Abstract
The Restriction-modification system AhdI contains two convergent transcription units, one with genes encoding methyltransferase subunits M and S and another with genes encoding the controller (C) protein and the restriction endonuclease (R). We show that AhdI transcription is controlled by two independent regulatory loops that are well-optimized to ensure successful establishment in a naïve bacterial host. Transcription from the strong MS promoter is attenuated by methylation of an AhdI site overlapping the -10 element of the promoter. Transcription from the weak CR promoter is regulated by the C protein interaction with two DNA-binding sites. The interaction with the promoter-distal high-affinity site activates transcription, while interaction with the weaker promoter-proximal site represses it. Because of high levels of cooperativity, both C protein-binding sites are always occupied in the absence of RNA polymerase, raising a question how activated transcription is achieved. We develop a mathematical model that is in quantitative agreement with the experiment and indicates that RNA polymerase outcompetes C protein from the promoter-proximal-binding site. Such an unusual mechanism leads to a very inefficient activation of the R gene transcription, which presumably helps control the level of the endonuclease in the cell.
INTRODUCTION
A type II restriction-modification (R-M) system consists of (i) a restriction endonuclease that recognizes a specific DNA sequence and introduces double-stranded breaks at or around the recognition site and (ii) a methyltransferase (methylase) that recognizes the same DNA sequence and methylates it. Methylation prevents site recognition by the endonuclease and thus protects the target DNA from cleavage. Type II R-M systems genes are often plasmid-encoded and can rapidly spread from one bacterial host to another, crossing species boundaries and impacting genome evolution on a global scale (1,2).
During cell entry and establishment of a plasmid containing R-M genes, unmodified host DNA can be cleaved by the endonuclease causing host cell death. Therefore, expression of R-M genes should be very tightly regulated to ensure that enough methylase is produced to methylate host DNA before active endonuclease is synthesized. In many R-M systems, such as AhdI (3), BamHI (4), BglII (5), Eco72I (6), EcoRV (7), Esp1396I (8), PvuII (9) and SmaI (10) the endonuclease gene expression is regulated by specialized Control (C) proteins (11). C proteins bind to palindromic DNA sequences called C-boxes and activate transcription of their own genes and endonuclease genes. Genes coding for C proteins are usually located upstream of, and partially overlap with the endonuclease gene (12). It is believed that a time delay in activation of endonuclease gene expression necessary for establishment of a C protein-regulated R-M system in a naïve host is achieved by (i) inefficient translation of the C protein ORF and (ii) the fact that C-proteins must dimerize in order to bind DNA. Indeed, cells overproducing a C protein cannot be transformed with a plasmid containing a cognate R-M system, presumably due to premature activation of endonuclease expression (13).
In all cases studied to date, C-boxes located in front of CR transcription units contain two sites for C-protein binding. It is believed that the presence of two binding sites allows a more precise control of the endonuclease gene transcription: binding of a C protein to the high-affinity promoter-distal site activates the endonuclease gene transcription, while binding to the intrinsically weaker promoter-proximal site represses it (3,14,15,17). Such a mechanism of regulation is similar to that described for the phage λ repressor (16); indeed C-proteins are evolutionary related to this well-studied transcription regulator. However, the exact mechanism(s) of transcription activation and repression by C proteins may be different from that used in the λ switch and may also differ in different R-M systems. For example, while the binding of the λ repressor to adjacent DNA-binding sites is cooperative, the interaction of C protein from AhdI system with its binding site is cooperative (3,17) but that from EcoRV system is not (14).
During establishment of an R-M system in a naïve host, efficient initial synthesis of the methylase is achieved due to high-level transcription. However, excessive methylase can be detrimental later on. For example, it may modify invading phage DNA before it is cleaved by the endonuclease, leading to the death of the host cell and therefore of the plasmid that harbours the R-M genes. The R-M systems appear to have evolved special mechanisms to decrease transcription of the methylase gene. Most C-protein dependent R-M systems are organized as two divergently transcribed transcription units, one containing the C and R genes, and another containing the M gene. The two transcription units are separated by a short intergenic region that contains divergent and partially overlapping promoters. C protein-dependent activation of the CR promoter simultaneously leads to decrease of the M promoter activity, either indirectly (through promoter competition) or in some systems such as Esp1396I or Kpn2I, directly (through occlusion of the M promoter by bound C protein). Overproduction of cognate C proteins in such systems was shown to repress expression of methylase genes in vivo (8,18).
In the object of this study, the R-M system AhdI, genes are organized as convergent transcription units, raising a question of how the regulation of the methylase synthesis is accomplished. Recent in vitro analysis of C.AhdI binding to its C-box indicates that because of the very high cooperativity, both C-protein-binding sites are occupied simultaneously at all protein concentrations tested (17), raising a question of how activated transcription of the CR transcription unit is accomplished. To answer these questions we here perform the in vivo and in vitro analysis of AhdI transcription and present a mathematical model that adequately describes control of the CR promoter, as well as the dynamics of the AhdI genetic switch.
MATERIALS AND METHODS
Bacteria strains, phages and media
Escherichia coli Z85 and HB101 (19), XL1-Blue (Stratagene) were used as host strains to study gene expression restriction of the λvir phage growth. Phage λvir was propagated as described (20). All bacterial strains were grown in standard Luria–Bertani (LB) medium at 37°C with appropriated antibiotics. XL10-Gold ultracompetent cells (Stratagene) were used for transformation during site-direct mutagenesis according to the manufacturer's protocol. E. coli BL21(DE3) strain was used for overexpression of recombinant proteins.
Plasmids and proteins
pAhdIMR, a pUC19-based plasmid containing the AhdI R-M genes cassette, was described earlier (3). A pAhdIMR derivative with a frame-shift mutation near the 5′ end of the ahdIC gene was prepared by site-directed PCR mutagenesis with QuikChange® Site-Direct mutagenesis kit (Stratagene). The mutation introduced an extra G/C base pair between base pairs 8 and 9 of the ahdIC gene. Sequences of primers used are available from the authors upon request. Plasmid pCBR was prepared by cloning a PCR fragment containing the ahdIC gene under control of its own promoter into the EcoRV site of pBR322. DNA fragments containing substitutions in the AhdI C-box were prepared by site-directed PCR mutagenesis. The fragments (extending from −214 tp +70 with respect to transcription start point at +1) were cloned into pT7Blue vector from the Perfectly Blunt Cloning Kit (Novagen) according to the manufacturer's protocol. The resulting plasmids were named pT7C1 (mutant OL), pT7C4 (mutant OR), and pT7C1C4 (a double mutant). pAhdsMR with a substitution in the target site (GACN5GTC → GACN5GAC) was created by site-directed mutagenesis using pAhdMR as a template. PCR fragments containing wt or mutant target sites were fused to the lacZ gene of the pSG206 (provided Sean Garrety, Harvard Medical School).
The AhdI methylase and C.AhdI were purified as described [refer (21) and (3), respectively].
Primer extension reactions
E.coli Z85 cells harbouring pAhdIMR or its derivative were grown to OD600 of ∼0.4 and total RNA was extracted using RNeasy Mini Kit (QIAGEN) according to the manufacturer's protocol with the inclusion of DNase I digestion step with RNase-Free DNase (QIAGEN). For a single primer extension reaction, 20 μg of total RNA was reverse-transcribed with 100 U of SuperScript III reverse transcriptase from the First-Strand Synthesis Kit for RT-PCR (Invitrogen) according to the manufacturer's protocol in the presence of 1 pmol of appropriate primers 32P-labelled at their 5′ ends. The reaction products were treated with RNase H, ethanol precipitated, dissolved in a loading buffer containing 7 M urea-formamide and resolved on 7% sequencing gels. The products of sequencing reactions performed with the same end-labelled primers and the pAhdIMR plasmid as a template using DNA Cycle Sequencing System (Promega) were run alongside the primer extension reactions. After electrophoresis, reaction products were revealed using PhosphorImager (Molecular Dynamics).
