantify changes in gene expression level of a cell population, they cannot be used for in vivo detection of mRNA. On the other hand, the use of reporter plasmids for fusion of GFP to RNA binding proteins requires the identification of unique proteins for each specific mRNA; it may not reflect the true expression level of the target mRNA. Significant challenges exist in detecting endogenous gene expression in vivo, including probe design and in vivo delivery, specific targeting and reporting, probe toxicity and the sensitivity of the imaging methods (1,2). In particular, it is necessary to design sophisticated probes in targeting intracellular disease markers so that high detection specificity, sensitivity and signal to background ratio can be achieved.
Of all candidate technologies for in vivo endogenous mRNA detection, the most promising one is molecular beacons (MBs) technology. MBs are dual-labeled antisense oligonucleotide (ODN) probes with a fluorophore at one end and a quencher at the other end (3,4). They are designed to form a stem–loop (hairpin) structure in the absence of a complementary target so that fluorescence of the fluorophore is quenched. Hybridization with the target mRNA opens the hairpin and physically separates the reporter from quencher, allowing a fluorescence signal to be emitted upon excitation. Although MBs have the potential to detect endogenous gene expression in living cells with high sensitivity, to realize this potential, many probe design issues need to be carefully addressed.
To establish the live-cell endogenous mRNA detection capability of MBs, efforts have been made to increase the signal-to-background (S/B) ratio by using dual MBs that form a FRET pair (5,6), to increase the MB stability by modifying the oligonucleotide probe backbone using 2′-O-methyl chemistry (7–9), and to use color-shifting MBs (10). More recently, fluorescent protein complementation has also been used to increase the S/B of the live-cell imaging assay (11,12). However, in all these MB-based methods, a major design issue is target accessibility. It is well known that an mRNA in a living cell has secondary structures and binds to RNA-binding proteins to form an RNP (ribonucleoprotein). Thus, if the target sequence for an MB is so chosen that it involves double-stranded portions of the mRNA or it is occupied by RNA-binding proteins, the beacon has to compete off the mRNA strand or RNA-binding protein(s) in order to hybridize to the target. This may significantly reduce the signal level resulting from MBs binding to their target mRNA, especially with DNA backbone MBs. Although MBs with 2′-O-methyl backbone have an increased affinity to target mRNAs (and an enhanced resistance to degradation by DNase or RNase-H), thus having a better target accessibility when competing with double-strand RNA or RNA-binding proteins, the beacon backbone is RNA-like, hence upon probe/target binding the resulting double-strand RNA may trigger RNA interference, or other unwanted cellular response. It is also possible that 2′-O-methyl MB could interfere with protein production (13,14).
In this work, we focused on addressing target accessibility issues with DNA backbone MBs. We chose bone morphogenic protein 4 (BMP-4, a member of the TGF-β superfamily) as the target mRNA since BMP-4 protein is involved in bone formation, stem cell differentiation (15–17) and plays a key role in proatherogenic inflammatory response and hypertension (18–21). We tested 10 different designs of MBs to identify the optimal beacon design, and performed both upregulation and downregulation of BMP-4 using, respectively, adenovirus infection and siRNA knockdown. Co-expressing GFP and the use of blocking RNA and in situ hybridization further confirmed the detection specificity. The results in this study provide important insights into target accessibility, and a basis for MB design rules. The ability to monitor BMP-4 mRNA expression and localization in living cells in real time may also provide a useful tool for specific studies of basic biology and disease states.
MATERIALS AND METHODS
Cell culture
Normal human dermal fibroblast (HDF) cells (Cambrex, NJ, USA) were grown in Clonetics fibroblast growth medium supplemented with 2% FBS, insulin, fibroblast growth factor, gentamicin sulfate and amphotericin-B (all from Cambrex, NJ, USA). HDF cells were used in our MB studies due to their low auto-fluorescence and ease for probe delivery.
MB design and synthesis
Ten different MBs (MB1-MB8, MB8a, Mb8b) targeting mouse BMP-4 mRNA and a ‘random’-sequence MB (‘random beacon’) were designed, synthesized and tested. The ‘random beacon’ for negative control assays has a specific 16-base target sequence that does not match with any mammalian gene. All MBs were designed to have DNA (2′-deoxy) backbone labeled with Cy3 fluorophore at the 5′ end and Black Hole quencher 2 (BHQ2) at the 3′ end; the specific sequences of these MBs are shown in Table 1, with the underlined sequences as stem sequences. The corresponding target sequences of mouse BMP-4 mRNA are shown in Figure 2. As shown in Table 1, all MBs have a stem length of 5 bases (with GC to AT ratio of 4) and, except MB5 which has the same stem sequence as MB3, all other beacons have different stem sequences. The calculated melting temperatures of MBs and MB/target duplexes are also shown in Table 1. The MB melting temperature was calculated using the program at: http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi, and that of MB/target duplexes was calculated using the Hyther Server at: http://ozone3.chem.wayne.edu/, with 200 nM concentration for beacons and DNA target, respectively. All calculates were using folding temperature of 37°C and ionic condition of 10 mM KCl and 5 mM MgCl2. All MBs were synthesized by MWG Biotech (High Point, NC).
Table 1.
