Abstract
Atopic dermatitis (AD) is characterized by a defective skin barrier which allows increased allergen and pathogen penetration. Loricrin (LOR) and involucrin (IVL) are proteins important for skin barrier formation and integrity. In this study, we demonstrate that the gene and protein expression of LOR and IVL is significantly decreased in acute (LOR: p<0.001; IVL: p<0.001) and non-lesional (LOR: p<0.001; IVL: p<0.001) skin of AD subjects, as compared to skin from healthy subjects. Using primary keratinocytes, we further demonstrate the down-regulatory effect of IL-4 and IL-13 – which are over-expressed in the skin of AD patients – on LOR and IVL expression in keratinocytes. Additionally, skin biopsies from signal transducer and activator of transcription (STAT)-6 transgenic mice were deficient in the expression and production of LOR and IVL. This study suggests that Th2 cytokines inhibit expression of LOR and IVL through a STAT-6 dependent mechanism.
Keywords: atopic dermatitis, loricrin, involucrin, Th2 cytokines, STAT-6
INTRODUCTION
Atopic dermatitis (AD) is a chronic inflammatory skin disease that affects up to 20% of children and significantly disrupts the quality of life for each individual affected by the disease [1]. In recent years, it has been suggested that the epidermal skin barrier plays a significant role in AD disease susceptibility and severity [2–5]. Using a murine model of AD, it has been shown that skin barrier dysfunction enhances allergen sensitization leading to increased IgE levels and airway hyper-reactivity [6]. This supports the notion that absorption of allergens through the skin may be the first step of the atopic march.
Loricrin (LOR) and involucrin (IVL) are important proteins that facilitate terminal differentiation of the epidermis and formation of the skin barrier [7–12]. Human LOR is an insoluble protein initially expressed in the granular layer of the epidermis during cornification, and comprises 80% of the total protein mass of the cornified envelope (CE) [7, 13–16]. Additionally, LOR functions as a main reinforcement protein for the CE and is deposited onto a scaffold of IVL and other calcium-binding proteins [17]. IVL is an also common component of the CE and provides a scaffold to which other proteins subsequently become cross-linked [8]. In the CE structure, IVL is adjacent to the cell membrane and form the exterior surface of the CE [18].
The current studies were therefore conducted to investigate the expression of LOR and IVL in eczematous skin lesions and non-lesional skin from AD subjects and investigate the role of IL-4 and IL-13, cytokines over-expressed in AD skin [19, 20], on the expression of LOR and IVL.
MATERIALS AND METHODS
Patients
Subjects included 13 healthy persons with no history of skin disease (mean age: 34.3 years) and 14 patients with moderate-to-severe atopic dermatitis (mean age: 33.6 years; 20%–60% skin involvement). None of the patients had previously received systemic corticosteroids or cyclosporine, and none had received topical corticosteroid or calcineurin inhibitors for at least one week before enrollment. This study was approved by the Institutional Review Board at National Jewish Medical and Research Center in Denver and conducted according to the Declaration of the Helsinki Guidelines. All subjects gave written informed consent prior to participation in this study.
Two millimeter punch biopsies were collected from erythematous lesions that were less than three days of old (involved atopic dermatitis) and uninvolved skin from the same AD patients, and normal healthy skin. The skin biopsies were immediately submerged in 1 mL Tri-Reagent (Molecular Research Center, Inc., Cincinnati, OH) and frozen at −80°C for future RNA isolation or immediately in 1 mL 10% buffered formalin for immunohistochemistry.
Mice
Heterozygous STAT-6 transgenic mice were obtained from Mark Kaplan (Indiana University, Indianapolis, IN). These mice constitutively express STAT-6. Upon arrival, heterozygous mice were bred with C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) to generate additional transgenic mice and wild type controls. All animal protocols were approved by the Institutional Animal Care and Use Committee at National Jewish Medical and Research Center. This institution has an animal welfare assurance number (A3026-1) on file with the Office of Protection and Research Risks.
The dorsal thoracic and lumbar regions of mice were clipped and treated with the depilatory agent Nair to remove hair. Seventy-two hours following hair removal, mice were euthanized via carbon dioxide asphyxiation. Four millimeter biopsies were collected and immediately submerged in 1 mL Tri-Reagent (Molecular Research Center, Inc., Cincinnati, OH) for RNA isolation.
RNA preparation and real-time RT-PCR
Total RNA was isolated from skin biopsies by chloroform:phenol extraction and isopropanol precipitation according to manufacturer’s guidelines (Molecular Research Center, Inc., Cincinnati, OH). RNeasy Mini Kits (Qiagen, Valencia, CA) were used according to the manufacturer’s protocol to further purify RNA from skin biopsies and to isolate RNA from cell cultures. One microgram RNA was reverse-transcribed in a 20-μl reaction containing Random Primers (Invitrogen, Carlsbad, CA), dNTP Mix (Invitrogen), 5 × First Strand Buffer (Invitrogen), RNase Inhibitor (Invitrogen) and Superscript III enzyme (Invitrogen) for 60 minutes at 42°C and then 70°C for 15 minutes. Real-time RT-PCR was performed and analyzed by the dual-labeled fluorogenic probe method using an ABI Prism 7300 sequence detector (Applied Biosystems, Foster City, CA) as previously described [20]. Primers and probes for human GAPDH, LOR and IVL were purchased from Applied Biosystems. Relative expression levels were calculated by the relative standard curve method as outlined in the manufacturer’s technical bulletin (Applied Biosystems). A standard curve was generated using the fluorescent data from 10-fold serial dilutions of total RNA of the highest expression sample. To allow for comparisons between samples and group, quantities of all targets in test samples were normalized to the corresponding GAPDH levels in the skin biopsies and cultured keratinocytes.
