Abstract
The Krüppel-like zinc finger protein Gli-similar 3 (Glis3) plays a critical role in pancreatic development and has been implicated in a syndrome with neonatal diabetes and hypothyroidism (NDH). In this study, we examine three steps critical in the mechanism of the transcriptional regulation by Glis3: its translocation to the nucleus, DNA binding and transcriptional activity. We demonstrate that the putative bipartite nuclear localization signal is not required, but the tetrahedral configuration of the fourth zinc finger is essential for the nuclear localization of Glis3. We identify (G/C)TGGGGGGT(A/C) as the consensus sequence of the optimal, high-affinity Glis3 DNA-binding site (Glis-BS). All five zinc finger motifs are critical for efficient binding of Glis3 to Glis-BS. We show that Glis3 functions as a potent inducer of (Glis-BS)-dependent transcription and contains a transactivation function at its C-terminus. A mutation in Glis3 observed in NDH1 patients results in a frameshift mutation and a C-terminal truncated Glis3. We demonstrate that this truncation does not effect the nuclear localization but results in the loss of Glis3 transactivating activity. The loss in Glis3 transactivating function may be responsible for the abnormalities observed in NDH1.
INTRODUCTION
Gli-similar (Glis) 1–3 constitute a subfamily of Krüppel-like zinc proteins that are related to members of the Gli and Zic family (1–6). These proteins share a highly conserved zinc finger domain consisting of five Cys2His2-type zinc finger motifs; however, they exhibit little homology outside their zinc finger domains. Gli and Zic proteins mediate their transcriptional regulation by binding to specific DNA elements, referred to as Gli-binding sites (GBS), in the promoter region of target genes (7,8).
During embryonic development Glis1–3 genes are expressed in a spatial and temporal manner and play a critical role in the regulation of several physiological processes (1–6,9). Glis1 is highly expressed in dermal papilla cells in the skin and is highly induced in the epidermis of psoriatic patients (10). Glis2 is expressed in the cranial and dorsal ganglia, neural tube and in the intermediate zone of the hindbrain in E9.5 mouse embryos (6). Recent studies showed that loss of Glis2 expression causes nephronophthisis, an autosomal recessive kidney disease and the most frequent genetic cause for end-stage renal failure in humans (11,12). Moreover, Glis2 has been implicated in the regulation of neuronal differentiation (3). Glis3 is highly expressed in the metanephric mesenchyme during embryonic development and in the uterus, pancreas and kidney of adult mice (9). Glis3 was shown to enhance osteoblast differentiation by inducing the expression of FGF18 (13). A recent study linked mutations in Glis3 to a human syndrome consisting of neonatal diabetes and congenital hypothyroidism (NDH) (14). NDH1-type patients exhibit the most severe effects and die between 10 days and 2 years after birth. These patients show a number of pathologies, including diabetes, polycystic kidney disease, glaucoma, hyperthyroidism, facial dysmorphology and liver fibrosis suggesting that Glis3 plays a critical role in the regulation of pancreatic development and in several other tissues.
Glis proteins control these physiological processes by regulating the transcription of specific genes in these target tissues. Changes in the function or activity of Glis3 proteins result in alterations in gene expression and, subsequently, abnormalities in cell and tissue functions. However, relatively little is known about the mechanisms by which Glis proteins regulate gene expression. Although Glis proteins have been reported to bind the GBS consensus (1–6), the sequence of their optimal DNA-binding site has not yet been determined.
To obtain greater insight into the physiological functions of Glis3 and its role in diseases, including NDH, it is important to obtain a better understanding of the various steps involved in the transcriptional regulation by Glis3, including its translocation to the nucleus, its interaction with specific DNA-binding sites and transcriptional activation through its activation domain. The objective of this study is to gain further insight into these three critical steps. We demonstrate that not the putative bipartite nuclear localization signal (bNLS) but ZF4 of Glis3 is required for its nuclear translocation. In addition, we define for the first time the consensus sequence of the optimal DNA-binding site of Glis3 (Glis-BS) and show that all five individual zinc finger motifs are required for optimal binding. Moreover, we show that the full-length Glis3 functions as an activator of transcription and that the activation domain resides at its C-terminus. We demonstrate that Glis3(NDH1), a C-terminal-truncated Glis3 associated with NDH1 patients, still localized to the nucleus but lost its transactivation function. This loss may be responsible for the abnormalities observed in NDH1 patients. Our study provides greater insight into the different steps required for the transcriptional regulation by Glis3 and will help in understanding its function in the regulation of pancreatic development and renal functions and its role in NDH.
MATERIALS AND METHODS
Plasmids
The reporter plasmid p(Glis-BS)6-LUC containing six copies of the consensus Glis-BS sequence, was generated by cloning two copies of 5′-CGTGGGGGGTCATGACATGTGGGGGGTCATGACATGTGGGGGGTCTCGA, containing three (Glis-BS) elements into pCRII-TOPO (Invitrogen, Carlsbad, CA, USA) and its subsequent insertion into the KpnI and BglII restriction sites of the luciferase reporter plasmid pTAL-LUC (Clontech, Palo Alto, CA, USA). The reporter plasmid p(Glis-BSmut)6-LUC was generated in the same way using 5′-CGTGGGAGGTCATGACATGTGGGAGGTCATGACATGTGGGAGGTCTCGA.
Transactivation, immuno-pulldown and quantitative real-time RT-PCR analysis
Electrophoretic mobility shift assay (EMSA) was carried out as described previously (15). EMSA, transactivation, and quantitative real-time RT-PCR (QRT-PCR) analyses are described in detail in Supplementary Data.
Selection of Glis3-binding sites
The consensus DNA-binding site of Glis3 was determined by a procedure similar to that reported previously (15). A mixture of 60 bp double-stranded DNA fragments was synthesized by PCR using the random oligomer 5′-CGAATTCGCTCAGCT-N22-GTACTGCAGGATCCA as template and 5′-CGAATTCGCTCAGCT-3′ (primer 1) and 5′-TGGATCCTGCAGTAC-3′ (primer 2) as the forward and reverse primers, respectively. PCR amplification was carried out using 20 pmol of oligomer, 100 pmol of 32P-end-labeled forward primer and 100 pmol of reverse primer for three cycles under the following conditions: 1 min at 94°C, 1 min at 55°C and 1 min at 72°C for each cycle. The double-stranded mixed DNA fragments generated were purified, incubated with Glis3(ZFD) protein and complexes analyzed by EMSA. A band corresponding to the Glis3/Glis-BS complex was excised and the DNA eluted in TE buffer. Recovered DNA was amplified by PCR for 15 cycles and used for another round of EMSA analysis as described above. This procedure was repeated five times. In the sixth round, PCR products were cloned into pCRII-TOPO (Invitrogen). The inserts from individual white colonies were amplified and used for EMSA with Glis3(ZFD). The amplified DNA inserts that bound Glis3(ZFD) as indicated by EMSA were subjected to sequence analysis. The sequences of 36 independent clones were analyzed.
