Abstract
The transcription factor NF-Y is a trimer with histone-like subunits that binds and activates CCAAT-containing promoters. NF-Y controls the expression of several key regulators of the cell cycle. In this study, we examined the functional and molecular effects of NF-YB knockdown. Cell cycle progression is affected with a G2/M-specific depletion. This is due to the inability of activation of G2/M-specific genes, as evidenced by expression profiling, RT-PCR and ChIP data. Surprisingly, apoptosis is also observed, with Caspase 3/7/8 cleavage. A role of p53 and Bcl-2 family members is important. NF-YB inactivation is sufficient to functionally activate p53, in the absence of DNA damage. Failure to maintain a physiologic level of CCAAT-dependent transcription of anti-apoptotic genes contributes to impairment of Bax/Bcl-2 and Bax/Bcl-XL ratios. Our data highlight the importance of fine balancing the NF-Y-p53 duo for cell survival by (i) maintaining transcription of anti-apoptotic genes and (ii) preventing p53 activation that triggers the apoptotic cascade.
INTRODUCTION
The CCAAT-binding factor NF-Y is a mammalian transcription factor that binds to CCAAT boxes in the promoters of a wide variety of genes. The CCAAT box is a common promoter element, and, in higher eukaryotes, it is found in ∼60% of tissue-specific, housekeeping and cell cycle-regulatory genes (1,2). In vitro and in vivo assays clearly demonstrated that NF-Y is the major CCAAT-binding activator (3,4). NF-Y is a heterotrimeric complex composed of three subunits, A, B and C, which are all essential for CCAAT binding (5,6). NF-YB and NF-YC contain histone fold motifs (HFMs) common to all core histones; NF-YB and NF-YC dimerization is essential for NF-YA association and sequence-specific DNA binding (7 and references therein). NF-Y is required to organize the chromatin in proximity of transcriptional start sites, thereby enabling recruitment of coactivators (8,9).
NF-Y controls the expression of several key regulators of the cell cycle (10–16). A bioinformatic analysis of cell cycle promoters showed a remarkable and specific abundance of CCAAT boxes in promoters regulated during the G2/M phase (17). Chip assays clearly demonstrated that NF-Y interactions with cell cycle regulated promoters, such as CDC2, CDC25A/B/C, cyclin A2, cyclin B1/B2 and E2F1, is highly dynamic through the cell cycle (18).
The three subunits are regulated at different levels. NF-YA has two major isoforms generated by alternative splicing, variously expressed in different cell types (19). NF-YC also has different isoforms that are regulated in a cell-specific way (Salvatoni,L. and Mantovani,R., unpublished data). The nuclear localization of NF-YC is cell cycle regulated and depends on the presence of its histone fold partner NF-YB (20). NF-YA is not ubiquitously expressed, being undetectable in adult skeletal muscle and heart tissues, whereas the NF-YB/NF-YC heterodimer is expressed in most tissues (21). Another level of regulation involves the acetylation of NF-YB (22) and the acetylation and phosphorylation of NF-YA [(22), Caretti,G. and Mantovani,R., unpublished data].
NF-Y is also involved in the modulation of the activity of cell cycle promoters in response to DNA damage (14,23), through wild-type p53-dependent transcriptional inhibition (16,24). p53 forms a complex with NF-Y on CCAAT box-containing promoters, and upon DNA damage, this complex can recruit histone-deacetylases (HDACs) and release acetyltransferases (HATs), causing a repression of cell cycle promoters (24). Interestingly, mutant p53/NF-Y complexes have the opposite effect on transcription upon DNA damage, that is, transactivation of proliferative genes, and aberrant cell cycle regulation (25,26). Finally, a recent genome-wide mRNA expression profiling of the transformation process verified that p53-mediated transcriptional repression of several targets is dependent on the activities of NF-Y, p21 and E2F (27).
Attempts to define the role of NF-Y in cell cycle regulation focused on NF-YA. A conditional deletion of both NF-YA alleles in primary cultures of mouse embryonic fibroblast cells led to a complete block in cell cycle progression (28). Expression of the dominant negative NF-YA mutant in mouse NIH 3T3 fibroblasts caused the cells to grow slower, with a modest increase in the time required for the cells to complete the S phase of the cell cycle (29).
We decided to sytematically examine the functional and transcriptional effects of abrogation of the NF-YB subunit by RNAi, using FACS, IF, gene expression profiling, RT-PCR and ChIP analysis in wt p53 HCT116 cells and in their E6 expressing counterpart, in which p53 is functionally inactivated.
MATERIALS AND METHODS
Cell lines and drugs
The human colon carcinoma HCT116 and HCT116/E6 were generously provided by Bert Volgestein (Johns Hopkins University School of Medicine, Baltimore, MD, USA). The cells were maintained as described previously (30). SW1353 human chondrosarcoma cells (ATCC, USA) were cultured in Leibovitz's L-15 medium containing 10% fetal bovine serum (Euroclone), 2 mM glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin according to the ATCC protocol.
For caspase inhibitor treatments, cells were treated with 25 μM ZVAD-fmk (Sigma-Aldrich) 1 h before and during the 48 h of siRNA transfection.
Small interfering RNA (siRNA)
HCT116 and HCT116/E6 cells were transfected (Lipofectamine 2000, Invitrogen) with 200 nM of paired NF-YB and non-targeting control small interfering RNA (siRNA) designed by Ambion, or with scramble and NF-YB shRNAs designed by Sigma-Aldrich.
NF-YB siRNAs
NF-YB1: sense-GGAAGUCAUUAUGUUAUACtt, antisense- GUAUAACAUAAUGACUUCCtc;
NF-YB2: sense-GGACAGCAUGAAGGAUCAUtt, antisense-AUGAUCAUUCAUGCUGUCCtc;
NF-YB3: sense- GGCAUUUACUAACCAGUUAtt, antisense-UAACUGGUUAGUAAAUGCCtc.
NF-YB shRNA
CCGGGCTATGTCTACTTTAGGCTTTCTCGAGAAAGCCTAAAGTAGACATAGCTTTTT;
CCGGGCAAGTGAAAGGTGCCATCAACTCGAGTTGATGGCACCTTTCACTTGCTTTTT.
Cells were monitored by microscopy at 24, 48 and 72 h. Total extracts and RNA were prepared after 48 h upon siRNA transfection.
Immunoblotting
For immunoblotting, equivalent amount of total extracts were resolved by SDS–PAGE, electrotransferred to nitrocellulose membrane and immunoblotted with NF-YB, NF-YA and NF-YC-purified rabbit polyclonal antibodies, anti-CycB1 and Cyc-A (Cell Signaling), anti-p53 (Active Motif), anti-phospo-Ser15 p53 (Cell Signaling), anti-Bax (N-20, S.Cruz); anti-CytC (S.Cruz) and anti-actin (Sigma).
Immunofluorescence
HCT116 cells were mock and NF-YB siRNA transfected for 48 h, then immunofluorescence assays were performed as described previously (30) with the following antibodies: anti-cleaved caspases 3-6-7-8 (Cell Signaling), anti-p53 Ab-7 (Calbiochem), anti-phospho histone H2AX (Ser-139) (Upstate) and Hoechst. Immunofluorescence was examined with a Zeiss AxioSkop 40 fluorescence microscope (Carl Zeiss, Germany) and the images collected with an AxioCam HRc camera and AxioVision version 3.1 software package.