Immunohistochemistry
Paraffin-embedded tissues were cut at 5 μm and placed on frosted microscope slides. Using xylene and a series of ethanol washes, slides were deparaffinized and then re-hydrated. Slides were then immersed in antigen retrieval solution (0.01 mol/L citric acid, 0.05 mol/L NaOH, pH 6.0) and microwaved for 7 minutes to retrieve masked antigen. Cell and Tissue Staining Kits (R&D Systems, Minneapolis, MN) were used according to the manufacturer’s protocol. Endogenous peroxidase was blocked with Peroxidase Blocking Reagent (R&D Systems) for 5 minutes followed by a block with Serum Blocking Reagent (R&D Systems) for 30 minutes at room temperature. The sections were stained with a polyclonal rabbit anti-human antibody directed against LOR (1:500 dilution; Abcam, Cambridge, MA) and a monoclonal mouse anti-human antibody directed against IVL (1:750 dilution; Abcam) at 4°C overnight. Slides were then washed and incubated for 60 minutes with a Biotinylated Secondary Antibody (R&D Systems). The antigen-antibody complex was detected using the avidin-biotin peroxidase complex method. Diaminobenzidine (R&D Systems) was used to visualize the antibody specific staining. Slides were counterstained with hematoxylin solution (Sigma-Aldrich, Inc., St. Louis, MO). Antibody specificity was determined by replacing the primary antibody with an isotype-matched control (purified non-immune rabbit or mouse IgG; Southern Biotechnology, Birmingham, AL). All slides were coded before the samples were evaluated so that the identity of the study subjects was not revealed. Images were collected at 40X magnification and the intensity of the immunostaining scored on a scale from 0 to 5, with 0 indicating no staining and 5 indicating the most intense staining.
Keratinocyte cell culture
Primary human keratinocytes derived from neonatal foreskin were grown in serum-free EpiLife cell culture medium (Cascade Biologics, Portland, OR) containing 0.06 mmol/L calcium chloride, 1% human keratinocytes growth supplement V2 (Cascade Biologics), and 1% of penicillin and streptomycin).
To investigate the effects of the IL-4 and IL-13, primary keratinocytes were seeded at 2 × 105 cells per well and then differentiated with 1.3 mmol/L CaCl2 for 5 days in the presence and absence of IL-4 (R&D Systems) and/or IL-13 (R&D Systems). Total RNA was isolated from keratinocytes by using RNeasy kits according to the manufacturer’s guidelines (Qiagen, Valencia, CA) for real-time RT-PCR.
Statistics
Statistical analysis was conducted using Graph Pad Prism, version 4.03 (San Diego, CA). Statistical differences between groups were determined using an unpaired T test with significant differences conferred when p<0.05. In cases where multiple groups were compared, data were analyzed by a one-way analysis of variance (ANOVA), and significant differences were determined by a Tukey-Kramer test [21].
RESULTS
Deficiency of LOR and IVL in AD Skin
As shown in Figure 1-A, LOR gene expression was significantly decreased in involved (mean: 4.36 ± 1.10 ng LOR/ng GAPDH, P < 0.01) and uninvolved (mean: 22.31 ± 6.24 ng, P < 0.05) skin from patients with AD compared with skin from normal subjects (mean: 82.82 ± 28.35 ng). In addition, LOR gene expression was significantly decreased in involved AD skins compared with uninvolved AD skin (P < 0.05). Similarly, IVL gene expression was significantly decreased in involved (mean: 2.92 ± 0.53 ng, P < 0.001) and uninvolved (mean: 5.85 ± 0.99 ng, P < 0.01) skin from patients with AD compared with skin from normal subjects (mean: 9.14 ± 0.93 ng) (Figure 1-B). Additionally, IVL gene expression was significantly decreased in involved AD skin compared with uninvolved AD skin (P < 0.05).
Figure 1.