The design of mouse BMP-4 and random molecular beacons and their computed melting temperature
| Beacon ID | Molecular Beacon Design | Melting temperature (°C) | |
|---|---|---|---|
| MB | Duplex | ||
| MB1 | 5′-Cy3-CCCGTTATAATAACAGTCCATACGGG-BHQ2-3′ | 56.0 | 45.4 |
| MB2 | 5′-Cy3-CGGACTCTGTAGAAGTGTCGCCGTCCG-BHQ2-3′ | 52.4 | 59.4 |
| MB3 | 5′-Cy3-CCCACGGAGCCGGTAAAGATGTGGG-BHQ2-3′ | 57.6 | 55.8 |
| MB4 | 5′-CY3-CCAGCCTGCTCTTCCTCCTCCTCGCTGG-BHQ2-3′ | 52.3 | 62.4 |
| MB5 | 5′-Cy3-CGCTCCCCGGTTCCCTGGCTCGAGCG-BHQ2-3′ | 54.8 | 66.3 |
| MB6 | 5′-CY3-CACCCTAACGATCGGCTGATGGGTG-BHQ2-3′ | 55.3 | 54.5 |
| MB7 | 5′-Cy3-CGCTCCCCAATCTCCACTCCGAGCG-BHQ2-3′ | 55.7 | 56.6 |
| MB8 | 5′-Cy3-CGCAGCCTCTACCACCATCTCCCTGCG-BHQ2-3′ | 55.4 | 57.5 |
| MB8a | 5′-Cy3-CGAGCCCATCTCCTGATAATTTGCTCG-BHQ2-3′ | 55.2 | 51.4 |
| MB8b | 5′-Cy3-CCGACCACCCCTCTACCACGTCGG-BHQ2-3′ | 55.0 | 55.3 |
| Random MB | 5′-Cy3-CGACGCGACAAGCGCACCGATACGTCG-BHQ2-3′ | 57.7 | NA |
Figure 2.
The cDNA sequence of mouse BMP-4 mRNA and the hybridization sites of BMP-4 targeting molecular beacons (MB1–MB8). The oligonucleotide sequences of the MBs are shown in Table 1.
MB delivery and imaging experiments
MBs were delivered into live HDF cells with a reversible permeabilization method using activated Streptolysin O (SLO) (6). Specifically, SLO was activated first by adding 5 mM of TCEP to 2 U/ml of SLO for 30 min at 37°C. Cells were incubated for 10 min in 250 μl of serum-free medium containing 0.1–0.2 U/ml of activated SLO (0.25–0.5 U SLO per 106 cells) and 0.5 μl (0.2 μM) or 2.5 μl (1 μM) of MB for cell permeabilization and beacon delivery. Cells were then resealed by adding 0.5 ml of the typical growth medium and incubated for 1 h at 37°C before performing fluorescence microscopy imaging. Each MB experiment was performed at least three times to ensure reproducibility.
The fluorescence imaging of live cells was performed using a Zeiss Axiovert 100 TV epifluorescence microscope coupled to a Cooke Sensicam SVGA cooled CCD camera. Zeiss 100× and 40× EC Plan-NEOFLUAR oil objectives with N.A. (Numerical Aperture) of 1.3 were used for the experiments. The fluorescence of Cy3-labeled beacons was imaged with excitation at 545 nm and emission detection at 570 nm, and GFP fluorescence was imaged with excitation at 470 nm and emission detection at 525 nm. Same exposure times (30 ms for GFP and 400 or 800 ms for Cy3) were used for the imaging experiments.
Real-time quantitative PCR
Total RNA was isolated from cells using Qiagen RNeasy Mini Kit. 100 ng of total RNA was used for cDNA synthesis by random hexamers with Invitrogen Thermoscript RT–PCR kit. For real-time PCR, the cDNA was amplified using a Stratagene Mx3005P (Stratagene) RT–PCR machine. The Ambion's 18S primers were used as an internal control for real-time PCR. PCR amplification was performed with the following primers with 60°C as the annealing temperature: 5′–TGGACTGTTATTATGCCTT-3′ and 5′-GGAGATCACCTCATTTTCTGG-3′ (22).
Adenovirus and siRNAs
To upregulate BMP-4 expression, HDF cells were infected for 2 days with adenovirus containing both ‘mouse’ BMP-4 and polycistronic GFP cDNAs (BMP-4 adenovirus). GFP was used as an independent marker for infection, but not tagged to BMP-4 protein. The BMP-4 siRNA (contains MB6 hybridization site) (sense: 5′-GUCAGAAUCAGCCGAUCGUUACCUCAA-3′, antisense: 5′-UUGAGGUAACGAUCGGCUGAUUCUGAC-3′) and nonsilencing siRNA (Qiagen) were transfected with Oligofectamine (Invitrogen) at the concentration of 100 nM. Four hours after siRNA transfection, HDF cells were infected by BMP-4 adenovirus in the growth medium and incubated for 2 days. BMP-4 mRNA knockdown experiments (n = 4) were also performed using siRNAs that target the hybridization sites of MB1 (siRNA-MB1) and MB8 (siRNA-MB8), respectively. The siRNA-MB1 (sense: 5′-AUGGACUGUUAUUAUAUGCCUUGUUUU-3′, antisense: 5′-AAAACAAGGCAUAUAAUAACAGUCCAU-3′) and the siRNA-MB8 (sense: 5′-UAUCAGGAGAUGGUGGUAGAGGGGUGU-3′, antisense: 5′-ACACCCCUCUACCACCAUCUCCUGAUA-3′) were transfected and the results quantified following the same procedure as described above for BMB-4 siRNA.
Blocking RNA experiment
To confirm detection specificity, we used blocking RNAs that can compete with MBs targeting BMP-4 mRNA, with targeting sequences of 5′-CAUAUAAUAACAGUCCAUGA-3′ (competing with MB1, see Table 1) and
5′-CCCCUCUACCACCAUCUCCUG-3′ (competing with MB8, see Table 1). Two days after adenovirus infection, these blocking RNAs were delivered into HDF cells by SLO with different concentrations of 0.2–40 μM, followed by 2-h incubation in normal media for recovery and hybridization of blocking RNA to its target. BMP-4 targeting MBs were then delivered into cells at a concentration of 100 nM and the resulting fluorescence signal was imaged by epifluorescence microscope 1 h after MB delivery.