Electrophoretic mobility shift assay
Plasmids pAhdMR, pT7C1, pT7C4, and pT7C1C4 were used as templates for PCR amplification of DNA fragments to be used for EMSA, in vitro transcription, and footprinting assays. Reactions contained, in 10 μl, 20 nM 32P-end-labelled wild-type or mutant PahdICR DNA fragments, 40 mM Tris–HCl (pH 8.0), 90 mM KCl, 10 mM MgCl2, 100 μg/ml BSA, 2 mM DTT, 5% glycerol. Reactions were incubated for 10 min at 37°C in the presence of varying concentrations of C.AhdI (from 0 to 188 nM). After the addition of 2 μl of loading buffer (30% glycerol, 10 mM EDTA, 0.05% bromphenol blue), the samples were loaded on a running 8% non-denaturing polyacrylamide gel. After electrophoresis, reaction products were revealed using PhosphorImager (Molecular Dynamics).
Footprinting and in vitro transcription
DNase I footprinting was carried out with reactions set up as described for EMSA. After a 10 min incubation at 37°C, 0.05 U DNase I (Worthington) was added and incubation was continued for another 45 s. Reactions were stopped by the addition of 20 μl stop buffer (1% SDS, 200 mM EDTA, pH 8.0, 50 μg/ml calf thymus DNA) (22) and ammonium acetate to the final concentration of 1 M. Samples were precipitated with ethanol, dried and resuspended in 8 μl of 7 M urea–formamide loading buffer. G+A sequencing reactions were carried out as described (22). Samples were applied on 6% sequencing gels and products were revealed using PhosphorImager (Molecular Dynamics).
Templates used in transcription assays with PahdICR and its derivatives were prepared by PCR. The endpoints of DNA fragments used were from +63 to −120 relative to transcription start point at +1. The methylated PahdIMS promoter fragment template was generated by BstEII/BsaI digestion of pAhdIMR plasmid. The resulting fragment extended from from positions +109 to −151 relative to transcription start point. The unmethylated template of the same size was prepared by PCR.
For in vitro transcription, promoter complexes were allowed to form for 10 min at 37°C in 10 μl reactions containing 40 mM Tris–HCl (pH 8.0), 90 mM KCl, 10 mM MgCl2, 2 mM DTT, 5% glycerol, 100 nM E. coli RNAP σ70 holoenzyme, and 8 nM of AhdI DNA fragments. Where indicated, the DNA fragments were incubated for 10 min at 37°C with varying concentrations of C.AhdI prior to the addition of RNAP. Transcription was initiated by the addition of a mixture of transcription substrates and heparin (30 μg/ml) to the final concentrations of 200 μM (ATP, CTP, UTP), 20 μM GTP, 10 μM [α-32P]GTP (3000 Ci/mmol). After a 15 min incubation, reactions were terminated by the addition of 15 μl 7 M urea–formamide loading buffer and resolved on 8% sequencing gel. After electrophoresis, transcription products were revealed using PhosphorImager (Molecular Dynamics).
KMnO4 probing was conducted under conditions used in in vitro transcription assays. Complexes were treated with 1 mM KMnO4 for 30 s at 37°C followed by the addition of β-mercaptoethanol and ammonium acetate to the final concentrations of 1 M and 0.3 M, respectively. Samples were precipitated by ethanol, dried and resuspended in 90 μl H2O. After the addition of 10 μl piperidine, samples were incubated at 90°C for 20 min, precipitated with ethanol, dried and resuspended in 8 μl of 7 M urea–formamide loading buffer. Samples were applied to 6% sequencing gel and revealed by PhosphorImager (Molecular Dynamics).
β-galactosidase assays
The activity of β-gal was determined as described by Miller (23) with modifications (24). Thirty microlitres of overnight bacterial cultures were inoculated into 3 ml LB containing appropriate antibiotics and grown at 37°C until OD600 reached 0.5–0.7. Total 200 μl of each culture was withdrawn, mixed with 800 μl of Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol, pH 7.0) and 200 μl of 4 mg/ml ONPG (Sigma) was added. Reactions were incubated for various periods of time, terminated by the addition of 500 μl 1 M Na2CO3 and OD420, OD550 was measured. The β-gal activity was determined using the following formula: Units activity=1000×[OD420 – (1.75×OD550)/t×v×OD600], where t is the time of the reaction in min, and v is the volume (in ml) of the culture used for the assay.
RESULTS
Mapping the ahdI promoters in vivo
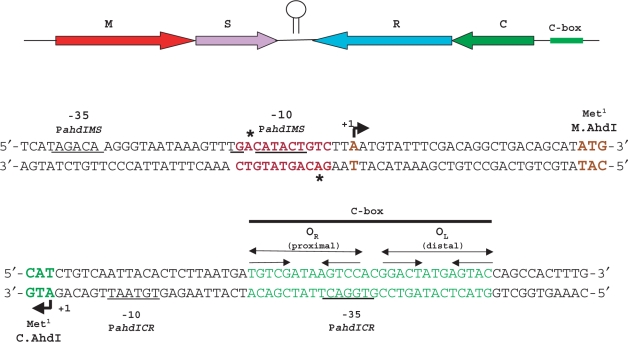
The genetic organization of the AhdI R-M system from Aeromonas hydrophila is schematically illustrated on Figure 1. The ahdIM and ahdIS genes, and the ahdIC and ahdIR genes are transcribed convergently. The ahdIM gene, which codes for the catalytic subunit of the AhdI methyltransferase, is immediately followed by the ahdIS gene, which codes for the specificity subunit of the enzyme. Thus, these genes are likely co-transcribed. The ahdIC gene is immediately followed by ahdIR and these genes are likely co-transcribed, as is also the case in other C protein-regulated R-M systems (14,25,26).
Figure 1.
Structural organization of the AhdI restriction-modification system. The ahdI genes are schematically shown on the top. Arrows show the direction of transcription. Two ahdI transcription units (MS and CR) are separated by a short self-complementary region that may act as a bidirectional transcription terminator (depicted here as a hairpin). DNA sequences around transcription start points of the ahdIMS and ahdICR promoters are expanded below. Beginnings of coding regions are indicated with colours that match those at the top of the figure. The PahdIMS and PahdICR start sites are shown, respectively, by rightward and leftward arrows above and below the sequence. Promoter elements are underlined and transcription starts are colour-coded. The ahdI C-box is indicated and two sets of inverted repeats are shown by convergent arrows. The AhdI site in front of PahdIMS is indicated in red; astericks indicate methylated adenine residues. The start codons of ahdC and ahdIM are indicated.