Confocal microscopy
HEK293T cells were plated in glass bottom culture dishes (Mattek Corp., Ashland, MA, USA) and the next day transfected with various p3×FLAG-CMV-Glis3 plasmids. After an additional 24 h incubation, cells were fixed in 4% paraformaldehyde followed by treatment with 0.2% Triton X-100 for 7 min. Cells were washed in PBS and subsequently incubated for 15 min in 1% BSA, then for 2 h with anti-FLAG M2 mouse monoclonal antibody (Sigma), and finally for 40 min with goat anti-mouse Alexa Fluor594 antibody (Molecular Probes, Eugene, OR, USA). Cells were washed with PBS and nuclei stained with 4′-6-diamidino-2-phenylindole (DAPI). Alternatively, cells were transfected with pEGFP-Glis3 or pEGFP-Glis3(ZF)1–5m. Fluorescence was observed 24 h later in a Zeiss confocal microscope LSM 510 NLO (Zeiss, Thornwood, NY, USA).
Electrophoretic Mobility Shift Assay
Escherichia coli BL21 (DE3) transformed with pQE32-Glis3(ZFD), pQE32-Glis3(ZFD) mutants and pQE32-Gli1(ZFD) were grown at 37°C to mid-log phase and then treated with isopropyl-β-d-thiogalactopyranoside (0.5 mM final concentration) for 3 h. (His)6-tagged proteins were purified using Ni-NTA+ resin (Qiagen) or Talon magnetic beads (Clontech). (His)6-tagged Glis3(ZFD) was further purified by FPLC using a S-200 column (Supplementary Figure 1S). For EMSA (His)6-Glis3(ZFD) (2.0 μg or as indicated otherwise) was incubated with 32P-labeled, double-stranded oligonucleotides (0.4 nM or as indicated otherwise) in binding buffer (20 mM Hepes, pH 8.0, 5% glycerol, 2.5 mM MgCl2, 1 mM DTT, 50 mM KCl, 10 mM ZnSO4, 300 μg poly(dI-dC) for 30 min at room temperature (15). The protein–DNA complexes were separated on a 10% native polyacrylamide gel (Hoefer 18 × 16 cm2) and visualized by autoradiography. Bound and free Glis-BS was measured using a Fluoskan 8900 densitometer (Alpha Innotech, San Leandro, CA, USA). Equilibrium dissociation constant (Kd) values were calculated from a Hill function fit to the bound and total concentration data.
RESULTS
Identification and expression of a longer variant of mouse Glis3
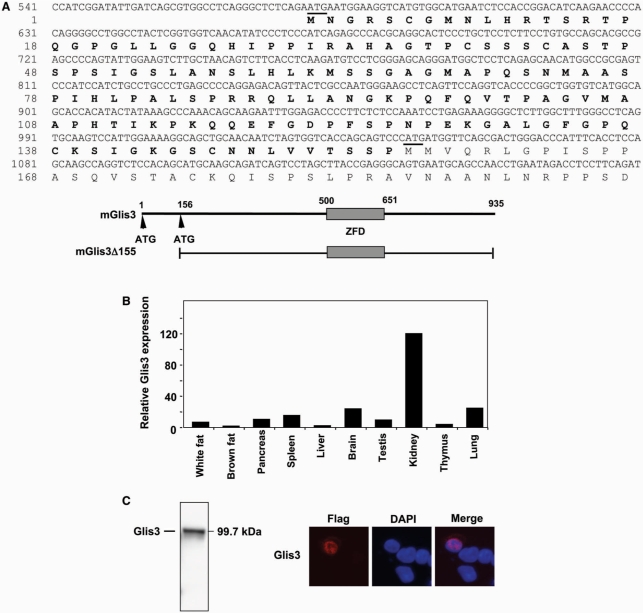
Comparison of the mouse Glis3 sequence (9) with sequences in Genbank identified a mouse sequence (AK054020) that was identical to Glis3 but extended the coding region another 465 nt towards the 5′-end. This sequence encoded a variant of Glis3 that is 155 amino acids longer at the amino-terminus. The nucleotide and deduced amino acid sequences of the extended region of this variant are shown in Figure 1A. In this study, we will refer to the full-length Glis3 and its shorter variant as Glis3 and Glis3(ΔN155), respectively. The open reading frame of Glis3 sequence encodes a protein of 935 amino acids with a calculated mass of 99.7 kDa and pI of 8.73. Pattern/profile searches did not reveal the presence of any specific motif in the extended 155 amino acid amino-terminus. QRT-PCR analysis with primers that specifically amplify a 5′ region of Glis3, demonstrated that the longer transcript was expressed in several tissues analyzed (Figure 1B) and was most highly expressed in kidney as reported previously for Glis3(ΔN155) expression (9).
Figure 1.
(A) Nucleotide and amino acid sequence of Glis3. The new extended N-terminal sequence of Glis3 is indicated in bold; only the 5′-end of Glis3 is shown. The start codons are underlined; the first start codon belongs to full-length Glis3, the second one to the previously reported Glis3(ΔN155) (9). A schematic view of Glis3 is shown below the sequence. ZFD indicates zinc finger domain. (B) Expression of full-length Glis3 mRNA in several mouse tissues. Total RNA isolated from various adult tissues was examined by real time QRT-PCR analysis using primers specific for the 5′-UTR of full-length Glis3. (C) The full-length Glis3 localizes to the nucleus. p3×FLAG-CMV-Glis3 plasmid was transfected in C3H10T1/2 cells and after 48 h incubation proteins were isolated and examined by western blot analysis using anti-FLAG M2 antibody. Subcellular localization of FLAG-Glis3 was examined by confocal microscopy with anti-FLAG M2 and Alexa Fluor 594 antibodies; nuclei were identified by DAPI staining.