Plasmids and transfections
A total of 2 × 105 HCT116 cells were transiently transfected with Lipofectamine 2000 (Invitrogen) using the indicated doses of DN-YA and NF-Y, 200 ng of BI-1 or Bcl-2 luciferase reporter, 50 ng of β-galactosidase and carrier plasmid to keep the total DNA concentration constant at 800 ng. Cells were recovered 24 h after transfection, resuspended in lysis buffer (1% Triton X-100, 25 mM glycil-glycine, 15 mM MgSO4, 4 mM EGTA) for luciferase activities. β-Galactosidase was assayed to control for transfection efficiency. Three independent transfections in duplicate were performed.
Chromatin immunoprecipitation
Chromatin immunoprecipitations were essentially performed as described previously (24,30). Chromatin was prepared upon 48 h of control siRNA or NF-YB siRNA transfection. CcnB2, Cdc2 and Cdc25C primers were described in Ref. (30). Oligonucleotide sequences are indicated in Supplementary Data.
Gene expression profiling and data analysis
Gene expression profiling was performed as described in Ref. (31). All expression values for the genes in the MAS 5.0 absolute analyses were determined using the global scaling option. MAS 5.0 comparison algorithm was used for pairwise analysis. Gene Ontology enrichment P-values were computed according to the hypergeometric distribution. GO terms have been ranked by increasing P-value; a ‘ratio’ has been calculated between the number of genes belonging to the GO term, in the sample-set versus the universe-set. Only terms with (i) P-value <1.00E−03 and (ii) ratio >2 and (iii) number of sample genes >4 have been displayed; in addition, redundant terms were manually removed, and terms were grouped by macro-categories (including terms from all the three GO partitions: MF, BP and CC). Differentially Expressed Genes (DEGs) were selected using SAM method (as provided by R package DEDS). The parameter ‘delta’ was set to 0.4, with an estimated FDR (False Discovery Rate) = 25% (32). A detailed list of all the genes identified in this study is available upon request.
Reverse Transcription (RT)-PCR analysis
RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany), according to the manufacturer's protocol, from HCT116 and HCT116/E6 control or NF-YB siRNA-transfected cells. For cDNA synthesis, 5 μg of RNA was retrotranscribed with a Moloney murine leukemia virus reverse transcriptase (Finzymes). Semiquantitative PCR analysis was performed with oligonucleotides indicated in Supplementary Data. CcnB2, Cdc25C and Bax primers were described previously (30). Glyceraldehyde 3-phosphate dehydrogenase control RT-PCR was performed with standard oligonucleotides. Oligonucleotide sequences are indicated in Supplementary Data.
Flow cytometric cell cycle analysis
Cells were harvested after 48 h of siRNA transfection and DNA distribution analysis of propidium–iodide-stained cells was performed by an Epics cytofluorimeter (Beckman Coulter). Apoptotic cells were identified by fluorescence-activated cell sorting (FACS) using Annexin V-PE conjugate (Invitrogen) following the protocol of the manufacturer.
Isolation of cytosolic fraction by digitonin lysis method
A total of 2 × 105 HCT116 cells were transiently transfected with 1 μg of expression plasmids for each NF-Y subunits. After 24 h the cells were incubated overnight with Adriamycin (0.2 and 0.5 μg/ml) and were then harvested. After two washes with PBS, the pellet was resuspended in digitonin lysis buffer (75 mM NaCl, 1 mM NaH2PO4, 8 mM Na2HPO4, 250 mM sucrose and 190 mg/ml of digitonin) containing protease inhibitors and incubated on ice for 5 min. The releasate was centrifuged at 15 000 r.p.m. at 4°C for 30 min and used for western blotting using mouse cytochrome c antibody (S. Cruz).
RESULTS
Silencing of NF-YB by RNA interference
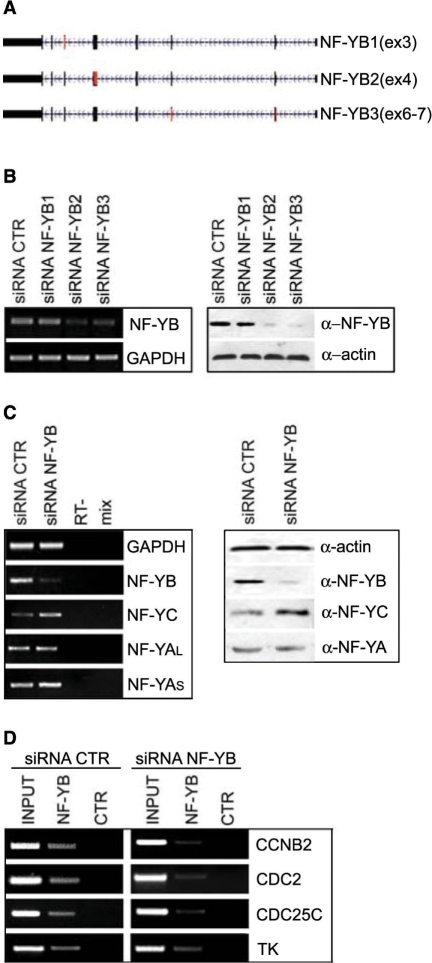
We designed three RNA-interference (RNAi) oligos to disrupt the function of the NF-YB subunit (Figure 1A); transfections of HCT116 cells with two of these siRNAs —NF-YB2 and NF-YB3— resulted in a significant and reproducible degradation of NF-YB mRNA, as shown by RT-PCR analysis (Figure 1B, left panel), and protein, as judged by western blotting (Figure 1B, right panel). The degree of knockdown was 75–95%. The actin protein and GAPDH mRNA were used as internal controls (Figure 1B, right and left panels, respectively). We analyzed the effect of NF-YB silencing on the transcription and expression of the other NF-Y subunits. We noticed increased transcription of NF-YC, and no changes in the two NF-YA isoforms, as shown by RT-PCR analysis (Figure 1C, left panel). Consistent with this, western blots showed increased NF-YC levels, and no change for NF-YA (Figure 1C, right panel and Supplementary Figure 1A). Finally, to ascertain whether NF-Y binding was affected in RNAi inactivated cells, ChIP assays with the anti-NF-YB antibody were performed. As shown in Figure 1D, NF-YB siRNA reduces considerably the binding to CCNB2, Cdc2, Cdc25C and TK promoters. The PCR of each ChIP were quantitatively compared to a Flag control antibody (Supplementary Figure 2A). We conclude that effective inactivation of NF-YB can be achieved in HCT116 cells.
Figure 1.
NF-YB RNAi in HCT116 cells. (A) Three siRNA were designed on human NF-YB gene. Black boxes represent exons; red boxes indicate targeted exons. (B) Left panel: RT-PCR analysis of NF-YB and GAPDH mRNA transcripts in negative control and NF-YB1-2-3-silenced cells. Right panel: western blot analysis of HCT116 total extracts with anti-NF-YB and anti-actin antibodies, transfected with control and NF-YB siRNAs. (C) RT-PCR (left panel) and western blot analysis (right panel) of the three NF-Y subunits in control or NF-YB2 transfected cells. (D) Chromatin immunoprecipitation analysis of NF-Y targets, using control and NF-YB-silenced HCT116 cells with NF-YB and Flag antibodies.