Fluorescence in situ hybridization (FISH)
Normal cells and adenoviral BMP-4 infected cells were fixed with 4% paraformaldehyde for 15 min at room temperature and then permeabilized with 0.2% Triton X-100 for 3 min. Blocking was performed with 0.1% Escherichia coli tRNAs and 1% RNase-free BSA in 2× SSC/25% formamide solution for 1 h at 50°C. Formamide was added to decrease the melting temperature between the probe and mRNA. After washing three times, cells were incubated with BMP-4-targeting FISH probe with 2′-O-methyl backbone, 5′-Cy3-ACUCCCUUGAGGUAACGAUCGGCUGAUUCUGACAUGCUGG-3′, at the concentration of 50 nM in 2× SSC/25% formamide overnight at 50°C. After washing with 2 × SSC/25% formamide for three times, cells were observed with epifluorescence microscope.
RESULTS AND DISCUSSION
Target accessibility
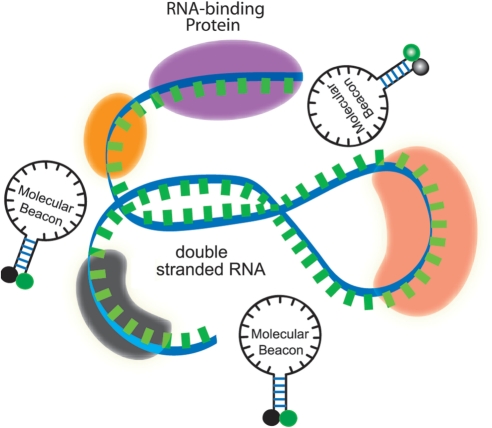
An important issue in live-cell mRNA detection is target accessibility. In contrast to in vitro (in solution) hybridization of MBs with target oligonucleotides, for a MB probe to hybridize to a target mRNA in a living cell, it has to overcome several obstacles, including the secondary structure of mRNAs, RNA-binding proteins and degradation of the probes due to endonucleases. As illustrated in Figure 1, the folded structure of an mRNA in a living cell is due to both the double-stranded segments of the mRNA and the RNA-binding proteins. If the target sequence is buried deep in the folded structure, a beacon has to penetrate into the folded mRNA. If the target sequence has a double-stranded portion, a beacon needs to compete with the RNA strand. More significantly, since an mRNA in a living cell always has RNA-binding proteins on it to form an RNP, if the target sequence is occupied by an RNA-binding protein (or proteins), the beacon has to compete off the RNA-binding protein(s) in order to hybridize to the target sequence. Thus, although an MB can be designed to have its probe sequence unique to the target mRNA, the target sequence may not be necessarily accessible in a living cell. Furthermore, although double-stranded portions of an mRNA can be predicted using, for example, mFOLD, predictions of the folded structure of an mRNA may not be accurate. More problematically, due to the very limited available information on RNA-binding proteins, it is not possible to predict, at any given stage of an mRNA, what proteins bind to it and where. Therefore, there is a lack of guidelines on how to optimize the MB design to have a better target RNA accessibility in a living cell. This severely limits the application of MBs to live-cell detection of endogenous mRNAs.
Figure 1.
A schematic illustration of a segment of the target mRNA with a double-stranded portion and RNA-binding proteins. A molecular beacon has to compete off an mRNA strand or RNA-binding protein(s) in order to hybridize to the target.
To address the target accessibility issues and to identify possible design rules for MBs, in this work we used mouse BMP-4 mRNA as a model system not only because of its biological significance in cardiovascular and cancer studies using animal models, but also because the availability of reagents in our group to use adenovirus for its upregulation and siRNAs probes for its downregulation. We have designed eight different BMP-4 targeting MBs to target different sites on the BMP-4 mRNA, including siRNA-binding sites, translation initiation site, FISH probe-binding sites and predicted antisense oligonucleotide-binding sites. Specifically, as shown in Figure 2 and Table 1, MB probe #1 (hereafter referred to as MB1) was designed to hybridize to the site containing the AUG translation initiation codon. This site was selected since previous studies in antisense research have suggested that the translation initiation site is a good site for generating an antisense effect, indicating that this site might be accessible for the antisense oligonucleotide probes. MB2 and MB6 were designed to hybridize respectively to the two siRNA-binding sites that have been effectively used by our group to downregulate the BMP-4 mRNA level. MB4 was designed based on a FISH probe sequence that has been previously reported for the in situ detection of BMP-4 mRNA (23). MB7 and MB8 were designed based on the sequences of antisense oligonucleotide probes recommended by mFOLD at the Integrate DNA Technology web site (http://www.idtdna.com/Scitools/Applications/mFold/). MB3 and MB5 were randomly chosen. The sequences of all eight probes were checked with BLAST search to make sure that they are unique to the BMP-4 mRNA.
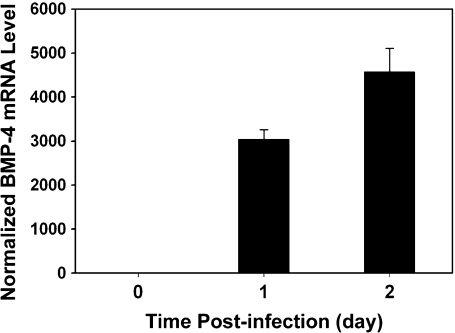
Shown in Figure 3 are the increases of BMP-4 mRNA 1 and 2 days after adenovirus infection, quantified using real-time PCR. Evidently, 2 days after infection, BMP-4 mRNA level increased by more than 4000-fold compared to that of control (uninfected) cells. Thus, in the subsequent target accessibility studies, MBs were delivered into cells 2 days after infection and incubated for another 1 h before fluorescence imaging. GFP signal from the same cells was used as an indication of the effect of infection, since the adenoviral constructs co-express GFP and mouse BMP-4. In most cases, HDF cells without viral infection (cells did not express mouse BMP-4 mRNA) did not show discernible fluorescence signal from MB delivery and hybridization, indicating that, without target mRNAs in HDF cells, MBs produced very low background signal due to nonspecific interaction and/or degradation of probes (data not shown).