The fourth ZF plays a critical role in the nuclear localization of Glis3
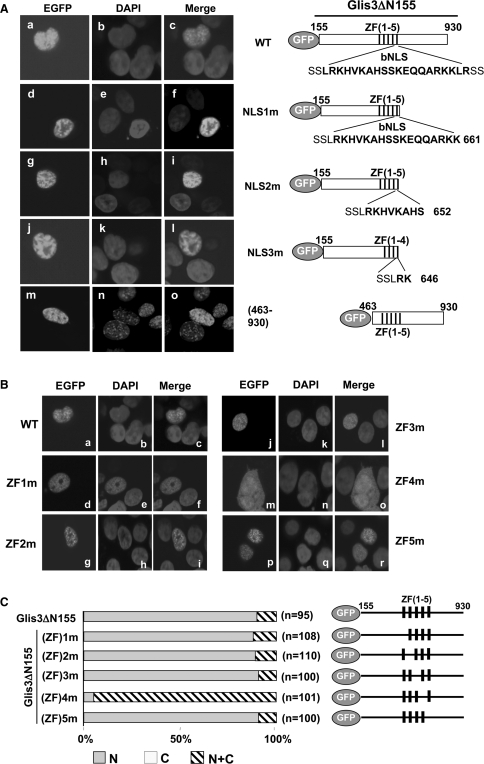
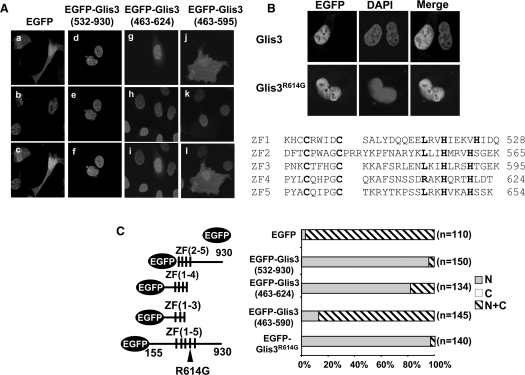
Translocation of Glis3 to the nucleus is necessary to enable it to regulate transcription. To investigate the subcellular localization of Glis3, murine pluripotent mesenchymal C3H10T1/2 cells were transfected with an expression vector encoding 3×FLAG-Glis3. The expression of Glis3 in C3H10T1/2 cells was first examined by western blot analysis. As shown in Figure 1C, the anti-FLAG antibody recognized a protein of the expected size. Analysis of the subcellular localization of Glis3 by confocal microscopy showed that Glis3 was localized predominantly to the nucleus (Figure 1C). Glis3 was localized to the nucleus in >95% of the cells in which Glis3 was detected. Similar results were obtained in CV-1 and HEK293 cells (data not shown). To determine what region is required for its nuclear localization, we examined the Glis3 sequence for the presence of potential nuclear localization signals using pattern/motif analysis. This analysis identified a potential bipartite nuclear localization signal (bNLS) between Arg645 and Lys661, a region that overlaps with ZF5 (Figure 2A). To determine whether this bNLS is required for the nuclear localization of Glis3, we examined the effect of several deletions within the bNLS on its nuclear localization in HEK293 cells. Deletion of the C-terminus up to Lys661, just up to the bNLS, did not affect the nuclear localization of Glis3. Further C-terminal deletions up to Ser652 and Lys646, which delete large portions of the bNLS, also did not affect the nuclear localization of Glis3. These data suggested that the bNLS is not required for the nuclear localization of Glis3 but that a different region of Glis3 is involved. In addition, deletion of the N-terminus up to Lys462 also did not affect the nuclear localization (Figure 2A). These data suggested a role for the zinc finger domain, rather than the N- and C-terminal regions, in the nuclear localization of Glis3. We, therefore, examined the effect of a Cys/Ala mutation within each single zinc finger on the subcellular localization of Glis3. The first Cys in each single Cys2His2-type zinc finger motif was mutated thereby destroying their tetrahydral configuration. As shown in Figure 2B, a mutation in either ZF1, ZF2, ZF3 or ZF5 did not affect the nuclear localization of Glis3 significantly. These Glis3 mutants were preferentially localized to the nucleus in >95% of the cells in which they were expressed. In contrast, Glis3 containing the mutation in ZF4 was distributed about equally between cytoplasm and nucleus in >90% of the cells. These results suggest that the tetrahedral configuration of ZF4 is critical in the nuclear localization of Glis3. Analysis of several additional mutants showed that EGFP-Glis3(532–930), containing an N-terminal deletion including ZF1 and EGFP-Glis3(463–624) containing ZF1–4, are still localized to the nucleus; however, Glis3(463–595) containing only ZF1–3, was equally distributed between nucleus and cytoplasm (Figure 3). These data support the critical role of ZF4 in the nuclear localization of Glis3. The nuclear localization of Zic3 has been reported to depend on a bNLS within ZF4 (16). Although the ZF4 of Glis3 does not contain a bNLS, it has two basic amino acids Arg614 and Lys616 in common with the bNLS of Zic3. However, an Arg614Gly mutation did not have any effect on the nuclear localization of Glis3 (Figure 3) suggesting that Arg614 is not essential.
Figure 2.
Nuclear localization of Glis3 does not require the putative bNLS but is dependent on ZF4. HEK293T cells were transfected with the pEGFP-Glis3 plasmid indicated and 30 h later examined by confocal microscopy for EGFP and DAPI fluorescence as described in ‘Materials and Methods’ section. EGFP was equally divided between cytoplasm and nucleus (data not shown). (A) The bNLS is not required for the nuclear localization of Glis3. Subcellular localization of wild-type (WT) EGFP-Glis3 (a–c), EGFP-Glis3-NLS1m (d–f), EGFP-Glis3-NLS2m (g–i), EGFP-Glis3-NLS3m (j–l), EGFP-Glis3(463–930) (m–o). A schematic view of each construct is shown on the right. The putative bNLS sequence is shown in bold. (B) The nuclear localization of Glis3 is dependent on the tetrahedral configuration of ZF4. Subcellular localization of EGFP-Glis3 (a–c), EGFP-Glis3(ZF)1m (d–f), EGFP-Glis3(ZF)2m (g–i), EGFP-Glis3(ZF)3m (j–l), EGFP-Glis3(ZF)4m (m–o) and EGFP-Glis3(ZF)5m (p–r). A schematic view of EGFP-Glis3(ZF)1–5m mutants is shown on the right in (C). The first Cys in each zinc finger motif is mutated to Ala causing loss of the tetrahedral zinc finger configuration as indicated. (C) Quantitation of the subcellular localization of Glis3 and Glis3 mutants. The percentage of cells in which Glis3 is largely restricted to the nucleus (N) or about equally distributed between the nucleus and cytoplasm (N+C) was calculated. ‘n’ indicates the number of Glis3 expressing cells counted.
Figure 3.
Subcellular localization of several Glis3 mutants. HEK293T cells were transfected with pEGFP-Glis3 or a pEGFP-Glis3 mutant as indicated. The subcellular localization of the fusion proteins was examined 30 h later by fluorescent confocal microscopy as described in ‘Materials and methods’ section. (A) Subcellular localization of EGFP (a–c), EGFP-Glis3(532–930) (d–f), EGFP-Glis3(463–624) (g–i) and EGFP-Glis3(463–595) (j–l). (B) Subcellular localization of Glis3 and Glis3(R614G) mutant in HEK293T cells. The sequences of the five zinc finger domains are shown below. (C) Quantitation of the subcellular localization of Glis3 mutants.