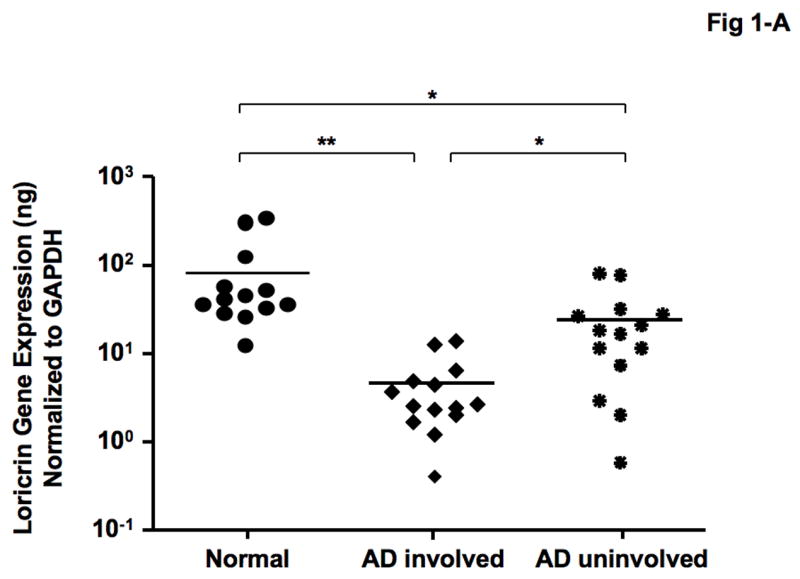
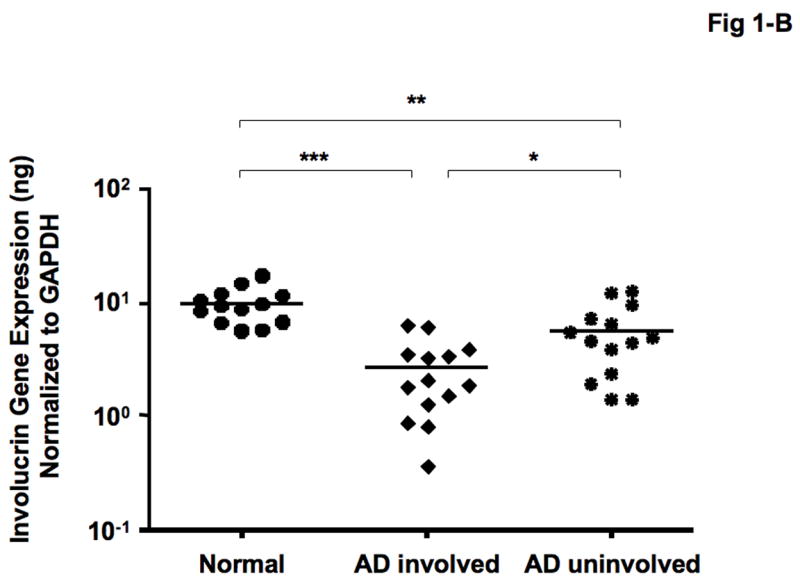
Gene expression of LOR and IVL in skin from normal subjects and AD patients. RNA was isolated from the skin of 13 normal subjects and 14 patients with AD, and the gene levels of LOR (A) and IVL (B) were evaluated by real-time RT-PCR. *P < 0.05; **P < 0.01; ***P < 0.001.
Immunohistochemical staining confirmed decreased expression of both LOR and IVL in involved and uninvolved skin from patients with AD compared with skin from normal subjects (Fig 2- A, B). The composite data for LOR and IVL immunostaining in all samples are shown in Fig 2-C and D. The staining intensity of LOR and IVL was significantly decreased in involved AD skin as compared to skin with normal subjects (LOR: P < 0.001; IVL: P < 0.001). And the staining intensity of LOR and IVL was significantly decreased in uninvolved AD skin as compared to skin from normal subjects (LOR: P < 0.05; IVL: P < 0.05). In addition, the staining intensity of LOR and IVL in involved AD skin was significantly decreased compared with uninvolved AD skin (LOR: P < 0.01; IVL: P < 0.05).
Figure 2.
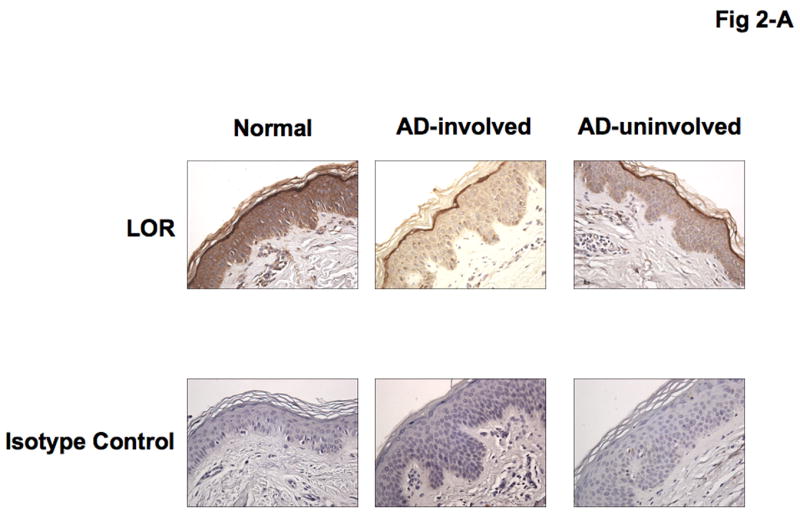
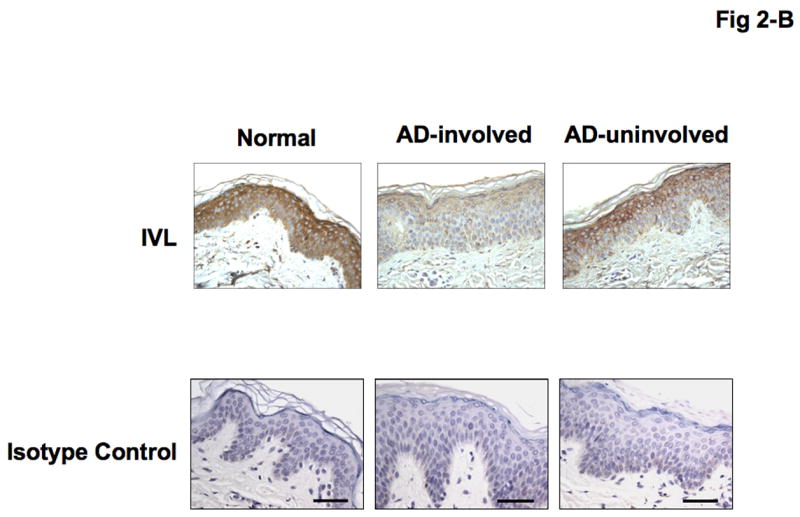