NF-YB silencing induces depletion in G2/M and apoptosis
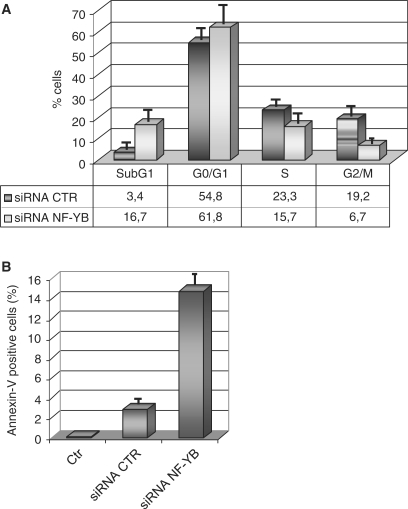
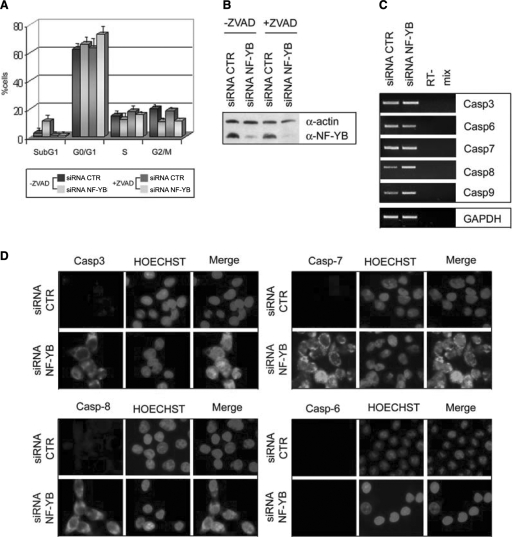
The effect of NF-Y inactivation on cellular physiology was investigated by flow cytometry after siRNA treatment: the most notable change was a decrease in cells in the G2/M phase, from 19.2% to 6.7% (Figure 2A). This is not accompanied by a concomitant increase of cells in G1. Unexpectedly, hyplodiploid sub-G1 events, evaluated after propidium iodide staining, increased from 3.4% to 16.7%. This might be due to induction of apoptosis: the annexin V staining of negative control and NF-YB siRNA-treated cells, followed by FACS analysis, indeed showed a parallel increase—from 3% to 15%—in annexin V positive cells, confirming that these cells are undergoing apoptosis (Figure 2B). A second siRNA (NF-YB3) has been used to confirm that NF-YB depletion impairs G2/M progression and induces apoptosis (Supplementary Figure 1B). In addition the same cell cycle alterations after NF-YB silencing has been observed in the human chondrosarcoma cell line SW1353 (Supplementary Figure 3A).
Figure 2.
Cell cycle progression in HCT116 NF-YB-silenced cells. (A) Cell cycle distribution analysis via flow cytometry of HCT116 cells, transfected NF-YB and non-targeting control siRNA. (B) Apoptosis was determined by staining with annexin V; the values represent the portion of annexin V staining cells. Error bars indicate standard deviations.
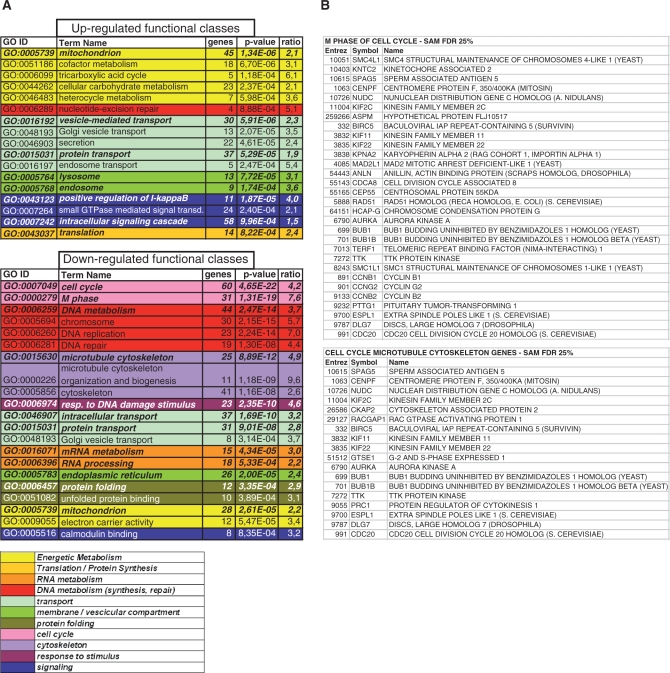
In parallel, we performed expression profiling analysis of cells transfected with negative control and NF-YB RNAi oligos, with the Affymetrix HG-U133 plus2.0 platform, containing 47 000 transcripts. Using relatively stringent criteria, 478 genes were down-regulated and 803 were up-regulated. For the repressed genes, we found that cell cycle, microtubule cytoskeleton and DNA metabolism are the statistically most important functions (Gene Ontology enrichment analysis) (Figure 3A). The list of cell cycle-microtubule cytoskeleton and G2/M genes is shown in Figure 3B. All of these categories are expected, given the functional role of NF-Y. On the other hand, apoptosis is not an over-represented category, with the only significant changes being an up-regulation of FADD, Caspase-9, IFM2 and FASTK. Furthermore, setting the FDR threshold to 12.3%, we found other potential candidates as anti-apoptotic genes regulated by NF-Y: BAG3, AERHGDIA, BRE, PIK3R2, BDNF, according to the expression profiling experiments of Figure 3.
Figure 3.
Microarray data analysis. (A) Gene Ontology terms displaying a statistically significant over-representation in the sample-set (up- and down-regulated genes, respectively). Parameter ‘genes’ is the number of genes annotated by the corresponding term in the sample set; parameter ‘P-value’ refers to the over-representation (see Methods section for statistical details); parameter ‘ratio’ is calculated as the ratio between term frequency in the sample set versus the universe set (all microarray genes). Terms have been pruned, and then grouped according to manually determined macro-categories, identified by different colors: see the legend for the categories names; categories with similar colors are functionally related. (B) Down-regulated genes, annotated by GO terms ‘M Phase’, both ‘Cell Cycle’ and ‘Microtubule Cytoskeleton’. IDs are expressed according to NCBI Entrez Gene.
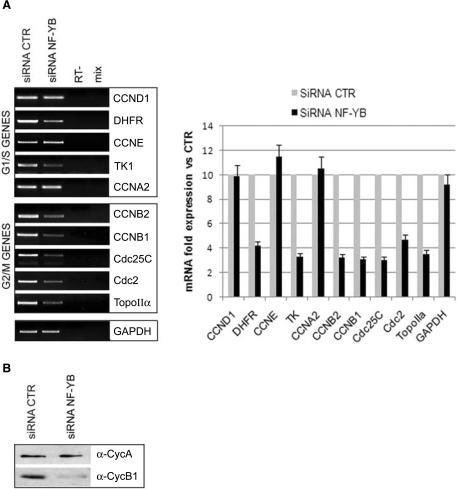
The profiling analysis was validated by RT-PCR, focusing on CCAAT-promoters regulated during the cell cycle. As shown in Figure 4A, all the G2/M promoters analyzed—CCNB2, CCNB1, Cdc25C, Cdc2, Topoisomerase IIα—revealed an evident decrease in transcription. Western blot analysis of whole cell extracts showed that CyclinB1, but not CyclinA protein levels, was drastically reduced upon NF-YB knockdown (Figure 4B). On the other hand, the transcription of many genes involved in G1/S transition was not impaired by NF-YB silencing: CCND1, CCNE, CCNA2; conversely TK1 and DHFR decreased, suggesting some differential behavior within this class. The validation by RT-PCR analysis was further performed by transfecting a second NF-YB siRNA (NF-YB3) (Supplementary Figure 1A, right panel). In conclusion, results from profiling data help us explain the G2/M depletion phenotype of NF-YB inactivated cells, indicating that a generalized defect in transcription of G2/M genes is responsible for it.