Identification of the consensus DNA-binding sequence for the Glis3
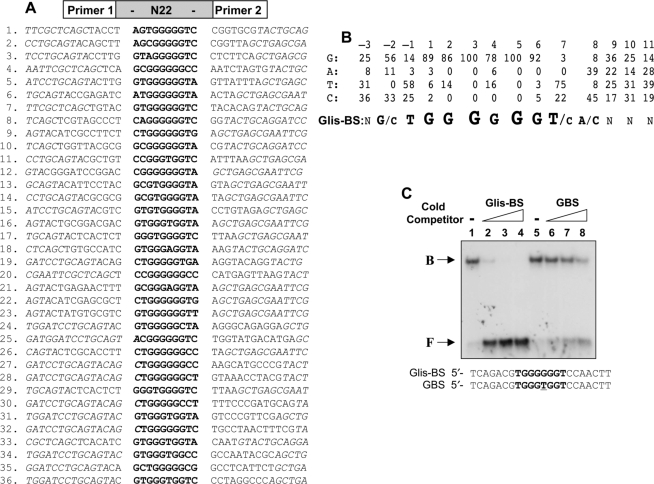
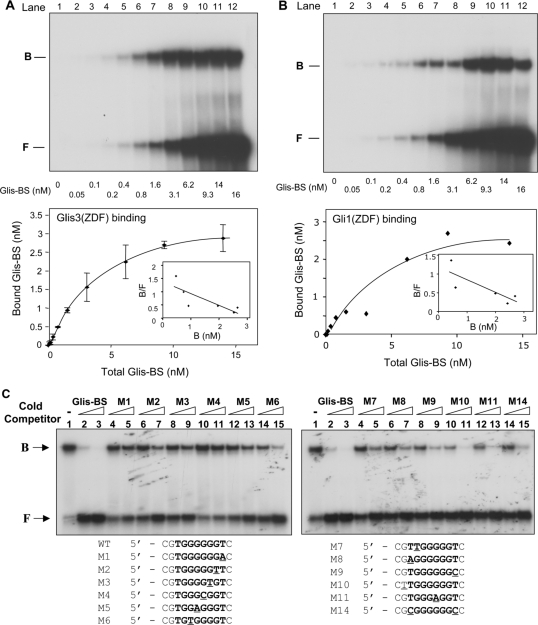
After localizing to the nucleus, Glis3 regulates transcription by interacting with specific DNA-binding sites in the promoter of target genes. Although members of the Gli and Zic subfamilies can bind the GBS consensus, these proteins exhibit distinct affinities for different binding sequences (8). Previous studies reported that Glis proteins can also bind the GBS consensus sequence (2,6,9); however, the sequence of the optimal Glis-BS has not been determined. To identify in an unbiased manner the consensus sequence that binds Glis3 with high affinity, we used a DNA-binding site selection strategy that is based on a combination of PCR and EMSA (15). This strategy used a mixture of 60 bp long oligonucleotides that contain 22 random nucleotides flanked on either side by a known sequence to facilitate PCR amplification and subsequent cloning of the selected DNA sequence. The PCR products generated after the sixth round were cloned into pCRII-TOPO. Inserts from individual clones were then analyzed by EMSA for Glis3(ZFD) binding and the inserts that bound Glis3(ZFD) strongly were sequenced. The sequences of 36 independent clones are shown in Figure 4A. The number of times that A, T, G and C appear at each position in these sequences was calculated and from this the Glis-BS consensus sequence was deduced (Figure 4B). This analysis indicated that Glis3 bound most effectively to a sequence consisting of the consensus (G/C)TGGGGGGT(A/C) in which the nucleotides shown in bold form the core motif.
Figure 4.
Identification of the consensus DNA-binding site (Glis-BS). To determine the consensus sequence of the Glis-BS that binds Glis3 with highest affinity, a DNA-binding site selection strategy was followed that was based on a combination of PCR and EMSA as described in ‘Materials and Methods’ section. A mixture of 60 bp oligonucleotides containing 22 random bases (N22) flanked on each side by a 20 bp primer (primer 1 and 2) was used as template. (A) Sequences of the 22 nt long fragments that bind Glis3 with high affinity identified in 36 independent clones. The Glis-BSs are indicated in bold. Italic nucleotides are part of the primer sequence. (B) Calculation of the consensus Glis-BS. Top line: numbers indicate the position of the nucleotide in the Glis-BS. The next four lines indicate the percentages by which G, A, T or C were present at each position. The bottom line shows the calculated consensus sequence of Glis-BS. The font size relates to the frequency by which the nucleotide is found at each position. N indicates no strong requirement for a specific nucleotide. (C) Binding of His6-Glis3(ZFD) (2 μg) to the 32P-labeled consensus Glis-BS (0.5 nM) was examined by EMSA in the absence (lane 1) and presence of a 5-, 25- and 50-fold excess of unlabeled consensus Glis-BS (lanes 2–4) or GBS (lanes 6–8). The sequence of Glis-BS and GBS is shown at the bottom. B, indicates bound Glis-BS or GBS; F, free Glis-BS or GBS.
EMSA analysis with 32P-labeled consensus Glis-BS (GTGGGGGGTC) and His6-Glis3(ZFD) confirmed binding of Glis3(ZFD) to the consensus sequence (Figure 3C). Addition of unlabeled Glis-BS competed effectively with 32P-Glis-BS for Glis3 binding. The unlabeled GBS consensus, consisting of CGTGGGTGGTC, also competed for Glis3 binding but less efficiently than the Glis-BS consensus. This was confirmed by experiments comparing the binding of His6-Glis3(ZFD) to the consensus Glis-BS or GBS as a function of increasing concentrations of His6-Glis3(ZFD) (Supplementary Figure 2S). These results are in agreement with the conclusion that Glis3 exhibits a much lower affinity for GBS than the Glis-BS consensus sequence.
To determine the affinity of Glis3 for the consensus Glis-BS, we examined the binding of His6-Glis3(ZFD) as a function of increasing concentrations of Glis-BS. Saturation analysis showed that optimal binding was reached at concentrations of about 10 nM Glis-BS (Figure 5A). From these data the apparent Kd was calculated by Scatchard plot analysis. The Kd of the binding of Glis-BS to His6-Glis3(ZFD) was 3.69 ± 0.26 nM.
Figure 5.
Analysis of the affinity and specificity of the binding of Glis3 and Gli1 to Glis-BS. (A) Binding of Glis3 to Glis-BS. The binding of His6-Glis3(ZFD) to radiolabeled Glis-BS was examined by EMSA as a function of increasing concentrations of Glis-BS as indicated (top panel). The amount of bound and free Glis-BS was determined and the Kd calculated by Scatchard plot analysis (lower panel). (B) Binding of Gli1 to Glis-BS. The binding of His6-Gli1(ZFD) to radiolabeled Glis-BS was examined by EMSA as a function of increasing concentrations of Glis-BS as indicated (top panel). The amount of bound and free Glis-BS was determined and the Kd calculated by Scatchard plot analysis (lower panel). (C) Effect of various mutations in Glis-BS on Glis3 binding. EMSA analysis was performed with His6-Glis3(ZFD) and the 32P-labeled consensus Glis-BS (0.5 nM) in the absence and presence of 5- or 25-fold excess of unlabeled Glis-BS or mutant Glis-BS (M1-M14) as indicated. The sequences of the M1-M14 mutants are shown beneath the EMSA. The mutated nucleotides are underlined.
To further analyze the binding specificity of Glis3, we examined the effect of a series of point mutations in Glis-BS on Glis3 binding. As shown in Figure 5C, the Glis-BS mutants M6, M8-M11 and M14 competed well with radiolabeled Glis-BS for Glis3(ZFD) binding, suggesting that these changes in nucleotides do not have a major impact on Glis3(ZFD) binding. The mutants M2, M3, M5 and M7, competed less efficiently, while M1 and M4 were poor competitors. These results were in good agreement with results obtained from experiments examining the ability of Glis3(ZFD) to bind 32P-labeled Glis-BS mutants (Supplementary Figure 3S).