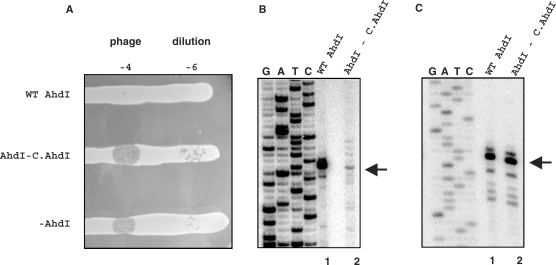
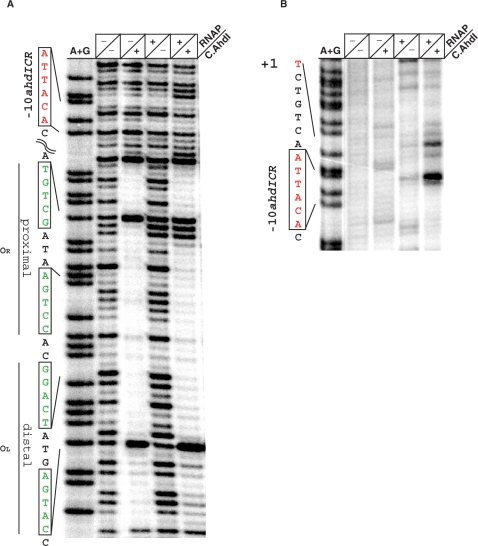
The start points of the ahdI promoters have not been experimentally identified. We performed primer extension with total RNA prepared from E. coli cells harbouring a plasmid containing the AhdI system. This pUC19-based plasmid was described previously (3); we call it pAhdIRM. E. coli cells harbouring pAhdIRM restricted the growth of phage λ (Figure 2A, labelled as ‘WT AhdI’), indicating that endonuclease (and therefore methyltransferase) is synthesized. Because the pAhdIRM plasmid is stably maintained in E. coli, the ahdI genes are apparently coordinately expressed in this heterologous host. In contrast, cells harbouring a derivative of pAhdIRM with a frame-shift mutation in the beginning of ahdIC did not restrict phage growth (Figure 2A, labelled as ‘AhdI—C.AhdI’), indicating that intact C.AhdI is required for endonuclease production. Total RNA from these cells was also used in primer extension reactions as a control.
Figure 2.
Expression of the ahdI genes in vivo. (A) The horizontal lines show overnight 37°C growth of E. coli Z85 strain harbouring indicated plasmids on an LB agar plate. Cells were spotted with indicated dilutions of λ-vir phage lysate. (B and C) Primer extension analysis of ahdI transcripts. RNA was purified from E. coli Z85 strain harbouring wild-type AhdI plasmid pAhdIRM (lanes 1) or pAhdIRM derivative with disrupted ahdIC (lanes 2) and primer extension reactions were performed with oligonucleotide primers complementary to the ahdIC (Figure 2B) and ahdIM (Figure 2C) genes. Sequencing reactions marker lanes were prepared with pAhdIRM and primers used for primer extension.
Extension of primers annealing to various places inside the ahdIM–ahdIS and ahdIC–ahdIR genes revealed, in each case, a single primer extension product corresponding to the same transcription start point in each transcription unit (data not shown). Thus, the ahdIM–ahdIS gene pair and the ahdIC–ahdIR gene pair are each transcribed from a single promoter and the genes in each pair form operons. Results of a representative primer extension experiment with one primer set are presented in Figures 2B and C. The primer extension product corresponding to the ahdICR transcription unit mapped to the first nucleotide of the ahdIC initiating codon. Thus, the ahdIC mRNA is leaderless, as is also the case in the PvuII R-M system (25). The experimentally determined transcription start site coincides with an earlier prediction (3); it is preceded by a TGTAAT sequence that is appropriately positioned to serve as a −10 promoter element (consensus sequence TATAAT). 18 bp upstream, a TGGACT sequence that is similar to the TTGACA –35 promoter element consensus sequence is located. The two C.AhdI-binding sites of the C-box (3) are centered on base pairs −29 and −44 with respect to transcription start point. Primer extension experiment with RNA prepared from non-restricting cells that contained the pAhdIRM derivative with non-functional ahdIC revealed that transcription of the ahdIC–ahdIR operon was strongly decreased (more than 10-fold, Figure 2B, compare lanes 1 and 2). Thus, the ahdICR promoter (PahdICR) requires C.AhdI for optimal transcription.
The 3′ ends of major extension products of primers annealing to ahdIM or ahdIS were located 26 bp upstream of the ahdIM start codon. An upstream CATACT sequence likely serves as a −10 promoter element for transcripts corresponding to major primer extension products. It is preceded by a TGA sequence, which may act as an extended −10 motif (consensus sequence TGn). 17 bp upstream of the −10 element there is a TAGACA sequence similar to the −35 element consensus sequence. Several minor primer extension products, most corresponding to transcripts whose 5′ ends we located 2–7 nucleotides downstream of the major transcript, were also reproducibly observed (Figure 2C). These primer extension products may correspond to minor downstream transcription initiation events or result from processing of the major transcript. The activity of the ahdIMS promoter (PahdIMS) did not depend on the presence of functional ahdIC (Figure 2C, compare lanes 1 and 2).
Regulation of transcription from PahdICR in vitro
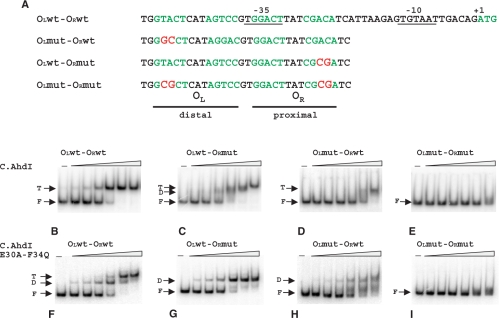
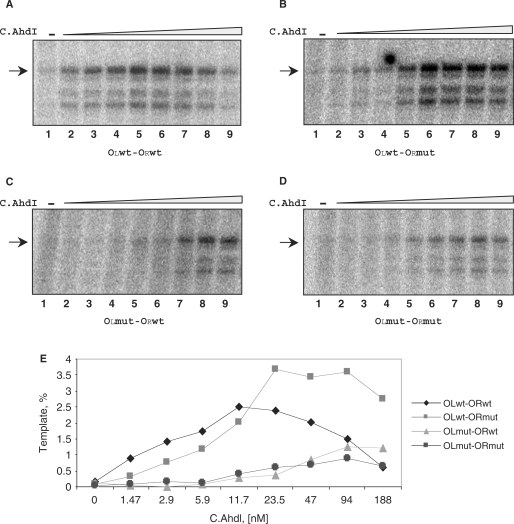
Previous work showed that promoter-distal C.AhdI-binding site has high affinity for C.AhdI, while promoter-proximal C.AhdI-binding site has lower affinity (17). However, the interaction of C.AhdI dimers with the C-box is highly cooperative and under no conditions could a complex with a C.AhdI dimer bound only at the promoter-proximal site be detected (17). We studied C.AhdI binding and C.AhdI-dependent in vitro transcription from wild-type PahdICR and from PahdICR-containing DNA fragments with mutations in promoter-proximal (OR), promoter-distal (OL), or both C.AhdI-binding sites. The mutations introduced double substitutions in each site (Figure 3A) that should abolish or strongly decrease the interaction with C.AhdI. In a gel retardation assay, the addition of increasing amounts of C.AhdI to the wild-type PahdICR fragment resulted in gradual disappearance of free DNA band and the appearance of a single sharp low-mobility band that persisted over the entire range of C.AhdI concentrations used (Figure 3B).
Figure 3.
Analysis of C.AhdI complexes on the wild-type and mutant ahdICR promoters using electrophoretic mobility shift assay. (A) Sequences of the wild-type and mutant C-boxes are presented. Two sets of inverted repeats that form promoter-proximal (OR) and promoter-distal (OL) C.AhdI-binding sites are highlighted in green colour. Promoter elements and transcription start site of the ahdICR promoter are also indicated. Substitutions introduced in mutant ahdI fragments are highlighted in red. (B–I) Increasing concentrations of the wild-type (panels B–E) or mutant (panels F–I) C.AhdI were combined with 20 nM of the indicated AhdI C-box DNA fragments, complexes were allowed to form and separated by EMSA. Results of electrophoretic separation of reaction products on an 8% native polyacrylamide gel are shown. F—free DNA, D—complexes containing bound C.AhdI dimer, T—complexes containing two C.AhdI dimers.