Figure 4.
Expression analysis of NF-Y targets in NF-YB-silenced HCT116 cells. (A) RNA expression levels of the indicated NF-Y target genes relative to RNA levels in control and NF-YB-silenced cells (left panel). Quantification of the RT-PCRs, relative to control siRNA expression levels, is plotted in the right panel. Error bars indicate standard deviations. (B) Cyclin A and Cyclin B1 expression analysis of total extracts of HCT116 non-targeting control or NF-YB siRNA transfected cells.
NF-YB silencing induces caspase dependent apoptosis
To explain the role of NF-Y inactivation in the induction of apoptosis, we turned our attention to caspases, because their cleavage and activation play a central role in the initiation and execution of this process. We first examined whether apoptosis is a caspase-dependent phenomenon in NF-YB-inactivated HCT116, treating them with the broad-spectrum caspase inhibitor Z-VAD. Figure 5A shows that apoptosis was significantly reduced in the presence of Z-VAD. A dramatic decrease in sub-G1 cells is indeed observed, associated to a modest increase in G1 cells. Western blot analysis confirmed that NF-YB protein levels were drastically reduced both in untreated and in Z-VAD-treated cells (Figure 5B), ruling out that the lack of apoptosis is due to inefficient NF-YB inactivation.
Figure 5.
NF-YB silencing induces apoptosis via caspases activation. (A) Cell cycle distribution analysis via flow cytometry of non-targeting control and NF-YB siRNA transfected cells, untreated or treated with ZVAD. (B) Western blot analysis of negative control and NF-YB-silenced HCT116 total extracts with anti-NF-YB and anti-actin antibodies, untreated or treated with ZVAD. (C) Expression analysis (RT-PCR) of the indicated caspase mRNA transcripts. (D) Immunofluorescence analysis of the indicated cleaved caspases and HOECHST.
We examined by RT-PCR the mRNA levels of caspase genes, before and after siRNA transfection. Figure 5C shows no change for caspase 6 and 7, and a modest increase for caspase 3, 8 and 9, the latter also emerging from the profiling data. Traditionally, caspases activity is determined by the cleavage of pro-caspases into active caspases. Thus, we performed immunofluorescence in HCT116 cells stained with antibodies specific for cleaved Caspase 3, 6, 7 and 8. Upon transfection of NF-YB siRNA, the cleavage of caspase 3, 7 and 8 was activated, while that of caspase 6 was not (Figure 5D). Thus, NF-YB inactivation leads to caspases cleavage, and the complete inhibition of caspase activity by Z-VAD prevent apoptosis induced by NF-YB depletion.
Role of p53 in apoptosis induction upon NF-YB silencing
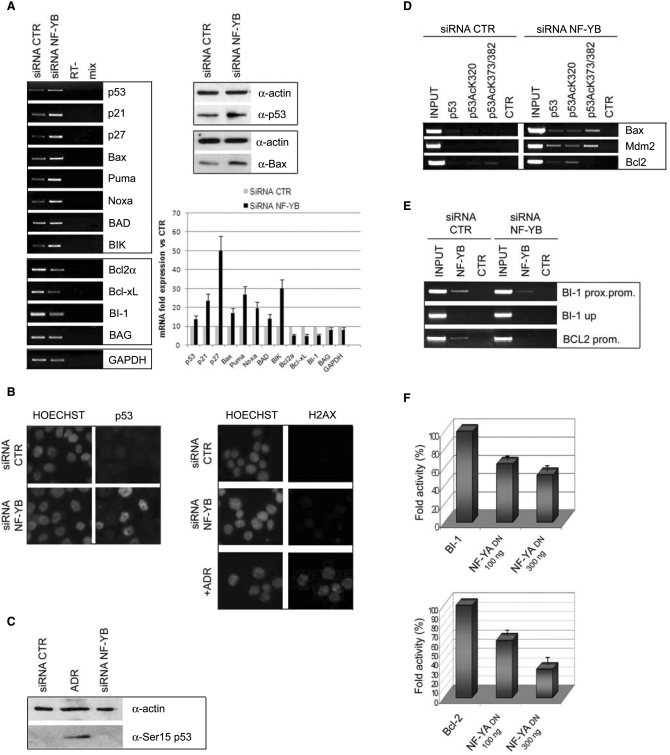
In vivo data indicate that NF-Y and p53 are strictly connected in the control of cell cycle progression upon DNA damage (24–26). A possible explanation for the apoptotic behavior of NF-YB inactivated cells would be activation of p53. To investigate this, we first performed RT-PCR analysis of the p53 gene: Figure 6A shows a modest, but reproducible, p53 induction both at the mRNA and protein levels. The increase of p53 expression was confirmed by immunofluorescence: Figure 6B (left panel) shows positive p53 staining in HCT116 NF-YB inactivated cells, compared to control cells. p53 activation was further detected by transfecting a second siRNA (NF-YB3) and two different shRNAs (shRNA NF-YB1 and NF-YB2) in HCT116 (Supplementary Figure 1A, right panel and Supplementary Figure 1C). In addition we confirmed p53 transcriptional activation in the human chondrosarcoma cell line SW1353 (Supplementary Figure 3B). As for p53 target genes, induction was seen for Bax and the BH3-only family members, such as Puma, Noxa and BIK, as well as the CDK inhibitors p21Waf1/Cip1 and p27 (Figure 6A, left panel). The expression level of BAD did not change. We also investigated Bcl-2 family members that inhibit apoptosis, Bcl-2 and Bcl-XL: as shown in Figure 6A, transcription of these genes was reduced. Another anti-apoptotic gene is Bax-inibitor 1 (BI-1), regulator of cell death pathways controlled by Bcl-2 and Bax. RT-PCR analysis showed a decrease of BI-1 transcription (Figure 6A).
Figure 6.
Activation of p53 and its target genes upon NF-YB silencing. (A) Left panel: RT-PCR analysis of the indicated mRNA transcripts in control and NF-YB-silenced cells. RNA expression levels of the indicated genes are quantitated relative to control siRNA transfected cells (lower right panel). Upper right panel: total extracts subjected to western immunoblotting using anti-p53, anti-bax and anti-actin antibodies. (B) Left panel: p53 and HOECHST staining of non-targeting control and NF-YB siRNA-transfected cells. Right panel: DNA damage was detected by H2AX staining of negative control and NF-YB siRNA transfected and Adriamycin-treated HCT116 cells. (C) Western blot analysis with anti-phospho Ser15 p53 and anti-actin antibodies of cell lysates from control siRNA, Adriamycin-treated and NF-YB siRNA-transfected cells. (D) ChIP assays of control and NF-YB-silenced cells of Bax, Mdm2 and Bcl-2 promoters, with the indicated antibodies. (E) Chromatin of control and NF-YB-silenced cells was immunoprecipitated with anti-NF-YB and Flag antibodies. PCR amplifications were performed with primers for BI-1 proximal promoter, BI-1 upstream promoter and Bcl-2. (F) Dose–response analysis (100–300 ng) of NF-YA DN in HCT116 cells with BI-1 and Bcl-2 reporters. Error bars indicate standard deviations.