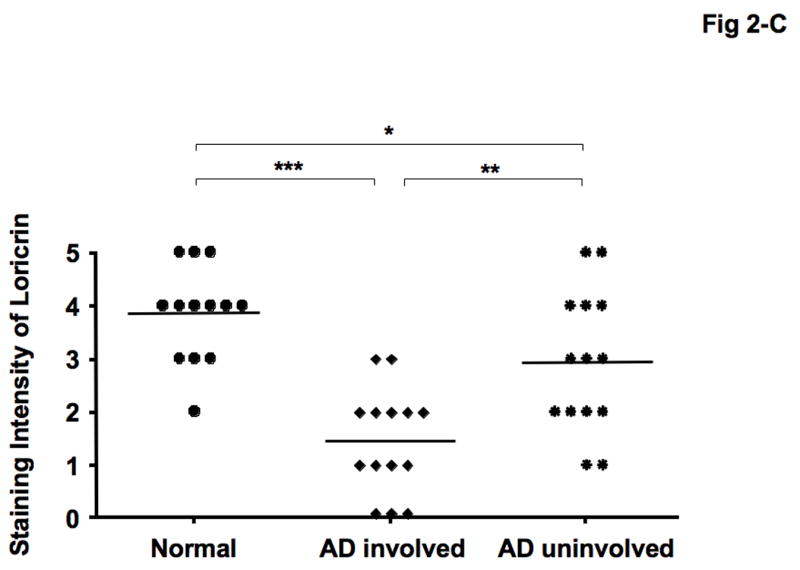
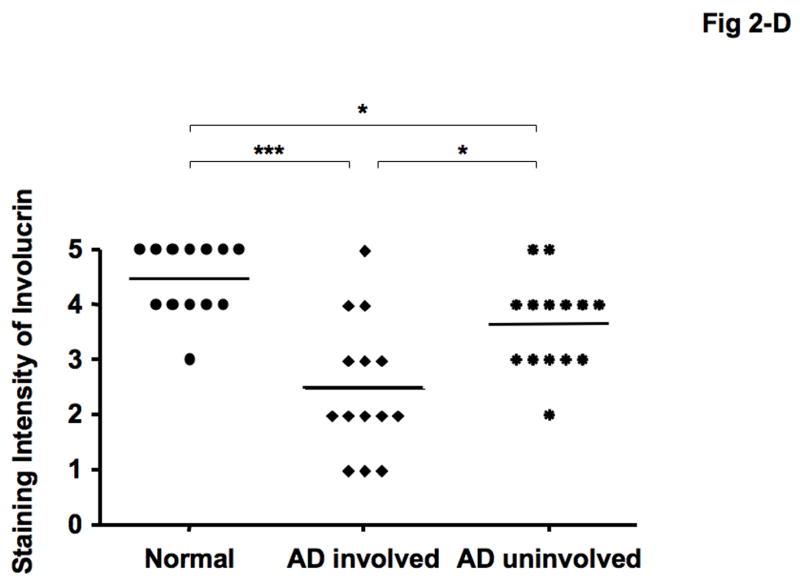
Decreased LOR and IVL staining in AD skin. (A, B) Representative paraffin embedded skin biopsies from normal subjects (n = 13), and patients with AD (n = 14) stained for LOR (A) and IVL (B) are shown. Images were collected at ×400 magnification. (C, D) The intensities of the staining for LOR (C) and IVL (D) were graded visually on a scale from 0 (no staining) to 5 (the most intense staining). *P < 0.05; **P < 0.01; ***P < 0.001.
Th2 cytokines inhibit gene expressions of LOR and IVL
IL-4 and IL-13 have previously been shown to be elevated in the skin from patients with AD [19, 20]. Therefore, we examined whether Th2 cytokines modulate the expression of LOR and IVL in human keratinocytes. Figures 3-A & B show that differentiating keratinocytes in the presence of IL-4 (LOR: 2.23 ± 0.81; IVL: 0.72 ± 0.05), IL-13 (LOR: 3.23 ± 0.54; IVL: 1.11 ± 0.06), or the combination of IL-4 and IL-13 (LOR: 2.80 ± 0.90; IVL: 0.88 ± 0.10) significantly inhibits the induction of LOR and IVL as compared with keratinocytes differentiated in media alone (LOR: 12.82 ± 2.15; IVL: 2.45 ± 0.26; P < 0.001 for all comparisons).