Comparison of Glis3 and Gli1 binding
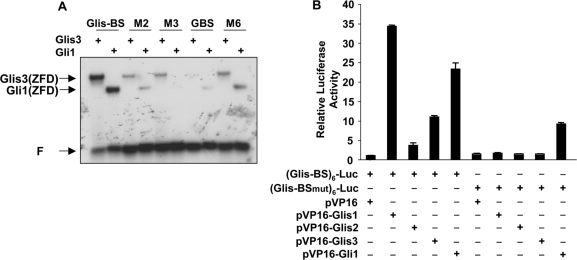
We next compared the binding of Glis3 to the Glis-BS consensus, the Glis-BS mutants M2, M3 and M6, and the GBS consensus with that of Gli1. Our EMSA showed that both Glis3 and Gli1 bound the consensus Glis-BS more effectively than GBS (Figure 6A). Moreover, Glis3 bound moderately well to the M3 mutant while Gli1 did not bind M3. These observations suggest that although Glis3 and Gli1 bind similar binding sites, they can exhibit different affinities for distinct sites.
Figure 6.
(A) Comparison of the binding specificity of His6-Glis3(ZFD) and His6-Gli1(ZFD) to 32P-labeled Glis-BS, GBS, and the Glis-BS mutants M2, M3 and M6. Binding was analyzed by EMSA. (B) Comparison of (Glis-BS)- and (Glis-BSmut)-mediated transcriptional activation by VP16-Glis1-3 and VP16-Gli1. HEK293T cells were co-transfected with pVP16-Glis1-3 or pVP16-Gli1 (0.1 μg), (Glis-BS)6-LUC, (Glis-BSmut)6-LUC and pCMVβ as indicated. After 30 h, cells were assayed for luciferase (LUC) and β-galactosidase activity as described in ‘Materials and methods’ section. The relative LUC reporter activity was calculated and plotted. The sequence of Glis-BSmut is CGTGGTGGGTC (M13 in Supplementary Figure 3S).
These results also showed that Gli1 bound Glis-BS more efficiently than GBS, which was reported to be the optimal binding sequence of Gli1. We, therefore, determined the Kd for the binding of Gli1 to the Glis-BS consensus by Scatchard plot analysis (Figure 5B). This analysis indicated that the apparent Kd for the interaction of Gli1 with Glis-BS was 3.32 ± 1.11 nM. These data demonstrate that both Glis3 and Gli1 exhibit a high and similar affinity for the consensus Glis-BS.
Next, we compared the ability of VP16-Glis1-3 proteins to induce (Glis-BS)6-dependent transcriptional activation of the LUC reporter gene (Glis-BS)6-LUC reporter, in which the luciferase reporter is under the control of six consensus Glis3 binding sites. This activation is largely a reflection of the ability of the fusion proteins to bind Glis-BS. The data in Figure 6B show that VP16-Glis1, VP16-Glis3 and VP16-Gli1 enhanced (Glis-BS)6-dependent reporter gene expression 34-, 11- and 23-fold, respectively. These results are in agreement with the conclusion that VP16-Glis1, -Glis3 and -Gli1 fusion proteins effectively bind Glis-BS. VP16-Glis1 and VP16-Glis3 did not significantly induce LUC reporter through Glis-BSmut (Figure 5B) in agreement with the observation that Glis3 has low affinity for Glis-BSmut (5′-CGTGGTGGGTC; M13 in Supplementary Figure 3S). In contrast, VP16-Gli1 was still able to increase LUC transcription supporting the conclusion that Glis3 and Gli1 have distinct but overlapping binding specificities. Similar results were obtained when this assay was performed in Chinese hamster ovary (CHO) cells (data not shown). VP16-Glis2 induced Glis-BS-dependent LUC reporter activity only 3-fold. This might be due to low affinity binding or, since Glis2 acts as a repressor, to the presence of a dominant Glis2 repressor function.
All five zinc finger domains are important in the binding of Glis3 to Glis-BS
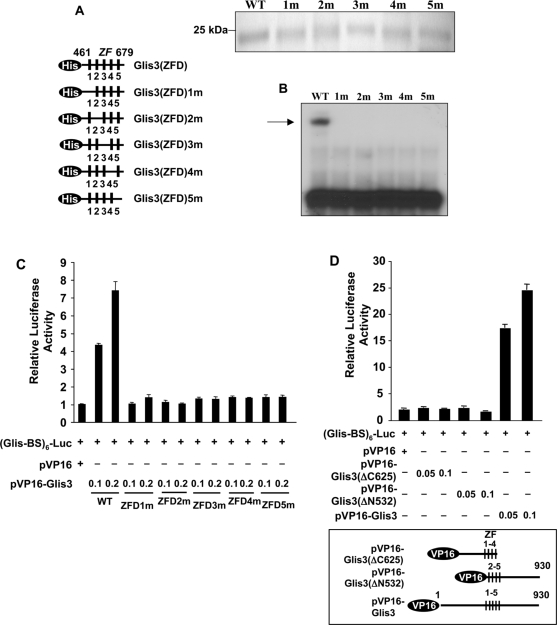
To examine the role of each individual zinc finger in the binding of Glis3(ZFD) to the Glis-BS consensus sequence, the effect of several mutations on the binding of Glis3 was determined by EMSA. The first Cys in each single Cys2His2-type zinc finger motif was mutated resulting in the loss of their tetrahedral configuration (Figure 7A). As shown in Figure 7B, a mutation in each single zinc finger motif totally abolished binding of Glis3 to the consensus Glis-BS suggesting that the tetrahedral conformation of each zinc finger motif is required for optimal binding of Glis3. This conclusion was supported by analysis of the activation of (Glis-BS)6-LUC reporter by VP16-Glis3 proteins (Figure 7C). The transcriptional activation by these fusion proteins is dependent on the binding of VP16-Glis3 to Glis-BS and the VP16 activation domain. The data showed that none of the VP16-Glis3 zinc finger mutants was able to induce LUC reporter activity suggesting that they did not bind Glis-BS in agreement with our EMSA results (Figure 7B and C). The importance of the first and fifth zinc finger motifs in the binding of Glis3 to Glis-BS was supported by the analysis of two additional mutants VP16-Glis3ΔC625, containing a C-terminal deletion up to ZF4, and VP16-Glis3ΔN532, containing an N-terminal deletion including ZF1. Both mutants were unable to induce LUC transcription (Figure 7D) in agreement with the conclusion that they do not bind Glis-BS.
Figure 7.
All five zinc finger motifs are required for optimal binding of Glis3 to the consensus Glis-BS. (A) A schematic view of His6-Glis3(ZFD)1–5m mutants in which the first Cys in each zinc finger motif is mutated to Ala (left panel). PAGE analysis of His6-Glis3(ZFD) proteins used in EMSA (upper panel). (B) EMSA using His6-Glis3(ZFD) and His6-Glis3(ZFD)1–5m mutant proteins and 32P-labeled consensus Glis-BS. The position of the Glis3/Glis-BS complex is indicated by the arrow. (C) The ability of VP16-Glis3 and VP16-Glis3(ZFD)1–5m mutants to induce (Glis-BS)6-dependent transactivation of the LUC reporter gene was examined in HEK293T cells. (D) The ability of VP16-Glis3 and mutants VP16-Glis3(ΔC625) and VP16-Glis3(ΔN532), encoding a C- and N-terminal deletion mutant of Glis3 as indicated in the lower panel, to induce (Glis-BS)6-dependent transactivation of the LUC reporter gene was examined. The relative luciferase activity was calculated and plotted. Numbers indicate amount of plasmid (μg) transfected.