It is well known that p53 activation follows DNA damage, as well as other noxious stimuli. It has been reported recently that NF-Y is involved in regulating many genes of the DNA damage response (33), a notion confirmed by our profiling experiments (Figure 3). Thus, one possible explanation for the above results would be that NF-YB inactivation leads to DNA damage. To investigate whether this is the case, we used a sensitive immunofluorescence assay with an antibody against H2AX, a sensitive marker of activation of the DNA-damage response: no staining could be detected after NF-YB inactivation (Figure 6B, right panel), whereas positivity was readily observed in cells treated with the DNA-damaging agent Adriamycin. Phosphorylation of p53 at residue Ser15 has been shown to occur in response to DNA damage (34,35). Such Ser15 phosphorylation is catalyzed by multiple protein kinases, including ATM, which may be activated by different types of damages. Western blot analysis of the cell lysates from control and NF-YB siRNA-transfected cells compared to Adriamycin-treated cells revealed that NF-YB depletion, unlike Adriamycin, did not cause p53 phosphorylation at Ser15 (Figure 6C). Therefore, NF-YB inactivation is not associated to an overt DNA-damage event.
p53 activation is accompanied by post-translational modifications; among others, acetylations are associated to p53 functional activation and DNA-binding. To ascertain whether this is the case in our system, we performed ChIP analysis with anti-p53, anti-acetyl-K320-p53, anti-acetyl-K373-382-p53 and control antibodies in HCT116 and NF-YB-inactivated cells. Figure 6D indeed shows p53 recruitment on Bax and Mdm2 upon NF-Y silencing; importantly, p53 is efficiently acetylated at K320 and K373-382. In addition to Bax and Mdm2, the Bcl-2 promoter also showed recruitment of p53, specifically acetylated at K320. Overall, these data prove that functional inactivation of NF-YB leads to p53 acetylation, increased DNA-binding to pro-apoptotic promoters and increased expression of these genes.
Role of NF-Y in the control of anti-apoptotic genes
Many anti-apoptotic genes contain CCAAT boxes. As a way to explain the anti-apoptotic behavior of NF-YB, ChIP analysis confirmed NF-Y binding to the core promoter of the anti-apoptotic BI-1 and Bcl-2 CCAAT promoters (Figure 6E). Control amplifications of upstream regions were negative. The PCR of each ChIP were quantitatively compared to the Flag control antibody (Supplementary Figure 2B). Indeed, NF-YB silencing decreased NF-Y binding, a result that parallels the reduced transcription observed in RT-PCR analysis (Figure 6A). To unambiguously prove the role of NF-Y in BI-1 and Bcl-2 transcription, we co-transfected HCT116 cells with the BI-1-luciferase and Bcl2-luciferase vectors, together with increasing amounts of the highly diagnostic dominant negative NF-YA vector YA13m29. This vector expresses a mutant NF-YA subunit that still enables NF-YB/NF-YC dimer interaction, but renders the resulting trimer inactive in CCAAT recognition (36). Figure 6F shows that the DN vector is indeed inhibitory on both promoters. These data indicate that NF-Y silencing leads to down-regulation of anti-apoptotic genes.
p53 is necessary for the apoptotic response upon NF-Y knockdown
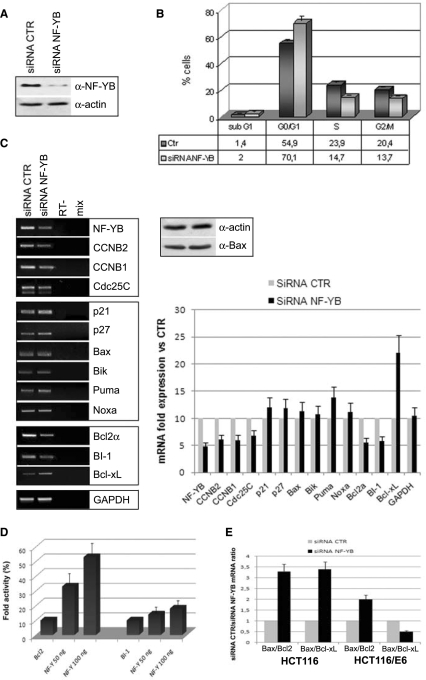
To detail the role of p53 and its target genes in apoptosis, we switched to HCT116/E6 cells, in which p53 is inactivated by expression of the E6 protein (37–40). A significant reduction of NF-YB levels was achieved (Figure 7A) and FACS analysis shows an increase in cells in G1, and a modest decrease in S and G2/M cells (Figure 7B). Comparison with FACS analysis of HCT116 cells in Figure 2A highlights that the decrease in G2/M is not as efficient as with p53-positive cells, confirming the role of p53 in G2/M depletion. Moreover, hyplodiploid sub-G1 events were essentially absent—from 1.4% to 2%—indicating that p53 is necessary to induce apoptosis by NF-YB inactivation. mRNA levels of cell-cycle regulated genes were evaluated by RT-PCR (Figure 7C, left panel). As expected, NF-YB siRNA transfection induced a reproducible decrease of NF-YB; G2/M promoters regulated by NF-Y were modestly decreased compared to HCT116. p21 and p27 did not increase and no change was seen for BH3-only Bik and Bax (Figure 7C), as confirmed by western blot analysis (Figure 7C, right panel); the anti-apoptotic Bcl-XL also showed no decrease. However, the transcription level of Bcl-2α and BI-1 were efficiently reduced, comparable to that of p53-positive cells (Figure 7C, left panel). To investigate whether this inhibition is a direct effect of NF-YB depletion on the promoter activity, we transiently co-transfected the Bcl-2α-luciferase vector together with increasing amounts of NF-Y trimer in HCT116/E6 cells. Figure 7D shows that the NF-Y expression activates the transcription of the Bcl-2α promoter.
Figure 7.
Role of p53 in apoptosis induction. (A) Expression levels of NF-YB and actin in HCT116/E6 upon NF-YB silencing. (B) Cell cycle progression analysis by FACS of control and NF-YB-silenced HCT116/E6 cells. Error bars indicate standard deviations. (C) Left panel: RT-PCR analysis of G2/M, pro- and anti-apoptotic genes upon NF-YB siRNA transfection. PCR amplified genes are indicated and their quantitation, relative to control expression levels, is plotted in the right lower panel. Right upper panel: total extracts subjected to western immunoblotting using anti-bax and anti-actin antibodies. (D) Dose–response analysis (50–100 ng) of NF-Y in HCT116/E6 cells with Bcl-2 reporter. Error bars indicate standard deviations (E) The histogram represents the Bax/Bcl-2 and Bax/Bcl-XL mRNA ratios in HCT116 and HCT116/E6 NF-YB-silenced cells.
What is particularly relevant are the mRNA ratios of pro- versus anti-apoptotic Bcl-2 family members, Bax to Bcl-2 and to Bcl-XL, in HCT116 and HCT116/E6 NF-YB-silenced cells, which is shown in Figure 7E. The Bax/Bcl-2 and Bax/Bcl-XL ratios are skewed toward Bax in HCT116 cells; interestingly, the former remains increased in HCT116/E6, by virtue of lack of Bax activation and Bcl-2 decrease, whereas the Bax/Bcl-XL ratio drops considerably in the absence of p53 and NF-YB, due to increase in Bcl-XL. These data provide evidence that p53 is required in the apoptotic response upon NF-Y RNAi, suggesting that decreased levels of anti-apoptotic genes are not sufficient to induce apoptosis, as pro-apoptotic members of the Bcl-2 family activated by p53 play a significant role in this phenomenon.