Two amino acid residues, Glu30 and Phe34, have been proposed to be important contacts for the interaction of C.AhdI with the sigma subunit of RNA polymerase (27) and they may also be involved in dimer–dimer contacts in the tetrameric complex. We constructed the mutant protein E30A-F34Q. As expected, the mutant was unable to activate transcription in in vitro assays (data not shown). Quantitative gel retardation experiments confirmed that the mutation effectively abolished the cooperativity of C.AhdI interaction with the C-box, leading to ∼360-fold decrease in the affinity for the proximal-binding site, but having no effect on the affinity for the distal site (27). When the E30A-F34Q C.AhdI mutant was used instead of the wild-type C.AhdI, an intermediate mobility band was present at lower concentrations of C.AhdI (Figure 3F). At higher C.AhdI concentrations this band was converted to the low-mobility band seen in reactions containing wild-type C.AhdI (compare Figures 3B and F). Thus, it is likely that the low-mobility band corresponds to two C.AhdI dimers bound to DNA, while the intermediate mobility band seen in reactions containing the mutant C.AhdI corresponds to a single C.AhdI dimer bound to the high-affinity promoter-distal-binding site.
A DNA fragment harbouring mutations in both binding sites did not interact with C.AhdI in a gel retardation assay (Figures 3E and I), indicating that mutations indeed abolished or strongly decreased the interaction. Addition of C.AhdI to a DNA fragment with a mutation in the strong promoter-distal C.AhdI-binding site led to formation of a complex containing C.AhdI tetramer (Figure 3D). However, higher concentrations of C.AhdI were required to obtain such a complex with the mutant fragment than with the wild-type PahdICR fragment (compare Figures 3B and D), as expected. The addition of C.AhdI to DNA fragment containing mutation in the weak promoter-proximal C.AhdI-binding site produced, at low C.AhdI concentrations, small amounts of a complex corresponding to C.AhdI dimer bound to DNA (Figure 3C). This complex was very transient; further increase in the concentration of C.AhdI reproducibly led to unusual ‘gradual’ conversion of the dimer band to that corresponding to C.AhdI tetramer-containing complex (Figure 3C). Only the dimer complex band was seen when tetramerisation-defective C.AhdI mutant was used with this DNA fragment (Figure 3G). Thus, formation of a complex containing wild-type C.AhdI tetramer on the PahdICR fragment with mutated promoter-proximal C.AhdI-binding site was driven by cooperative interactions between C.AhdI dimers. The reason(s) for unusual gradual conversion of the dimer band to the tetramer band observed with this fragment are unknown.
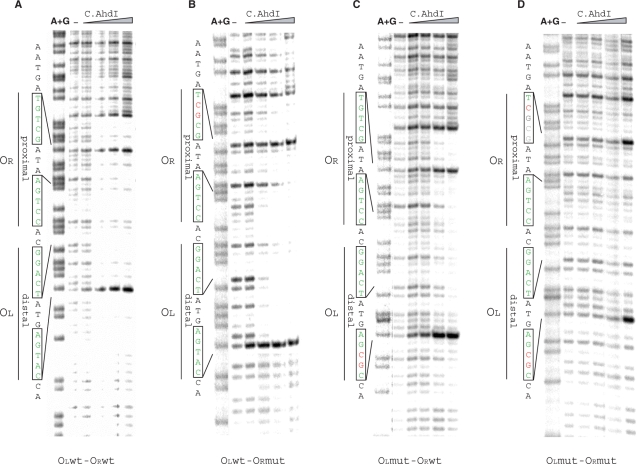
In agreement with results of gel retardation analysis, DNase I footprinting revealed that both C.AhdI-binding sites were always protected to the same degree on both the wild-type and mutant PahdICR fragments (Figure 4), suggesting that complexes containing C.AhdI tetramers were formed over the entire range of C.AhdI concentrations used. Interestingly, specific binding of C.AhdI to the double mutant fragment was detected at high concentrations of C.AhdI (Figure 4D, right lanes), even though this fragment did not appear to interact with C.AhdI in gel retardation assays. The residual-specific binding explains the observed low-level activated transcription from this fragment (see below).
Figure 4.
DNase I footprinting of C.AhdI complexes on wild-type and mutant ahdICR promoters. 32P-end labelled (bottom strand) ahdICR promoter-containing fragments (22.5 nM) were combined with increasing concentrations (0–188 nM) of C.AhdI and treated with DNase I. Two sets of inverted repeats are indicated at the left of each panel.
The results of in vitro transcription by the E. coli σ70 RNA polymerase (RNAP) holoenzyme from PahdICR-containing DNA fragments in the presence of increasing concentrations of C.AhdI are presented in Figure 5. Wild-type PahdICR was virtually inactive in the absence of C.AhdI (Figure 5A, lane 1) but was activated upon the addition of C.AhdI (compare lane 1 with lanes 2–5). Further increase in the amount of C.AhdI repressed transcription (Figure 5A, compare lane 5 with lanes 7–9). Primer extension experiments revealed that the in vitro transcription start point was the same as that observed in vivo (data not shown). Several minor transcripts seen correspond to minor primer extension products observed when analysing total cellular RNA; apparently they are generated when RNA polymerase initiates transcription several bases up- or downstream of the main transcription initiation start point.
Figure 5.
In vitro transcription from wild-type and mutant ahdICR promoters. (A–D) E. coli RNAP σ70 holoenzyme (100 nM) was combined with DNA fragments containing wild-type or mutant PahdICR (8 nM) in the presence of increasing concentrations of C.AhdI (0–500 nM) and a single-round transcription reaction was performed. Reaction products were separated by denaturing gel electrophoreses. (E) Quantification of results presented in (A).
Transcription from a fragment containing mutations in both C.AhdI-binding sites was several-fold less efficient than that from the wild-type fragment and required higher concentrations of C.AhdI (Figure 5D). The activation of transcription from this fragment at high concentrations of C.AhdI is a result of residual-specific interaction of C.AhdI with mutant operators, seen on Figure 4D. A fragment with mutated promoter-distal OL site behaved similarly (Figure 5C), underscoring the importance of C.AhdI binding to this site for transcription activation. Transcription from a fragment containing mutated promoter-proximal OR site was activated by C.AhdI as efficiently as from the wild-type fragment; however, only weak repression of transcription from the mutant template was observed and only at high concentrations of C.AhdI (Figure 5B, compare lanes 8 and 9). This result is intuitively expected since C.AhdI interaction with the promoter-proximal site, which presumably is required for transcription repression, is weakened by the mutation.
Quantification of the amount of PahdICR transcripts produced in this single-round in vitro transcription experiments at optimal C.AhdI concentrations indicated that less than 2.5% of templates were transcriptionally active (data not shown), even though the amount of RNAP used in the experiment exceeded the amount of promoter DNA several-fold and so RNAP was not limiting. Indeed, DNase I footprinting of reactions containing both C.AhdI and RNAP holoenzyme revealed no extra protection compared to reactions containing C.AhdI only (Figure 6A, compare lanes 2 and 4). The only difference was the appearance of two DNase I sensitive bands at the downstream boundary of promoter-proximal C.AhdI-binding site. These bands, located at positions −25/−26 with respect to transcription start point likely correspond to DNase I hypersensitive bands commonly seen in open promoter complexes footprints. However, since no protection around the −10 promoter element and the transcription start site is seen in the presence of C.AhdI and RNAP, the amount of open complexes must be very low. On the other hand, the results of KMnO4 probing, which reports the presence of unpaired thymines in transcription-competent open promoter complex, revealed that thymines in the −10 element of PahdICR reacted poorly with KMnO4 in the presence of RNAP alone (Figure 6B, lane 3), but became strongly reactive when both RNAP and C.AhdI were present together (Figure 6B, lane 4).