Figure 3.
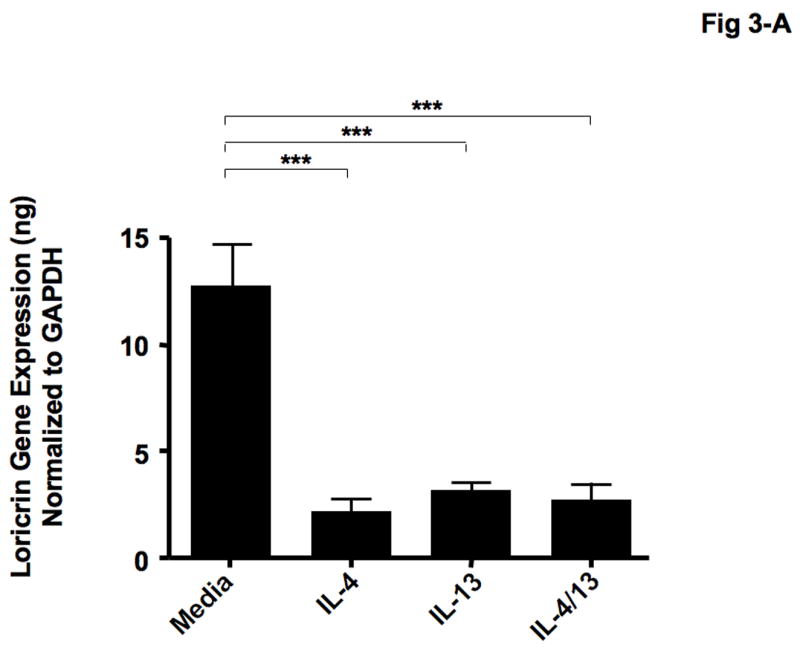
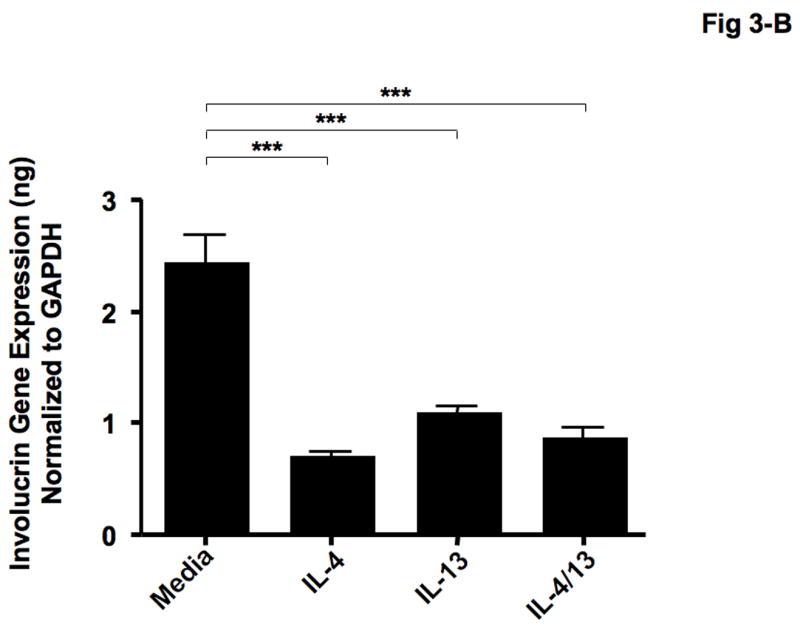
Th2 cytokines down-regulate the gene expressions of LOR and IVL. Primary human keratinocytes were differentiated in the absence and presence of IL-4, IL-13 or combination of IL-4 and IL-13 for 5 days. Then RNA was isolated from the cells, and the gene expressions of LOR (A) and IVL (B) were examined by real-time RT-PCR. ***P < 0.001.
LOR and IVL gene expression is down-regulated in STAT-6 transgenic mice
IL-4 and IL-13 have been shown to activate the STAT-6 signaling pathway [22, 23], therefore we investigated the role of the STAT-6 signaling pathway on LOR and IVL expression. Skin biopsies were collected from STAT-6 transgenic mice and their wild type controls (C57BL/6) and evaluated for LOR and IVL gene expression. LOR and IVL gene expression was significantly decreased in skin from STAT-6 transgenic mice (LOR: 0.0049 ± 0.0013, P = 0.037; IVL: 0.0014 ± 0.0004, P = 0.034) as compared to skin of wild type mice (LOR: 0.0102 ± 0.0032; IVL: 0.0035 ± 0.0011) (Fig 4-A, B).
Figure 4.