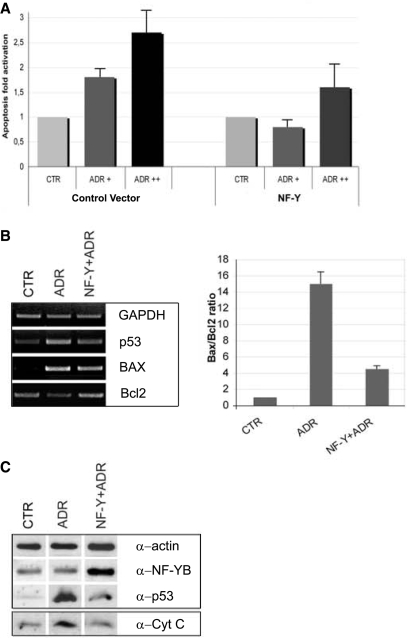
To futher investigate the role of NF-Y in preventing apoptosis, we overexpressed the NF-Y trimer in HCT116 cells and we activated p53 by Adriamycin treatment after 24 h. As shown in Figure 8A, NF-Y reduces apoptosis activation mediated by p53. RT-PCR and western Blot analysis confirm an increase in p53 mRNA and protein levels, which is reverted by NF-Y overexpression (Figure 8B). In addition, Bcl2 repression after Adriamycin treatment is reversed by transfecting NF-Y, and Bax activation is—modestly—reduced. NF-Y overexpression correlates with decreasing ratios of Bax to Bcl2 (Figure 8C). Finally, the increase in cytochrome C release into the cytoplasm after ADR is obliterated by NF-Y overexpression (Figure 8D). We conclude that increasing the levels of NF-Y protects cells from entering a p53-mediated pathway of apoptosis.
Figure 8.
Overexpression of NF-Y prevents p53-mediated apoptosis. (A) Apoptosis fold activation in control cells and in NF-Y overexpressing cells concurrently with Adriamycin treatment. Error bars indicate standard deviations. (B) Left panel: RT-PCR analysis of p53, Bax and Bcl-2 genes upon Adriamycin and NF-Y overexpression concurrently with Adriamycin treatment. Right panel: the histogram represents the Bax/Bcl-2 mRNA ratios in Adriamycin and NF-Y overexpressing/Adriamycin-treated cells. Error bars indicate standard deviations. (C) Upper panel: total extracts subjected to western immunoblotting using anti-actin, anti-NF-YB and anti-p53 antibodies. Lower panel: isolated cytosolic fractions of control, Adriamycin and NF-Y overexpressing/Adriamycin-treated cells, subjected to western blot analysis with anti-cytochrome C antibody.
DISCUSSION
NF-Y targets include regulatory genes that ensure orderly progression of the cell cycle. Over the last years, many efforts have been made to establish the role of NF-Y in controlling cell proliferation in vivo (18,28,29). In this study, we examined the effects of knocking down the NF-YB subunit for cell cycle progression. Three relevant findings are presented: (i) NF-Y silencing impairs G2/M progression and induces apoptosis; (ii) widespread activation of G2/M and anti-apoptotic genes requires NF-Y; (iii) p53 is activated by a decrease in NF-YB and it is involved in cell cycle alteration and programmed cell death.
NF-Y silencing impairs G2/M progression and induces apoptosis
We have determined that RNAi-mediated depletion of the NF-YB subunit can induce cell cycle progression failure, resulting in impairment of the G2/M transition (Figure 2A). Our results showed a massive delay in mitotic progression. Importantly, >60% of cells failed to progress through G2/M during a 48 h observation period, whereas only a small proportion of cells is impaired in progression to S phase. RT-PCR analysis confirmed a decrease in transcription of G2/M-regulated genes after NF-YB knockdown (Figure 4), nevertheless we cannot exclude that factors outside of the transcriptional apparatus contribute significantly to the G2/M depletion effect.
Two bioinformatic studies on CCAAT boxes link them to cell cycle regulation: the analysis of cell cycle promoters showed a remarkable and specific abundance of CCAAT boxes in promoters regulated during the G2/M phase (17) and the recent development of a computational algorithm to identify transcription factors that preferentially act together, identified the NF-Y-CDE-CHR module as predictive of G2/M genes (41). CDE-CHR elements are cis-regulatory sequences originally characterized in genetic experiments for their importance in restricting expression in G2 and M (42,43). Our profiling analysis indicates that genes of G2 and M phases are hit, as well as those required for microtubule, cytoskeleton and spindle formation, occurring during G2, and, in general, required for mitotic entry. Therefore, these experiments represent the molecular basis for the bioinformatic predictions. They also support the conclusion that the cell cycle depletion is due to a generalized requirement for NF-Y in genes expressed in G2 and M, rather than to specific G2/M regulators, such as CDC25C, Cyclin B1, Cdc2 and AuroraB.
Two different approaches were used to gather functional information about pathways regulated by NF-Y/CBF, by targeting NF-YA/CBF-B, a different subunit from the one analyzed here. (i) The CBF-B/NF-YA gene was inactivated in ES cells and KO mice derived: they die early in embryogenesis and inactivation in primary mouse embryonic fibroblasts results in a block in cell proliferation and inhibition of S phase or DNA synthesis, followed by induction of apoptosis. This latter event, however, was a very late phenomenon, being manifest after 5 days (28). (ii) Overexpression of an inducible Dominant Negative CBF-B/NF-YA lacking the transcription activation domain (Bdbd) showed a dramatic arrest in G2/M and lack of expression of G2/M genes (29,44). No apoptosis was observed. The results presented for NF-YB inactivation are quite different and there are two possible scenarios.
One possibility focuses on the special relationship between NF-Y and p53 as a transcriptional G2/M checkpoint ‘duo’, described with a different NF-YA Dominant Negative mutated in the DNA-binding subdomain (YAm29). This DN inhibited the p53 response, notably a G2/M block, both in cells with wt p53 and with tumor derived p53 mutants (24,25). The YAm29 and Bdbd act in fundamentally different ways: the latter associates with the NF-YB/NF-YC heterotrimer, binds DNA, but it does not activate transcription, due to lack of the Q-rich activation domain. The mechanism is most likely related to the lack of recruitment of general transcription factors (44). The former inhibits DNA binding of the trimer, by forming a defective heterotrimer, which is incapable to associate with DNA. This removes p53 from G2/M promoters, possibly unlike Dbdb. In this scheme, apoptosis would result from the transcriptional block and the release of p53 from G2/M promoters; with Bdbd, instead, NF-Y stays on the promoter and might keep p53 bound: only a transcriptional block would ensue, without the accompanying p53 activation.
Alternatively, it is possible that the three different subunits have somewhat specialized roles in different sets of genes. NF-YA has no histone-like structure, unlike NF-YB-NF-YC, whose resemblance to H2A-H2B has been detailed by chrystallography: it is possible that the elimination—or unbalance—of the histone-fold dimer might bring a more serious threat to cell survival and trigger a prompt apoptotic response. Indeed, there are tissues in vivo in which NF-YA, but not NF-YB, is apparently absent (21,45), suggesting a more ‘structural’ role for the heterodimer. We also note that the NF-YB/NF-YC heterodimer is 2/3-fold in excess with respect of NF-YA in growing cells (Salvatoni,S. and Mantovani,R., unpublished data). The dimer might thus be involved in connections other than CCAAT binding and transcriptional regulation.
In agreement with previous studies, there is no G1/S block upon NF-YB inactivation. In general, this is surprising, considering the number of G1/S genes that are functioning through NF-Y, including some of the key regulators of S phase entry (11,18,46,47). The profiling analysis supports the idea that transcription of G1/S genes is less dependent on NF-Y. It is possible that the complete elimination of NF-YB, as opposed to the 80–90% reduction of protein levels reported here, could cause additional G1/S phenotypes not detected in this study.
p53-NF-Y: a special connection ?