Figure 3.
Real-time PCR result of BMP-4 mRNA before and after viral infection in HDF cells. The BMP-4 mRNA levels were normalized against the 18S rRNA levels. Bar graph shows the upregulated BMP-4 mRNA levels divided by the level of uninfected control cells.
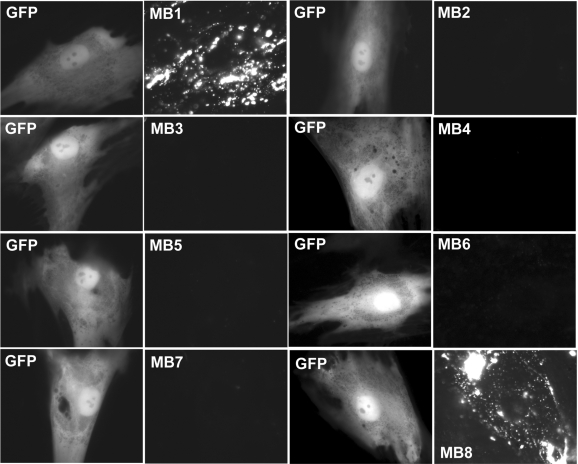
Figure 4 shows the fluorescence signal of 8 BMP-4 targeting MBs (MB1-MB8) in infected HDF cells. Specifically, MB1 and MB8 produced intense punctate fluorescence signals. However, only very weak fluorescence signals were produced by MB2, MB3, MB4, MB5, MB6 and MB7, while the same cells showed a high level of GFP expression, indicating that these beacons have limited or no accessibility to BMP-4 mRNA targets. Note the punctate MB signal shown in Figure 4 (and Figures 5 and 6), which appears to be an indication of specific subcellular localization of BMP-4 mRNA. The subcellular localization of other mRNAs has been reported elsewhere (6,24–26).
Figure 4.
Fluorescence signal from nine different MBs targeting BMP-4 mRNA and from GFP HDF cells 2 days after adenovirus infection. Left and right panels display respectively the epifluorescence images of GFP expression and signal from MBs targeting BMP-4 mRNA in the same cell. The same exposure times were used for GFP and MB imaging. The results showed clearly that only MB1 and MB8 gave high signal level, indicating good target accessibility, whereas all other MBs (MB2–MB7) have poor target accessibility, as indicated by the very weak signal levels.
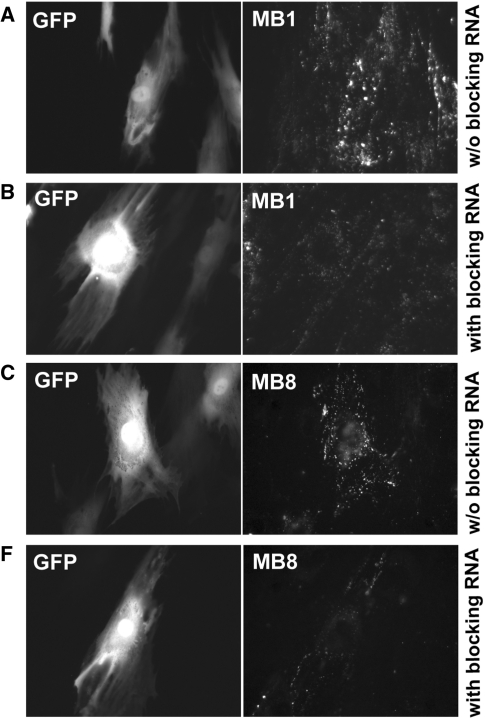
Figure 5.
Evaluation of the specificity of MB1 and MB8 in targeting BMP-4 using blocking RNAs. Left and right panels display the epifluorescence images of GFP expression and MB signal respectively in the same cells. Two hours after delivery of 40 μM blocking RNAs into infected HDF cells, 0.2 μM of MB1 (A, B) and MB8 (C, D) were delivered into HDF cells with and without blocking RNAs. For comparison, images in (A, C) and (B, D) show respectively fluorescence signal in HDF cells without and with blocking RNAs, indicating a reduced level of MB signal due to blocking, whereas the GFP signal remained the same.
Figure 6.
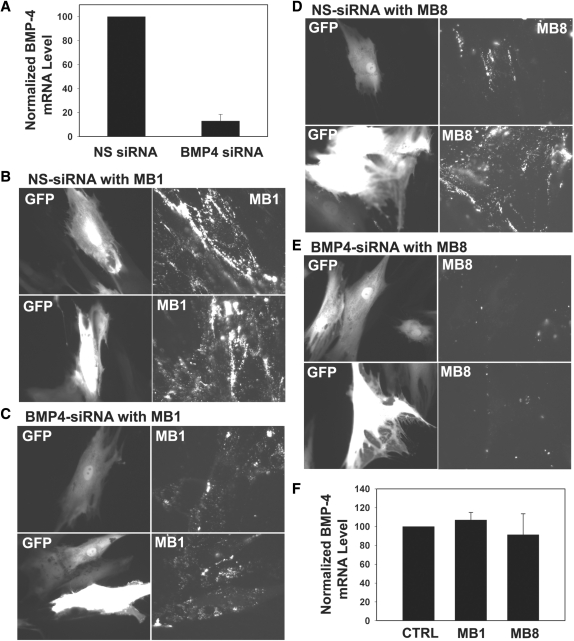
The effect of siRNA knockdown in conforming the specificity of MB1 (B, C) and MB8 (D, E). HDF cells with upregulated BMP-4 level were transfected with either nonsilencing siRNA or BMP-4 siRNA 4 h before adenovirus infection. (A) The effect of siRNA in knocking down BMP-4 mRNA level as measured by RT–PCR. The bar graph shows an ∼90% knockdown in the BMP-4 mRNA level as compared with that of control cells transfected with nonsilencing siRNA. The BMP-4 mRNA levels were normalized by the 18S rRNA level. (B, D) Images of GFP (left panel) and BMP-4-targeting molecular beacon (right panel) signals in control cells transfected with 100 nM of nonsilencing siRNA followed by adenovirus infection. The signal levels of MB1 (B) and MB8 (D) in the control cells corresponded well with the GFP intensity. (C, E) Images of GFP (left panel) and BMP-4-targeting molecular beacon (right panel) signals in cells transfected with 100 nM of BMP-4 siRNA followed by adenovirus infection. The same exposure times for imaging GFP and MB signals were used for all the experiments. Regardless of the GFP signal level, fluorescence signals from MB1 (C) and MB8 (E) almost diminished completely as a result of siRNA knockdown, indicating excellent beacon signal specificity. (F) Real-time PCR analysis suggesting that there is no antisense effect of BMP-4 targeting molecular beacons (MB1 and MB8) in live HDF cells. Cells were infected with virus for 2 days and then delivered with 1 μM of MB1 and MB8, respectively. Twenty-four hours after delivery, RNA was isolated from the cells. Bar graph shows the BMP-4 mRNA levels divided by that of control cells without any MB.