Figure 6.
Footprinting of RNA polymerase complexes PahdICR. The indicated proteins were combined with the wild-type PahdICR DNA fragment, complexes were allowed to form and footprinted with DNase I (A) or probed with KMnO4 (B). C.AhdI binding sites and the −10 element of the promoter are indicated.
Modelling transcription activity of PahdICR
The in vitro transcription results described above show that the level of transcription from PahdICR is low in the absence of C.AhdI, increases upon the addition of C.AhdI and, after reaching a maximum, gradually decreases. Presumably, activated transcription is due to the recruitment of RNAP through favorable protein-protein interactions between a C.AhdI dimer bound to promoter-distal-binding site and RNAP bound at the promoter, as is common for other activated promoter complexes. On the other hand, in vitro binding of C.AhdI to the wild-type PahdICR fragment reveals only complexes containing the C.AhdI tetramer, raising a question of how transcription from PahdICR is activated. We here propose a model, by which transcription from PahdICR is activated through RNAP ability to passively outcompete C.AhdI in the binding to the promoter-proximal site. In the absence of RNAP, binding of C.AhdI dimers to the two binding sites is highly cooperative, so that a C.AhdI dimer that is bound to the promoter-distal (high affinity) binding site recruits the second dimer to the promoter-proximal (low affinity) binding site. Therefore, in the absence of RNAP, only tetramer complexes are observed. However, when RNAP is also present in solution, it competes with C.AhdI dimers for binding to the lower affinity promoter-proximal-binding site. As a consequence, in addition to the C.AhdI tetramer complexes, C.AhdI dimer–RNAP–DNA complexes can also be formed. Our experiments demonstrate that in vitro such complexes are present in low abundance.
To determine if the regulatory mechanism described above can explain the experimentally measured dependence of PahdICR activity from the concentration of C.AhdI, we developed a quantitative model that is based on the following reactions:
| 2.1 |
| 2.2 |
| 2.3 |
| 2.4 |
| 2.5 |
In the equations above, the following notations are used: M and D denote C.AhdI monomers and dimers, respectively; D-DNA denotes C.AhdI dimer bound to promoter-distal (high affinity)-binding site; RNAP-DNA and T-DNA denote, respectively, complexes of promoter DNA with RNAP and the C.AhdI tetramer; D-DNA-RNAP is a complex consisting of the C.AhdI dimer, promoter DNA and RNAP. Equilibrium constants of these reactions are denoted by K1–K5. Equation (2.1) describes dimerization of C.AhdI. Equation (2.2) describes the binding of RNAP to PahdICR in the absence of C.AhdI. Equation (2.3) describes the binding of a C.AhdI dimer to promoter-distal (high affinity)-binding site. Equation (2.4) describes recruitment of a C.AhdI dimer to promoter-proximal (low affinity)-binding site through cooperative interactions with the dimer that is bound at the promoter-distal (high affinity) site (2.4). Finally, Equation (2.5) describes the competing reaction of RNAP recruitment to promoter facilitated by protein–protein interactions with the C.AhdI dimer bound at promoter-distal-binding site.
We further assume that transcription from PahdICR is proportional to its equilibrium occupancy by RNAP, which is a standard assumption (28,29). We also note that the published value of C.AhdI dimerization constant K1 (2.5 μM) (3) is significantly higher than the range of C.AhdI concentrations used in the experiment (from 0 to 188 nM), so under our conditions most C.AhdI molecules in solution exist as monomers. In Supplement A, we show that these assumptions together with Equations (2.1)–(2.5) lead to the following dependence of transcription activity of PahdICR from C.AhdI concentration:
| 2.6 |
where φ is transcription activity and [C] is the concentration of C.AhdI. Constants a, b and c depend on the concentration of RNAP and on the reaction equilibrium constants as given in Supplement A.
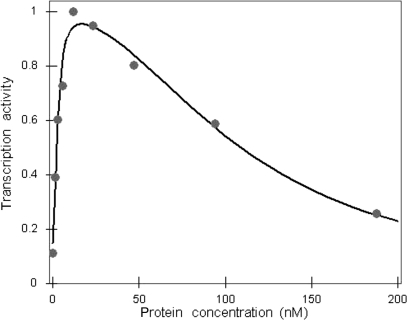
In Figure 7, the fit of Equation (2.6) to experimentally measured transcription activity levels at various C.AhdI concentrations is shown. As can be seen, there is a very good quantitative agreement between the model (grey line) and the experimental data (grey dots). It should be noted that several alternative models tested, for example one that assumed that C.AhdI binds DNA as a monomer (3), led to poor agreement with the experiment. Since alternative models have the same number of free parameters as the model given by Equation (2.6) (i.e. three parameters), the excellent agreement between our model and the experiment is not due to over-fitting.
Figure 7.
Modelling transcription activity of PahdICR versus C.AhdI protein concentration. Experimentally measured values of the transcription activity for the wild-type operator sequence are given by the grey circles. The transcription activities are measured in arbitrary units, so the transcription activity values are normalized such that the maximal value corresponds to one. The curve obtained by fitting the quantitative model to the experimental points is shown by the black line.
Equations (2.1)–(2.5) should also adequately describe data obtained using promoter templates with mutations in the C.AhdI-binding sites, since the only difference between the wild-type and mutant templates is that some of the values of the equilibrium binding constants should change (decrease) due to mutations. Indeed, similarly to the wild-type promoter case, there is a very good agreement between our quantitative model [Equation (2.6)] and the experimental results (Figure S1). Furthermore, changes in model parameters, which can be inferred from the fits described above, should be consistent with the nature of mutations in PahdICR. To intuitively better understand the meaning of parameters b and c, one should note that parameter b is related to the probability of activated (i.e. C.AhdI dimer-RNAP-DNA) complex formation, while parameter c is related to the probability of repressed (C.AhdI tetramer) complex formation (see Supplement A). Analysis presented in Supplement B shows that compared to the wild-type: (i) mutations introduced in either the promoter-distal C.AhdI-binding site alone or in both promoter-distal and promoter-proximal-binding sites should decrease the value of parameter b by approximately the same amount; (ii) mutations in the promoter-proximal C.AhdI-binding site alone should not affect the value of b and (iii) mutations in either of the two C.AhdI-binding sites (or in both of them) should decrease the value of c. This decrease should be smaller when a mutation is introduced in promoter-proximal C.AhdI-binding site.
Changes of the parameters determined from the fits to experimental data agree with the model predictions: the value of parameter b decreases ca. two orders of magnitude in the presence of mutations in either promoter-distal C.AhdI-binding site or in both sites but does not change significantly (only ∼20%) when a mutation in the promoter-proximal site is introduced. The value of parameter c decreases for all three mutants; the extent of the decrease can not be accurately determined in the case of mutation in the promoter-distal site, which is due to the fact that repression of transcription is not observed for this mutant at the measured range of C.AhdI concentrations. For the other two mutants, the decrease in c is smaller for the template with a mutated promoter-proximal C.AhdI-binding site than for the template with both sites mutated (two and three orders of magnitude, respectively). In summary, the quantitative model is in a very good agreement with experimental measurements, both in terms of the fits to experimentally measured data points and in terms of predicted changes of model parameters upon introduction of mutations in C.AhdI-binding sites. Therefore, we conclude that activated transcription from PahdICR is caused by the RNAP ability to compete with the strongly cooperative binding of C.AhdI to the promoter-proximal-binding site, and that control of PahdICR can be appropriately described by the mathematical relationship given above.