The connections between p53 and NF-Y were known before. In fact, it has been shown that the two factors interact directly, both with wt p53 and in tumor cells with mutant p53 alleles (24,25). Regions within the two proteins have been pinpointed that mediate interactions: the C-terminus of p53 and the αC of NF-YC. Under unstressed conditions, p53 is bound to G2/M promoters and the importance of this G2/M transcriptional ‘checkpoint’ is highlighted by the findings that gain-of-function p53 mutants subvert it completely, promoting transcription of key cell cycle regulators. Indeed, such p53 mutants show a stronger inherent interaction with NF-Y (25).
p53 activation occurs normally under stressed conditions, specifically after DNA damage, and modulation of p53 activity is exerted via post-translational modifications (48). The sensitive H2AX assay, which is commonly used to monitor cells that undergo DNA lesions, and the lack of p53-Ser15 phosphorylation, suggest that removal of NF-YB does not cause DNA damage. Thus, activation of p53 is an apparently surprising result. Our ChIP assays further indicate that it is an acetylated, ‘active’ p53 that binds to the target promoters. There are two possibilities. (i) The p53-NF-Y duo, positioned on G2/M genes might be a sensor of stress (see above). The link between the two proteins could be direct, as revealed by protein–protein interaction assays, or mediated by other factors. Removal of p53 from its ‘niche’ G2/M promoters by NF-Y depletion, might trigger exposure of specific p53 residues and activation of the protein. An interacting partner might mask p53 residues, as long as the NF-Y-p53 interaction is intact. (ii) Alternatively, one should consider that NF-Y is a key factor in the transcriptional response to numerous stress signals (8,49). NF-YB inactivation could be ‘sensed’ as a potentially dangerous situation simply because stress responses, which play a fundamental role in cell survival, would be highly diminished. Two groups of genes come to mind: the first are genes coding for proteins required in various DNA-damage responses, which are under NF-Y control. This has been detailed in ChIP on chip experiments with an oligo array enriched in this category (33). Most of these genes are actually not induced by the damage, and they are expressed at low, constant levels. The second group is genes that mediate the response to Endoplasmic Reticulum stress: most of these genes are strongly dependent on NF-Y (49 and references therein). Falling short on a minimum of expression of these genes, or closing down their promoter accessibility, as it is the case when NF-Y is not bound, could be sensed as a stress condition, triggering an indirect p53 activation.
Mechanisms of activation of apoptosis
The p53-inducible proapoptotic genes are involved in several death pathways (50). Activated p53 induces the transcription of apoptotic genes such as Bax (51), Puma (52), Noxa (53), p53AIP (54), FAS/APO1 (55) and represses the anti-apoptotic Bcl-2 (56). CDK inhibitors p21 and p27 are transcriptionally activated by NF-YB knockdown, as well as Bax and the BH3-only Bik, Puma and Noxa (Figure 6A). The analysis of both cell cycle progression and of the activation of p53 target genes in HCT116/E6 cells indicates that p53 is required for the apoptotic response. Our results imply that p53 is involved in activating pro-apoptotic target genes by directly binding to their promoters. Interestingly, the analysis of p53 targets—Bax, Mdm2 and Bcl2—by ChIP assays showed that p53 is differently acetylated. The activated promoters, Bax and Mdm2, are positive for p53 acetylation at both Lys320 and Lys 373-382, while the repressed Bcl2 promoter showed a recruitment of p53 specifically acetylated at Lys320.
Downstream of p53, the Bcl-2 family members play a role in apoptosis. The translocation of p53 to the mitochondria and the formation of complexes with the anti-apoptotic Bcl-2 family proteins induce the release of sequestered pro-apoptotic proteins and subsequent permeabilization of the outer mitochondrial membrane (53–57). NF-YB inactivation correlates with the cleavage of procaspase-3, 7 and 8 and an increase in Bax/Bcl-2 and Bax/Bcl-XL ratios. The overexpression of anti-apoptotic Bcl-2 family proteins, such as Bcl-2 and Bcl-XL, blocks the release of cytochrome c in response to a variety of apoptotic signals (58). On the contrary, the relocalization of Bax to mitochondria is linked to increased cytochrome c levels in the cytosol (58–60). The comparison between HCT116 and HCT116/E6 data clearly suggests that inhibition of the anti-apoptotic Bcl-2α and BI-1 is due to NF-Y inactivation (Figure 7). This highlights the importance of a fine balance between p53 and NF-Y-regulated genes (Figure 9). NF-Y silencing increases the Bax/Bcl-2 ratio both in HCT116 and in HCT116/E6. In p53 positive cells, the level of Bax is induced, in the absence of p53 it is not. Thus, the increase in HCT116/E6 cells is caused by the failure to activate Bcl-2 as a consequence of NF-Y knockdown. Conversely, the Bax/Bcl-XL ratio is 3-fold increased only in HCT116, pointing out that Bcl-XL is a direct target of p53, but not of NF-Y. Therefore, it seems reasonable to postulate that apoptosis may be caused by a reduction in Bcl-XL/Bax heterodimerization. Disruption of this balance can occur either following a loss NF-Y activity, or by p53 activation, through DNA damage. The role of NF-Y in preventing apoptosis has been clearly demonstrated by comparing the Bax to Bcl2 ratios, after Adriamycin treatment or NF-Y overexpression concurrently with Adriamycin. NF-Y overexpression causes a decrease in the Bax to Bcl2 ratio, and abrogates the cytochrome C release into the cytoplasm.
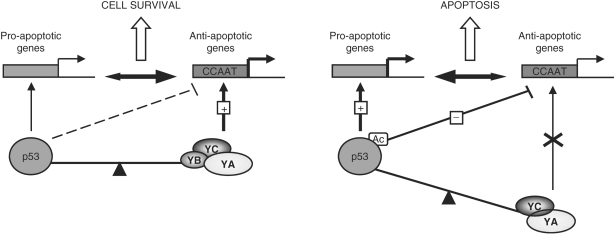
Figure 9.
A model of the fine balance between p53 and NF-Y regulated genes.