The observation shown in Figure 4 has significant implications. For example, MB2 and MB6 were designed based on the validated siRNA sites; but these sites were clearly not accessible to beacons, possibly due to the differences between siRNAs and MBs in probe–target hybridization process. For siRNAs, their hybridization to target mRNA involves the RNA-induced silencing complex (RISC), which may remove RNA-binding proteins and unwind the secondary structure of mRNA, thereby enabling the siRNAs to hybridize readily to the target mRNA. However, there is no data to suggest a similar protein complex that facilitates the hybridization of a beacon to its mRNA target. Based on our limited observation, the effective sites for MBs to detect mRNA in living cells may be different from that of siRNA-binding sites.
The fact that MB3, MB4, MB5 and MB7 showed very low signal from probe–target hybridization is also noteworthy. It is understood that randomly chosen target sequences, although unique, may not be accessible in live-cell mRNA detection using MBs, as demonstrated by MB3 and MB5. In situ hybridization assays could increase the accessibility of oligonucleotide probes to target mRNA by employing high temperature and low salt concentration to unfold the mRNA secondary structures and remove the RNA-binding proteins. However, these conditions cannot be used in live-cell imaging studies and therefore, the FISH probe-binding site does not ensure the accessibility of beacons, as is the case for MB4. It is clear from Table 1 that the melting temperature for MB and beacon/target hybrids is not a factor in the extent of MB binding.
Interestingly, MB7 and MB8 showed very different signal levels (Figure 4), although both beacons were designed based on antisense oligonucleotide probe-binding sites. As shown in Figure 4, MB8, which was designed to hybridize to the coding region of BMP-4 mRNA near the stop codon, exhibited very high fluorescence signals in cells that expressed high levels of GFP, whereas cells having MB7 did not show much signal from BMP-4 mRNA targeting (even though they had high GFP signal). This indicates that an antisense oligonucleotide probe-binding site may or may not be accessible for MBs in living cells.
Signal specificity
Another important issue in live-cell mRNA detection is signal specificity. To determine whether the fluorescence signal from cells containing MB1 or MB8 is specific, we performed several control studies. We first ruled out the possibility of high background signal due to nonspecific interaction and/or probe degradation by endonucleases in living cells. If the high fluorescence signal from MB1 and MB8 were due to these false-positive events, then other beacon designs (MB2–MB7) would show similar levels of signal, which clearly is not the case (Figure 4). To drive this point home, we delivered random-sequence MBs (random beacons) into infected cells and observed very weak signal level (data not shown), confirming the signal specificity of MB1 and MB8, since random beacons do not have any complementary mRNA target in human and mouse cells, and therefore the very weak signal from random beacons indicates a low level of probe degradation and nonspecific interaction in live HDF cells. We also delivered MB1 and MB8 respectively into cells without viral infection and found that very weak fluorescence signal was emitted from the MBs (data not shown), again indicating signal specificity, since these cells do not express mouse BMP-4 mRNA.
To further confirm the signal specificity of MB1 and MB8, we performed two additional experiments, one using blocking RNA and the other siRNA, combined with adenovirus infection. The blocking RNA approach is similar to the use of blocking peptides (that mimic the antigen-binding site of an antibody) to prevent the binding between antigen and antibody in determining the specificity of an antibody. Specifically, blocking RNAs of 20mer (5′-CAUAUAAUAACAGUCCAUGA-3′) and 21mer (5′-CCCCUCUACCACCAUCUCCUG-3′) were designed to hybridize to the same targeting sequences of MB1 and MB8, respectively, thus competing with MB1 and MB8 for binding to BMP-4 mRNA. For each beacon, we first delivered blocking RNAs with 0.2–40-μM concentration, then the corresponding MBs with 100-nM concentration into infected cells that have upregulated BMP-4 expression. Infected cells without blocking RNAs were used as a control for each MB (MB1 and MB8) in detecting BMP-4 mRNA.
As shown in Figure 5, cells incubated with blocking RNAs prior to beacon delivery exhibited reduced intensity of fluorescence signal (Figure 5B andD) compared with cells without blocking RNAs (Figure 5A and C), although these cells showed the same level of GFP intensity (an indication of having roughly the same level of BMP-4 mRNA level). This result suggests that MB1 and MB8 have good target specificity to BMP-4 mRNA, since the blocking RNAs appeared to reduce the amount of MBs binding to BMP-4 mRNA. We observed that the blocking effect was small when low concentrations (<10 μM) of blocking RNAs were used (data not shown). This is likely due to the difference in hybridization kinetics compared to MBs, and the fact that short blocking RNAs can be readily degraded in living cells.