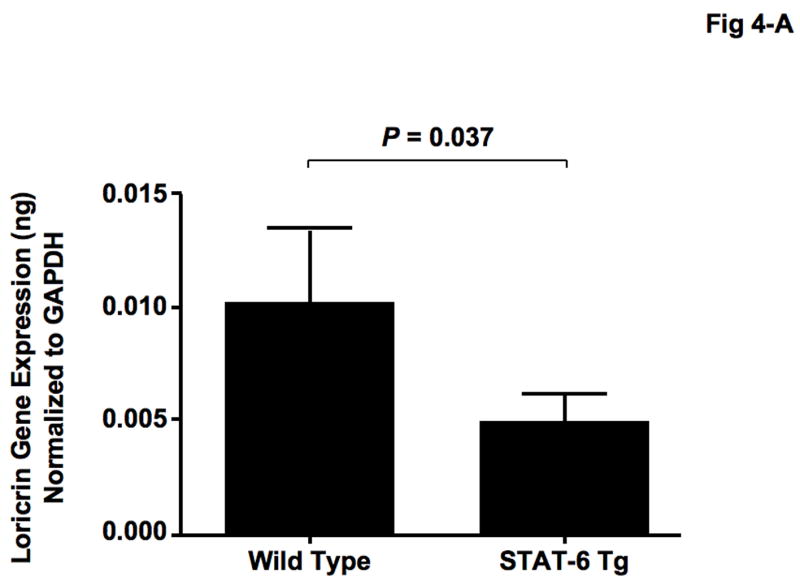
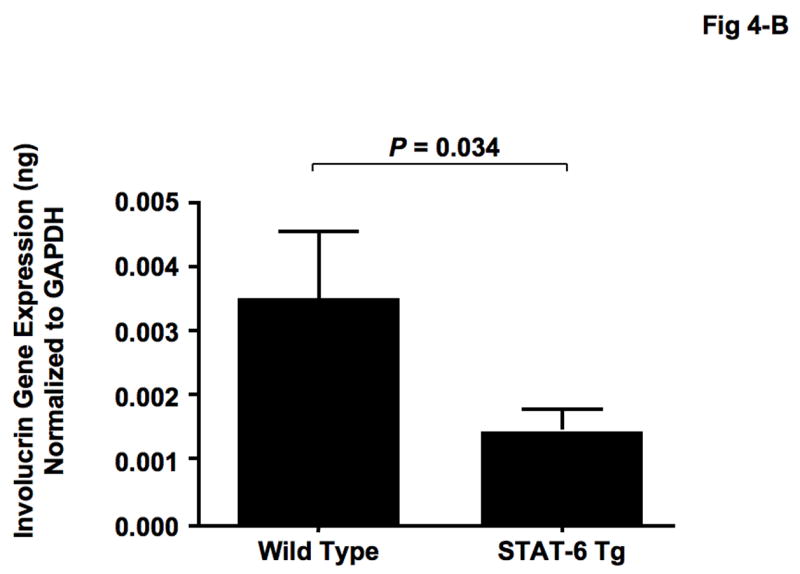
Gene expression of LOR and IVL was decreased in STAT-6 transgenic mice. Skin biopsies were obtained from 15 wild type (C57BL/6) and 15 STAT-6 transgenic mice, and gene expression of LOR (A) and IVL (B) was evaluated using real-time RT-PCR.
DISCUSSION
The stratum corneum and epidermis serve as a protective physical barrier against invading allergens and pathogens. This physical barrier is significantly compromised in the skin of AD patients; resulting in increased allergen sensitization, as well as viral and bacterial infections [24, 25]. Previous studies have shown that increased penetration of allergens and human pathogens augment the atopic inflammatory response which can exacerbate the disease [6, 26]. It is not fully understood how this additionally affects the epidermal barrier. The epidermal differentiation complex (EDC) is a cluster of genes located on chromosome 1q21 that encode a number of proteins important for barrier function [27]. LOR and IVL are two proteins encoded within the EDC, therefore these studies evaluated the relationship between their expression and the atopic inflammatory response.
In our current study, we demonstrate that both LOR and IVL expression are down-regulated in lesional and non-lesional AD skin as compared to skin from normal subjects. This is contradictory to previous studies by Sugiara et al [28] and Jensen et al. [29]. Using gene microarrays, Sugiara et al. [28] observed increased IVL and decreased LOR gene expression in lesional AD skin. Jensen et al. [29] observed increased LOR and decreased IVL expression using immunohistochemistry and western blotting. Important differences in experimental design may explain the discrepancies between our observations and those previously published: 1) while gene microarrays provide a significant amount of information, they are known to have limitations and can be falsely positive or negative [30, 31]. Our study used real-time RT-PCR, which has a higher degree of sensitivity, to examine the genetic expression of LOR and IVL and immunohistochemistry to confirm the observation on the protein level. 2) The lesional skin used in our current study is collected solely from acute atopic lesions. This factor is not clearly defined in either of the previous studies [28, 29]. AD lesions are bi-phasic with acute lesions having more of a Th2 cytokine milieu and chronic lesions having more of a Th1 cytokine milieu [19]. The relationship between cytokine milieu and barrier integrity is not completely understood. In fact, our current study is the first to investigate the effect of the cytokine milieu on LOR and IVL expression.
Keratinocytes are the primary cell of the epidermis and migrate through the basal, spinous, and granular regions to differentiate terminally into cornified cells and form the CE [15, 32]. For that reason, we used primary human keratinocytes to investigate why LOR and IVL expression is decreased in AD skin. Acute AD skin is characterized by the over-expression of IL-4 and IL-13 [19]. Therefore, we differentiated normal primary keratinocytes in the presence and absence of IL-4 and/or IL-13 for 5 days and evaluated the gene expression of LOR and IVL. The induction and expression of LOR and IVL was significantly inhibited by IL-4 and IL-13, suggesting that expression of LOR and IVL is down-regulated in AD skin due to the over-expression of Th2 cytokines in AD skin.