As the expression changes of many of the anti-apoptotic genes are below the stringent criteria used here in the profiling analysis, much work lies ahead in the determination of the complete picture of pro-survival genes regulated by NF-Y. Finally, the behavior of gain-of-function p53 mutants should be re-evaluated, in terms of the enhancement of NF-Y anti-apoptotic and pro-survival function, which might impact in their pro-growth strategy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
C.I. thanks R. Mantovani for many helpful discussion and comments on the article. We thank L.M. Boxer and M. J-Brady for Bcl-2 and BI-1 luciferase plasmids; V. Zappavigna for anti-CyclinA and anti-CyclinB1 antibodies. C.I. is supported by Ministero dell'Istruzione dell'Università e della Ricerca grant (PRIN). We thank Associazione Italiana per la Ricerca sul Cancro (AIRC) for support. V.B. is supported by a Federazione Italina per la Ricerca sul Cancro (FIRC) fellowship. Funding to pay the Open Access publication charges for this article was provided by Ministero dell'I;struzione dell'U;niversità e della Ricerca grant (PRIN).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 5.McNabb DS, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 6.Sinha S, Maity SN, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY,allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl Acad. Sci. USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caretti G, Motta MC, Mantovani R. NF-Y associates with H3-H4 tetramers and octamers by multiple mechanisms. Mol. Cell Biol. 1999;19:8591–8603. doi: 10.1128/mcb.19.12.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huet X, Rech J, Plet A, Vie A, Blanchard JM. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol. Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Ginkel PR, Hsiao KM, Schjerven H, Farnham PJ. E2F-mediated growth regulation requires transcription factor cooperation. J. Biol. Chem. 1997;272:18367–18374. doi: 10.1074/jbc.272.29.18367. [DOI] [PubMed] [Google Scholar]
- 12.Bolognese F, Wasner M, Dohna CL, Gurtner A, Ronchi A, Muller H, Manni I, Mossner J, Piaggio G, et al. The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT-binding activity is cell-cycle regulated. Oncogene. 1999;18:1845–1853. doi: 10.1038/sj.onc.1202494. [DOI] [PubMed] [Google Scholar]
- 13.Farina A, Manni I, Fontemaggi G, Tiainen M, Cenciarelli C, Bellorini M, Mantovani R, Piaggio G. Down-regulation of cyclin B1 gene transcription in terminally 26 differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18:2818–2817. doi: 10.1038/sj.onc.1202472. [DOI] [PubMed] [Google Scholar]
- 14.Yun J, Chae HD, Choy HE, Chung J, Yoo HS, Han MH, Shin DY. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J. Biol. Chem. 1999;274:29677–29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- 15.Jung MS, Yun J, Chae HD, Kim JM, Kim SC, Choi TS, Shin DY. p53 and its homologues, p63 and p73, induce a replicative senescence through inactivation of NF-Y transcription factor. Oncogene. 2001;20:5818–5825. doi: 10.1038/sj.onc.1204748. [DOI] [PubMed] [Google Scholar]
- 16.Manni I, Mazzaro G, Gurtner A, Mantovani R, Haugwitz U, Krause K, Engeland K, Sacchi A, Soddu S, et al. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J.Biol. Chem. 2001;276:5570–5576. doi: 10.1074/jbc.M006052200. [DOI] [PubMed] [Google Scholar]
- 17.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulatros controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J. Biol. Chem. 2003;278:30435–30440. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]
- 19.Li XY, Hooft van Huijsduijnen R, Mantovani R, Benoist C, Mathis D. Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J. Biol. Chem. 1992;267:8984–8990. [PubMed] [Google Scholar]
- 20.Frontini M, Imbriano C, Manni I, Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle. 2004;3:217–222. [PubMed] [Google Scholar]
- 21.Gurtner A, Manni I, Fuschi P, Mantovani R, Guadagni F, Sacchi A, Piaggio G. Requirement for down-regulation of the CCAAT binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol. Biol. Cell. 2003;14:12706–12715. doi: 10.1091/mbc.E02-09-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yun J, Chae HD, Choi TS, Kim EH, Bang YJ, Chung J, Choi KS, Mantovani R, Shin DY. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J. Biol. Chem. 2003;278:36966–36972. doi: 10.1074/jbc.M305178200. [DOI] [PubMed] [Google Scholar]
- 23.Adachi N, Nomoto M, Kohno K, Koyama H. Cell-cycle regulation of the DNA topoisomerase IIalpha promoter is mediated by proximal CCAAT boxes: possible involvement of acetylation. Gene. 2000;245:49–57. doi: 10.1016/s0378-1119(00)00040-8. [DOI] [PubMed] [Google Scholar]
- 24.Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, et al. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Peart MJ, Prives C. Mutant p53 gain of function: the NF-Y connection. Cancer Cell. 2006;10:173–174. doi: 10.1016/j.ccr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Tabach Y, Milyavsky M, Shats I, Brosh R, Zuk O, Yitzhaky A, Mantovani R, Domany E, Rotter V, et al. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol. Syst. Biol. 2005;1:1–15. doi: 10.1038/msb4100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- 29.Hu Q, Maity SN. Stable expression of a dominant negative mutant of CBF/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J. Biol. Chem. 2000;275:4435–4444. doi: 10.1074/jbc.275.6.4435. [DOI] [PubMed] [Google Scholar]
- 30.Basile V, Mantovani R, Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J. Biol. Chem. 2006;281:2347–2357. doi: 10.1074/jbc.M507712200. [DOI] [PubMed] [Google Scholar]
- 31.Tagliafico E, Tenedini E, Manfredini R, Grande A, Ferrari F, Roncaglia E, Bicciato S, Zini R, Salati S, et al. Identification of a molecular signature predictive of sensitivity to differentiation induction in acute myeloid leukemia. Leukemia. 2006;20:1751–1758. doi: 10.1038/sj.leu.2404358. [DOI] [PubMed] [Google Scholar]
- 32.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceribelli M, Alcalay M, Viganò MA, Mantovani R. Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle. 2006;5:1102–1110. doi: 10.4161/cc.5.10.2777. [DOI] [PubMed] [Google Scholar]
- 34.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 35.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh S, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J. Biol. Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 37.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 38.Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell Biol. 1993a;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell Biol. 1993b;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Z, Shendure J, Church GM. Discovering functional transcription factors combinations in the human cell cycle. Genome Res. 2006;15:848–855. doi: 10.1101/gr.3394405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zwicker J, Gross C, Lucibello FC, Truss M, Ehlert F, Engeland K, Muller R. Cell cycle regulation of cdc25C transcription is mediated by the periodic repression of the glutamine-rich activators NF-Y and Sp1. Nucleic Acid Res. 1995;23:3822–3830. doi: 10.1093/nar/23.19.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, Muller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Q, Lu JF, Luo R, sen S, Maity SN. Inhibition of CBF/NF-Y mediated transcription activation arrests cells at G/M phase and suppresses expression of genes activated at G2/M phase of the cell cycle. Nucleic Acid Res. 2006;34:6272–6285. doi: 10.1093/nar/gkl801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marziali G, Perrotti E, Ilari R, Coccia EM, Mantovani R, Testa U, Battistini A. The activity of the CCAAT-box binding factor NF-Y is modulated through the regulated expression of its A subunit during monocyte to macrophage differentiation: regulation of tissue-specific genes through a ubiquitous transcription factor. Blood. 1999;93:519–526. [PubMed] [Google Scholar]
- 46.Chang ZF, Huang DY, Hu SF. NF-Y-mediated trans-activation of the human thymidine kinase promoter is closely linked to activation of cyclin-dependent kinase. J. Cell Biochem. 1999;75:300–309. [PubMed] [Google Scholar]
- 47.Good LF, Chen KY. Cell cycle- and age-dependent transcriptional regulation of human thymidine kinase gene: the role of NF-Y in the CBP/tk binding complex. Biol. Signals. 1996;5:163–169. doi: 10.1159/000109214. [DOI] [PubMed] [Google Scholar]
- 48.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 49.Donati G, Imbriano C, Mantovani R. Dynamic recruitment of transcription factors and epigenetic changes on the ER stress response gene promoters. Nucleic Acids Res. 2006;34:3116–3127. doi: 10.1093/nar/gkl304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourdon JC, De Laurenzi V, Melino G, Lane D. p53: 25 years of research and more questions to answer. Cell Death Diff. 2003;10:397–399. doi: 10.1038/sj.cdd.4401243. [DOI] [PubMed] [Google Scholar]
- 51.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 52.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 53.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000a;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 54.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 55.Owen-Schaub LB, Zhang W, Cusack JC, Angelo LS, Santee SM, Fujiwara T, Roth JA, Deisseroth AB, Zhang WW, et al. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol. Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 57.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 58.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 59.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 60.Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, et al. p73 induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.