As yet another confirmation of signal specificity, we used the specific siRNA to downregulate the expression of BMP-4, and visualized the BMP-4 mRNA level using MB1 and MB8. As a control, nonsilencing siRNA was delivered into cells to confirm that RNA-interference induced BMP-4 knockdown was not due to nonspecific oligonucleotides. Specifically, MB1 and MB8 were respectively delivered into HDF cells containing either nonsilencing or BMP-4 siRNA 2 days after siRNA transfection and viral infection, and the resulting fluorescence signal was visualized using an epifluorescence microscope. Real-time PCR analysis of BMP-4 mRNA level was carried out for HDF cells after 2 days incubation following siRNA transfection and BMP-4 adenovirus infection. The RT–PCR result shown in Figure 6A indicates that the siRNA knocked down 90% of BMP-4 mRNA compared to that of control cells (cells with nonsilencing siRNA). In the imaging studies of siRNA knockdown, we again used GFP level as an independent indicator, since the adenovirus encodes BMP-4 and GFP from the same bicistronic vector, and the BMP-4 siRNA is expected to knock down BMP-4 without affecting GFP levels in the virus-infected cells.
As can be seen from Figure 6B, with nonsilencing siRNA, the fluorescence signal from MB1 was quite high, consistent with the GFP fluorescence signal level. When siRNA was used, the fluorescence signal from MB1 was deceased significantly (Figure 6C), although the GFP signal remained high, reflecting the knockdown effect of siRNA. Similarly, with nonsilencing siRNA, the GFP signal correlated well with BMP-4 mRNA levels detected by MB8 (Figure 6D). However, transfection of BMP-4 siRNA diminished most of the fluorescence signals from MB8, while GFP signals from the same cells were high (Figure 6E). The results in Figure 6 strongly suggest that the signals from MB1 and MB8 are very specific in detecting BMP-4 mRNAs in live HDF cells.
We next determined whether the BMP-4 targeting MBs would induce BMP-4 mRNA degradation like the siRNAs. It is well established that antisense oligonucleotides with DNA backbone may induce RNase H activity to degrade target mRNA in the DNA–RNA duplex by hydrolysis (27). To determine if the reduced level of BMP-4 mRNA in the siRNA experiment was due to unwanted cleavage of BMP-4 mRNAs triggered by the beacons rather than siRNA, RT–PCR assays was performed to quantify the BMP-4 mRNA level using RNA samples isolated from cells that were incubated with 1 μM of MB1 or MB8 2 days after infection. As shown in Figure 6F, BMP-4 mRNA levels maintained the same even when cells contained MBs (MB1 or MB8). These results indicate that the MB did not induce antisense effects on target mRNAs, therefore the reduction in beacon signal shown in Figure 6C and E were entirely due to siRNA.
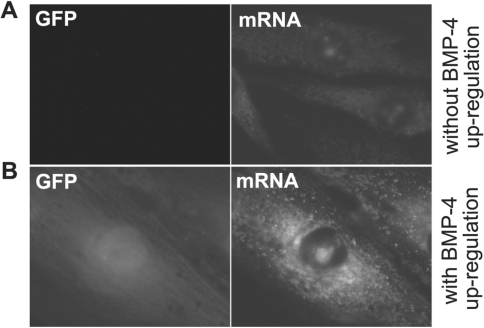
As the final check of signal specificity, we carried out a FISH assay to visualize the BMP-4 mRNA in fixed and permeabilized cells. Both control cells and cells with upregulation of BMP-4 via adenovirus infection were fixed and incubated with the FISH probes that were designed to hybridize with BMP-4 mRNA; the resulting fluorescence images are shown in Figure 7A and B for control and infected cells, respectively. We found that only cells with high GFP signals exhibited high fluorescence signal from the FISH probes; control cells showed very low fluorescence signal, as expected. The localization pattern of BMP-4 mRNA in fixed cells was essentially the same as that in live cells. The only discernible difference observed in the FISH assay was that MB signals from BMP-4 mRNA were dispersed instead of punctated in the perinuclear region seen in living cell assays. This difference can be attributed to changes in cell structure and mRNAs localization (28) due to the fixation process. Nevertheless, in situ hybridization of BMP-4 mRNA with fluorescent probes confirmed the results of BMP-4 mRNA detection in live cells.
Figure 7.
BMP-4 mRNA detection using FISH probes in HDF cells. FISH analysis of BMP-4 mRNA in cells without (A) and with (B) viral infection induced upregulation. Only cells with GFP expression showed an increased fluorescence signal from BMP-4 FISH probe. Control cells without infection showed very low fluorescence signal level, consistent with the low endogenous expression level of BMP-4 in HDF.
Target sequence sensitivity
Taken together, the results shown in Figures 4–6 indicated that, of the eight MBs designed to target BMP-4 mRNA (Table 1), MB1 and MB8 produced robust and specific signal above background, suggesting that the BMP-4 sequences targeted by MB1 and MB8 are structurally accessible. Other beacons (MB2–MB7) that designed to target different sites on BMP-4 mRNA all gave a very low level of fluorescence signal, suggesting that these sites are not readily accessible. This is most likely due to the secondary structure of BMP-4 mRNA and the RNA-binding proteins that occupy the specific sequences targeted by MB2-MB7. It is not clear if there is a large segment on the BMP-4 mRNA that has probe accessibility, or there is only a ‘narrow’ region where MB could hybridize to the target mRNA.