Glis3 is a potent activator of Glis-BS-dependent transcription
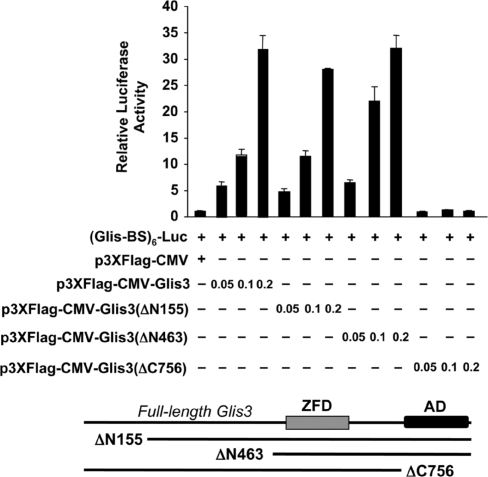
The aforementioned results characterized the nuclear translocation and DNA binding of Glis3, we next determined whether the full-length Glis3 was able to activate Glis-BS-dependent transcription and function as a transcriptional activator. For this purpose HEK293T cells were transiently transfected with a Glis3 expression and a (Glis-BS)6-LUC reporter plasmids and the transcriptional activity of Glis3 analyzed. As shown in Figure 8, Glis3 induced (Glis-BS)-dependent transcription 32-fold suggesting that it functions as a transcriptional activator. Glis3(ΔN155) and Glis3(ΔN463) containing amino-terminal deletions, were almost as active as full-length Glis3 suggesting that the amino-terminus does not appear to contain a repressor or activation function. In contrast, the carboxyl-terminal mutant Glis3(ΔC756) totally lacked transcriptional activity suggesting the loss of an activation function at the C-terminus.
Figure 8.
Full-length Glis3 functions as a potent transcriptional activator. HEK293T cells were co-transfected with p3×FLAG-CMV-Glis3, p3×FLAG-CMV-Glis3(ΔN155), p3×FLAG-CMV-Glis3(ΔN463), p3×FLAG-CMV-Glis3(ΔC756), pCMVβ and (Glis-BS)6-LUC as indicated. After 30 h, cells were assayed for luciferase (LUC) and β-galactosidase activity. The relative LUC reporter activity was calculated and plotted. A schematic view of the deletion mutants is shown below the graph. AD, activation domain; ZDF, zinc finger domain.
NDH1 mutation results in the loss of Glis3 transactivating function
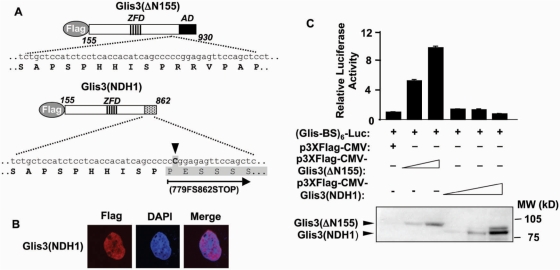
A recent genetic study implicated Glis3 in a syndrome with NDH (14). One family of patients (NDH1) with the most severe NDH contained a homozygous insertion (2997insC) leading to a frameshift and a truncated Glis3 protein, referred to as Glis3(NDH1) (Figure 9A). To examine the effect of this truncation on the subcellular localization and transcriptional activity of Glis3, we generated a mutant Glis3(NDH1) expression plasmid that contains this NDH1 truncation (Figure 9A). As WT Glis3, Glis3(NDH1) still localized to the nucleus (Figure 9B). However, in contrast to WT Glis3, Glis3(NDH1) did not induce Glis-BS-dependent transcription of the LUC reporter in HEK293T (Figure 9C). These data are in agreement with our obervations that the transactivating domain is localized at the C-terminus of Glis3 (Figure 8). Thus, the NDH1 mutation induces loss of the Glis3 transactivating function. This loss might be responsible for the abnormalities observed in NDH1 patients.
Figure 9.
Glis3(NDH1) localized to the nucleus, but lost its transactivation activity. (A) Schematic view of Glis3(Δ155) and Glis3(NDH1). ZDF and AD are indicated. An insertion of C (arrow head) in the Glis3 gene (NDH1) is linked to the most severe type of NDH. This insertion results in a frame shift at 779 and a stop codon at 862 (779FS862STOP) as indicated (shaded box). (B) Glis3(NDH1) localizes to the nucleus. HEK293T cells were transfected with p3×FLAG-CMV-Glis3(NDH1) and subcellular localization was examined by confocal microscopy with anti-FLAG M2 and Alexa Fluor 594 antibodies; nuclei were identified by DAPI staining. (C) Glis3(NDH1) lacks transcriptional activity. HEK293T cells were co-transfected with p3×FLAG-CMV-Glis3(ΔN155), p3×FLAG-CMV-Glis3(NDH1), pCMVβ and (Glis-BS)6-LUC as indicated. After 30 h, cells were assayed for luciferase (LUC) and β-galactosidase activity. The relative LUC reporter activity was calculated and plotted. The expression of Glis3(ΔN155) and Glis3(NDH1) proteins was examined by western blot analysis using anti-FLAG antibody (lower panel).
DISCUSSION
Glis3 is a member of the Glis Krüppel-like zinc finger subfamily of transcription factors that are related to members of the Gli and Zic families (5,6,9). In this study, we describe a sequence that extends the previously reported mouse Glis3 protein by an additional 155 amino acids (9) at the N-terminus and further characterize several steps (nuclear localization, DNA binding and transcriptional activity) that are critical in the mechanism by which Glis3 regulates transcription. Translocation of the Glis3 to the nucleus is an essential step in the pathway by which it regulates transcription. Regulation of nuclear import and export provides an important mechanism by which the cell can control the transcriptional activity of many transcription factors (17). The trafficking of Gli and Zic proteins between cytoplasm and nucleus is controlled at multiple levels and provides a mechanism to regulate their transcriptional activity (16,18). And although Gli and Zic proteins are related, the mechanism involved in their nucleocytoplasmic trafficking are very different (16,19). We show that, in number of cell types, full-length Glis3 localized preferentially to the nucleus. Nuclear import and export can occur by passive diffusion of proteins smaller than ∼40 kDa (20) while larger proteins are translocated by active transport or in complex with actively transported proteins. Because of its size (97 kDa), the nuclear localization of Glis3 likely involves an active transport mechanism. The classical import mechanism involves binding of the importin-α/β heterodimer to a nuclear localization signal consisting of one or two basic regions thereby targeting it for transport through nuclear pore complexes. The energy for this transport is provided by the GTPase Ran (17). Pattern/motif analysis showed that Glis3 contains one putative bipartite nuclear localization signal (bNLS) that overlaps with ZF5. Our data demonstrate that this motif is not required but that ZF4 is essential for the nuclear localization of Glis3. These observations suggest that the nuclear translocation of Glis3 is not mediated by a direct interaction with importins but by a different mechanism.