Regulation of transcription from the PahdIMS promoter
Inspection of the PahdIMS sequence revealed the presence of an AhdI site that partially overlaps with the −10 element of the promoter (Figure 1). The subunit composition and the sequence of the AhdI methyltransferase are highly similar to enzymes from Type I R-M systems, which invariably methylate the sixth position of an adenine in the target sequence (30,31). Based on this similarity, it is highly likely that the AhdI methylase also methylates an adenine. In the case of the AhdI site embedded in PahdIMS, a non-template strand A between the extended −10 TG motif and the CATACT −10 element and the template strand A two bases downstream of the −10 element should be methylated (Figure 1). To determine whether methylation of the AhdI site affects transcription from PahdIMS, two PahdIMS-containing fragments, identical except for the methylation state of the AhdI site, were prepared and used in in vitro transcription (Figure 8A). The results of multiple experiments consistently showed that the methylated template was utilized slightly less (∼50%) efficiently than the unmodified template. The effect was highly reproducible with different preparations of PahdIMS DNA. In vitro transcription from both fragments was independent of the presence of C.AhdI and initiated from the same start site as that determined in vivo (data not shown).
Figure 8.
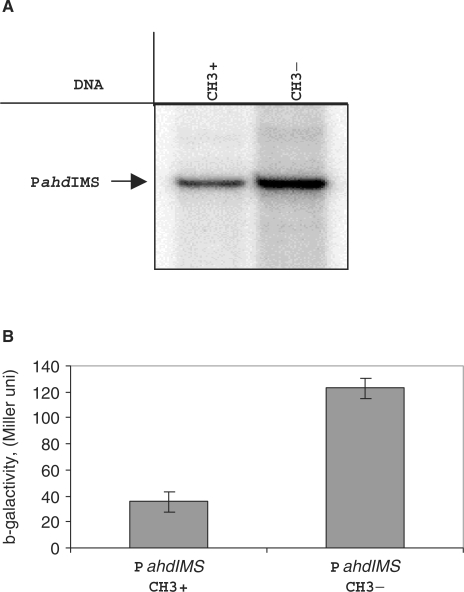
Regulation the PahdIMS activity in vitro and in vivo. (A) A single-round transcription in vitro by E.coli RNAP σ70 holoenzyme (100 nM) from regulatory region DNA fragment (13 nM) containing methylated and unmethylated promoter region were performed and products were separated by gel electrophoreses. Autoradiographs of 8% denaturing polyacrylamide gels are shown. (B) Effect of methylation on the expression of ahdIM gene. Plasmid pPM containing transcriptional fusion PahdIM’::’lacZ were co-transformed with the pAhdIMRinC plasmid (only methylase gene is active), bacterial cultures were grown until OD600 0.5 and β-galactosidase activity was measured in three independent sets of experiments.
The effect of methylation on PahdIMS expression was also studied in vivo (Figure 8B). To this end, a plasmid containing PahdIMS::lacZ transcription-translation fusion was created. The plasmid afforded robust expression of β-gal activity in lacZ− cells, as expected. We next introduced in these cells a compatible plasmid harbouring the entire AhdI R-M system with interrupted ahdIC (above). This plasmid was meant to serve as a source of the AhdI methylase and indeed control experiments showed that DNA from cells harbouring this plasmid was resistant to digestion with AhdI (data not shown). Quantitative β-gal measurements revealed that the presence of the AhdI R-M plasmid resulted in ∼4-fold decrease in β-gal activity, supporting the in vitro results and suggesting that there is a negative autoregulatory loop that controls the steady-state level of transcription from PahdIMS through the catalytic function of the AhdI methylase.
Modelling the dynamics of the AhdI system
We next wanted to determine whether the architecture of the ahdICR and ahdIMS loops, as well as the experimentally measured dependence of the ahdICR promoter activity from C.AhdI concentration allows the establishment of equilibrium states (in an equilibrium state, the values of transcript and protein concentrations are such that the system can indefinitely stay in this state) and whether such equilibrium states, if they exist, are stable, i.e. whether upon small perturbations of proteins and transcripts concentrations from the equilibrium value the system is able to return to the equilibrium state. To address these questions, a dynamical model of the two loops was made. First, we modelled the in vivo dynamics of the ahdICR loop. The model takes in account that the ahdIC and ahdIR genes are transcribed from the same promoter and assumes that the ahdIC ORF is translated less efficiently than the ahdIR ORF (32). The following equations describe how the amounts of ahdIC and ahdIR transcripts and products change with time upon the entry of the ahdICR loop in the cell:
| 2.7 |
| 2.8 |
| 2.9 |
where c is the concentration of the ahdICR transcripts, while CC and CR are concentrations of the C.AhdI and R.AhdI proteins, respectively. Constants λ and β denote, respectively, transcript and protein decay rates (the same decay rate for C.AhdI and R.AhdI is assumed). Constants αC and αR denote the rates of translation of C.AhdI and R.AhdI, respectively. The dependence of the PahdICR transcription rate from the concentration of C.AhdI is denoted as φ(CC) and was experimentally measured and theoretically modelled above. The first term on the right-hand side of Equation (2.7) describes the generation of the ahdICR transcripts, while the second term describes their decay. The first terms on the right-hand sides of Equations (2.8) and (2.9) represent transcript translation, while the second terms represent protein decay.
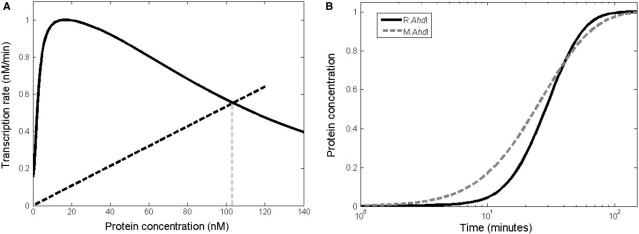
From the above equations it is straightforward to see that CR(t) = αR/αCCC(t), so in the further analysis we can concentrate only on Equations (2.7) and (2.8). In Supplement C, we show that these two equations have an equilibrium state, which is given by the intersection of the φ(CC) curve and a straight line whose slope is equal to the product of protein and transcript decay rates divided by protein translation rate (βλ/αC). The position of the equilibrium value of protein concentration is graphically illustrated in Figure 9A. In Supplement D we show that this equilibrium state is stable as long as the slope of the straight line has a larger value than the slope of the φ(CC) curve at the point of intersection (i.e. at the equilibrium point). From Figure 9A it follows that the stability condition is satisfied for virtually any parameter values, i.e. for any value of the equilibrium protein concentration. We therefore conclude that the ahdICR loop architecture and the experimentally measured curve of PahdICR activity versus C.AhdI concentration are consistent with the biological expectation that the system should have a stable equilibrium point.
Figure 9.