IL-4 and IL-13 signal through IL-4 receptor α1 and α2 to activate the STAT-6 signaling pathway [22, 23]. This pathway has previously been shown to down-regulate important components of the innate immune response [33], therefore we investigated the role of STAT-6 in the Th2 mediated down-regulation of LOR and IVL. Skin biopsies from STAT-6 transgenic mice exhibited significantly lower levels of both LOR and IVL as compare to their wild type controls. This suggests that IL-4 and IL-13 modulation of LOR and IVL is mediated by STAT-6 activation.
The current study demonstrates that gene and protein expression of LOR and IVL are decreased in the skin from AD patients. In addition, we demonstrate that gene expression of LOR and IVL is down-regulated by IL-4 and IL-13 through STAT-6. Taken together, our data suggest that decreased expression of LOR and IVL is caused, in part, by the over-expression of Th2 cytokines that down-regulate LOR and IVL expression during the differentiation into CE. Therefore, our current study suggests that cutaneous sensitization by various allergens and pathogens in AD is due, in part, to defect in epidermal skin barrier and may result in frequent skin inflammation and infection in AD skin.
Acknowledgments
This work was supported in part by Korea Research Foundation-KRF-2007-611-E00010 (BEK); NIH grants NIH/NIAID contracts N01 AI40029 and N01 AI40030, AR41256, R21 AR051634-01, General Clinical Research Center grant MO1 RR00051 from the Division of Research Resources, the Edelstein Family Chair in Pediatric Allergy and Immunology, and the University of Colorado Cancer Center (D.Y.M.L.).
The authors are additionally indebted to the nursing staff in the General Clinical Research Center for the recruitment of patients and collection of specimens. The authors also thank Maureen Sandoval for her help in preparing this manuscript.
ABBREVIATIONS
- AD
Atopic dermatitis
- CE
Cornified envelope
- EDC
Epidermal differentiation complex
- IL
Interleukin
- IVL
Involucrin
- LOR
Loricrin
- STAT
Signal transducer and activator of transcription
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leung DY, Bieber T. Atopic dermatitis. Lancet. 2003;361:151–160. doi: 10.1016/S0140-6736(03)12193-9. [DOI] [PubMed] [Google Scholar]
- 2.Taieb A, Hanifin J, Cooper K, Bos JD, Imokawa G, David TJ, Ring J, Gelmetti C, Kapp A, Furue M, de Prost Y, Darsow U, Werfel T, Atherton D, Oranje AP. Proceedings of the 4th Georg Rajka International Symposium on Atopic Dermatitis, Arcachon, France, September 15–17, 2005. J Allergy Clin Immunol. 2006;117:378–390. doi: 10.1016/j.jaci.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 3.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, Debenedetto A, Schneider L, Beck LA, Barnes KC, Leung DY. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2007;120:150–155. doi: 10.1016/j.jaci.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsson M, Broberg A, Jernas M, Carlsson L, Rudemo M, Suurkula M, Svensson PA, Benson M. Increased expression of aquaporin 3 in atopic eczema. Allergy. 2006;61:1132–1137. doi: 10.1111/j.1398-9995.2006.01151.x. [DOI] [PubMed] [Google Scholar]
- 5.Sator PG, Schmidt JB, Honigsmann H. Comparison of epidermal hydration and skin surface lipids in healthy individuals and in patients with atopic dermatitis. J Am Acad Dermatol. 2003;48:352–358. doi: 10.1067/mjd.2003.105. [DOI] [PubMed] [Google Scholar]
- 6.Spergel JM, Mizoguchi E, Brewer JP, Martin TR, Bhan AK, Geha RS. Epicutaneous sensitization with protein antigen induces localized allergic dermatitis and hyperresponsiveness to methacholine after single exposure to aerosolized antigen in mice. J Clin Invest. 1998;101:1614–1622. doi: 10.1172/JCI1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinertd PM, Marekov LN. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem. 1995;270:17702–17711. doi: 10.1074/jbc.270.30.17702. [DOI] [PubMed] [Google Scholar]
- 8.Bennett HS, Dienstfrey A, Hudson LT, Oreskovic T, Fuerst T, Shepherd J. Standards and measurements for assessing bone health-workshop report co-sponsored by the International Society for Clinical Densitometry (ISCD) and the National Institute of Standards and Technology (NIST) J Clin Densitom. 2006;9:399–405. doi: 10.1016/j.jocd.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Hudson TJ. Skin barrier function and allergic risk. Nat Genet. 2006;38:399–400. doi: 10.1038/ng0406-399. [DOI] [PubMed] [Google Scholar]
- 10.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O’Regan GM, Watson RM, Cecil JE, Bale SJ, Compton JG, DiGiovanna JJ, Fleckman P, Lewis-Jones S, Arseculeratne G, Sergeant A, Munro CS, El Houate B, McElreavey K, Halkjaer LB, Bisgaard H, Mukhopadhyay S, McLean WH. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 11.Candi E, Melino G, Mei G, Tarcsa E, Chung SI, Marekov LN, Steinert PM. Biochemical, structural, and transglutaminase substrate properties of human loricrin, the major epidermal cornified cell envelope protein. J Biol Chem. 1995;270:26382–26390. doi: 10.1074/jbc.270.44.26382. [DOI] [PubMed] [Google Scholar]