The nuclear localization of several other proteins, including Wilm's tumor 1 (WT1) (21), the nuclear receptor NR2C1 (22), early growth response 1 (Egr1) (23), human Snail (24), JAZ (25) and erythroid Krüppel-like factor (EKLF) (26), Zic3 (16) are determined by their zinc finger motifs. However, the sequence requirements for nuclear localization are very different between these proteins. Our data show that the nuclear localization of Glis3 is dependent on the tertiary structure of ZF4 as has been reported for Egr1, JAZ and human Snail (23–25). In contrast, the nuclear localization of EKLF1 is independent of the tertiary structure of its zinc fingers (26). The loss of nuclear retention was not due to the inability of Glis3 to bind DNA since mutations in other zinc finger motifs, which also abolish DNA binding, did not effect nuclear localization. The nuclear localization of Zic3 has been reported to depend on a bNLS within ZF4 (16). Although the ZF4 of Glis3 shows 58% sequence homology with that of Zic3, this bNLS is not conserved. Moreover, mutation of Arg614 within ZF4 into Gly, which further abolishes this site, did not have any effect on the nuclear localization of Glis3. Zic3 also contains a nuclear export signal comprising the second and third zinc finger motif. However, Glis3 does not appear to contain any nuclear export signal.
After entering the nucleus, Glis3 functions as a transcriptional regulator by binding to specific DNA elements (Glis-BS) in the promoter of target genes. In this study, we determined for the first time the consensus sequence required for optimal Glis3 binding using a PCR/EMSA-based site selection strategy. This strategy identified 5′-(G/C)TGGGGGGT(A/C) as the optimal binding sequence of Glis3. Glis1, in which ZFD exhibits 93% sequence homology to that of Glis3, also binds effectively to the consensus Glis-BS. Analysis of various point mutations in the Glis-BS consensus on Glis3 binding confirmed the specificity of this interaction and showed that a single nucleotide change in the consensus sequence can result in the total loss of Glis3 binding. Glis3 binds with high affinity (an apparent Kd of 3.69 ± 0.26 nM) to the Glis-BS consensus. The consensus Glis-BS shows a great similarity with the binding sites of Gli and Zic proteins (7,8,27,28). However, as observed for Gli and Zic proteins (8), differences in the binding to distinct DNA-binding sites were observed between Gli1 and Glis3. Glis3 bound the Glis-BS mutant M3 moderately well while Gli1 did not bind. Our data also demonstrated that Glis3 bound Glis-BS more effectively than to the GBS consensus. Unexpectedly, Gli1 also interacted more effectively with the Glis-BS than with the GBS consensus which is considered to be the optimal binding site of Gli1 (7,27). We show that Gli1 exhibited high affinity for the Glis-BS consensus (an apparent Kd of 3.32 ± 1.11 nM) similar to that of Glis3. The observed overlap in binding specificity suggests that Glis and Gli proteins can compete for binding to the same DNA binding sequence in the promoter of target genes (9). The latter provide a mechanism by which the Glis and Gli signaling pathways might interact or interfere with each other.
To determine the requirements of the interaction of Glis3 with Glis-BS, we examined the effect of several point mutations on this binding. Mutation of the first cysteine in each individual zinc finger motif to alanine, which demolishes the tetrahedral configuration of the zinc finger, destroyed the ability of Glis3 to bind Glis-BS (Figure 7A and B). These observations indicate that each zinc finger motif is critical for the interaction of Glis3 with Glis-BS. The requirement for ZF1 and ZF5 was supported by the inability of VP16-Glis3 mutants, lacking ZF1 or ZF5, to induce (Glis-BS)-dependent transcription (Figure 7C and D). Previously, crystal structure analysis of Gli1–DNA complexes and analysis of Gli3 DNA binding have shown that ZF2-5 bind in the major groove and wrap around the DNA and that ZF4 and ZF5 make extensive base contacts (27,29). Although these studies indicated that ZF1 does not contact the DNA, the inability of ZF1 mutants of Glis3 to bind Glis-BS suggests that ZF1 might be important for maintaining the proper structure of the zinc finger domain required for optimal Glis3 binding. This concept is supported by a recent study showing that mutations within ZF1 of ZIC3 can influence the nuclear localization, protein stability, and activity of ZIC3 indicating the importance of ZF1 in ZIC3 activity (30).
After binding Glis-BS, does full-length Glis3 function as a transcriptional activator? We demonstrated that the full-length Glis3 induces the transcription of (Glis-BS)6-dependent reporter suggesting that it functions as a transcriptional activator. Deletion of a 155- or 463-amino acid region at the amino-terminus did not affect the transcriptional activity of Glis3 suggesting that this region does not appear to contain a repressor or activation function. In contrast, deletion of the C-terminus totally abolished the transcriptional activity of Glis3 in agreement with the conclusion that the deleted region contains an activation domain (9). As reported for other transcription factors, transcriptional repression or activation by Glis proteins is likely mediated through a recruitment of intermediary proteins that function, respectively, as co-repressors or co-activators (31). This is supported by previous findings showing that C-terminal binding protein 1 (CtBP1) and HDAC3 function as co-repressors of Glis2 (1); however, no co-activators or co-repressors have yet been identified for Glis3. In addition to the sequence of the DNA-binding site and binding affinity, the context of the binding site within the promoter regulatory region has been found to be an important determinant in which transcription factor and intermediary proteins are recruited to the site (32). This may also apply to the binding of Gli, Zic and Glis proteins to GBS in target genes.
Recently, mutations in the Glis3 gene have been implicated in a syndrome with neonatal diabetes mellitus, polycystic kidney disease and NDH (14). These studies have indicated that Glis3 plays a critical role in pancreatic development and the maintenance of normal kidney functions. NDH1-type patients exhibit the most severe effects and die between 10 days and 2 years after birth. These patients contain a frameshift mutation in the Glis3 gene that yields a truncated Glis3 protein [Glis3(NDH1); Figure 9A]. In our study, we show that Glis3(NDH1) still localized to the nucleus but was unable to induce (Glis-BS)-dependent transactivation. These findings are in agreement with our observations (Figure 8) that the C-terminus of Glis3 contains a transactivation function. The loss of this transactivation function appears at least in part responsible for the abnormalities observed in NDH1 patients. However, we cannot rule out that in vivo the Glis3(NDH1) protein is unstable and rapidly destroyed by the proteasome system resulting in decreased levels of Glis3(NDH1) protein.