Modelling of AhdI system dynamics. (A) Equilibrium position for C.AhdI. The black line presents transcription activity versus protein concentration. The equilibrium is determined by the intersection of the black dashed line with the transcription activity curve, and the equilibrium protein concentration is indicated by the vertical grey dashed line. Parameter values used for C.AhdI are: the maximal value of the transcription rate for PahdICR is 1 nM/min, protein and transcript half-lives are 30 min and 5 min, respectively, and C.AhdI transcript is translated three times during its lifetime. (B) Dynamics of equilibrium establishment for R.AhdI and M.AhdI. Values on the vertical axis give protein concentration scaled by the equilibrium value, while the horizontal axis corresponds to time post-plasmid entry. The full and dashed lines correspond, respectively, to the change of the protein concentrations with time for R.AhdI and M.AhdI. For R.AhdI we use five times larger translation rate compared to C.AhdI, while other parameters are the same. For M.AhdI KD = 650 nM, while the other parameters are the same as for R.AhdI.
The above analysis can be directly extended to the ahdIMS loop since Equations (2.7) and (2.8) also hold for this case with the following modifications. First, the decay rates and the translation rates associated with C.AhdI now become associated with M.AhdI. Second, φ(CC), the transcription activity versus C.AhdI concentration for PahdICR should be changed to φ(CM), transcription activity of PahdIMS versus AhdI methylase (M.AhdI) concentration. Since we established above that AhdI methylation attenuates the activity of PahdIMS, it is plausible to assume that φ(CM) has a Michaelis–Menten form, i.e. φ(CM) = KD/(CM + KD), where CM is the concentration of the AhdI methylase and KD determines the concentration of the methylase in the presence of which the transcription activity of PahdIMS drops to half-maximal value. By following arguments similar to those presented in the case of the ahdICR loop, we again conclude that consistent with biological expectations, the ahdIMS loop has a stable equilibrium point (determined by the intersection of the φ(CM) curve and the straight line described above).
We were next interested in determining how the concentrations of AhdI methyltransferase and restriction endonuclease approach the equilibrium states, i.e. how these concentrations change upon entry of an AhdI R-M plasmid in the cell. To this end, we solved Equations (2.7)–(2.9) and the equivalents of Equations (2.7) and (2.8) for the AhdI methylase with the initial condition that the transcript and protein levels corresponding to all genes equal zero at the time of plasmid entry, which is taken as a zero time point. We solve Equations (2.7)–(2.9) as described in Supplement C, using parameter values typical for bacteria (see the legend of Figure 9A). The value of KD (see above) is chosen such that the equilibrium protein concentrations are the same for C.AhdI and M.AhdI, allowing direct comparisons of dynamics of the approach to equilibrium for both loops. The obtained dependence of the endonuclease and methylase concentrations from time is shown in Figure 9B. As can be seen, the concentrations of both R.AhdI and M.AhdI increase and then saturate at stable equilibrium values but the dynamics of establishment of the two equilibria is significantly different. That is, the rate of increase of the R.AhdI protein concentration is very small for early times post-plasmid entry but increases rapidly at later times, so that the transition between the OFF and the ON states of the ahdICR loop is (i) delayed and (ii) happens in a narrow time interval. On the other hand, the concentration of the AhdI methylase increases rapidly at early times post-plasmid entry but the rate of protein accumulation decreases at later times.
DISCUSSION
In this work, we map the AhdI system promoters and demonstrate that transcription of both the restriction endonuclease and methyltransferase genes is controlled by independent autoregulatory loops, each leading to stable equilibrium states. We present experimental measurements of the wild-type and mutant ahdICR promoters activities as a function of C.AhdI concentration along with a quantitative model that explains the experimentally observed behaviors. We show that the architecture of the two loops is such that when a plasmid carrying the ahdI genes enters a naïve host (i) the restriction endonuclease is synthesized with a delay with respect to the methyltransferase and (ii) the transition from the ON to the OFF state of ahdICR transcription occurs in a narrow time interval. These dynamical properties of the system are largely due to the mechanism of transcription regulation by C.AhdI.
Previous work as well as results presented here demonstrates that when C.AhdI is combined with the wild-type ahdICR promoter DNA, only free DNA or complexes containing C.AhdI tetramers can be detected (3,17). Either of these templates are transcriptionally inactive, the former due to the weakness of the promoter and the latter due to steric hindrance from the C.AhdI dimer bound to promoter-proximal-binding site. Therefore, a question of how transcription from PahdICR is activated arises. We show here, that the experimentally observed profile of C.AhdI-dependent transcription from the ahdICR promoter can be quantitatively explained by a model in which RNAP outcompetes the C.AhdI dimer that would bind to the promoter-proximal-binding site in the absence of RNAP. This competition occurs due to favorable interactions between the C.AhdI dimer bound at promoter-distal site and RNAP, most likely its σ70 subunit region 4 domain (27). Therefore, the mechanism of transcription regulation by C.AhdI is characterized by the mutually opposing effects of (i) strong cooperativity in the binding of C.AhdI dimers to promoter DNA and (ii) the ability of RNAP to outcompete the dimer bound at the promoter-proximal position. The mechanism of ahdICR promoter activation by C.AhdI is quite inefficient, since in vitro transcription indicates that only a small fraction of templates becomes transcriptionally active even at optimal conditions. However, such inefficiency is likely to be biologically significant as it decreases the steady-state level of expression of restriction endonuclease, a highly toxic protein.
With regard to the ahdIMS loop, the methyltransferase activity is expected to be high in the beginning of the establishment of ahdI genes in a naïve host, so that the host DNA becomes protected from the endonuclease cleavage early on. This is ensured by the high basal transcription rate of the ahdIMS promoter. Since high levels of methyltransferase activity can prevent the R-M system's effectiveness in excluding foreign DNA from the host, a mechanism must exist that attenuates the initial strong synthesis of methyltransferase so that optimal steady-state concentration of methyltransferase is established in the cell. In C protein dependent R-M systems with divergently transcribed M and CR genes, such regulation can occur through direct coupling of transcription activities of divergent promoters. Indeed, in such systems overproduction of C protein was shown to decrease the level of M promoter transcription in vivo (6,8,18). Obviously, such mechanisms cannot operate in C protein-dependent systems with convergently transcribed genes. We here show that the AhdI methylase attenuates transcription of its own genes by methylating a site that partially overlaps with the -10 element of its promoter. This mechanism of negative autoregulation is similar to that we previously described for CfrBI, a C-protein-independent R-M system whose divergent transcription is controlled by methylation of a CfrBI site overlapping with the -35 element of the methyltransferase promoter (19). The ‘engineering’ solution used in AhdI for attenuation of the synthesis of methyltransferase, is clearly not the only one possible in convergently-transcribed C-protein-dependent systems. For example, in the Esp1396I system, the promoter of the standalone methyltranferase gene contains a strong C-protein-binding site that couples methyltranferase gene transcription to CR operon transcription [(8) and our unpublished data]. The existence of such arrangements further underscores the importance of controlling the expression of methyltransferase for the host cell.
The in vitro analysis of the transcription control of the ahdICR promoter by C.AhdI protein has allowed us to model the in vivo dynamics of the system during establishment in a naïve host. The dynamics of changes in the AhdI products concentrations during approach to equlibrium (Figure 9B) is biologically reasonable and can be intuitively understood in terms of the properties of the system architecture and the mechanisms of transcriptional control. First, the small rate of R.AhdI accumulation at early times post-plasmid entry is a consequence of low basal transcription rate from the ahdICR promoter that leads to the absence of significant transcription until sufficient amounts of C.AhdI are synthesized. Furthermore, the relatively high dimerization dissociation constant of C.AhdI ensures an additional delay, since only C.AhdI dimers can bind DNA and activate transcription. The likely low rates of translation of the leaderless C.AhdI transcript should also contribute to the delay. Therefore, there are multiple features behind the loop design that ensure that the synthesis of the endonuclease occurs with a delay and in a highly cooperative manner, arguing that this is a major constraint exhibited on the loop architecture and transcriptional control.