- 12.Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch PJ, de Viragh PA, Scharer E, Bundman D, Longley MA, Bickenbach J, Kawachi Y, Suga Y, Zhou Z, Huber M, Hohl D, Kartasova T, Jarnik M, Steven AC, Roop DR. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J Cell Biol. 2000;151:389–400. doi: 10.1083/jcb.151.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneda K, Steinert PM. Overexpression of human loricrin in transgenic mice produces a normal phenotype. Proc Natl Acad Sci USA. 1993;90:10754–10758. doi: 10.1073/pnas.90.22.10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalinin A, Marekov LN, Steinert PM. Assembly of the epidermal cornified cell envelope. J Cell Sci. 2001;114:3069–3070. doi: 10.1242/jcs.114.17.3069. [DOI] [PubMed] [Google Scholar]
- 16.Steinert PM, Kartasova T, Marekov LN. Biochemical evidence that small proline-rich proteins and trichohyalin function in epithelia by modulation of the biomechanical properties of their cornified cell envelopes. J Biol Chem. 1998;273:11758–11769. doi: 10.1074/jbc.273.19.11758. [DOI] [PubMed] [Google Scholar]
- 17.Robinson NA, Lapic S, Welter JF, Eckert RL. S100A11, S100A10, annexin I, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J Biol Chem. 1997;272:12035–12046. doi: 10.1074/jbc.272.18.12035. [DOI] [PubMed] [Google Scholar]
- 18.Nemes Z, Marekov LN, Fesus L, Steinert PM. A novel function for transglutaminase 1: attachment of long-chain omega-hydroxyceramides to involucrin by ester bond formation. Proc Natl Acad Sci USA. 1999;96:8402–8407. doi: 10.1073/pnas.96.15.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamid Q, Boguniewicz M, Leung DY. Differential in situ cytokine gene expression in acute versus chronic atopic dermatitis. J Clin Invest. 1994;94:870–876. doi: 10.1172/JCI117408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, Darst MA, Gao B, Boguniewicz M, Travers JB, Leung DY. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–3269. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 21.Tukey JW. Some thoughts on clinical trials, especially problems of multiplicity. Science. 1977;198:679–684. doi: 10.1126/science.333584. [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 23.Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S. Impaired IL-13-mediated functions of macrophages in STAT6-deficient mice. J Immunol. 1996;157:3220–3222. [PubMed] [Google Scholar]
- 24.Boguniewicz M, Leung DY. Atopic dermatitis. J Allergy Clin Immunol. 2006;117:S475–480. doi: 10.1016/j.jaci.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004;113:651–657. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laouini D, Kawamoto S, Yalcindag A, Bryce P, Mizoguchi E, Oettgen H, Geha RS. Epicutaneous sensitization with superantigen induces allergic skin inflammation. J Allergy Clin Immunol. 2003;112:981–987. doi: 10.1016/j.jaci.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Volz A, Korge BP, Compton JG, Ziegler A, Steinert PM, Mischke D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993;18:92–99. doi: 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- 28.Sugiura H, Ebise H, Tazawa T, Tanaka K, Sugiura Y, Uehara M, Kikuchi K, Kimura T. Large-scale DNA microarray analysis of atopic skin lesions shows overexpression of an epidermal differentiation gene cluster in the alternative pathway and lack of protective gene expression in the cornified envelope. Br J Dermatol. 2005;152:146–149. doi: 10.1111/j.1365-2133.2005.06352.x. [DOI] [PubMed] [Google Scholar]
- 29.Jensen JM, Folster-Holst R, Baranowsky A, Schunck M, Winoto-Morbach S, Neumann C, Schutze S, Proksch E. Impaired sphingomyelinase activity and epidermal differentiation in atopic dermatitis. J Invest Dermatol. 2004;122:1423–1431. doi: 10.1111/j.0022-202X.2004.22621.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Hessner MJ, Wu Y, Pati N, Ghosh S. Quantitative quality control in microarray experiments and the application in data filtering, normalization and false positive rate prediction. Bioinformatics. 2003;19:1341–1347. doi: 10.1093/bioinformatics/btg154. [DOI] [PubMed] [Google Scholar]
- 31.Liao JG, Lin Y, Selvanayagam ZE, Shih WJ. A mixture model for estimating the local false discovery rate in DNA microarray analysis. Bioinformatics. 2004;20:2694–2701. doi: 10.1093/bioinformatics/bth310. [DOI] [PubMed] [Google Scholar]
- 32.Watt FM. Terminal differentiation of epidermal keratinocytes. Curr Opin Cell Biol. 1989;1:1107–1115. doi: 10.1016/s0955-0674(89)80058-4. [DOI] [PubMed] [Google Scholar]
- 33.Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, Leung DY. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–348. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]