In summary, in this study we demonstrate that full-length Glis3 localizes to the nucleus and that its putative bNLS is not required but that the tetrahedral configuration of ZF4 is essential for the nuclear localization of Glis3. These observations suggest that Glis3 is imported into the nucleus by a nonclassical mechanism. In addition, we identify the consensus, high affinity Glis-BS sequence required for optimal Glis3 binding and show that all five Glis3 zinc finger motifs are critical for optimal binding. We further show that full-length Glis3 functions as a potent activator of (Glis-BS)-dependent transcription and contains an activation domain at its C-terminus. This transactivation function is lost in NDH1 patients and offers a mechanism explaining the abnormalities observed in these patients. Our study provides greater insight into three critical steps involved in the mechanism by which Glis3 controls transcription. These observations help us to understand the role of Glis3 in human disease, such as NDH, and in the regulation of pancreatic development and renal functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Dr C. Teng and Dr John Roberts for their valuable comments on the manuscript. This research was supported by the Intramural Research Program of the NIEHS, NIH. Funding to pay the Open Access publication charges for this article was provided by the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kim SC, Kim YS, Jetten AM. Kruppel-like zinc finger protein Gli-similar 2 (Glis2) represses transcription through interaction with C-terminal binding protein 1 (CtBP1) Nucleic Acids Res. 2005;33:6805–6815. doi: 10.1093/nar/gki985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim YS, Lewandoski M, Perantoni AO, Kurebayashi S, Nakanishi G, Jetten AM. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem. 2002;277:30901–30913. doi: 10.1074/jbc.M203563200. [DOI] [PubMed] [Google Scholar]
- 3.Lamar E, Kintner C, Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–1346. doi: 10.1242/dev.128.8.1335. [DOI] [PubMed] [Google Scholar]
- 4.Nakashima M, Tanese N, Ito M, Auerbach W, Bai C, Furukawa T, Toyono T, Akamine A, Joyner AL. A novel gene, GliH1, with homology to the Gli zinc finger domain not required for mouse development. Mech. Dev. 2002;119:21. doi: 10.1016/s0925-4773(02)00291-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F, Jetten AM. Genomic structure of the gene encoding the human GLI-related, Kruppel- like zinc finger protein GLIS2. Gene. 2001;280:49–57. doi: 10.1016/s0378-1119(01)00764-8. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Nakanishi G, Kurebayashi S, Yoshino K, Perantoni A, Kim YS, Jetten AM. Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcriptional factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. J. Biol. Chem. 2001;12:10139–10149. doi: 10.1074/jbc.M108062200. [DOI] [PubMed] [Google Scholar]
- 7.Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizugishi K, Aruga J, Nakata K, Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J. Biol. Chem. 2001;276:2180–2188. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- 9.Kim YS, Nakanishi G, Lewandoski M, Jetten AM. GLIS3, a novel member of the GLIS subfamily of Kruppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003;31:5513–5525. doi: 10.1093/nar/gkg776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakanishi G, Kim YS, Nakajima T, Jetten AM. Regulatory role for Kruppel-like zinc-finger protein Gli-similar 1 (Glis1) in PMA-treated and psoriatic epidermis. J. Invest. Dermatol. 2006;126:49–60. doi: 10.1038/sj.jid.5700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attanasio M, Uhlenhaut NH, Sousa VH, O'Toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nature Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y-S, Kang HS, Herbert R, Beak JY, Collins JB, Grissom SF, Jetten AM. Krüppel-like zinc finger protein Glis2 is essential for the maintenance of normal renal functions. Mol. Cell. Biol. 2007 doi: 10.1128/MCB.01722-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beak JY, Kang HS, Kim YS, Jetten AM. Kruppel-like zinc finger protein Glis3 promotes osteoblast differentiation by regulating FGF18 expression. J. Bone Miner. Res. 2007;22:1234–1244. doi: 10.1359/jbmr.070503. [DOI] [PubMed] [Google Scholar]
- 14.Senee V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, Charon C, Nicolino M, Boileau P, Cavener DR, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nature Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 15.Yan ZH, Medvedev A, Hirose T, Gotoh H, Jetten AM. Characterization of the response element and DNA binding properties of the nuclear orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor. J. Biol. Chem. 1997;272:10565–10572. doi: 10.1074/jbc.272.16.10565. [DOI] [PubMed] [Google Scholar]
- 16.Bedard JE, Purnell JD, Ware SM. Nuclear import and export signals are essential for proper cellular trafficking and function of ZIC3. Hum. Mol. Genet. 2007;16:187–198. doi: 10.1093/hmg/ddl461. [DOI] [PubMed] [Google Scholar]
- 17.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell. Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 19.Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic hedgehog signalling. Eur. J. Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 20.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 21.Bruening W, Moffett P, Chia S, Heinrich G, Pelletier J. Identification of nuclear localization signals within the zinc fingers of the WT1 tumor suppressor gene product. FEBS Lett. 1996;393:41–47. doi: 10.1016/0014-5793(96)00853-8. [DOI] [PubMed] [Google Scholar]
- 22.Yu Z, Lee CH, Chinpaisal C, Wei LN. A constitutive nuclear localization signal from the second zinc-finger of orphan nuclear receptor TR2. J. Endocrinol. 1998;159:53–60. doi: 10.1677/joe.0.1590053. [DOI] [PubMed] [Google Scholar]
- 23.Matheny C, Day ML, Milbrandt J. The nuclear localization signal of NGFI-A is located within the zinc finger DNA binding domain. J. Biol. Chem. 1994;269:8176–8181. [PubMed] [Google Scholar]
- 24.Yamasaki H, Sekimoto T, Ohkubo T, Douchi T, Nagata Y, Ozawa M, Yoneda Y. Zinc finger domain of Snail functions as a nuclear localization signal for importin beta-mediated nuclear import pathway. Genes Cells. 2005;10:455–464. doi: 10.1111/j.1365-2443.2005.00850.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang M, May WS, Ito T. JAZ requires the double-stranded RNA-binding zinc finger motifs for nuclear localization. J. Biol. Chem. 1999;274:27399–27406. doi: 10.1074/jbc.274.39.27399. [DOI] [PubMed] [Google Scholar]
- 26.Pandya K, Townes TM. Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J. Biol. Chem. 2002;277:16304–16312. doi: 10.1074/jbc.M200866200. [DOI] [PubMed] [Google Scholar]
- 27.Vortkamp A, Gessler M, Grzeschik KH. Identification of optimized target sequences for the GLI3 zinc finger protein. DNA Cell Biol. 1995;14:629–634. doi: 10.1089/dna.1995.14.629. [DOI] [PubMed] [Google Scholar]
- 28.Ishiguro A, Inoue T, Mikoshiba K, Aruga J. Molecular properties of Zic4 and Zic5 proteins: functional diversity within Zic family. Bioch. Biophys. Res. Comm. 2004;324:302–307. doi: 10.1016/j.bbrc.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 29.Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 30.Chhin B, Hatayama M, Bozon D, Ogawa M, Schön P, Tohmonda T, Sassolas F, Aruga J, Valard AG, Chen SC, et al. Elucidation of penetrance variability of a ZIC3 mutation in a family with complex heart defects and functional analysis of ZIC3 mutations in the first zinc finger domain. Hum. Mutat. 2007;28:563–570. doi: 10.1002/humu.20480. [DOI] [PubMed] [Google Scholar]
- 31.Jetten AM, Joo JH. Retinoid-related orphan receptors (RORs): roles in cellular differentiation and development. Adv. Devel. Biol. 2006;16:314–354. doi: 10.1016/S1574-3349(06)16010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Rosenfeld MG, Hamilton BA. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.