On the other hand, at later times, accumulated C.AhdI starts to significantly enhance transcription due to the fast (quadratic) increase of ahdICR promoter activity with C.AhdI concentration. We note that such a fast increase is a direct consequence of cooperativity (3), i.e. of the fact that most of C.AhdI is present in solution as monomer and that a dimer (two monomers) enters the activation complex (C.AhdI dimer–RNAP–DNA). For example, if only C.AhdI monomers were binding to promoter-distal-binding site, the transcription activity of the promoter would increase linearly instead of quadratically and the switch from the OFF to the ON state would be exhibited over a broader C.AhdI concentration (and therefore, time) interval. When the amount of C.AhdI increases further, the repression complex starts to dominate over the activation complex and transcription of ahdICR is decreased. As a consequence, a certain equilibrium concentration of the regulator and endonuclease transcripts is established, and we show that this equilibrium is stable with respect to small perturbations.
In summary, while AhdI presents a relatively simple system, its properties appear well optimized to perform the desired biological function. Study of other R-M systems should more fully map different mechanistic solutions through which the design principles, discussed above in the context of the AhdI system, can be efficiently implemented.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
We thank Dr Simon Streeter for providing purified C.AhdI protein. This work was supported by an NIH RO1 grant GM59295, NIH FIRCA research grant R03 TW07145, RFBR grant 060449667A and Russian Academy of Sciences Presidium Program in Molecular and Cellular Biology New Groups Grant to K.S. M.D. acknowledges support from NSF under Agreement No. 0112050 and NSF grant MCB-0418891. G.K. acknowledges the support from BBSRC (UK). Funding to pay the Open Access publication charges for this article was provided by NIH RO1 grant GM 9295.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 1996;42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Streeter SD, Papapanagiotou I, McGeehan JE, Kneale GG. DNA footprinting and biophysical characterization of the controller protein C.AhdI suggests the basis of a genetic switch. Nucleic Acids Res. 2004;32:6445–6453. doi: 10.1093/nar/gkh975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ives CL, Nathan PD, Brooks JE. Regulation of the BamHI restriction-modification system by a small intergenic open reading frame, bamHIC, in both Escherichia coli and Bacillus subtilis. J. Bacteriol. 1992;174:7194–7201. doi: 10.1128/jb.174.22.7194-7201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anton BP, Heiter DF, Benner JS, Hess EJ, Greenough L, Moran LS, Slatko BE, Brooks JE. Cloning and characterization of the BglII restriction-modification system reveals a possible evolutionary footprint. Gene. 1997;187:19–27. doi: 10.1016/s0378-1119(96)00638-5. [DOI] [PubMed] [Google Scholar]
- 6.Rimseliene R, Vaisvila R, Janulaitis A. The eco72IC gene specifies a trans-acting factor which influences expression of both DNA methyltransferase and endonuclease from the Eco72I restriction-modification system. Gene. 1995;157:217–219. doi: 10.1016/0378-1119(94)00794-s. [DOI] [PubMed] [Google Scholar]
- 7.Zheleznaya LA, Kainov DE, Yunusova AK, Matvienko NI. Regulatory C protein of the EcoRV modification-restriction system. Biochemistry. 2003;68:105–110. doi: 10.1023/a:1022105804578. [DOI] [PubMed] [Google Scholar]
- 8.Cesnaviciene E, Mitkaite G, Stankevicius K, Janulaitis A, Lubys A. Esp1396I restriction-modification system:structural organization and mode of regulation. Nucleic Acids Res. 2003;31:743–749. doi: 10.1093/nar/gkg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijesurier RM, Carlock L, Blumenthal RM, Dunbar JC. Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2000;182:477–487. doi: 10.1128/jb.182.2.477-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidmann S, Seifert W, Kessler C, Domdey H. Cloning, characterization and heterologous expression of the SmaI restriction-modification system. Nucleic Acids Res. 1989;17:9783–9796. doi: 10.1093/nar/17.23.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bart A, Dankert J, van der Ende A. Operator sequences for the regulatory proteins of restriction modification systems. Mol. Microbiol. 1999;31:1277–1278. doi: 10.1046/j.1365-2958.1999.01253.x. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl Acad. Sci. USA. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semenova E, Minakhin L, Bogdanova E, Nagornykh M, Vasilov A, Heyduk T, Solonin A, Zakharova M, Severinov K. Transcription regulation of the EcoRV restriction-modification system. Nucleic Acids Res. 2005;33:6942–6951. doi: 10.1093/nar/gki998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mruk I, Rajesh P, Blumenthal R. Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm837. Advance Access published on 11 October 2007; doi:10.1093/nar/gkm837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 2005;30:275–279. doi: 10.1016/j.tibs.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 17.McGeehan JE, Papapanagiotou I, Streeter SD, Kneale GG. Cooperative binding of the C.AhdI controller protein to the C/R promoter and its role in endonuclease gene expression. J. Mol. Biol. 2006;358:523–531. doi: 10.1016/j.jmb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Lubys A, Jurenaite S, Janulaitis A. Structural organization of the plasmid-borne restriction-modification system type II Kpn2I from Klebsiella Pneumoniae RLF2. Nucleic Acids Res. 1999;27:4228–4234. doi: 10.1093/nar/27.21.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakharova M, Minakhin L, Solonin A, Severinov K. Regulation of RNA polymerase promoter selectivity by covalent modification of DNA. J. Mol. Biol. 2004;335:103–111. doi: 10.1016/j.jmb.2003.09.081. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Marks P, McGeehan J, Kneale G. A novel strategy for expression and purification of the DNA methyltransferase, M.AhdI. Prot. Expres. Purif. 2004;37:236–242. doi: 10.1016/j.pep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Metzger W, Heumann H. Footprinting with Exonuclease III. In: Kneale GG, editor. DNA–Protein Interactions: Principles and Protocols. 1994. Vol. 30. Methods in Molecular Biology. Humana Press Inc., Totowa, NJ, pp. 11–20. [Google Scholar]
- 23.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 24.Dove SL, Hochschild A. A bacterial two-hybrid system based on transcription activation. In: Fu H, editor. Protein-Protein Interaction: Principles and Protocols. 2004. Vol. 261. Methods in Molecular Biology. Humana Press Inc., Totowa, NJ, pp. 231–250. [DOI] [PubMed] [Google Scholar]
- 25.Knowle D, Lintner RE, Touma YM, Blumenthal RM. Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2005;187:488–497. doi: 10.1128/JB.187.2.488-497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kita K, Tsuda J, Nakai SY. C.EcoO109I, a regulatory protein for production of EcoO109I restriction endonuclease, specifically binds to and bends DNA upstream of its translational start site. Nucleic Acids Res. 2002;30:3558–3565. doi: 10.1093/nar/gkf477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGeehan JE, Streeter SD, Papapanagiotou I, Fox GC, Kneale GG. High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila. J. Mol. Biol. 2005;346:689–701. doi: 10.1016/j.jmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Buchler NE, Gerland NE, Hwa N. On schemes of combinatorial transcription logic. Proc. Natl. Acad Sci. USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J. Mol. Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- 30.Marks P, McGeehan J, Wilson G, Errington N, Kneale G. Purification and characterisation of a novel DNA methyltransferase, M.AhdI. Nucleic Acids Res. 2003;31:2803–2810. doi: 10.1093/nar/gkg399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray N. Type I restriction systems: sophisticated molecular machines (a legacy of Bertrani and Weigle) Microbiol. Mol. Rew. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rew. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.