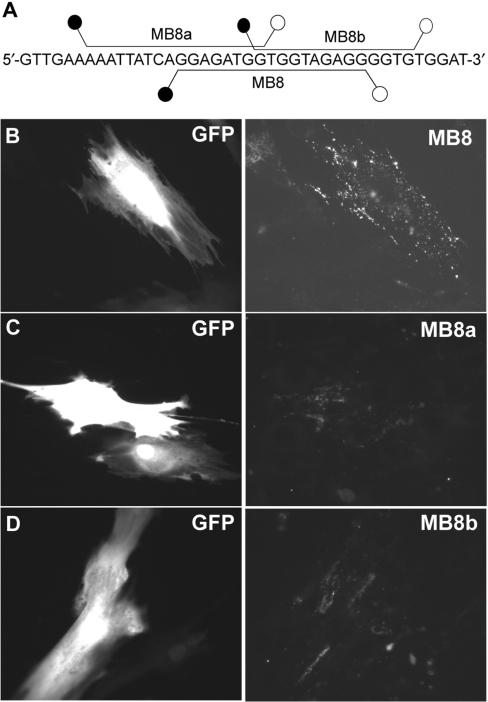
To address this issue, we evaluated the accessibility of MBs that target sequences adjacent to MB8-targeting sequence on BMP-4 mRNA. As shown in Figure 8A (also Table 1), two MBs, MB8a and MB8b, were designed so that their target sequences share respectively 8 and 10 overlapping bases with the target sequence of MB8. Specifically, MB8a has 8 of its 17-base sequence overlapping with the 5′ end of the MB8 sequence, and MB8b has 10 of its 14-base sequence overlapping with the 3′ end of the MB8-targting sequence (Figure 8A). As shown in Figure 8C and D, MB8a and MB8b produced much lower fluorescence signal compared with that of MB8 (Figure 8B), although the GFP signal remained the same. This result further demonstrates that the fluorescence signal from MB8 was critically dependent on the specific target sequences; any change to the target sequence, even a few bases, could significantly reduce the target accessibility. Therefore, MBs need to be designed properly with very specific sequences in order to have good target accessibility in live-cell mRNA detection.
Figure 8.
Effect of target-sequence shifting of MB8 on BMP-4 mRNA detection. (A) Hybridization sites of MB8, MB8a and MB8b. Two molecular beacons, MB8a and MB8b were designed so that their hybridization domains are shifted to the left and right, respectively from that of MB8. (B–D) BMP-4 mRNA detection in HDF cells using respectively 0.2 μM of MB8 (B), 1 μM of MB8a (C) and 1 μM MB8b (D). Note that higher (5×) concentrations of MB8a and MB8b were used in this experiment as it was difficult to detect any signal with the same exposure time when a low concentration (0.2 μM) of MB8a or MB8b was used.
Working toward beacon design rules
The ability to perform sensitive live-cell RNA detection is important for biological studies of mRNA expression, localization and transport, and for clinical studies such as disease detection and diagnosis. Although FISH analysis has been used extensively for mRNA detection in intact cells, it cannot be applied to track the dynamics of changes in the amount and localization of mRNA in living cells, and the fixation and washing steps are not only time consuming but prone to artifacts. Live-cell visualization of mRNA using MBs has the potential to address all these issues and provide a powerful tool for gene expression dynamics studies. However, target accessibility and signal specificity remain major challenges in using MBs to detect endogenous mRNA in living cells.
As demonstrated in this study, due to the lack of target accessibility, MBs designed to target a specific mRNA (such as BMP-4 mRNA) may have very low signal, rendering these MBs useless. Since MBs with 2′-O-methyl backbone have an increased affinity to mRNAs compared with DNA beacons (9), we tested if the use of 2′-O-methyl MBs could lead to better target accessibility. We synthesized a beacon that has exactly the same design as of MB6 but with 2′-O-methyl backbone chemistry for its loop sequence and targeted BMP-4 mRNA in infected cells (with BMP-4 upregulation). However, we did not see any signal increase compared with MB6 that used DNA backbone, although the GFP level was very high (data not shown). Therefore, 2′-O-methyl modification of MBs may result in higher affinity to target mRNA, but not necessarily a better target accessibility.
A specific challenge is that accurate prediction of the structure(s) of an mRNP in a living cell is very difficult, if not impossible, mainly due to the very limited biological knowledge of RNA-binding proteins. As a result, no computational models or general rules are available at the present time to identify, at any stage of an mRNA, the RNA-binding proteins bound, and the corresponding binding sites. An alternative method is to identify accessible sites on an mRNA at which MBs could hybridize and give a high fluorescence signal. As candidate sites, in this study we tested siRNA-binding sites, antisense oligonucleotide-binding sites, FISH probe-binding sites and other sites that are located near the translation start codon or termination codon regions. Interestingly, MBs (MB2 and MB6) targeting siRNA-binding sites that have previously been used to downregulate the BMP-4 mRNA level only exhibited a weak fluorescence signal compared with that from MB1 and MB8.
To confirm that MB hybridization and siRNA knockdown do not correlate well, we designed two additional siRNAs, siRNA-MB1 and siRNA-MB8, that target the hybridization sites of MB1 and MB8, respectively. The siRNA-MB1 and siRNA-MB8 were transfected using the same procedure as with BMP-4 siRNA and NS-siRNA, followed by BMP-4 adenovirus infection. We found that BMP-4 siRNA targeting the MB1 hybridization site only gave a moderate knockdown effect (44%), while siRNA targeting the MB8 hybridization site exhibited a high knockdown effect (83%), comparable to the siRNA that targets the BMP-4 mRNA sequence containing MB6 hybridization site (90%). Therefore, it appears that the siRNA-binding sites are not necessarily accessible targets for MB hybridization, and sites that are accessible for MB hybridization in living cells are not necessarily good siRNA-binding sites.
Among all the hybridization sites tested, we found that sequences located near the start or termination codon are more accessible for beacons to hybridize to its target mRNA. Clearly, more experimental studies are required in order to verify that the start or termination codon regions are indeed accessible for a large group of mRNA molecules. In our previous studies (6,9), we targeted exon–exon junctions of Survivin and GAPDH genes and found that they are accessible. Although more studies are needed to confirm that these are general design rules, we believe the work performed in and insight gained from the current study on MB design is the first step toward the development of robust beacon design rules.
ACKNOWLEDGEMENTS
This work was supported by the National Heart Lung and Blood Institute of the NIH (HL80711) as a Program of Excellence in Nanotechnology (G.B.), by the National Cancer Institute of the NIH (CA119338) as a Center of Cancer Nanotechnology Excellence (G.B.), and by NIH HL70531 (H.J.). Funding to pay the Open Access publication charges for this article was provided by NIH (HL80711).
Conflict of interest statement. None declared.
REFERENCES
- 1.Tsien RY. Imagining imaging's future. Nat. Rev. Mol. Cell Biol. 2003;4:ss16–ss21. [PubMed] [Google Scholar]
- 2.Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH. Visualization of single molecules of mRNA in situ. Methods Enzymol. 2003;361:245–304. doi: 10.1016/s0076-6879(03)61015-3. [DOI] [PubMed] [Google Scholar]
- 3.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 5.Tsourkas A, Behlke MA, Xu Y, Bao G. Spectroscopic features of dual fluorescence/luminescence resonance energy-transfer molecular beacons. Anal. Chem. 2003;75:3697–3703. doi: 10.1021/ac034295l. [DOI] [PubMed] [Google Scholar]
- 6.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majlessi M, Nelson NC, Becker MM. Advantages of 2′-O-methyl oligonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998;26:2224–2229. doi: 10.1093/nar/26.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molenaar C, Marras SA, Slats JC, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:E89–E89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsourkas A, Behlke MA, Bao G. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002;30:5168–5175. [PMC free article] [PubMed] [Google Scholar]
- 10.Tyagi S, Marras SA, Kramer FR. Wavelength-shifting molecular beacons. Nat. Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi S. Splitting or stacking fluorescent proteins to visualize mRNA in living cells. Nat. Method. 2007;4:391–392. doi: 10.1038/nmeth0507-391. [DOI] [PubMed] [Google Scholar]
- 12.Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Method. 2007;4:421–427. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- 13.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecular 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 14.Prakash TP, Johnston JF, Graham MJ, Condon TP, Manoharan M. 2′-O-[2-[(N,N-dimethylamino)oxy]ethyl]-modified oligonucleotides inhibit expression of mRNA in vitro and in vivo. Nucleic Acids Res. 2004;32:828–833. doi: 10.1093/nar/gkh220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li RH, Wozney JM. Delivering on the promise of bone morphogenetic proteins. Trends Biotechnol. 2001;19:255–265. doi: 10.1016/s0167-7799(01)01665-1. [DOI] [PubMed] [Google Scholar]
- 16.Massague J. How cells read TGF-b signals. Nat. Rev. Mol. Cell. Biol. 2002;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 17.Hogan BL. Bone morphogenetic proteins in development. Curr. Opin. Genet. Dev. 1996;6:432–438. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- 18.Sorescu GP, Song H, Tressel SL, Hwang J, Dikalov S, Smith DA, Boyd NL, Platt MO, Lassegue B, Griendling KK, et al. Bone morphogenic protein 4 produced in endothelial cells by oscillatory shear stress induces monocyte adhesion by stimulating reactive oxygen species production from a nox1-based NADPH oxidase. Circ. Res. 2004;95:773–779. doi: 10.1161/01.RES.0000145728.22878.45. [DOI] [PubMed] [Google Scholar]
- 19.Miriyala S, Nieto MCG, Mingone C, Smith D, Dikalov S, Harrison DG, Jo H. Bone morphogenic protein-4 induces hypertension in mice: role of noggin, vascular NADPH oxidases, and impaired vasorelaxation. Circulation. 2006;113:2818–2825. doi: 10.1161/CIRCULATIONAHA.106.611822. [DOI] [PubMed] [Google Scholar]
- 20.Chang K, Weiss D, Suo J, Vega JD, Giddens D, Taylor WR, Jo H. Bone morphogenic protein antagonists are coexpressed with bone morphogenic protein 4 in endothelial cells exposed to unstable flow in vitro in mouse aortas and in human coronary arteries: role of bone morphogenic protein antagonists in inflammation and atherosclerosis. Circulation. 2007;116:1258–1266. doi: 10.1161/CIRCULATIONAHA.106.683227. [DOI] [PubMed] [Google Scholar]
- 21.Jo H, Song H, Mowbray A. Role of NADPH oxidases in disturbed flow- and BMP4- induced inflammation and atherosclerosis. Antioxid. Redox Signal. 2006;8:1609–1619. doi: 10.1089/ars.2006.8.1609. [DOI] [PubMed] [Google Scholar]
- 22.Soares ML, Haraguchi S, Torres-Padilla M-E, Kalmar T, Carpenter L, Bell G, Morrison A, Ring CJA, Clarke NJ, Glover DM, et al. Functional studies of signaling pathways in peri-implantation development of the mouse embryo by RNAi. BMC Dev. Biol. 2005;5:28. doi: 10.1186/1471-213X-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh PCH, Kenagy RD, Mulvihill ER, Jeanette PJ, Wang X, Chang CMC, Yao Z, Ruzzo WL, Justice S, Hudkins K, et al. Bone morphogenetic protein 4: potential regulator of shear stress-induced graft neointimal atrophy. J. Vasc. Surg. 2006;43:150–158. doi: 10.1016/j.jvs.2005.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martone ME, Pollock JA, Ellisman MH. Subcellular localization of mRNA in neuronal cells. Contributions of high-resolution in situ hybridization techniques. Mol. Neurobiol. 1998;18:227–246. doi: 10.1007/BF02741301. [DOI] [PubMed] [Google Scholar]
- 25.Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J. Cell. Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoharan M. 2′-Carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim. Biophys. Acta. 1999;1489:117–130. doi: 10.1016/s0167-4781(99)00138-4. [DOI] [PubMed] [Google Scholar]
- 28.Dirks RW, Molenaar C, Tanke HJ. Methods for visualizing RNA processing and transport pathways in living cells. Histochem. Cell Biol. 2001;115:3–11. doi: 10.1007/s004180000214. [DOI] [PubMed] [Google Scholar]